Abstract
Molecular typing of 246 Staphylococcus aureus isolates from unselected patients in Thailand showed that 10 (4.1%) were actually Staphylococcus argenteus. Contrary to the suggestion that S. argenteus is less virulent than S. aureus, we demonstrated comparable rates of morbidity, death, and health care-associated infection in patients infected with either of these two species.
TEXT
The population genetic structure of Staphylococcus aureus has been extensively described using multilocus sequence typing (MLST) (1). The original primers failed to amplify all 7 MLST loci for a small proportion of presumptive S. aureus isolates, resulting in the delayed description of an early branching lineage that was initially referred to as clonal complex 75 (CC75). Sequence type 1223 (ST1223), which belongs to the same lineage but does not cluster within CC75, was subsequently described (2, 3). The new species name Staphylococcus argenteus was proposed for CC75 and related STs (4), followed by formal taxonomic classification as S. argenteus sp. nov. (5).
Using a single-nucleotide polymorphism (SNP) genotyping method based on MLST, staphylococci belonging to CC75 were found to account for 71% of community-associated methicillin-resistant S. aureus (MRSA) skin and soft tissue infections in indigenous communities in the Northern Territory of Australia (6). Further isolation of CC75 from these communities (3) was followed by reports of its isolation in Cambodia (2), Indonesia (2), Fiji (7), and Trinidad and Tobago (8) and from members of an isolated village in the Amazonian forest in French Guinea (9). Information contained in the MLST database suggests that S. argenteus is also present in Europe and Africa. Overall, this indicates widespread dissemination of S. argenteus, and a report of its isolation from the feces of the straw-colored fruit bat in Nigeria (10) indicates that it may be carried by other animal species.
Isolation of S. argenteus in Thailand has not been reported, but based on isolation elsewhere in Southeast Asia we presumed that it was present. This is made more plausible by the fact that S. argenteus cannot be distinguished from S. aureus using routine diagnostic microbiology identification (2, 6, 7, 9). We investigated the presence of S. argenteus in Thailand through a reevaluation of S. aureus isolates obtained during a prospective study of 270 consecutive patients presenting with invasive infection at Sappasithiprasong Hospital in Northeast Thailand between November 2006 and November 2007 (11). All the isolates were originally identified based on Gram staining and the coagulase test and further evaluated for methicillin susceptibility and the presence of pvl and mecA and stored at −80°C (11).
Of the 270 isolates originally stored, 246 (91.1%) were available for genotyping using a combination of PCR and multilocus sequence typing (MLST). Of these isolates, 37 were MRSA. As ST239 is the predominant MRSA lineage in Thailand (12), MRSA isolates were initially screened using an ST239-specific PCR assay (12). All non-ST239 isolates were then typed using MLST as described previously (1), except for modifications in primer design to improve PCR amplification (see Table S1 in the supplemental material). Allele and ST assignments were performed using the MLST database (http://saureus.mlst.net). S. argenteus isolates in the study collection were identified based on phylogenetic clustering. To enrich the population for relevant genotypes, 38 STs were downloaded from the MLST database (http://www.mlst.net) as a reference collection. These were selected following an initial analysis in which phylogenetic trees were drawn for each S. aureus MLST locus using all of the data held in the MLST database (not shown). This demonstrated that several arcC alleles (36, 151, 207, and 271) and pta alleles (39, 107, 145, 175, 198, 256, 268, and 287) clustered in a single branch containing known S. argenteus isolates and were not shared with S. aureus (see Fig. S1 and S2 in the supplemental material). The remaining five alleles were shared between the two species. Based on this, we selected all STs that contained these particular arcC and pta alleles. The 38 STs were visualized using eBURST with a cutoff of 4 or more shared loci, which demonstrated that 33 STs belonged to one of four CCs: CC75 (n = 4 STs), CC2198 (n = 8 STs), CC2483 (n = 10 STs), or CC1594 (n = 11 STs). The remaining 5 STs were either singletons (ST2225, ST2351, or ST2470) or paired (ST2596 and ST2793). A maximum-likelihood tree was reconstructed from concatenated sequences of 7 MLST loci using PhyML version 3.0.1 (13). The CLC Main Workbench version 7.0 (Qiagen, USA) was used to edit and display the tree.
