Abstract
We describe here 35 animal cases of tuberculosis due to Mycobacterium microti in France (2002–2014). Recently, molecular tools that overcome the difficulty of confirming infection by this potentially zoonotic agent have revealed an increasing number of cases, suggesting that its prevalence may have been underestimated.
INTRODUCTION
Mycobacterium microti belongs to the Mycobacterium tuberculosis complex (MTC), which also includes M. tuberculosis, Mycobacterium bovis, Mycobacterium caprae, Mycobacterium africanum, and Mycobacterium pinnipedii. This species was originally described as the cause of tuberculosis (TB) in wild rodents. Indeed, field voles (Microtus agrestis), bank voles (Myodes glareolus), wood mice (Apodemus sylvaticus), and shrews (Sorex araneus), which are particularly susceptible to M. microti infection, constitute its natural reservoirs (1). However, an increasing number of cases have also been reported in domestic and wild mammals (2), such as cats (3, 4), pigs (5), European wild boar (Sus scrofa) (6), ferrets (Mustela putorius), badgers (Meles meles) (4), New World camelids (Lama glama and Vicugna pacos) (4), squirrel monkeys (Saimiri sciureus) (7), meerkats (Suricata suricatta) (8), and a dog (9). Until now, 27 cases of M. microti infection in both immunocompetent (10) and immunocompromised human patients (11) have been described (12), demonstrating its capacity for causing clinical illness and thus for being a potential zoonotic agent.
In the first described animal cases with M. microti, the infection was due to bacilli of limited genotypic diversity, as determined by spoligotyping, with the vole-type SB0118 and the llama-types SB0112 and SB0423 (13, 14) being dominant. However, Smith et al. (15) recently showed that this species is more diverse than was expected, and they described 15 new spoligotype patterns for M. microti. Moreover, a number of spoligopatterns described in M. microti infection cases in humans have also been reported, although they were not submitted to the spoligotype database, confirming an even larger diversity for this mycobacterium (4, 12).
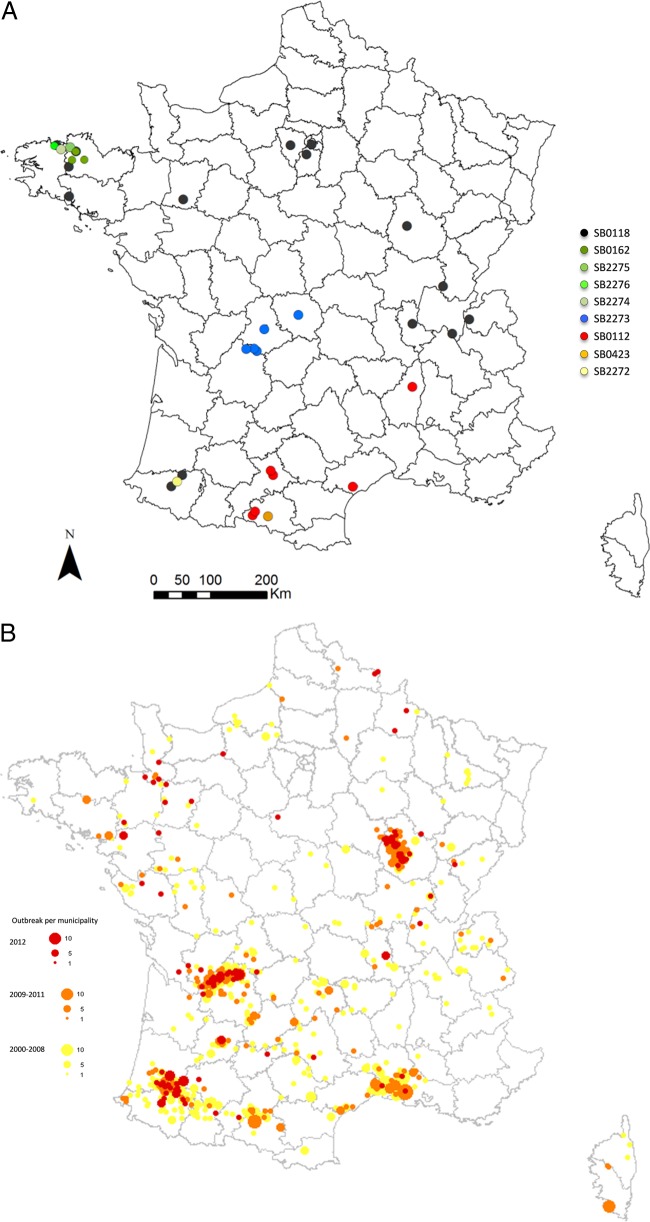
The present study, which is the first retrospective analysis of M. microti infection in animals in France, describes the animal M. microti infection cases identified by the Tuberculosis National Reference Laboratory in France from May 2002 to April 2014, with a particular focus on genotypic diversity by spoligotyping and the geographic localization of the pathogen. Here, we describe 35 cases in a large variety of domestic and wild animal species. The individuals presenting with TB-like lesions had tissues analyzed by bacteriology and, from 2005, also by molecular technologies; spoligotyping was performed by Luminex either on MTC isolates or directly onto PCR-positive samples (16, 17) (Table 1). With the PCR-positive samples, in which the DNA concentration