Abstract
Bacterial profiles of saliva in subjects with periodontitis and dental caries have been demonstrated to differ from that of oral health. The aim of this comparative analysis of existing data generated by the Human Oral Microbe Identification Microarray (HOMIM) from 293 stimulated saliva samples was to compare bacterial profiles of saliva in subjects with periodontitis and dental caries.
Keywords: saliva, HOMIM, bacteria, periodontitis, dental caries
The bacterial profile of saliva in oral health has been investigated (1, 2), and has been shown to be most closely associated with the bacterial profile of the throat, the tonsils, and the dorsum of the tongue (3). However, our group has recently demonstrated that bacterial profiles of saliva in samples from subjects with periodontitis or dental caries differ significantly from that of oral health (4, 5). Thus, the purpose of the present analysis was to directly compare existing data on bacterial profiles of saliva from subjects with periodontitis with those from individuals with dental caries.
The present comparative analysis is part of a group of investigations analyzing the microbial profiles of the same batch of stimulated saliva samples from the Danish Health Examination Survey (DANHES) (4–6). Thus, in this analysis, we used existing data generated by the Human Oral Microbe Identification Microarray (HOMIM) from a periodontitis group and from a dental caries group that have previously been compared against a control cohort (4, 5). Periodontitis was defined as bleeding on probing (BOP) at ≥25% of total sites, minimum of two teeth with clinical attachment level ≥4 mm, and minimum two teeth with probing depth ≥6 mm. Dental caries was defined as untreated caries lesions on at least three surfaces. We excluded data from 10 subjects who had both periodontitis and dental caries. Therefore, in this analysis, data from a total of 129 periodontitis subjects were compared with data from 164 subjects with dental caries. Because we previously demonstrated that smoking has a profound effect on microbial profiles (6), we also performed a separate comparative analysis of data from non-smoking subjects (periodontitis n=98, dental caries n=139).
The cohort of DANHES and the subpopulation participating in the oral part of DANHES have previously been presented (7, 8), and the protocols describing clinical examination and collection of stimulated saliva samples have been outlined in detail (7). Briefly, the periodontal examination was performed in two randomly selected quadrants (one in the upper and one in the lower jaw), except third molars, including gingival margin position, probing pocket depth and BOP recorded at six sites on each tooth. Full mouth was screened for caries. Caries was recorded as a clinically cavitated lesion present on surface and tooth levels. No information from dental radiographs was included in this study. The laboratory procedure of DNA isolation (6) and subsequent DNA–DNA hybridization to a HOMIM (9, 10) has been described previously. In brief, HOMIM is a culture-independent technique that uses two separate PCR amplifications and subsequent DNA–DNA hybridization for identification of around 300 bacterial species, as single-stranded fluorescent-labeled PCR products are captured by highly specific bacterial 16S rRNA probes (18–20 bases) printed on a customized aldehyde-coated glass slide. Based on a qualitative HOMIM value of each target, further analysis was performed with the HOMIM online tool (http//:bioinformatics.forsyth.org/homim/).
In this present analysis, comparisons between groups at taxon/cluster level were performed using information about frequency and mean HOMIM values from 327 probes as end points by means of Mann–Whitney test with Benjamini–Hochberg correction for multiple dependent assumptions (11). An adjusted p-value <0.01 was considered statistically significant. In addition, data reduction and graphic presentation were performed using principal component analysis. We used GraphPad Prism 5 (San Diego, CA) and MeV 4.8.1 (12) to analyze and plot our data.
The present comparative analysis of the periodontitis group and the dental caries group revealed a significant higher mean age (56.8 years) in the periodontitis group than in the dental caries group (50.1 years; p<0.0001). In addition, the periodontitis group tended to comprise a higher proportion of daily smokers (24%) as compared to the dental caries group (15%; p=0.06).