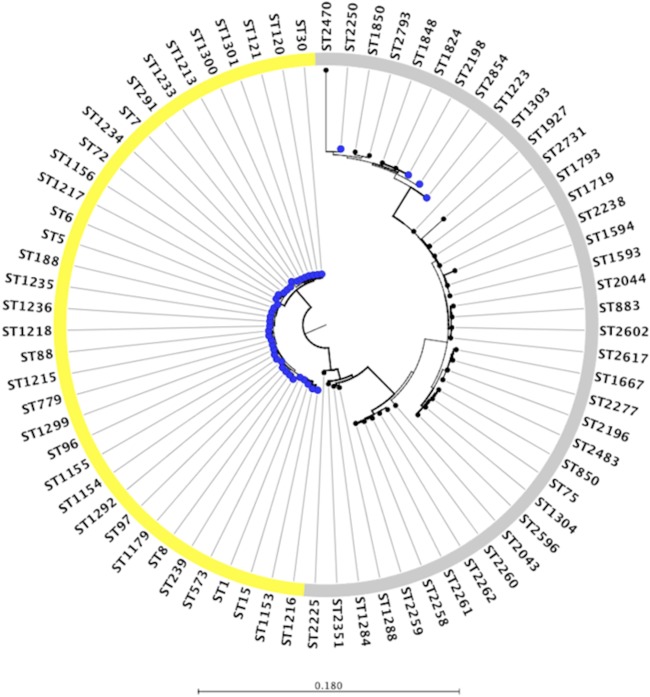
A total of 40 STs were identified in the 246 Thai isolates. The predominant genotype in the collection was ST121 (95/246, 38.6%); the frequency of STs for the collection is summarized in Fig. S3 in the supplemental material. A maximum-likelihood phylogenetic tree was reconstructed based on concatenated sequences of 7 MLST loci for these STs, together with the 38 reference S. argenteus STs from the MLST website (Fig. 1). The phylogenetic tree divided the Thai collection into two major branches containing 36 STs that were S. aureus and 4 STs that clustered with the S. argenteus reference sequences. Three groups were observed for all of the S. argenteus data, one of which contained all four STs (representing 10 isolates) from the Thai collection. Three of these four STs were identified previously (ST1223 reported from Cambodia, Australia, and French Guiana, ST2250 from the United Kingdom, and ST2198 from Australia and Germany) (2, 3, 4, 9). The fourth ST (ST2854) is novel. Based on this phylogenetic analysis, we concluded that 10/246 isolates (4.1%) from an unselected consecutive collection of S. aureus isolates from Thailand should be recategorized as S. argenteus. Our findings confirm that this species is endemic in Thailand and provides additional evidence to suggest that S. argenteus may be more widely distributed across Southeast Asia.
FIG 1.
Phylogenetic tree of 246 putative S. aureus isolates based on concatenated sequences of 7 MLST loci using the maximum-likelihood method. Blue dots represent STs identified for the 246 study isolates, and black dots represent 38 STs downloaded from the MLST website (http://www.mlst.net). The colored ring denotes known or proposed species for each ST (yellow, S. aureus; gray, S. argenteus).
S. argenteus isolates identified in Australia are predominantly resistant to methicillin, but those identified elsewhere are susceptible to methicillin; isolates tested to date are universally pvl negative (6–9, 14). All 10 Thai S. argenteus isolates were methicillin susceptible, mecA negative, and pvl negative. The frequencies of methicillin resistance and pvl positivity for the entire Thai collection are summarized in Fig. S3 in the supplemental material.
