Abstract
The epidemiological and bacteriological investigations on four foodborne outbreaks caused by a new type of enterotoxin-producing Clostridium perfringens are described. C. perfringens isolated from patients of these outbreaks did not produce any known enterotoxin and did not carry the C. perfringens enterotoxin gene. However, the culture filtrates of these isolates induced the accumulation of fluid in rabbit ileal loop tests. The molecular weight of the new enterotoxin may be between 50,000 and 100,000, although the known C. perfringens enterotoxin is ca. 35,000. This new enterotoxin was heat labile, and its biological activities were inactivated by heating for 5 min at 60°C. The new enterotoxin was sensitive to pH values higher than 11.0 and protease treatment but was resistant to trypsin treatment. These results suggest that the new enterotoxin may be a protein. Although C. perfringens enterotoxin induced morphological changes in Vero cells, the changes induced by the new enterotoxin differed from those by the known C. perfringens enterotoxin. The new enterotoxin also induced morphological changes in L929 cells, whereas the known C. perfringens enterotoxin did not, because L929 cells lacked an appropriate enterotoxin receptor. Although C. perfringens enterotoxin is recognized as the only diarrheagenic toxin responsible for C. perfringens foodborne outbreaks, the results of the present study indicate that C. perfringens isolated from these four outbreaks produced a new type of enterotoxin.
INTRODUCTION
Clostridium perfringens gastroenteritis is characterized mainly by diarrhea and abdominal pain. C. perfringens enterotoxin (CPE) is considered to be the only virulence factor responsible for the gastrointestinal symptoms reported in C. perfringens type A foodborne outbreaks (1–4). The diarrheagenicity of CPE was clarified in oral administration experiments performed on volunteers and monkeys (5–8). Previous studies have examined the characteristics of CPE, methods to detect it, its production, the structures of its protein and gene, and the whole sequence of the cpe gene (9–11). Thus, C. perfringens isolated from foodborne outbreaks caused by C. perfringens has been confirmed to produce CPE (6, 12). On the other hand, non-CPE-producing C. perfringens is considered to be nondiarrheagenic and is a normal inhabitant in humans and animals.
In Japan, about 30 C. perfringens foodborne outbreaks occur every year. We encountered four unusual foodborne outbreaks from 1997 to 2010 in Japan. Although they might be caused by C. perfringens, C. perfringens isolates taken from patients did not produce CPE and also did not carry the cpe gene. Therefore, we suspected those isolates might produce novel enterotoxin. In the present study, we epidemiologically and bacteriologically investigate these four outbreaks.
MATERIALS AND METHODS
Samples. (i) Outbreak 1.
An outbreak occurred in October 1997 in Tokyo, Japan. The causal food was a boxed lunch. The fecal specimens of 29 patients, 4 healthy people who ate the same food as the patients, and 3 food handlers, as well as 9 foods, including 2 sets of leftover food and 13 swab samples from the cooking utensils of the responsible food facilities, were examined.
(ii) Outbreak 2.
An outbreak occurred in June 2003 in Tokyo. The fecal specimens of 4 patients and 12 food handlers as well as 26 foods, including 12 sets of leftover food that the patients had taken home from the restaurant and 4 sets of leftover food at the restaurant, were examined.
(iii) Outbreak 3.
An outbreak occurred in August 2009 in Osaka, Japan. Samples were tested at the Osaka City Institute of Public Health and Environmental Sciences, and 18 C. perfringens strains isolated from the outbreak were examined in our laboratory.
(iv) Outbreak 4.
An outbreak occurred in January 2010 in Tochigi, Japan. Samples were tested mainly at the Tochigi Prefectural Institute of Public Health and Environmental Science and our laboratory. Eleven fecal specimens from patients were tested in our laboratory, and 14 C. perfringens strains isolated at Tochigi Prefectural Institute of Public Health were also examined in our laboratory.
In addition, a total of 20 strains isolated from another 4 outbreaks, including 10 CPE-positive and 10 CPE-negative strains, were used for reference. Those strains were confirmed to be negative for new enterotoxin productivity.
Isolation and identification of Clostridium perfringens.
Samples were examined for enteric pathogens, including C. perfringens, Salmonella, Campylobacter, Shigella, Yersinia, enteropathogenic Escherichia coli, pathogenic Vibrio, Aeromonas, Plesiomonas, and Norovirus. To isolate C. perfringens, CW agar plates (Nissui Pharmaceutical, Tokyo, Japan) supplemented with kanamycin and egg yolk (5.0 g heart extract, 10.0 g proteose peptone, 10.0 g peptone, 5.0 g sodium chloride, 10.0 g lactose, 0.05 g phenol red, 20.0 g agar, 0.2 g kanamycin sulfate, 20 g egg yolk, and 1 liter distilled water, pH 7.2 ± 0.1) were used for direct plating, and fluid thioglycolate medium (5 g yeast, 15 g tryptone, 5 g glucose, 0.5 g l-cystine, 0.5 g sodium thioglycolate, 2.5 g sodium chloride, 0.001 g resazurin, 0.75 g agar, and 1 liter distilled water, pH 7.1 ± 0.1 [TGC medium; Eiken Chemical, Tokyo, Japan]) was used as the enrichment broth. Fecal specimens or food homogenates were directly streaked onto CW agar plates and were cultivated under anaerobic conditions at 37°C overnight. In the enrichment culture, fecal specimens, which had been inoculated into TGC medium and heated at 100°C for 10 min, were cultured at 37°C overnight. Food and swab samples were not heated before culturing. The cultured TGC medium was spread onto CW agar plates and cultured overnight under anaerobic conditions.
The identification of isolates was performed using biochemical tests such as anaerobic growth, spore formation, the neutralization of lecithinase activity with anti-alpha-toxin serum, and gelation hydrolysis. In addition, these isolates were examined using the PCR method with 2 kinds of species-specific primers for the 16S rRNA genes of C. perfringens (13, 14).
To count the number of C. perfringens CFU in samples, fecal specimens or food samples were serially diluted 10 times with sterile saline, spread onto CW agar plates, and then cultured at 37°C overnight under anaerobic conditions. Typical C. perfringens colonies were counted, and serotyping tests were performed.
Detection of Clostridium perfringens enterotoxin.