of the pathogen can be low, partial profiles were sometimes obtained. Bacteriology was positive for only eight of the cases (Table 1). The majority of the cases were in cats (n = 19 [54%]). The other infected animal species were wild boar (n = 7 [20%]), dogs (n = 3 [8%]), marmosets (Callithrix jacchus) (n = 2 [6%]), llamas or alpacas (n = 2 [6%]), a pig (n = 1 [3%]), and an otter (Lutra lutra) (n = 1 [3%]). The spatial distribution of the cases was obtained with the ArcGIS 9 software (Fig. 1A). As it is present in nine out of 22 French mainland regions, M. microti infection seems to be evenly distributed throughout the country.
TABLE 1.
List of M. microti infection cases in various hosts reported in 9 regions of France between 2002 and 2014 and the associated spoligopatterns
| Yr | Host | Region | Spoligopattern | Identification method |
|---|---|---|---|---|
| 2002 | Dog | Limousin | SB2273 | Bacteriology |
| 2003 | Cat | Brittany | SB0162 | Molecular biology |
| 2004 | Cat | Île de France | SB0118 | Bacteriology |
| 2005 | Llama | Midi-Pyrénées | SB0423 | Bacteriology |
| 2006 | Cat | Languedoc-Roussillon | SB0112 | Molecular biology |
| 2007 | Cat | Brittany | SB0118 | Bacteriology |
| 2007 | Otter | Brittany | SB0162 | Bacteriology |
| 2007 | Cat | Île de France | SB0118 | Molecular biology |
| 2008 | Marmosets | Brittany | SB0118 | Bacteriology |
| 2009 | Marmosets | Brittany | SB0118 | Bacteriology |
| 2009 | Cat | Brittany | SB0118 | Molecular biology |
| 2010 | Cat | Rhône-Alpes | SB0118 | Molecular biology |
| 2010 | Cat | Midi-Pyrénées | SB0112 | Molecular biology |
| 2010 | Cat | Brittany | SB2274 | Molecular biology |
| 2011 | Cat | Rhône-Alpes | SB0118 | Molecular biology |
| 2011 | Cat | Rhône-Alpes | SB0118 | Molecular biology |
| 2011 | Dog | Midi-Pyrénées | SB0112 | Bacteriology |
| 2011 | Pig | Brittany | SB2275 | Molecular biology |
| 2011 | Cat | Rhône-Alpes | SB0112 | Molecular biology |
| 2012 | Cat | Burgundy | SB0118 | Molecular biology |
| 2012 | Cat | Limousin | SB2273 partial 1 | Molecular biology |
| 2012 | Cat | Midi-Pyrénées | SB0112 | Molecular biology |
| 2013 | Cat | Brittany | SB0162 | Molecular biology |
| 2013 | Cat | Île de France | SB0118 | Molecular biology |
| 2013 | Dog | Midi-Pyrénées | SB0112 | Molecular biology |
| 2013 | Wild boar | Burgundy | SB0118 | Molecular biology |
| 2013 | Cat | Brittany | SB0118 | Molecular biology |
| 2013 | Wild boar | Aquitaine | SB2273 partial 2 | Molecular biology |
| 2013 | Wild boar | Aquitaine | SB2273 | Molecular biology |
| 2013 | Wild boar | Aquitaine | SB0118 | Molecular biology |
| 2013 | Wild boar | Aquitaine | SB0118 | Molecular biology |
| 2013 | Alpaca | Pays de Loire | SB0118 | Molecular biology |
| 2014 | Wild boar | Aquitaine | SB2273 partial 3 | Molecular biology |
| 2014 | Wild boar | Aquitaine | SB2272 | Molecular biology |
| 2014 | Cat | Brittany | SB2276 | Molecular biology |
FIG 1.