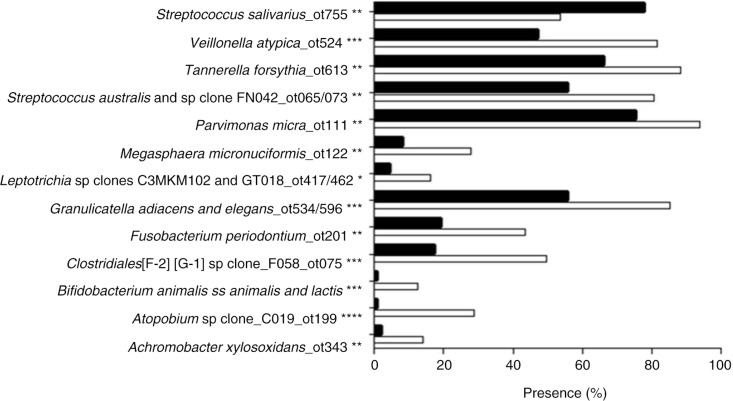
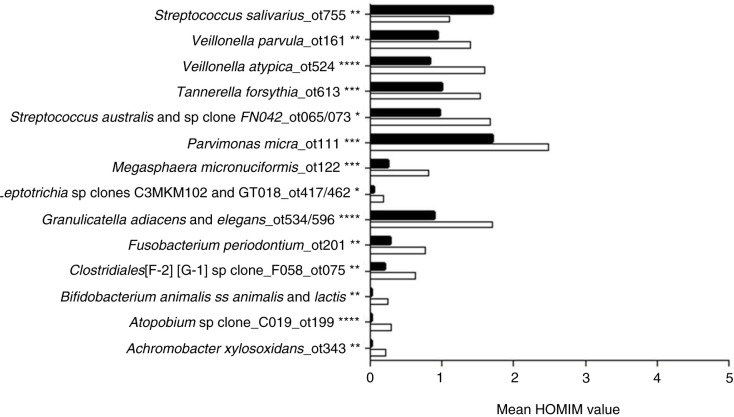
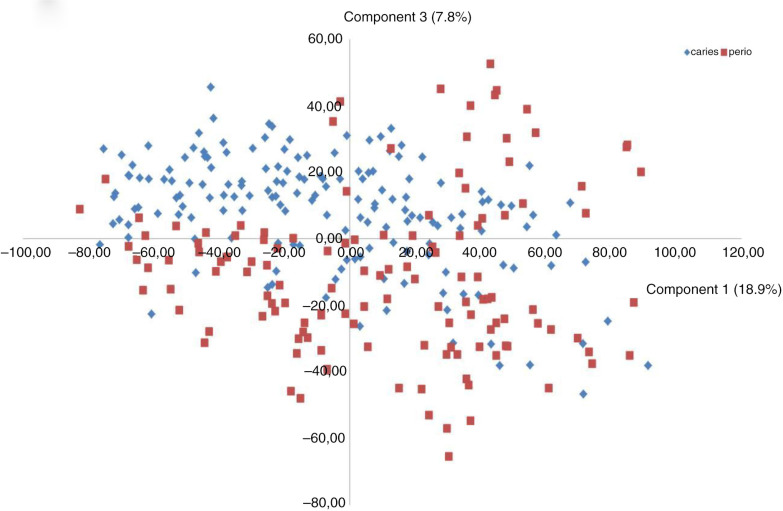
Targets of 12 probes (recognizing eight bacterial taxa and four bacterial clusters) were present significantly more often and at significantly higher levels (mean HOMIM value) in samples from periodontitis subjects (adjusted p-value<0.01) as compared to samples from subjects with dental caries. In contrast, Streptococcus salivarius was the only bacterial target detected significantly more often, and at significantly higher levels, in samples from subjects with dental caries (adjusted p-value<0.01) (Figs. 1 and 2). A separate analysis of samples from non-smoking subjects revealed that the same bacterial taxa and clusters were significantly associated with samples from the periodontitis group and dental caries group, as in the analysis comprising all subjects (data not shown). Data reduction through principal component analysis was performed to provide two-dimensional presentation of differences observed at taxon/cluster level, as mean HOMIM values of all probes identified were used as the end point to calculate the principal components of this n-dimensional data set. Based on the third most decisive component (component 3), accounting for 7.8% of the total variation of the data set, separation of samples from periodontitis and dental caries patients was evident (Fig. 3).
Fig. 1.