Isolates were cultured in modified Duncan-Strong medium (15) (m-DS; 4.0 g yeast, 15.0 g peptone, 4.0 g soluble starch, 1.0 g sodium thioglycolate, 5.0 g sodium dihydrogen phosphate dihydrate, 900 ml distilled water, pH 7.6 ± 0.1, with the addition of a 1/10 volume of 5% sodium bicarbonate just before use) at 37°C for 20 h (16), and spore formation was observed in the culture by phase-contrast microscopy. The supernatant of the cultures was examined for CPE with a reverse passive latex agglutination test using the CPE toxin detection kit (PET-RPLA; Denka Seiken, Niigata, Japan) in accordance with the instructions of the manufacturer. The detection of CPE in the patients' feces within 3 days of the onset was also performed using the same detection kit.
The presence of the cpe gene was examined by a PCR assay using the following three types of primers: commercial primers CPE-1 and -2 (TaKaRa Bio, Shiga, Japan), the primers 5′-GGAGATGGTTGGATATTAGG-3′ and 5′-GGACCAGCAGTTGTAGATA-3′, reported by Meer and Songer (17), and the primers 5′-ATGCTTAGTAACAAT TTAAATCC-3 and 5′-TTTTTGAAATAATATTGAATAAG-3′, reported by Ishimura et al. (18). To extract the DNA of C. perfringens isolates, colonies on the CW agar plate were suspended in sterilized water. Equal volumes of 50 mM NaOH solution were added to the colony suspensions, and the mixtures were then heated at 100°C for 10 min. After being heated, the suspensions were neutralized with equal volumes of Tris-HCl (pH 7.5) and were then centrifuged at 12,000 rpm for 10 min. The supernatants were used as the template DNA in the PCR assay.
Epidemiological analysis of isolates.
The serotypes of the isolates were examined with commercial antisera containing 17 serotypes (Hobbs1 to Hobbs17; Denka Seiken, Niigata, Japan) and a Japanese serological typing system (TW1 to TW74) prepared at our laboratory (19, 20). These isolates were also analyzed by pulsed-field gel electrophoresis (PFGE). Chromosomal DNA was prepared in gel blocks and digested with three types of restriction enzymes, such as NruI, SmaI, and SacII. After digestion, electrophoresis was performed using CHEF-Mapper systems (Bio-Rad Laboratories, California, USA).
Toxin typing tests of isolates.
Neutralization tests were performed as follows. The strains isolated from the feces of patients were inoculated into chopped meat broth with 1% fructose and cultured at 37°C for 8 h under anaerobic conditions. The supernatants of these cultures were used in toxin typing tests. The supernatant was mixed with each antitoxic serum of alpha-, beta-, epsilon-, and iota-toxin (Wellcome Research Laboratory, Beckenham, United Kingdom) and incubated at 37°C for 30 min. After being incubated, 0.5 ml of each mixture was injected into the caudal veins of three mice. The survival of mice after the injection was observed for 3 days (21). The animal experiments performed in this study were approved by the Institutional Review Board at the Tokyo Metropolitan Institute of Public Health.
Toxin genes were detected by PCR (17, 22–24). C. perfringens strain W4232, which was isolated from patients from a typical C. perfringens type A foodborne outbreak, was used as an alpha-toxin gene-positive strain. C. perfringens type C KZMT 49 (NCTC 3227), C. perfringens type D KZMT 82 (L9), and C. perfringens type E KZ 316 (E6 Leeds) were kindly provided by Takayoshi Yamagishi in the Department of Laboratory Science, School of Health Science, Faculty of Medicine, Kanazawa University, and were used as beta-, epsilon-, and iota-toxin gene-positive strains, respectively.
Rabbit ileal loop tests.
The isolates from four outbreaks were cultured in m-DS medium at 37°C for 20 h. The filtered supernatants of the cultures were used as samples. A total of 0.5 ml from each of the samples was injected into the ileal loops of Japanese White rabbits weighing 1.6 to 2.2 kg and under anesthesia with pentobarbital and medetomidine hydrochloride. The loops were examined by inducing the fluid accumulation approximately 15 h after the injection (25). We used a purified cholera toxin (Wako Pure Chemical Industries, Osaka, Japan) as the positive control and m-DS medium as the negative control.
Cytotoxicity assay.
Cytotoxicity was examined using Vero cells and L929 cells. Cells suspended in Eagle's modified essential medium (MEM) containing 5% fetal calf serum (FCS) and kanamycin (Nikken Bio Medical Laboratory, Kyoto, Japan) were distributed onto microplates (48- or 96-well plates) at a density of approximately 10 (4) cells/ml and cultured at 37°C for 20 h in a 5% CO2 incubator. Three kinds of media, including m-DS, cooked meat broth (BD Difco, New Jersey, USA) supplemented with 0.5% soluble starch and 0.5% glucose, and TGC II broth (5 g yeast, 15 g tryptone, 5 g glucose, 0.5 g l-cystine, 0.5 g sodium thioglycolate, 2.5 g sodium chloride, and 1 liter distilled water, pH 7.1 ± 0.1; Eiken Chemical, Tokyo, Japan), were used to prepare the samples. The isolates were cultured in these media for approximately 20 h, and the formation of spores in the media was assessed under a phase-contrast microscope. After the formation of spores in these media was confirmed, the supernatants of the cultures were filtered and used as sample solutions. Each sample solution was subjected to a 2-fold serial dilution with sterile phosphate buffer containing 0.85% NaCl.
Vero and L929 cell monolayers grown on microplates were washed with Dulbecco's phosphate buffer (Nissui Pharmaceutical, Tokyo, Japan), 1% FCS-Eagle MEM was then added at 0.5 ml/well (48-well plates) or 0.1 ml/well (96-well plates), and a 1/10 volume of the sample solution was added. The plates were then incubated at 37°C for 24 h, and cytotoxicity was observed under an inverted microscope. The crude antitoxin serum was prepared as follows. C. perfringens strain w5052, which was isolated from a patient in outbreak 1, was cultured in m-DS medium and was prepared as an immunogen with the ammonium sulfate precipitation of the culture filtrate and dialysis. The immunogen and Freund's complete adjuvant (BD Difco, New Jersey, USA) were mixed and intramuscularly injected into Japanese White rabbits. Blood was collected after the evaluated antibody titer was confirmed, and serum was separated. The neutralization of cytotoxic effects was examined using the filtrates of m-DS cultures that had reacted with the crude antitoxin serum at 37°C for 30 min.
Examinations of other related toxins.
Toxin A of Clostridium difficile and the toxin A and B genes of C. difficile were examined using the commercial kit (Shionogi Pharmaceutical, Osaka, Japan) and PCR, respectively (26–28). The verotoxin (VT) of enterohemorrhagic Escherichia coli and heat-labile toxin (LT) of enterotoxigenic E. coli were examined using the commercial kit (Denka Seiken, Niigata, Japan), and VT, LT, and heat-stable toxin (ST) genes were detected by PCR (29–31).