(A) Distribution of M. microti cases in France and their associated spoligopatterns. (B) Distribution by municipality of M. bovis outbreaks (adapted from reference 25).
The first case of M. microti infection described in France was identified by isolating this agent from a dog (9). Some years later, a study of six human cases of pulmonary TB due to M. microti was published (12). Recently, a case of feline cutaneous TB was described in Burgundy, eastern central France (18), but molecular characterization for this case was not performed. The number of cases reported in this study increased in the last few years. This increasing recognition is the result of the improved sensitivities of the diagnostic tests employed (mainly molecular typing tools), increased awareness about the infection, and the inclusion of wildlife cases studied using molecular biology techniques since 2013. This suggests that M. microti is circulating both in domestic and wild populations and that the prevalence of this pathogen may have been underestimated before the introduction of molecular techniques. Indeed, until recently, wildlife TB cases were confirmed by bacteriology only, which has a very poor sensitivity for diagnosing M. microti, even compared to that for M. bovis (14, 15), which is also a quite culture-recalcitrant bacterium.
Genetic diversity.
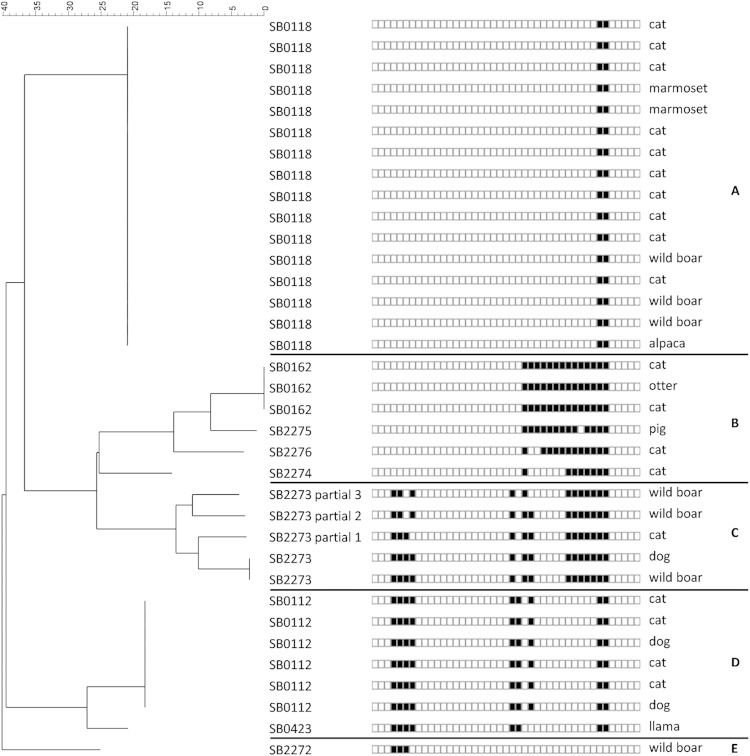
The spoligopatterns were determined for all cases in the current study and appear to form 5 groups on the neighbor-joining tree built with the BioNumerics software (Fig. 2). The authoritative names for the spoligopatterns (prefix of SB, followed by four digits) were obtained from the M. bovis Spoligotype Database website (http://www.Mbovis.org) (19). As suggested in the literature, the genetic diversity of M. microti seems to be greater than was previously thought (15). Until now, 28 different spoligopatterns of this MTC species (Table 2) were described in the literature, but only 24 were submitted to the M. bovis database. All spoligopatterns of M. microti are supposed to derive from a putative ancestral pattern, SB0155, also described in M. pinnipedii (15, 20). Spoligopatterns result from the unidirectional evolution of the direct repeat region, with accumulations of deletions leading to new patterns. The most interesting case is SB2277, recently described in an Italian wild boar, for which the direct repeat (DR) region lacks the classic 43 spacers (6). Here, we describe nine different spoligopatterns of M. microti for only 35 cases, four of which had already been described in the past, and five of which are new patterns. SB0118 (vole type), which is one of the main spoligopatterns described in the literature (13, 14), is also the most common (16 cases) and the most widespread in France (group A). Another well-described spoligopattern (4, 13, 15), SB0112 (llama type), with six identified cases, is the second most important pattern in France. This pattern has only a one-spacer difference with SB0423, which was described in French human cases (12, 13). These close patterns form a clear group on the neighbor-joining analysis and, moreover, seem to be colocalized in the south of France (group C). SB0162, reported previously in a badger (15), is described here in three cases. This pattern forms group B, with three new spoligotypes, SB2274 to SB2276, and has the same geographical localization, i.e., Brittany, in the northwest France. A last cluster, group D, was identified on the neighbor-joining tree and is composed of cases with a new pattern, SB2273, either complete or partial. This group seems to be clearly localized in the center of France. A new spoligotype, SB2272, is described here for the first time in a wild boar. This pattern is very close to the SB1403 pattern described in a cat in Britain (15).