S. argenteus isolates characteristically lack staphyloxanthin, a carotenoid pigment responsible for the characteristic golden colony color (4). All 10 Thai S. argenteus grew on blood agar as white colonies, which is consistent with this. Staphyloxanthin confers resistance against oxidative stress and neutrophil killing (15), leading to the hypothesis that S. argenteus may be less virulent than S. aureus (4). This is supported by the findings of a study showing that an S. argenteus isolate (ST1850) was associated with lower virulence than S. aureus in murine sepsis and skin infection models (16). Transformation of this isolate with a plasmid carrying the gene encoding staphyloxanthin led to increased resistance to oxidative stress in vitro but no change in resistance to neutrophil killing or in vivo virulence. A study that defined the frequency of S. argenteus in three different clinical collections in northern Australia reported that this species was predominantly associated with skin and soft tissue infections but rarely with bacteremia (1% [3/220] of methicillin-susceptible S. aureus [MSSA] bacteremias and 0% [0/47] of MRSA bacteremias) (16).
We tested the hypothesis that S. argenteus isolates were not associated with severe disease in our Thai cohort. Clinical details and outcomes from S. argenteus infection are shown in Table 1. Most patients (8/10, 80%) had community-acquired infection, but two had health care-associated infection, indicating that S. argenteus may cause nosocomial infection. We noted that S. argenteus was strongly associated with skin and soft tissue infections or superficial abscesses (8/10 cases), which is consistent with previous reports. However, three patients with S. argenteus infection had bacteremia, and two patients had bone and joint infections, indicating the ability of S. argenteus to cause invasive disease (see Table S2 in the supplemental material). Two patients infected with S. argenteus died, one as a direct result of the infection. In general, disease characteristics and outcomes were not different between the patients infected with S. argenteus and those infected with MSSA. However, the median age of the patients infected with S. argenteus was higher than that of patients infected with MSSA (56.5 versus 38; P = 0.04), and patients infected with S. argenteus were more likely to have diabetes and renal disease than patients infected with MSSA (Table 1).
TABLE 1.
Clinical manifestations and outcomes for patients infected with MSSA or S. argenteus
| Clinical parameter | Data for patients witha: |
P valueb | |
|---|---|---|---|
| MSSA | S. argenteus | ||
| Total cases | 199 | 10 | |
| Age (yr) | 38 (14–59, 1–84) | 56.5 (41–66, 11–73) | 0.04 |
| Sex (male) | 118 (59) | 7 (70) | 0.74 |
| Blood culture-positive isolates | 56 (28) | 3 (30) | >0.99 |
| Place of acquisition | |||
| Community acquired | 172 (86) | 8 (80) | 0.63 |
| Health care associatedc | 27 (14) | 2 (20) | |
| Underlying medical conditiond | 65 (33) | 7 (70) | 0.03 |
| Cardiac diseasee | 12 (6) | 1 (10) | 0.48 |
| Diabetes mellitus | 29 (15) | 6 (60) | 0.002 |
| Renal diseasef | 15 (8) | 6 (60) | <0.001 |
| Immunosuppressiong | 25 (13) | 2 (20) | 0.62 |
| Lung diseaseh | 9 (5) | 1 (10) | 0.39 |
| Pattern of disease | |||
| Identified site, bacteremia only | 17 (9) | 2 (20) | 0.19 |
| 1 identified site | 155 (78) | 6 (60) | |
| >1 identified site | 27 (13) | 2 (20) | |
| Other identifiable sites of infection | |||
| Superficial abscess | 81 (41) | 4 (40) | 0.15 |
| Deep abscess | 45 (23) | 0 | |
| Other skin and soft tissue infections | 21 (11) | 4 (40) | |
| Bone and joint infections | 24 (12) | 2 (20) | |
| Prosthetic material infection | 13 (7) | 0 | |
| Respiratory infection | 14 (7) | 0 | |
| Endocarditis | 6 (3) | 0 | |
| Other infection | 7 (4) | 0 | |
| Outcome | |||
| Cured | 150 (75) | 7 (70) | 0.58 |
| Treatment failure | 11 (6) | 1 (10) | |
| Death due to S. aureus | 26 (13) | 1 (10) | |
| Death due to other causes | 12 (6) | 1 (10) | |
Data shown are number (%) or median (interquartile range, range).