Characterization of the diarrheagenic toxin.
After the isolates had been cultured in m-DS medium at 37°C for approximately 20 h under anaerobic conditions, the cultures were centrifuged at 12,000 rpm for 10 min. These filtered supernatants were used as samples for the diarrheagenic characterization of the toxin. Samples were filtered to estimate the molecular weights using ultrafiltration membranes (Millipore, Tokyo, Japan). In the heat stability test, samples were heated at 60°C for 5 and 10 min and at 100°C for 10 min and then rapidly cooled. In the test against the acid and alkaline treatments, samples were adjusted to pH 4.0 with 0.1 M hydrochloric acid and pH 11.0 with ammonia solution and were incubated at 37°C for 2 h. After the reaction, the samples were neutralized with ammonia solution or diluted hydrochloric acid. Samples in which 0.1% (wt/vol) trypsin or pronase (guaranteed grade; Wako Pure Chemical Industries, Osaka, Japan) had been added were kept at 37°C for 2 h and immediately cooled on ice. The biochemical activities of these treated samples were examined using the accumulation of fluid in rabbit ileal loop test and cytotoxicity assay.
RESULTS
Epidemiological investigations.
An outline of the outbreaks is shown in Table 1. In outbreak 1 on 12 October 1997, 39 of 160 boys (4 teams) who had participated in baseball games had gastrointestinal symptoms approximately 15 h after eating a boxed lunch. The clinical symptoms of the patients included diarrhea (97.4%), abdominal pain (43.5%), nausea (7.9%), and vomiting (5.1%). No patients were hospitalized. The mean incubation period was 15.5 h. The foods in the boxed lunch that had been eaten by the patients included boiled and seasoned beef with stringy konnyaku (devil's tongue), rolled egg, fried lotus, grilled salmon, broiled eel, lettuce leaves, orange, and a Japanese rice ball, called an “onigiri.” An epidemiological investigation revealed that the incriminating foods may have been cooked on the previous day and left at room temperature overnight. The average attack rate of 4 teams was 24.3%, with each attack rate being 63.3%, 34.3%, 19.5%, and 5.9%. The attack rate was higher in team members who ate the boxed lunch which was prepared earlier and was left for a longer time after being cooked. The boiled and seasoned beef with stringy konnyaku was suspected as the incriminating food, but the chi-square test result was not significant, which did not allow for any definitive conclusions.
TABLE 1.
Outline of four outbreaks caused by new enterotoxin-producing Clostridium perfringens
| Outbreak | Date of occurrence | Place of occurrence | No. of patients/no. of people at risk (%) | Preparing facility | Incriminated food | % of patients with diarrhea, abdominal pain (main clinical symptoms) | Incubation period (mean) in h | Serotype of causal bacteria |
|---|---|---|---|---|---|---|---|---|
| 1 | 12 October 1997 | Tokyo | 39/160 (24.3) | Restaurant | Boxed luncha | 97.4, 43.5 | 4–28 (15.5) | TW27 |
| 2 | 16 June 2003 | Tokyo | 9/11 (81.8) | Restaurant | Stewed lamb | 100.0, 33.3 | 7–20 (10.0) | TW27 |
| 3 | 15 August 2009 | Osaka | 84/420 (20.0) | Restaurant | Roast beef | 100.0, 74.7 | 2.5–26 (12.5) | TW27 |
| 4 | 2 January 2010 | Tochigi | 79/171(46.2) | Restaurant | Roast beef | 98.7, 67.1 | 3–24 (9.5) | TW21 |
Included rolled egg, fried lotus, boiled and seasoned beef with stringy konnyaku (devil's tongue), grilled salmon, broiled eel, lettuce leaves, orange, and a Japanese rice ball (Onigiri), prepared on 11 October 1997.
Outbreak 2 occurred on 16 June 2003. Nine of 11 people had diarrhea (97.4%), abdominal pain (33.3%), and malaise (22.2%) approximately 10 h after eating stewed lamb. The stewed lamb had been cooked on the previous day and was kept at room temperature overnight.
In outbreak 3 on 15 August 2009, 84 of 420 people had diarrhea (100.0%), abdominal pain (74.7%), tenesmus (9.5%), nausea (7.1%), malaise(6.0%), and fever (6.0%) approximately 12.5 h after eating roast beef from a smorgasbord for lunch. The chi-square test revealed that roast beef was suspected of being the incriminating food.
In outbreak 4 on 2 January 2010, 79 of 171 people had diarrhea (98.7%) and abdominal pain (67.1%) approximately 9.5 h after eating roast beef for dinner.
The clinical symptoms of patients in these four outbreaks were similar to those from typical C. perfringens outbreaks.
Clostridium perfringens isolated from samples.
The results of the examination are shown in Table 2. Although results for the pathogenic bacteria for gastroenteritis were negative in fecal and foods samples, C. perfringens was isolated from 14 out of 29 fecal samples collected from patients, and serotype TW27 was isolated from 11 out of 14 patients, or 42 (80.8%) out of 52 isolates, in outbreak 1. Among healthy people, C. perfringens was isolated from 3 out of 4 people, and serotype TW27 was isolated only from 1 person or only 3 (25.0%) out of 12 isolates. The numbers of C. perfringens serotype TW27 CFU/g in the feces of 5 patients within 3 days of the onset were 2.8 × 106, 8.3 × 105, 4.9 × 104, 2.3 × 104, and 5.0 × 103.
TABLE 2.
Clostridium perfringens isolated from samples from three outbreaks
| Outbreak | Sample type | Sample source | Sampling day(s) from the onset | No. of samples examined | No. of C. perfringens-positive samples | No. of patients with serotype TW27-or TW21-positive samplesa,b | CFU/g of serotype TW27 or TW21 in samplesa |
|---|---|---|---|---|---|---|---|
| 1 | Fecal specimen | Patients | Day 3 | 29 | 14 | 11 (42/52) | 5.0 × 103–2.8 ×106 |
| Healthy persons who had eaten incriminated food | Day 3 | 4 | 3 | 1 (3/12) | 3.0 ×103 | ||
| Food handlers | Day 3 | 3 | 0 | 0 | |||
| 2 | Fecal specimen | Patients | Days 0–2 | 4 | 4 | 4 (44/44) | 8.3 × 103–2.6 × 106 |
| Food handlers | Days 1 and 2 | 12 | 7 | 1 (6/13) | 5.0 ×103 | ||
| Food | Food at the patient's home | 12 | 7 | 5 (44/64) | 1.1 × 107,c <3.0 × 102d | ||
| 4 | Fecal specimen | Patients in Tokyo | Days 3–5 | 11 | 11 | 10 (79/107) | 6.3 × 103 –2.1 × 105 |
TW27 in outbreaks 1 and 2 and TW21 in outbreak 4.
