Abstract
Clostridium difficile PCR ribotype 033 (RT033) is found in the gastrointestinal tracts of production animals and, occasionally, humans. The illumigene C. difficile assay (Meridian Bioscience, Inc.) failed to detect any of 52 C. difficile RT033 isolates, while all strains signaled positive for the binary toxin genes but were reported as negative for C. difficile by the Xpert C. difficile/Epi assay (Cepheid).
TEXT
Clostridium difficile is the most common cause of infection associated with health care in the United States (1). The diagnosis of C. difficile infection (CDI) remains a vexing problem in hospitals and laboratories. Most current methods of laboratory diagnosis involve detecting tcdA and/or tcdB, two genes located in the C. difficile pathogenicity locus (PaLoc) that encode the two main virulence factors of the bacterium, the large clostridial toxins A (TcdA) and B (TcdB). The detection of a third, binary toxin (CDT), an actin-specific ADP-ribosyltransferase also produced by some strains of C. difficile and not encoded in the PaLoc, has been largely overlooked (2). This has been attributed to the difficulty of detecting CDT by nonmolecular means and the fact that it is usually produced by strains that also produce toxins A and B (A+ B+) (3). An exception is the real-time PCR-based Xpert C. difficile/Epi system (Cepheid, Sunnyvale, CA). This U.S. Food and Drug Administration (FDA)-approved assay detects CDT gene sequences but only as a way of identifying putative ribotype 027 (RT027)/NAP1 C. difficile strains in fecal specimens. The assay also detects tcdB and the single base pair deletion at nucleotide position 117 of tcdC found in epidemic fluoroquinolone-resistant RT027/NAP1 strains (http://www.cepheid.com/us/cepheid-solutions/clinical-ivd-tests/healthcare-associated-infections/xpert-c-difficile-epi). The illumigene C. difficile assay (Meridian Bioscience, Inc.) is also approved by the FDA, but it instead targets a highly conserved region at the 5′ end of tcdA and uses loop-mediated isothermal DNA amplification (LAMP) technology (http://www.meridianbioscience.com/diagnostic-products/c-difficile/illumigene-molecular-diagnostic-system/illumigene-c-difficile.aspx).
C. difficile RT033 belongs to toxinotype XIa/b and has a truncated PaLoc but an intact CDT locus (4, 5). It is an emerging strain of clinical and veterinary importance that has been detected in cattle, veal calves, piglets, horses, and soil from various geographical locations worldwide (6–8). Its exact role in human CDI has yet to be determined; however, enterotoxic effects have been observed in rabbit ileal loop tests with toxin A-negative, toxin B-negative, CDT-positive (A− B− CDT+) strains of C. difficile, indicating that CDT may be a virulence factor in these strains (9). In addition, CDT has been linked to an increase in the prevalence of community-acquired CDI (CA-CDI), a high rate of recurrent CDI, and the need for hospital admission due to reinfection (10, 11). A− B− CDT+ strains of C. difficile, such as RT033, are not detected by most current diagnostic methods and hence may be missed by clinical laboratories.
Meridian Bioscience Inc. claims that the tcdA target sequence of their illumigene C. difficile assay allows for detection of the PaLoc of toxigenic C. difficile regardless of the expression phenotype and that the assay was designed to detect all known A+ B+ and A− B+ toxinotypes (http://www.meridianbioscience.com/diagnostic-products/c-difficile/illumigene-molecular-diagnostic-system/illumigene-c-difficile.aspx). This means that PaLoc-positive and phenotypically A− B− CDT+ C. difficile strains such as RT033 may be detected by the illumigene C. difficile assay and signal as CDT positive by the Xpert C. difficile/Epi assay. In this study, we tested C. difficile RT033 isolates for their ability to signal in the illumigene C. difficile and Xpert C. difficile/Epi assays.