P values were estimated using Fisher's exact test. Values for age were estimated using the Mann-Whitney test.
Health care-associated infections included nosocomial and nonnosocomial health care-associated infections (12).
Past medical history of any underlying chronic medical conditions reported by the patients and/or relatives or recorded in the medical notes.
“Cardiac disease” comprised congenital heart disease, valvular heart disease (including rheumatic heart disease), ischemic disease, or arrhythmias (including heart block requiring pacemaker).
“Renal disease” included end-stage renal failure with long-term dialysis and chronic renal failure (not on dialysis) due to diabetes mellitus, systematic lupus erythematous, multiple myeloma, glomerulonephritis, or an unknown etiology.
Immunosuppression included that from HIV, chemotherapy, untreated leukemia, radiotherapy, or immunosuppressive medications, including >30 mg/day of prednisolone for >1 week.
“Lung disease” comprised previously treated tuberculosis, previous empyema, lung cancer, long-term tracheostomy, chronic obstructive pulmonary disease, or asthma.
A limitation of this work is that the study cohort included only patients with S. aureus isolated from a normally sterile site. This is likely to have led to an underrepresentation of minor skin and soft tissue infections, which may have resulted in an underestimation of the true prevalence of S. argenteus. Our data demonstrate, however, that S. argenteus may be associated with serious morbidity, death, and nosocomial infection, particularly in patients with underlying diseases, such as diabetes mellitus and renal diseases.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Wellcome Trust (grant 089275/Z/09/Z).
We thank the medical and nursing staff at Sappasithiprasong Hospital and Mahidol-Oxford Tropical Medicine Research Unit for their support.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03049-14.
REFERENCES
- 1.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 38:1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruimy R, Armand-Lefevre L, Barbier F, Ruppe E, Cocojaru R, Mesli Y, Maiga A, Benkalfat M, Benchouk S, Hassaine H, Dufourcq JB, Nareth C, Sarthou JL, Andremont A, Feil EJ. 2009. Comparisons between geographically diverse samples of carried Staphylococcus aureus. J Bacteriol 191:5577–5583. doi: 10.1128/JB.00493-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ng JW, Holt DC, Lilliebridge RA, Stephens AJ, Huygens F, Tong SY, Currie BJ, Giffard PM. 2009. Phylogenetically distinct Staphylococcus aureus lineage prevalent among indigenous communities in northern Australia. J Clin Microbiol 47:2295–2300. doi: 10.1128/JCM.00122-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holt DC, Holden MT, Tong SY, Castillo-Ramirez S, Clarke L, Quail MA, Currie BJ, Parkhill J, Bentley SD, Feil EJ, Giffard PM. 2011. A very early-branching Staphylococcus aureus lineage lacking the carotenoid pigment staphyloxanthin. Genome Biol Evol 3:881–895. doi: 10.1093/gbe/evr078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tong SY, Schaumburg F, Ellington MJ, Corander J, Pichon B, Leendertz F, Bentley SD, Parkhill J, Holt DC, Peters G, Giffard PM. 30 September 2014. Novel staphylococcal species that form part of a Staphylococcus aureus related complex: the non-pigmented S. argenteus sp. nov. and the non-human primate associated S. schweitzeri sp. nov. Int J Syst Evol Microbiol doi: 10.1099/ijs.0.062752-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McDonald