Numbers in parentheses are the no. of TW27-positive isolates/no. of isolates examined.
Stewed lamb.
Foods included smoked salmon salad, marinated seafood, yellow fin tuna fish in olive oil, and rillettes.
In outbreak 2, C. perfringens was isolated from the feces of all 4 patients examined, and all were serotype TW27. The numbers of C. perfringens serotype TW27 CFU/g in the feces of 4 patients within 3 days of the onset were 2.6 × 106, 6.6 × 105, 1.8 × 105, and 8.3 × 103. Stewed lamb was suspected as the incriminating food. The number of C. perfringens serotype TW27 CFU/g in the stewed lamb taken to the patients' homes as leftovers was 1.1 × 107. C. perfringens serotype TW27 was isolated from 4 other foods at the patients' homes, and the number of CFU/g for this serotype was lower than 3 × 102.
In outbreak 3, C. perfringens was isolated from 29 out of 54 fecal samples collected from patients and 6 out of 12 fecal samples collected from the food handlers at laboratory in the Osaka City Institute of Public Health and Environmental Sciences. Eighteen strains, including 13 strains from the patients and 5 strains from the food handlers, were serotype TW27.
In outbreak 4, bacteriological examination was performed at both laboratories in Tochigi Prefecture and Tokyo. The results of the examination conducted at our laboratory revealed that C. perfringens serotype TW21 was isolated from 10 out of 11 patients, and 79 (73.8%) out of 107 isolates from these 10 patients were serotype TW21. The numbers of serotype TW21 CFU/g isolated from 5 patients within 3 to 5 days of the onset were 2.1 × 105, 8.3 × 104, 4.9 × 104, 2.3 ×104, and 6.3 × 103. In addition, a total of 11 strains isolated from 6 patients, 4 food handlers, and 1 roast beef were received from the Tochigi Prefectural Institute of Public Health and Environmental Science, and they were also confirmed as serotype TW21. The roast beef was from the same lot as the roast beef that was served, and the number of C. perfringens CFU/g in the roast beef was 9.0 × 102.
The isolated C. perfringens strains were subterminal spore-forming anaerobic bacilli, positive for lecithinase, which is neutralized by anti-alpha-toxin, and showed stormy fermentation (Table 3). These characteristics indicated that the isolates were typical C. perfringens. The isolates were also confirmed to be C. perfringens by PCR by using 2 kinds of primers.
TABLE 3.
Characteristics of new enterotoxin-producing Clostridium perfringens isolates from four outbreaks
| Characteristic | Resultk by outbreak/strain (no. of strains examined) |
|||||
|---|---|---|---|---|---|---|
| Outbreak 1 (5) | Outbreak 2 (5) | Outbreak 3 (5) | Outbreak 4 (5) | CPE-positivea strains (10) | CPE-negativeb strains (10) | |
| C. perfringens 16S rRNAc | + | + | + | + | + | + |
| C. perfringens enterotoxin (CPE) | ||||||
| RPLA | − | − | − | − | + | − |
| PCR (TaKaRa)d | − | − | − | − | + | − |
| PCR (Meer and Songer)e | − | − | − | − | + | − |
| PCR (Ishimura et al.)f | − | − | − | − | + | − |
| Other diarrheagenic toxinsg | − | − | − | − | − | − |
| Rabbit ileal loop tests | + | + | + | + | NT | − |
| Cytotoxic effects to Vero cells | +h | +h | +h | +h | + | − |
| Cytotoxic effects to L929 cells | + | + | + | + | − | − |
| Lecithinase production | + | + | + | + | + | + |
| Neutralization by anti-alpha-toxin | + | + | + | + | + | + |
| Alpha-toxin: mouse method and PCRi | + | + | + | + | + | + |
| Beta-toxin: mouse method and PCRj | − | − | − | − | − | − |
| Epsilon-toxin: mouse method and PCRj | − | − | − | − | − | − |
| Iota-toxin: mouse method and PCRj | − | − | − | − | − | − |
| Growth on aerobic blood agar | − | − | − | − | − | − |
| Motility | − | − | − | − | − | − |
| Spores (subterminal) | + | + | + | + | + | + |
| Gelatin hydrolysis | ||||||
| 2% | + | + | + | + | + | + |
| 10% | + | + | + | + | + | + |
| Milk digestion (stormy fermentation) | + | + | + | + | + | + |
| Indole production | − | − | − | − | − | − |
| Hemolysis | ||||||
| Horse | + | + | + | + | + | + |
| Sheep | + | + | + | + | + | + |
| Rabbit | + | + | + | + | + | + |
| Fermentation of: | ||||||
| Maltose | + | + | + | + | + | + |
| Lactose | + | + | + | + | + | + |
| Sucrose | + | + | + | + | + | + |
| Salicin | − | − | − | − | − | − |
| Mannitol | − | − | − | − | − | − |
| Raffinose | + | + | + | + | + | + |
| Cellobiose | − | − | − | − | − | − |
| Inulin | − | − | − | − | − | − |
| Inositol | + | + | + | + | v | v |
| Trehalose | − | − | − | − | v | v |
CPE-producing C. perfringens.
Non-CPE-producing C. perfringens.
Assayed by commercial PCR kit (TaKaRa).
Assayed by reference 17.
Assayed by reference 18.
C. difficile toxin A and B and E. coli VT, LT, and ST were tested by PCR and RPLA or ELISA, respectively.
The morphological changes were different from the change by the known CPE-producing isolates.
NT, not tested; v, valiable reaction; +, positive; −, negative.
Toxin type of the isolates.
C. perfringens serotype TW27, which was isolated from the first three outbreaks, and serotype TW21, which was isolated from outbreak 4, were identified as C. perfringens type A, because the lethal activity was neutralized with anti-alpha-toxin serum in a neutralization test using mice, and the alpha-toxin gene, but not the beta-, epsilon-, or iota-toxin gene, was detected from the isolates by PCR (Table 3).
Epidemiological analysis of the isolates with pulsed-field gel electrophoresis.