Probe targets identified to be present with different frequency in saliva samples from subjects with periodontitis and dental caries. Taxa and clusters identified at significantly different frequencies in samples from periodontitis and caries subjects. Binary values (presence/absence) of each probe target were compared between the periodontitis group and the caries group. The presence of each target as % of total samples is displayed. White bars: periodontitis group. Black bars: dental caries group. *: adjusted p-value<0.01, *: adjusted p-value<0.001, ***: adjusted p-value<1.0×10−5, ****: adjusted p-value<1.0×10−9.
Fig. 2.
Probe targets observed at different levels in saliva samples from subjects with periodontitis and dental caries. Taxa and clusters identified at significantly different levels (mean Human Oral Microbe Identification Microarray [HOMIM] value) in samples from subjects with periodontitis and subjects with dental caries. Mean HOMIM value of each taxon/cluster was compared between the periodontitis group and the caries group. The mean level of each target is displayed. White bars: periodontitis group. Black bars: dental caries group. *: adjusted p-value<0.01, **: adjusted p-value<0.001, ***: adjusted p-value<1.0×10−5, ****: adjusted p-value<1.0×10−8.
Fig. 3.
Principal component analysis. Principal component analysis was used for exploration of mathematical patterns, as mean HOMIM values (level) from each probe, were used as parameters for investigation.
It is noteworthy that we, indeed, determined two putative periodontal pathogens, Parvimonas micra and Tannerella forsythia, to be more associated with samples from subjects with periodontitis than from dental caries subjects. These results are in line with previous studies analyzing local bacterial community profiles associated with periodontitis, demonstrating that P. micra, T. forsythia, Megasphaera micronuciformis, Veillonella parvula, and Leptotrichia species were more associated with local periodontal lesions than with healthy control sites (9, 13). Furthermore, S. salivarius has been shown previously to be more associated with orthodontic-induced caries lesions than with healthy control sites (14) and also with caries in the permanent dentition (15). Interestingly, the different bacterial profiles between subjects with periodontitis and subjects with dental caries appeared to be independent of smoking status. This suggests that the impact of oral disease status on bacterial profiles of saliva may be more significant than that of smoking status.
Recent studies of stimulated saliva samples, performed by our group, on the same HOMIM-generated data set, have only sporadically identified suggested periodontal pathogens and cariogenic bacteria as Porphyromonas gingivalis, Aggregatibacter actinomycetemcomitans, and Streptococcus mutans ( 4–6). In line with this observation, this subsequent comparative analysis found none of these organisms to be predominantly associated with periodontitis or dental caries. This finding is conflicting with earlier reports, demonstrating that presence of P. gingivalis, A. actinomycetemcomitans, and other bacteria in saliva samples was associated with periodontitis (16, 17). Likewise, the presence of S. mutans in saliva has been suggested as a biomarker of early childhood caries (18). A possible explanation for these conflicting findings could be that these organisms were actually present in the samples, but in quantities below the detection limit of HOMIM. However, the association of P. micra and T. forsythia with saliva samples from subjects with periodontitis in this analysis suggests that local bacterial alterations in relation to periodontitis, and possibly dental caries, may be detected in saliva samples.
In conclusion, these comparative analyses suggest that bacterial salivary profiles of subjects with periodontitis and dental caries are differentiated from each other and that these differences are independent of smoking status. Obviously, longitudinal and interventional studies are warranted to further evaluate if bacterial profiles of saliva have the potential to serve as a biomarker of periodontitis and dental caries.
Acknowledgements
The authors would like to thank Dr. Morten Grønbæk and the National Institute of Public Health, University of Southern Denmark, Copenhagen, Denmark, for accessibility to DANHES-related data; Dr. Mogens Kilian for helpful advice with analysis of microbiological data; and Sean Cotton, Alexis Kokaras, and Christina Murphy for laboratory assistance at the Forsyth Institute.
Conflict of interest and funding
The authors declare no conflict of interest in this study. This study was supported financially by: the Danish Dental Association, the Danish Foundation for Mutual Efforts in Dental Care, TrygFonden, the Health Insurance Foundation, and the Simon Spies Foundation.