M, Dougall A, Holt D, Huygens F, Oppedisano F, Giffard PM, Inman-Bamber J, Stephens AJ, Towers R, Carapetis JR, Currie BJ. 2006. Use of a single-nucleotide polymorphism genotyping system to demonstrate the unique epidemiology of methicillin-resistant Staphylococcus aureus in remote aboriginal communities. J Clin Microbiol 44:3720–3727. doi: 10.1128/JCM.00836-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenney A, Holt D, Ritika R, Southwell P, Pravin S, Buadromo E, Carapetis J, Tong S, Steer A. 2014. The clinical and molecular epidemiology of Staphylococcus aureus infections in Fiji. BMC Infect Dis 14:160. doi: 10.1186/1471-2334-14-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monecke S, Stieber B, Roberts R, Akpaka PE, Slickers P, Ehricht R. 2014. Population structure of Staphylococcus aureus from Trinidad & Tobago. PLoS One 9:e89120. doi: 10.1371/journal.pone.0089120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruimy R, Angebault C, Djossou F, Dupont C, Epelboin L, Jarraud S, Lefevre LA, Bes M, Lixandru BE, Bertine M, El Miniai A, Renard M, Bettinger RM, Lescat M, Clermont O, Peroz G, Lina G, Tavakol M, Vandenesch F, van Belkum A, Rousset F, Andremont A. 2010. Are host genetics the predominant determinant of persistent nasal Staphylococcus aureus carriage in humans? J Infect Dis 202:924–934. doi: 10.1086/655901. [DOI] [PubMed] [Google Scholar]
- 10.Akobi B, Aboderin O, Sasaki T, Shittu A. 2012. Characterization of Staphylococcus aureus isolates from faecal samples of the straw-coloured fruit bat (Eidolon helvum) in Obafemi Awolowo University (OAU), Nigeria. BMC Microbiol 12:279. doi: 10.1186/1471-2180-12-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nickerson EK, Wuthiekanun V, Wongsuvan G, Limmathurosakul D, Srisamang P, Mahavanakul W, Thaipadungpanit J, Shah KR, Arayawichanont A, Amornchai P, Thanwisai A, Day NP, Peacock SJ. 2009. Factors predicting and reducing mortality in patients with invasive Staphylococcus aureus disease in a developing country. PLoS One 4:e6512. doi: 10.1371/journal.pone.0006512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feil EJ, Nickerson EK, Chantratita N, Wuthiekanun V, Srisomang P, Cousins R, Pan W, Zhang G, Xu B, Day NP, Peacock SJ. 2008. Rapid detection of the pandemic methicillin-resistant Staphylococcus aureus clone ST 239, a dominant strain in Asian hospitals. J Clin Microbiol 46:1520–1522. doi: 10.1128/JCM.02238-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 14.Tong SY, Lilliebridge RA, Bishop EJ, Cheng AC, Holt DC, McDonald MI, Giffard PM, Currie BJ, Boutlis CS. 2010. Clinical correlates of Panton-Valentine leukocidin (PVL), PVL isoforms, and clonal complex in the Staphylococcus aureus population of northern Australia. J Infect Dis 202:760–769. doi: 10.1086/655396. [DOI] [PubMed] [Google Scholar]
- 15.Liu GY, Essex A, Buchanan JT, Datta V, Hoffman HM, Bastian JF, Fierer J, Nizet V. 2005. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J Exp Med 202:209–215. doi: 10.1084/jem.20050846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tong SY, Sharma-Kuinkel BK, Thaden JT, Whitney AR, Yang SJ, Mishra NN, Rude T, Lilliebridge RA, Selim MA, Ahn SH, Holt DC, Giffard PM, Bayer AS, Deleo FR, Fowler VG Jr. 2013. Virulence of endemic nonpigmented northern Australian Staphylococcus aureus clone (clonal complex 75, S. argenteus) is not augmented by staphyloxanthin. J Infect Dis 208:520–527. doi: 10.1093/infdis/jit173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.