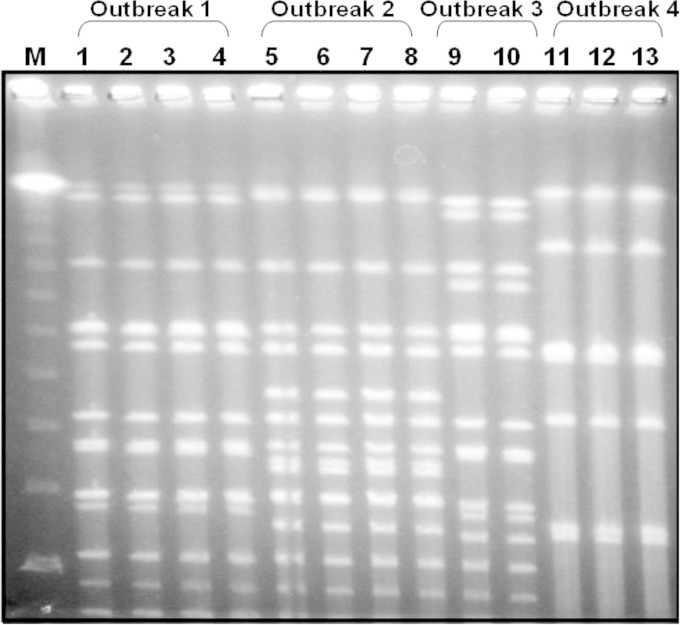
The PFGE patterns of the isolates from an outbreak were indistinguishable. In outbreaks 2 and 4, the PFGE patterns of the isolates from the patients and incriminating foods were also indistinguishable from each other. The isolates of the four outbreaks showed different PFGE patterns from each other (Fig. 1).
FIG 1.
Pulsed-field gel electrophoresis patterns of the isolates from four outbreaks. M, λ ladder; lanes 1 to 4, patients from outbreak 1; lanes 5 to 7, patients from outbreak 2; lane 8, stewed lamb from outbreak 2; lanes 9 and 10, patients from outbreak 3; lanes 11 and 12, patients from outbreak 4; lane 13, roast beef from outbreak 4. Restriction enzyme, NruI, pulse time of 0.47 to 30.82 s, 6.0 V/cm at 14°C for 20.18 h.
Productivity of the known enterotoxins.
The direct detection of CPE in the patients' feces in outbreaks 1, 2, and 4 was unsuccessful. C. perfringens serotype TW27, which was isolated from outbreaks 1, 2, and 3, and serotype TW21, which was isolated from outbreak 4, did not produce CPE by RPLA. In these C. perfringens isolates, the cpe gene was not detected by PCR using three kinds of primer sets (Table 3). RPLA and PCR did not detect the production of toxins A and B of Clostridium difficile or LT, ST, and VT of diarrheagenic Escherichia coli (Table 3).
Enteropathogenicity of the isolates by rabbit ileal loop tests.
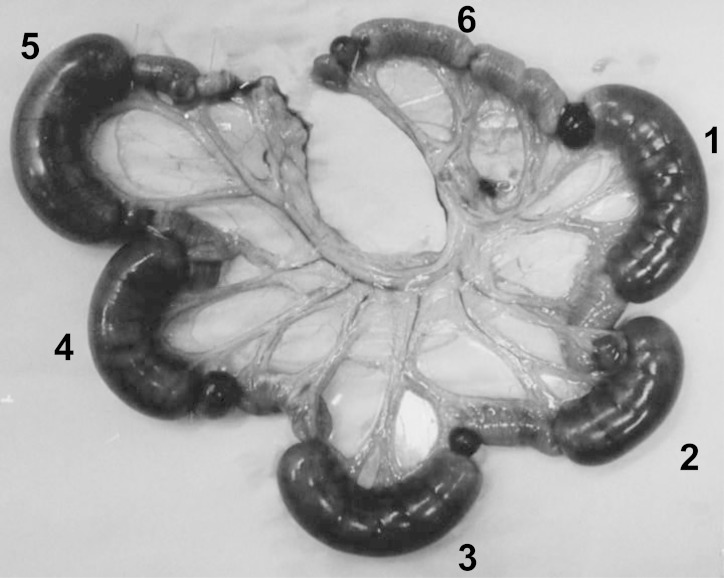
In order to clarify whether isolated C. perfringens produced diarrheagenic toxins, rabbit ileal loop tests were performed using culture filtrates of isolates. As shown in Fig. 2, fluid accumulation (FA) in the rabbit ileal loop was clearly observed in the filtered supernatants of the 4 strains isolated from patients as well as the cholera toxin as a positive control. These FA activities were detected in C. perfringens isolates from the four outbreaks. The FA ratios (ml/cm) of the isolates from outbreaks 1, 2, 3, and 4 were 1.3 to 2.0, 1.2 to 1.8, 0.9 to 1.6, and 1.1 to 1.6, respectively. FA was even observed in culture filtrates diluted 10 times with m-DS medium. The FA ratios of the original culture filtrate, filtrate that had been diluted 2 times, and filtrate diluted 10 times were 1.40, 0.97, and 0.54, respectively. The activity of FA was not neutralized by anti-alpha-toxin serum.
FIG 2.
The results of rabbit ileum loop tests of the isolates from outbreak 1. Each sample of 0.5 ml was injected into each ileum loop. Loop 1, culture filtrate (strain W5052); loop 2, culture filtrate (strain W5043); loop 3, culture filtrate (strain W5040); loop 4, culture filtrate (strain W5036); loop 5, cholera toxin at 1 μg (positive control); loop 6, m-DS medium (negative control).
Results of the cytotoxicity assay.
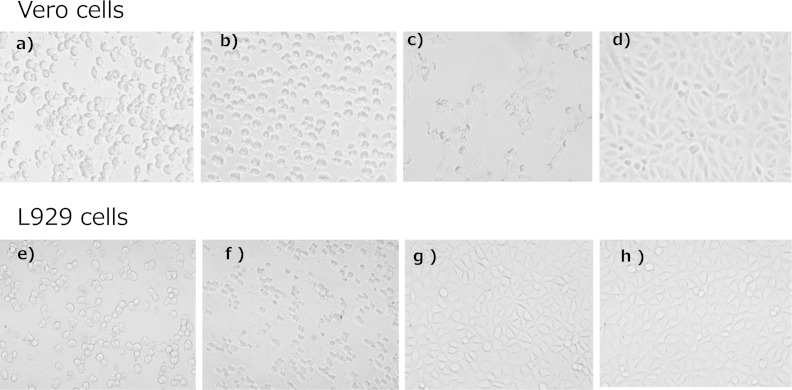
In Vero cells, the cytotoxic effects of isolates from the four outbreaks differed from the effects of the known CPE. Moreover, CPE was not cytotoxic to L929 cells, whereas the cultures of the isolates from four outbreaks were cytotoxic to L929 cells (Fig. 3, Table 4). Although cytotoxic effects were neutralized in Vero cells by the antitoxin serum, the cytotoxic effects induced by the known CPE were not neutralized by the antitoxin serum (Table 5).