RT033 isolates from human (n = 15), porcine (n = 16), and bovine (n = 21) sources were used in this study (Table 1). The animal isolates were collected as part of Australian livestock prevalence studies (6, 8). The clinical isolates were mostly from pathology laboratories around Australia. Frederick Barbut (National Reference Laboratory for C. difficile, France) kindly contributed 6 of the clinical RT033 isolates (12). For comparison, 11 non-RT033 isolates of various origins and 5 reference strains were also tested (Table 1).
TABLE 1.
Comparison of Meridian illumigene C. difficile and Cepheid Xpert C. difficile/Epi assays for detection of C. difficile RT033a
| Ribotype (strain) | No. tested | Toxin profile | Toxinotype(s) | Source (no. of isolates) | Xpert detection of: |
Xpert results | illumigene result | ||
|---|---|---|---|---|---|---|---|---|---|
| tcdB | CDT | tcdC | |||||||
| Test strains | |||||||||
| RT033 | 52 | A− B− CDT+ | Xla, Xlb | Human (15) | NEG | POS | NEG | Toxigenic C. difficile NEG; presumptive RT027 NEG | NEG |
| Bovine (21) | |||||||||
| Porcine (16) | |||||||||
| RT017 | 4 | A− B+ CDT− | VIII | Human | POS | NEG | NEG | Toxigenic C. difficile POS; presumptive RT027 NEG | POS |
| RT237 | 4 | A− B+ CDT+ | XXXI | Human (2) | POS | POS | NEG | Toxigenic C. difficile POS; presumptive RT027 NEG | NEG |
| Porcine (2) | |||||||||
| RT239 | 1 | A− B− CDT+ | NA | Human | NEG | POS | NEG | Toxigenic C. difficile NEG; presumptive RT027 NEG | NEG |
| RT280 | 1 | A− B+ CDT+ | XXX | Human | POS | POS | NEG | Toxigenic C. difficile POS; presumptive RT027 NEG | NEG |
| RT281 | 1 | A− B+ CDT+ | XXX | Human | POS | POS | NEG | Toxigenic C. difficile POS; presumptive RT027 NEG | NEG |
| Reference strains | |||||||||
| RT087 (VPI 10463) | 1 | A+ B+ CDT− | 0 | Human | POS | NEG | NEG | Toxigenic C. difficile POS; presumptive RT027 NEG | POS |
| RT033 (IS 58) | 1 | A− B− CDT+ | Xla | Human | NEG | POS | NEG | Toxigenic C. difficile NEG; presumptive RT027 NEG | NEG |
| RT012 (CD630) | 1 | A+ B+ CDT− | 0 | Human | POS | NEG | NEG | Toxigenic C. difficile POS; presumptive RT027 NEG | POS |
| RT027 (R 20291) | 1 | A+ B+ CDT+ | III | Human | POS | POS | POS | Toxigenic C. difficile POS; presumptive RT027 POS | POS |
| RT010 (CD062) | 1 | A− B− CDT− | NA | Human | NEG | NEG | NEG | Toxigenic C. difficile NEG; presumptive RT027 NEG | NEG |
POS, positive; NEG, negative; NA, not applicable (no pathogenicity locus present).
The identities of the C. difficile isolates were confirmed by their distinct colony morphology, yellow-green fluorescence under 360-nm UV light, and horse-dung odor when grown on blood agar for 48 h under anaerobic conditions (A35 anaerobic workstation; Don Whitley Scientific, Shipley, West Yorkshire, UK). Toxin gene profiling and PCR ribotyping of all isolates were performed using previously described methods (6). The illumigene and Xpert tests were performed according to the manufacturers' instructions, except that 0.5 McFarland standard saline suspensions of C. difficile from 48-h blood agar cultures were used instead of the recommended stool samples.