FIG 2.
Neighbor-joining tree based on spoligotype patterns identified in France. The letters on the right indicate the groups A to E.
TABLE 2.
Spoligopatterns of M. microti described in the literature and in this study
| SB no.a | Spoligopattern | Source(s)a | Reference(s) |
|---|---|---|---|
| SB0155 | ․․․◆◆◆◆․․․․․․․․․․․․․․․◆◆◆◆◆◆◆◆◆◆◆◆◆◆◆◆․․․․․ | Cat, llama, alpaca, badger | 15 |
| SB1510 | ․․․◆◆◆◆․․․․․․․․․․․․․․․◆◆◆◆◆◆◆◆◆◆◆◆◆◆․․․․․․․ | Pig | 15 |
| SB0326 | ․․․◆◆◆◆․․․․․․․․․․․․․․․․․◆◆◆․․◆◆◆◆◆◆◆․◆․․․․․ | Alpaca, cow | 15 |
| SB1514 | ․․․◆◆◆◆․․․․․․․․․․․․․․․․․◆◆◆․◆◆◆◆◆◆․․◆◆․․․․․ | Cat | 15 |
| SB0988 | ․․․․․․․․․․․․․․․․․․․․․․◆․◆◆◆◆◆◆◆◆◆◆◆◆◆◆․․․․․ | Cow | 15 |
| SB0162 | ․․․․․․․․․․․․․․․․․․․․․․․․◆◆◆◆◆◆◆◆◆◆◆◆◆◆․․․․․ | Badger, cat, otter | 15 |
| SB2275 | ․․․․․․․․․․․․․․․․․․․․․․․․◆◆◆◆◆◆◆◆◆․◆◆◆◆․․․․․ | Pig | |
| SB2276 | ․․․․․․․․․․․․․․․․․․․․․․․․◆․․◆◆◆◆◆◆◆◆◆◆◆․․․․․ | Cat | |
| SB2274 | ․․․․․․․․․․․․․․․․․․․․․․․․◆․․․․․․◆◆◆◆◆◆◆․․․․․ | Cat | |
| SB2273 | ․․․◆◆◆◆․․․․․․․․․․․․․․․◆․◆◆․․․․․◆◆◆◆◆◆◆․․․․․ | Wild boar, dog, cat | |
| ND | ․․․◆◆◆◆․․․․․․․․․․․․․․․◆․◆․․․․․․◆◆◆◆◆◆◆․․․․․ | Human | 12 |
| ND | ․․․◆◆◆◆․․․․․․․․․․․․․․․◆․◆․․․․․․◆․․◆◆◆◆․․․․․ | Human | 12 |
| SB0655 | ․․․◆◆◆․․․․․․․․․․․․․․․․․․․․․․․․․․․․◆◆◆◆․․․․․ | Badger | 15 |
| SB1403 | ․․․◆◆◆◆․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․ | Cat | 15 |
| SB2272 | ․․․◆◆◆․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․ | Wild boar | |
| SB0112 | ․․․◆◆◆◆․․․․․․․․․․․․․․․◆◆․◆․․․․․․․․․․◆◆․․․․․ | Alpaca, llama, pig, human, otter, cat, dog | 9–11, 13–15 |
| SB1505 | ․․․◆◆◆․․․․․․․․․․․․․․․․◆◆․◆․․․․․․․․․․◆◆․․․․․ | Llama | 15 |
| SB1509 | ․․․◆․◆◆․․․․․․․․․․․․․․․◆◆․◆․․․․․․․․․․◆◆․․․․․ | Alpaca | 15 |
| SB1511 | ․․․․◆◆◆․․․․․․․․․․․․․․․◆◆․◆․․․․․․․․․․◆◆․․․․․ | Cat | 15 |
| SB0657 | ․․․◆◆◆◆․․․․․․․․․․․․․․․◆◆․◆․․․․․․․․․․◆․․․․․․ | Cat | 13, 15 |
| SB0423 | ․․․◆◆◆◆․․․․․․․․․․․․․․․◆◆․․․․․․․․․․․․◆◆․․․․․ | Cat, human, llama | 4, 12, 13 |
| ND | ․․․◆◆◆◆․․․․․․․․․․․․․․․◆◆․․․․․․․․․․․․․․․․․․․ | Human | 4 |
| SB0654 | ․․․◆․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․◆◆․․․․․ | Cat, llama, badger, cow | 13, 15 |
| SB1513 | ․․․◆․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․◆․․․․․ | Cat | 15 |
| SB0118 | ․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․◆◆․․․․․ | Vole, ferret, llama, badger, pig, human, meerkat, squirrel monkey, cat, wild boar, alpaca, marmoset | 3, 4, 7, 8, 10, 12–15 |
| ND | ․․․․․․․․․․․․․․․․․․․․․․․․◆․․․․․․․․․․․◆․․․․․․ | Human | 12 |
| SB1512 | ․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․◆․․․․․․ | Cat | 15 |
| SB2277 | ․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․․ | Wild boar | 6 |