FIG 3.
Comparison of cytotoxicity to Vero cells and L929 cells. The cells were treated with each filtrate of m-DS cultures. (a, e) Isolate from a patient from outbreak 1; (b, f) isolate from a patient from outbreak 2; (c, g) enterotoxin-producing C. perfringens; (d, h) non-enterotoxin-producing C. perfringens.
TABLE 4.
The spore formation and the cytotoxic effects of the isolates in each medium
| Culture medium | Cell type | Resultd by outbreak/strain (no. of strains examined) |
|||||
|---|---|---|---|---|---|---|---|
| Outbreak 1 (5) | Outbreak 2 (5) | Outbreak 3 (5) | Outbreak 4 (5) | CPE-positivea strains (10) | CPE-negativeb strains (10) | ||
| Modified DS medium | |||||||
| Spore forming | +++ | +++ | +++ | +++ | +∼++ | +∼++ | |
| Cytotoxic effects | Vero | +c | +c | +c | +c | + | − |
| Cytotoxic effects | L929 | + | + | + | + | − | − |
| Cooked meat broth | |||||||
| Spore forming | − | − | − | − | − | − | |
| Cytotoxic effects | Vero | − | − | − | − | − | − |
| Cytotoxic effects | L929 | − | − | − | − | − | − |
| TGCII broth | |||||||
| Spore forming | − | − | − | − | − | − | |
| Cytotoxic effects | Vero | − | − | − | − | − | − |
| Cytotoxic effects | L929 | − | − | − | − | − | − |
CPE-producing C. perfringens.
Non-CPE-producing C. perfringens.
Morphological changes were different from the change by the known enterotoxin-producing isolates.
+, positive; −, negative; +++, spore-forming rate approximating >20%; +~++, spore-forming rate approximating <20%.
TABLE 5.
Characteristics of the diarrheagenic toxin produced by Clostridium perfringens isolated from four outbreaks
| Characteristic | Resultf by test and outbreak/strain (no. of strains examined) |
||||||
|---|---|---|---|---|---|---|---|
| Rabbit ileal loop test (FA)a for outbreak 1 (3) | Cytotoxicity (Vero cells) |
||||||
| Outbreak 1 (5) | Outbreak 2 (5) | Outbreak 3 (5) | Outbreak 4 (5) | CPE-positiveb strains (10) | CPE-negativec strains (10) | ||
| Crude toxin | + (1.3–2.0) | + | + | + | + | + | − |
| Mol wt | |||||||
| <30,000 | − | − | − | − | − | − | − |
| 30,000–50,000 | − | − | − | − | − | + | − |
| 50,000–100,000 | + (1.3–1.6) | +d | +d | +d | +d | − | − |
| >100,000 | − | − | − | − | − | − | − |
| Treatment with: | |||||||
| Heating at: | |||||||
| 60°C for 5 min | − | − | − | − | − | − | − |
| 60°C for 10 min | − | − | − | − | − | − | − |
| 100°C for 10 min | − | − | − | − | − | − | − |
| Acid (pH 4.0) | ± (0–0.6) | NT | NT | NT | NT | NT | NT |
| Alkali (pH 11.0) | − | NT | NT | NT | NT | NT | NT |
| Trypsin | + (1.0–1.9) | NT | NT | NT | NT | NT | NT |
| Pronase | − | NT | NT | NT | NT | NT | NT |
| Neutralization with antitoxine | NT | + | + | + | + | − | NT |
FA, fluid accumulation ratio. The strains from outbreaks 2, 3, and 4 are all positive on the rabbit ileal loop tests. FA ratios of the strain from outbreaks 2, 3, and 4 are 1.2 to 1.8, 0.9 to 1.6, and 1.1 to 1.6, respectively.
CPE-producing C. perfringens.
Non-CPE-producing C. perfringens.
The morphological changes were different from the change by the known CPE-producing isolates.
Antitoxin against the crude toxin prepared from the isolate on outbreak 1.
NT, not tested; +, positive; −, negative.
Sporulation and cytotoxic effects of isolates were observed in their culture of m-DS medium but not in cooked meat and TGC II broths (Table 4).
Characteristics of the diarrheagenic toxin.
The characteristics of the crude diarrheagenic toxin produced by the isolates from four outbreaks are summarized in Table 5. The crude toxin had a molecular weight of 50,000 to 100,000 and was assumed to be a thermolabile toxin that had been inactivated by heating at 60°C for 5 min, and also by acid, alkaline, and pronase treatments, but was not inactivated by the trypsin treatment.
DISCUSSION
CPE-producing C. perfringens is known as a major cause of foodborne outbreaks. Between 1997 and 2010, we identified four foodborne outbreaks of gastroenteritis caused by a new type of C. perfringens in Japan. In these outbreaks, the symptoms of the patients included mainly diarrhea and abdominal pain and appeared after median incubation periods of 9.5 to 15.5 h. These clinical features and the results of epidemiological investigations suggested that four foodborne outbreaks might be caused by C. perfringens. The results of bacteriological examinations corresponded to the bacteriological criteria used to confirm a diagnosis of C. perfringens food poisoning, including the presence of either (i) 105 C. perfringens organisms/g of stool from two or more ill people or (ii) 105 C. perfringens organisms/g of epidemiologically implicated food (32). More than 106 C. perfringens organisms/g of fecal specimen from patients was isolated in these outbreaks, and the same serotypes, such as serotype TW27 from outbreaks 1, 2, and 3 and serotype TW21 from outbreak 4, were isolated in each outbreak. These results indicated that the outbreaks may have been caused by C. perfringens.
An important point for the diagnosis of C. perfringens food poisoning is that CPE were detected in feces of patients within 3 days of the onset in many food poisoning outbreaks (16). However, CPE was not detected in all patients' feces within 3 days of the onset. The absence of CPE in patients' feces suggests that these outbreaks differed from typical C. perfringens food poisoning. The isolates formed spores at a high rate in m-DS medium, whereas CPE was not detected in the cultures. In addition, PCR using 3 types of primer sets revealed that cpe genes were not carried in the isolates. These results demonstrated that isolates from these four outbreaks did not produce the known CPE. We also investigated other known diarrheagenic factors. The isolates did not produce any diarrheagenic toxins: toxins A and B of C. difficile, LT and ST of enterotoxigenic E. coli, or VT of enterohemorrhagic E. coli. However, the culture filtrates of isolates from the four outbreaks induced the fluid accumulation in rabbit ileal loops. This result suggested that these isolates produced a new type of diarrheagenic enterotoxin.