All 52 human, bovine, and porcine C. difficile RT033 isolates failed to signal with the illumigene assay, as did the two A− B− reference strains (RT033 and RT010) and four non-RT033 test ribotypes (the only other A− B− ribotype [RT239] and 3 of 4 A− B+ ribotypes). The four RT017 strains were the only A− B+ isolates tested that signaled in the illumigene assay. The Xpert assay detected the CDT analyte in all 52 RT033 strains, all CDT+ non-RT033 test strains, and each of the CDT+ reference strains. Only the tcdB-positive strains were identified as toxigenic C. difficile, and only the RT027 reference strain was identified as a presumptive RT027/NAP1 strain (Table 1).
RT033 characteristically contains a truncated PaLoc region that retains only the putative translocation domain fragment (A2) and parts of the repetitive domain fragment (A3) of the tcdA gene (Fig. 1); hence, neither of the large clostridial toxins is produced (4, 5). The illumigene C. difficile assay did not detect the remnant fragments of the tcdA gene in any of the RT033 isolates tested, consistent with a deletion in the 5′ end region (4, 5). The illumigene C. difficile assay was designed to target a “conserved” region at the 5′ end of tcdA present in all known A+ B+ and A− B+ toxinotypes (13). This was supported by Couturier et al., who reported detection of the tcdA target sequence in A− B+ strains belonging to toxinotypes VIII and X using the illumigene C. difficile assay (13), and in the present study for RT017 (toxinotype VIII) strains which have a truncated tcdA gene retaining only the A1 fragment of the gene (4). Other A− B+ ribotypes tested (RT237, RT280, and RT281) that were known to contain a truncated tcdA region (14, 15) were not detected by the illumigene C. difficile assay. Therefore, in addition to PaLoc-negative C. difficile, the target region of the illumigene C. difficile assay is not conserved in the PaLocs of RT033, RT237, RT280, and RT281 strains.
FIG 1.
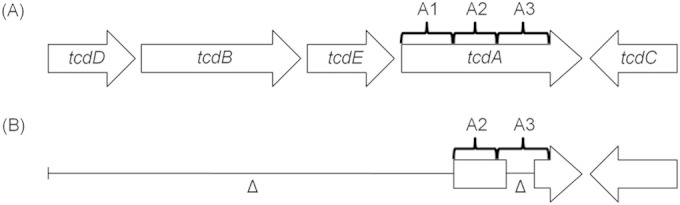
Comparison of a complete PaLoc with the virulence genes (tcdA, tcdB) and accessory genes (tcdD, tcdE, tcdC) (A) and the truncated RT033 PaLoc containing parts of tcdA (fragments A2 and A3) and tcdC (B). tcdD, positive regulator gene; tcdE, holin-like-protein-encoding gene; tcdC, negative regulator; tcdA, toxin A gene; tcdB, toxin B gene. (Based on data from reference 5.)
The Xpert C. difficile/Epi assay successfully detected CDT gene sequences in all RT033 isolates as expected. However, it was necessary to view the raw data obtained by the assay to identify a CDT-positive result because the assay does not recognize a test as positive for toxigenic C. difficile based on this result alone. The CDT target is only used to give a presumptive positive report of C. difficile RT027/NAP1 strains, and this is only when tcdB and the tcdC Δ117 deletion are detected also. Furthermore, the assay does not distinguish between the two CDT genes cdtA and cdtB (2). Variability and truncations within the CDT locus have been reported; however, the 5′ end region of cdtA and the 3′ end region of cdtB are usually highly conserved (2).
A limitation of this study was the use of pure cultures of C. difficile. Each assay has been validated for stool specimens only; however, this should not have reduced the sensitivities or specificities of the assays. As a common ribotype in veal calves and piglets in some parts of the world, the presence of RT033 isolates should be a concern for the veterinary industry (6, 16). While the role of C. difficile in idiopathic diarrhea in calves is still unclear, C. difficile is the most frequent cause of piglet neonatal diarrhea in the United States (15). The RT033 strain has also been isolated from symptomatic human patients, suggesting possible zoonotic transmission (12). However, our study highlights a problem when attempting to undertake surveillance for RT033 C. difficile strains using current molecular methods, making it difficult to assess the true scale and significance of such strains in CDI in humans and production animals. The Xpert C. difficile/Epi assay detects CDT gene sequences and hence may play a role in surveillance programs. There are other commercial diagnostic PCR kits, such as the EasyScreen C. difficile Reflex kit and the Verigene C. difficile test, capable of detecting CDT gene sequences, but they were not evaluated in this study. Ultimately, the development of a multiplex PCR assay that targets all four C. difficile toxin genes (tcdA, tcdB, cdtA, cdtB) is essential for maximizing C. difficile detection in clinical laboratories.