The spoligotype and sources of French cases are indicated in bold; ND, not determined.
Epidemiology of M. microti.
As previously demonstrated in Britain (15), this study suggests that M. microti is endemic in certain areas in France and absent in other areas; however, the number of cases in our study is less than that in the British study. Some authors hypothesize that in regions where M. microti infection is endemic in badgers or cats, it might provide some protection to cattle against M. bovis infection and thus explain the low incidence of bovine TB (15, 21). This protective immunity to M. bovis appears not to apply to the wild boar, which seem to be involved in the maintenance of M. bovis in some of the described regions, or for cattle, as new bovine TB outbreaks are discovered every year in these same regions. Indeed, M. microti and M. bovis infections overlap in some areas (Fig. 1A and B), especially in those regions where the wild boar was found to be infected by M. microti, i.e., in Aquitaine (Southwest France) and Burgundy (eastern central France). The prevalences of M. bovis and M. microti in wild boar of Aquitaine, through the surveillance of wildlife in the last 2 years, have been estimated to be 2.5% and 0.5%, respectively. A serological survey of wild boar conducted in France highlighted the distribution of animals exposed to MTC (mainly M. bovis and M. microti), which overlapped the distribution of bovine TB (bTB) outbreaks in cattle (22) and correspond to the same regions where we found wild boar infected with M. microti. These results do not support the hypothesis of competition between M. microti and M. bovis and suggest that these mycobacteria do not completely exclude each other geographically in France.
Zoonotic transmission might occur from rodents to humans via spillover hosts, such as cats, South American camelids, or dogs, or via environmental contamination environments. Indeed, small rodent populations are widely distributed not only in rural habitats but also in periurban regions. Potential zoonotic infections have been suggested to involve cats (23), mice (14), and either a raccoon or raccoon dogs (24). Even if M. microti is considered a rare human pathogen, its potential to cause clinical illness in humans has been demonstrated. Therefore, the accurate surveillance of TB using molecular tools that overcome the particular difficulty of confirming M. microti by bacteriology would help characterize the epidemiology of this pathogen.
ACKNOWLEDGMENTS
We thank Victoria Boschiroli and Edouard Reveillaud for their valuable comments on the manuscript.