McClane and McDonel (33, 34) reported that CPE induced morphological changes in Vero cells; therefore, we here examined the effects of isolates from the four outbreaks to Vero cells. The culture filtrates of the isolates from both outbreaks caused morphological changes in Vero cells; however, the morphological changes observed differed from those induced by the known CPE. Although the known CPE has been reported to induce no morphological changes in L929 cells (35), the culture filtrates of these isolates did. These results strongly suggested that the pathogenic factor produced by the isolates was a new type of enterotoxin that differed from the known CPE. The cytotoxic effects induced by the new type of enterotoxin were detected in the culture supernatant during sporulation, which is similar to the known CPE (36, 37).
The known CPE is a thermolabile protein that consists of 319 amino acids with a molecular weight of 35,517 and isoelectric point of 4.3 (2, 4, 38, 39). The molecular weight of the novel enterotoxin was estimated to be 50,000 to 100,000, which differed from that of the known CPE. The new enterotoxin was inactivated by heating at 60°C for 5 min and alkaline and pronase treatments but not by a trypsin treatment. Those characteristics suggest that the novel enterotoxin is also a protein, like CPE.
No other enteropathogenic factor, except for CPE, has previously been confirmed in C. perfringens, unlike E. coli enterotoxins. However, the results of the present study strongly suggest that the isolates from the four outbreaks produced a new type of diarrheagenic enterotoxin. We identified the novel diarrheagenic enterotoxin in the present study as C. perfringens iota-like enterotoxin (CPILE), a binary toxin similar to Clostridium spiroforme iota-like toxin (presented at the 87th Annual Meeting of Japanese Society for Bacteriology in 2013 and the 8th International Conference on the Molecular Biology and Pathogenesis of the Clostridia-ClostPath 2013, Palm-Cove, Australia, 2013), and isolates from the four outbreaks harbored and expressed the cpiles gene. Patients with diarrhea, but in whom any causative bacteria are not detected, should be tested for CPILE-producing C. perfringens, and further investigations on CPILE-producing C. perfringens and CPILE are necessary.
ACKNOWLEDGMENTS
We thank Hiromi Nakamura of the Osaka City Institute of Public Health and Environmental Sciences for the isolates from outbreak 3 and Hideki Uchida of Tochigi Prefectural Institute of Public Health and Environmental Science for the isolates from outbreak 4.
We are grateful to the members of the Health and Safety Division, Bureau of Social Welfare and Public Health, Tokyo Metropolitan Government, Adachi Public Health Center, Chiyoda Public Health Center, Osaka City, and Tochigi Prefecture for the helpful epidemiological investigation.
REFERENCES
- 1.Hauschild AHW, Nillo L, Dorward WJ. 1970. Response of ligated intestinal loops in lambs to an enteropathogenic factor of Clostridium perfringens type A. Can J Microbiol 16:339–343. [DOI] [PubMed] [Google Scholar]
- 2.Hauschild AHW, Hilsheimer R. 1971. Purification and characteristics of the enterotoxin of Clostridium perfringens type A. Can J Microbiol 17:1425–1433. [DOI] [PubMed] [Google Scholar]
- 3.Stark RL, Duncan CL. 1971. Biological characteristics of Clostridium perfringens type A enterotoxin. Infect Immun 4:89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stark RL, Duncan CL. 1972. Purification and biochemical properties of Clostridium perfringens type A enterotoxin. Infect Immun 6:662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duncan CL, Strong DH. 1971. Clostridium perfringens type A food poisoning. I. Response of the rabbit ileum as an indication of enteropathogenicity of strains of Clostridium perfringens on monkeys. Infect Immun 3:167–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strong DH, Dunkan CL, Perna G. 1971. Clostridium perfringens type A food poisoning. II. Response of the rabbit ileum as an indication of enteropathogenicity of strains of Clostridium perfringens in human being. Infect Immun 3:171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skjelkvåle R, Uemura T. 1977. Experimental diarrhea in human volunteers following oral administration of Clostridium perfringens enterotoxin. J Appl Bacteriol 43:281–286. doi: 10.1111/j.1365-2672.1977.tb00752.x. [DOI] [PubMed] [Google Scholar]
- 8.Hauschild AHW, Walcroft MJ, Campbell W. 1971. Emesis and diarrhea induced by enterotoxin of Clostridium perfringens type A in monkeys. Can J Microbiol 17:1141–1143. doi: 10.1139/m71-180. [DOI] [PubMed] [Google Scholar]
- 9.Van Damme-Jongsten M, Wernars K, Notermans S. 1989. Cloning and sequencing of the Clostridium perfringens enterotoxin gene. Antonie Van Leeuwenhoek 56:181–190. doi: 10.1007/BF00399981. [DOI] [PubMed] [Google Scholar]
- 10.Iwanejko LA, Routledge MN, Stewart GSAB. 1989. Cloning in Escherichia coli of the enterotoxin gene from Clostridium perfringens type A. J Gen Microbiol 135:903–909. [DOI] [PubMed] [Google Scholar]
- 11.Czeczulin JR, Hannna PC, McClane BA. 1993. Cloning, nucleotide sequencing, and expression of Clostridium perfringens enterotoxin gene in Escherichia coli. Infect Immun 61:3429–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haushild AHW, Nillo L, Dorward WJ. 1971. The role of enterotoxin in Clostridium perfringens type A enteritis. Can J Microbiol 17:987–991. doi: 10.1139/m71-156. [DOI] [PubMed] [Google Scholar]
- 13.Wang RF, Cao WW, Franklin W, Cambpbell W, Cerniglia CEA. 1994. 16S rDNA-based PCR method for rapid and specific detection of Clostridium perfringens in food. Mol Cell Probes 8:131–137. doi: 10.1006/mcpr.1994.1018. [DOI] [PubMed] [Google Scholar]
- 14.Kikuchi E, Miyamoto Y, Narushima S, Itoh K. 2002. Design of species-specific primers to identify 13 species of Clostridium harbored in human intestinal tracts. Microbiol Immunol 46:353–358. doi: 10.1111/j.1348-0421.2002.tb02706.x. [DOI] [PubMed] [Google Scholar]