References
- 1.Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4:962–74. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nasidze I, Li J, Quinque D, Tang K, Stoneking M. Global diversity in the human salivary microbiome. Genome Res. 2009;19:636–43. doi: 10.1101/gr.084616.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13:R42. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belstrom D, Fiehn NE, Nielsen CH, Kirkby N, Twetman S, Klepac-Ceraj V, et al. Differences in bacterial saliva profile between periodontitis patients and a control cohort. J Clin Periodontol. 2014;41:104–12. doi: 10.1111/jcpe.12190. [DOI] [PubMed] [Google Scholar]
- 5.Belstrom D, Fiehn NE, Nielsen CH, Holmstrup P, Kirkby N, Klepac-Ceraj V, et al. Altered bacterial profiles in saliva from adults with caries lesions: a case-cohort study. Caries Res. 2014;48:368–75. doi: 10.1159/000357502. [DOI] [PubMed] [Google Scholar]
- 6.Belstrom D, Holmstrup P, Nielsen CH, Kirkby N, Twetman S, Heitmann BL, et al. Bacterial profiles of saliva in relation to diet, lifestyle factors, and socioeconomic status. J Oral Microbiol. 2014;6:23609. doi: 10.3402/jom.v6.23609. doi: http://dx.doi.org/10.3402/jom.v6.23609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eriksen L, Gronbaek M, Helge JW, Tolstrup JS, Curtis T. The Danish Health Examination Survey 2007–2008 (DANHES 2007–2008) Scand J Public Health. 2011;39:203–11. doi: 10.1177/1403494810393557. [DOI] [PubMed] [Google Scholar]
- 8.Kongstad J, Ekstrand K, Qvist V, Christensen LB, Cortzen B, Grønbæk M, et al. Findings from the oral health study of the Danish Health Examination Survey 2007–2008. Acta Odontol Scand. 2013;71:1560–9. doi: 10.3109/00016357.2013.776701. [DOI] [PubMed] [Google Scholar]
- 9.Colombo AP, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80:1421–32. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Preza D, Olsen I, Willumsen T, Borches SK, Cotton CL, Grinde B, et al. Microarray analysis of the microflora of root caries in elderly. Eur J Clin Microbiol Infect Dis. 2009;28:509–17. doi: 10.1007/s10096-008-0662-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–18. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 12.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, et al. TM4 microarray software suite. Methods Enzymol. 2006;411:134–93. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 13.Colombo AP, Bennet S, Cotton SL, Goodson JM, Kent R, Haffajee AD, et al. Impact of periodontal therapy on the subgingival microbiota of severe periodontitis: comparison between good responders and “Refractory” subjects by the Human Oral Microbe Identification Microarray (HOMIM) J Periodontol. 2012;83:1279–87. doi: 10.1902/jop.2012.110566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torlakovic L, Klepac-Ceraj V, Ogaard B, Cotton SL, Paster BJ, Olsen I. Microbial community succession on developing lesions on human enamel. J Oral Microbiol. 2012;4:16125. doi: 10.3402/jom.v4i0.16125. doi: http://dx.doi.org/10.3402/jom.v4.16125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aas JA, Griffen AL, Dardis SR, Lee AM, Olsen I, Dewhirst FE, et al. Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol. 2008;46:1407–17. doi: 10.1128/JCM.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paju S, Pussinen PJ, Suominen-Taipale L, Hyvonen M, Knuuttila M, Kononen E. Detection of multiple pathogenic species in saliva is associated with periodontal infection in adults. J Clin Microbiol. 2009;47:235–8. doi: 10.1128/JCM.01824-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kononen E, Paju S, Pussinen PJ, Hyvönen M, Di Tella P, Suominen-Taipale L, et al. Population-based study of salivary carriage of periodontal pathogens in adults. J Clin Microbiol. 2007;45:2446–51. doi: 10.1128/JCM.02560-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parisotto TM, Steiner-Oliveira C, Silva CM, Rodrigues LK, Nobre-dos-Santos M. Early childhood caries and mutans streptococci: a systematic review. Oral Health Prev Dent. 2010;8:59–70. [PubMed] [Google Scholar]