REFERENCES
- 1.Cavanagh R, Begon M, Bennett M, Ergon T, Graham IM, De Haas PE, Hart CA, Koedam M, Kremer K, Lambin X, Roholl P, van Soolingen D. 2002. Mycobacterium microti infection (vole tuberculosis) in wild rodent populations. J Clin Microbiol 40:3281–3285. doi: 10.1128/JCM.40.9.3281-3285.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodriguez-Campos S, Smith NH, Boniotti MB, Aranaz A. 2014. Overview and phylogeny of Mycobacterium tuberculosis complex organisms: implications for diagnostics and legislation of bovine tuberculosis. Res Vet Sci 97:S5–S19. doi: 10.1016/j.rvsc.2014.02.009 [DOI] [PubMed] [Google Scholar]
- 3.Rüfenacht S, Bogli-Stuber K, Bodmer T, Jaunin VF, Jmaa DC, Gunn-Moore DA. 2011. Mycobacterium microti infection in the cat: a case report, literature review and recent clinical experience. J Feline Med Surg 13:195–204. doi: 10.1016/j.jfms.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xavier Emmanuel F, Seagar AL, Doig C, Rayner A, Claxton P, Laurenson I. 2007. Human and animal infections with Mycobacterium microti, Scotland. Emerg Infect Dis 13:1924-1927. doi: 10.3201/eid1312.061536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor C, Jahans K, Palmer S, Okker M, Brown J, Steer K. 2006. Mycobacterium microti isolated from two pigs. Vet Rec 159:59–60. doi: 10.1136/vr.159.2.59-a. [DOI] [PubMed] [Google Scholar]
- 6.Boniotti MB, Gaffuri A, Gelmetti D, Tagliabue S, Chiari M, Mangeli A, Spisani M, Nassuato C, Gibelli L, Sacchi C, Zanoni M, Pacciarini ML. 2014. Detection and molecular characterization of Mycobacterium microti in wild boar from northern Italy. J Clin Microbiol 52:2834–2843. doi: 10.1128/JCM.00440-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henrich M, Moser I, Weiss A, Reinacher M. 2007. Multiple granulomas in three squirrel monkeys (Saimiri sciureus) caused by Mycobacterium microti. J Comp Pathol 137:245–248. doi: 10.1016/j.jcpa.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 8.Palgrave CJ, Benato L, Eatwell K, Laurenson IF, Smith NH. 2012. Mycobacterium microti infection in two meerkats (Suricata suricatta). J Comp Pathol 146:278–282. doi: 10.1016/j.jcpa.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Deforges L, Boulouis HJ, Thibaud JL, Boulouha L, Sougakoff W, Blot S, Hewinson G, Truffot-Pernot C, Haddad N. 2004. First isolation of Mycobacterium microti (llama-type) from a dog. Vet Microbiol 103:249–253. doi: 10.1016/j.vetmic.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 10.Niemann S, Richter E, Dalugge-Tamm H, Schlesinger H, Graupner D, Konigstein B, Gurath G, Greinert U, Rusch-Gerdes S. 2000. Two cases of Mycobacterium microti derived tuberculosis in HIV-negative immunocompetent patients. Emerg Infect Dis 6:539–542. doi: 10.3201/eid0605.000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horstkotte MA, Sobottka I, Schewe CK, Schafer P, Laufs R, Rusch-Gerdes S, Niemann S. 2001. Mycobacterium microti llama-type infection presenting as pulmonary tuberculosis in a human immunodeficiency virus-positive patient. J Clin Microbiol 39:406–407. doi: 10.1128/JCM.39.1.406-407.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panteix G, Gutierrez MC, Boschiroli ML, Rouviere M, Plaidy A, Pressac D, Porcheret H, Chyderiotis G, Ponsada M, Van Oortegem K, Salloum S, Cabuzel S, Banuls AL, Van de Perre P, Godreuil S. 2010. Pulmonary tuberculosis due to Mycobacterium microti: a study of six recent cases in France. J Med Microbiol 59:984–989. doi: 10.1099/jmm.0.019372-0. [DOI] [PubMed] [Google Scholar]