- 15.Ohtani Y, Ujiiye A. 1987. Sporulation and enterotoxin production of Clostridium perfringens in a modified DS medium. Food Hyg Saf Sci 28:281–285. doi: 10.3358/shokueishi.28.281. [DOI] [Google Scholar]
- 16.Itoh T, Inaba M, Saito K, Sakai S, Uemura T, Sakaguchi G. 1979. Assay for enterotoxin in fecal specimens of Clostridium perfringens food poisoning. Kansenshogaku Zasshi 53:409–416. doi: 10.11150/kansenshogakuzasshi1970.53.409. [DOI] [PubMed] [Google Scholar]
- 17.Meer RR, Songer JG. 1997. Multiplex polymerase chain reaction assay for genotyping Clostridium perfringens. Am J Vet Res 587:702–705. [PubMed] [Google Scholar]
- 18.Ishimura K, Ito F, Kodama M, Kayashima T, Kasama Y, Nakano K, Yamaoka H, Ogino T. 1997. Availability of PCR-based detection and fingerprinting method for molecular epidemiologic analysis of enterotoxigenic Clostridium perfringens. Jpn J Food Microbiol 14:35–42. doi: 10.5803/jsfm.14.35. [DOI] [Google Scholar]
- 19.Itoh T. 1972. Incidence of heat resistant Clostridium perfringens in feces of healthy subjects, serotyping of isolates, and food poisoning caused by new serotype organism. Ann Rep Tokyo Metr Res Lab PH 24:347–339. [Google Scholar]
- 20.Monma C, Yanagawa Y, Kusunoki J, Kai A, Shingaki M, Obata H, Takahashi M, Itoh T, Ohta K, Kudoh H. 1995. Bacterial investigation of two outbreaks caused by Clostridium perfringens of new serotype. Ann Rep Tokyo Metr Res Lab PH 46:8–11. [Google Scholar]
- 21.Nishida S, Murakami M, Yanagishi T. 1962. Chopped meat broth as a medium for the production of toxins by Clostridia. 1. Production of toxins by Clostridium welchii. Jap J Microbiol 6:33–40. [Google Scholar]
- 22.Uzal FA, Plumb JJ, Blackall LL, Kelly WR. 1997. PCR detection of Clostridium perfringens different toxins in faces of goats. Lett Appl Microbiol 25:339–344. doi: 10.1046/j.1472-765X.1997.00247.x. [DOI] [PubMed] [Google Scholar]
- 23.Yoo HS, Lee SU, Park KY, Park YH. 1997. Molecular typing and epidemiological survey of prevalence of Clostridium perfringens types by multiplex PCR. J Clin Microbiol 35:228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamagishi T, Sugitani K, Tanishima K, Nakamura S. 1997. Polymerase chain reaction test for differentiation of five toxin types of Clostridium perfringens. Microbiol Immunol 41:295–299. doi: 10.1111/j.1348-0421.1997.tb01204.x. [DOI] [PubMed] [Google Scholar]
- 25.De SN. 1959. Enterotoxicity of bacteria-free culture-filtrate of Vibrio cholerae. Nature 183:1533–1534. doi: 10.1038/1831533a0. [DOI] [PubMed] [Google Scholar]
- 26.Wren BW, Heard SR, Al-Saleh AI, Tabaqchali S. 1993. Characterisation of Clostridium difficile strains by polymerase chain reaction with toxin A- and B-specific primers. J Med Microbiol 38:109–113. doi: 10.1099/00222615-38-2-109. [DOI] [PubMed] [Google Scholar]
- 27.Kato N, Ou CY, Kato H, Bartley SL, Brown VK, Dowell VR Jr, Ueno K. 1991. Identification of toxigenic Clostridium difficile by the polymerase chain reaction. J Clin Microbiol 291:33–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuhl SJ, Tang YJ, Navarro L, Gumerlock PH, Silva J Jr. 1993. Diagnosis and monitoring of Clostridium difficile infections with the polymerase chain reaction. Clin Infect Dis 16(Suppl 4):S234–S238. doi: 10.1093/clinids/16.Supplement_4.S234. [DOI] [PubMed] [Google Scholar]
- 29.Karch H, Meyer T. 1989. Single printer pair for amplifying segments of district Shiga-like-toxin genes by polymerase chain reaction. J Clin Microbiol 27:2751–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abe A, Obata H, Matsushita S, Yamada S, Kudoh Y, Bangtrakulnonth A, Ratchtrachenchat OA, Danbara H. 1992. A sensitive method for the detection of enterotoxigenic Escherichia coli by the polymerase chain reaction using multiple primer pairs. Zentralbl Bakteriol 277:170–178. doi: 10.1016/S0934-8840(11)80610-9. [DOI] [PubMed] [Google Scholar]
- 31.Victer T, Du Toit R, Van Zyl Bester AJ, Van Helden PD. 1991. Improved method for the routine identification of toxigenic Escherichia coli by DNA amplification of a conserved region of the heat-labile toxin a subunit. J Clin Microbiol 29:158–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Labbe R. 1989. Clostridium perfringens, p 191–234. In Doyle MP. (ed), Foodborne bacterial pathogens. Marcel Dekker, New York, NY. [Google Scholar]
- 33.McDonel JL, McClane BA. 1979. Binding versus biological activity of Clostridium perfringens enterotoxin in Vero cells. Biochem Biophys Res Commun 87:497–504. doi: 10.1016/0006-291X(79)91823-0. [DOI] [PubMed] [Google Scholar]
- 34.McClane BA, McDonel JL. 1979. The effects of Clostridium perfringens enterotoxin on morphology, viability, and macromolecular synthesis in Vero cells. J Cell Physiol 99:191–200. doi: 10.1002/jcp.1040990205. [DOI] [PubMed] [Google Scholar]
- 35.Katahira J, Onoue N, Horiguchi Y, Matsuda M, Sugimoto N. 1997. Molecular cloning and functional characterization of the receptor for Clostridium perfringens enterotoxin. J Cell Biol 136:1239–1247. doi: 10.1083/jcb.136.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duncan CL, Strong DH, Sebald M, Sebald M. 1972. Sporulation and enterotoxin production by mutants of Clostridium perfringens. J Bacteriol 110:378–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duncan CL. 1973. Time of enterotoxin formation, release during sporulation of Clostridium perfringens type A. J Bacteriol 13:932–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uemura T, Sakaguchi G, Itoh T, Okazawa K, Sakai S. 1975. Experimental diarrhea in cynomolgus monkeys by oral administration with Clostridium perfringens type A available cellar enterotoxin. J Med Sci Biol 28:165–177. doi: 10.7883/yoken1952.28.165. [DOI] [PubMed] [Google Scholar]
- 39.Sakaguchi G, Uemura T, Riemann HP. 1973. Simplified method for purification of Clostridium perfringens type A enterotoxin. Appl Microbiol 26:762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]