- 13.Kremer K, van Soolingen D, van Embden J, Hughes S, Inwald J, Hewinson G. 1998. Mycobacterium microti: more widespread than previously thought. J Clin Microbiol 36:2793–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Soolingen D, van der Zanden AG, de Haas PE, Noordhoek GT, Kiers A, Foudraine NA, Portaels F, Kolk AH, Kremer K, van Embden JD. 1998. Diagnosis of Mycobacterium microti infections among humans by using novel genetic markers. J Clin Microbiol 36:1840–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith NH, Crawshaw T, Parry J, Birtles RJ. 2009. Mycobacterium microti: more diverse than previously thought. J Clin Microbiol 47:2551–2559. doi: 10.1128/JCM.00638-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cowan LS, Diem L, Brake MC, Crawford JT. 2004. Transfer of a Mycobacterium tuberculosis genotyping method, spoligotyping, from a reverse line-blot hybridization, membrane-based assay to the Luminex multianalyte profiling system. J Clin Microbiol 42:474–477. doi: 10.1128/JCM.42.1.474-477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Abadia E, Refregier G, Tafaj S, Boschiroli ML, Guillard B, Andremont A, Ruimy R, Sola C. 2010. Mycobacterium tuberculosis complex CRISPR genotyping: improving efficiency, throughput and discriminative power of ‘spoligotyping' with new spacers and a microbead-based hybridization assay. J Med Microbiol 59:285–294. doi: 10.1099/jmm.0.016949-0. [DOI] [PubMed] [Google Scholar]
- 18.Laprie C, Duboy J, Malik R, Fyfe J. 2013. Feline cutaneous mycobacteriosis: a review of clinical, pathological and molecular characterization of one case of Mycobacterium microti skin infection and nine cases of feline leprosy syndrome from France and New Caledonia. Vet Dermatol 24:561–569. e133–e134. doi: 10.1111/vde.12066. [DOI] [PubMed] [Google Scholar]
- 19.Smith NH, Upton P. 2012. Naming spoligotype patterns for the RD9-deleted lineage of the Mycobacterium tuberculosis complex; www.Mbovis.org Infect Genet Evol 12:873–876. doi: 10.1016/j.meegid.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 20.Cousins DV, Bastida R, Cataldi A, Quse V, Redrobe S, Dow S, Duignan P, Murray A, Dupont C, Ahmed N, Collins DM, Butler WR, Dawson D, Rodriguez D, Loureiro J, Romano MI, Alito A, Zumarraga M, Bernardelli A. 2003. Tuberculosis in seals caused by a novel member of the Mycobacterium tuberculosis complex: Mycobacterium pinnipediisp. nov. Int J Syst Evol Microbiol 53:1305–1314. doi: 10.1099/ijs.0.02401-0. [DOI] [PubMed] [Google Scholar]
- 21.Gunn-Moore DA, McFarland SE, Brewer JI, Crawshaw TR, Clifton-Hadley RS, Kovalik M, Shaw DJ. 2011. Mycobacterial disease in cats in Great Britain: I. Culture results, geographical distribution and clinical presentation of 339 cases. J Feline Med Surg 13:934–944. doi: 10.1016/j.jfms.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richomme C, Boadella M, Courcoul A, Durand B, Drapeau A, Corde Y, Hars J, Payne A, Fediaevsky A, Boschiroli ML. 2013. Exposure of wild boar to Mycobacterium tuberculosis complex in France since 2000 is consistent with the distribution of bovine tuberculosis outbreaks in cattle. PLoS One 8:e77842. doi: 10.1371/journal.pone.0077842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGoldrick C, Coghlin C, Seagar AL, Laurenson I, Smith NH, Stewart WC, Kerr KM, Douglas JG. 2010. Mycobacterium microti infection associated with spindle cell pseudotumour and hypercalcaemia: a possible link with an infected alpaca. BMJ Case Rep 2010:pii:bcr1120092484. doi: 10.1136/bcr.11.2009.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frank W, Reisinger EC, Brandt-Hamerla W, Schwede I, Handrick W. 2009. Mycobacterium microti–pulmonary tuberculosis in an immunocompetent patient. Wien Klin Wochenschr 121:282–286. doi: 10.1007/s00508-009-1164-0. [DOI] [PubMed] [Google Scholar]
- 25.Fediaevsky A, Courcoul A, Boschiroli ML, Reveillaud E. 2013. Tuberculose bovine en France en 2012: des signaux favorable mais une situation toujours complexe dans certaines zones. Bull Epid Santé Anim Alim 59:4–10. [Google Scholar]