ACKNOWLEDGMENTS
We thank Cepheid Australia and Meridian (Bioline Australia) for the provision of materials for the study.
REFERENCES
- 1.Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Naddle J, Ray SM, Thompson DL, Wilson LE, Fridkin SK. 2014. Multistate point-prevalence survey of health care-associated infections. N Engl J Med 370:1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stare BG, Delmée M, Rupnik M. 2007. Variant forms of the binary toxin CDT locus and tcdC gene in Clostridium difficile strains. J Med Microbiol 56:329–335. doi: 10.1099/jmm.0.46931-0. [DOI] [PubMed] [Google Scholar]
- 3.Geric B, Johnson S, Gerding DN, Grabnar M, Rupnik M. 2003. Frequency of binary toxin genes among Clostridium difficile strains that do not produce large clostridial toxins. J Clin Microbiol 41:5227–5232. doi: 10.1128/JCM.41.11.5227-5232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rupnik M. 2008. Heterogeneity of large clostridial toxins: importance of Clostridium difficile toxinotypes. FEMS Microbiol Rev 32:541–555. doi: 10.1111/j.1574-6976.2008.00110.x. [DOI] [PubMed] [Google Scholar]
- 5.Rupnik M, Brazier JS, Duerden BI, Grabnar M, Stubbs SL. 2001. Comparison of toxinotyping and PCR ribotyping of Clostridium difficile strains and description of novel toxinotypes. Microbiology 147:439–447. [DOI] [PubMed] [Google Scholar]
- 6.Knight DR, Thean S, Putsathit P, Fenwick S, Riley TV. 2013. Cross-sectional study reveals high prevalence of Clostridium difficile non-PCR ribotype 078 strains in Australian veal calves at slaughter. Appl Environ Microbiol 79:2630–2635. doi: 10.1128/AEM.03951-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janezic S, Zidaric V, Pardon B, Indra A, Kokotovic B, Blanco JL, Seyboldt C, Diaz CR, Poxton IR, Perreten V, Drigo I, Jiraskova A, Ocepek M, Weese JS, Songer JG, Wilcox MH, Rupnik M. 2014. International Clostridium difficile animal strain collection and large diversity of animal associated strains. BMC Microbiol 14:173. doi: 10.1186/1471-2180-14-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knight DR, Squire MM, Riley TV. 2015. Nationwide surveillance study of Clostridium difficile in Australian neonatal pigs shows high prevalence and heterogeneity of PCR ribotypes. Appl Environ Microbiol 81:119–123. doi: 10.1128/AEM.03032-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geric B, Carman RJ, Rupnik M, Genheimer CW, Sambol SP, Lyerly DM, Gerding DN, Johnson S. 2006. Binary toxin-producing, large clostridial toxin-negative Clostridium difficile strains are enterotoxic but do not cause disease in hamsters. J Infect Dis 193:1143–1140. doi: 10.1086/501368. [DOI] [PubMed] [Google Scholar]
- 10.Stewart DB, Berg A, Hegarty J. 2013. Predicting recurrence of C. difficile colitis using bacterial virulence factors: binary toxin is the key. J Gastrointest Surg 17:118–124. doi: 10.1007/s11605-012-2056-6. [DOI] [PubMed] [Google Scholar]
- 11.Barbut F, Decré D, Lalande V, Burghoffer B, Noussair L, Gigandon A, Espinasse F, Raskine L, Robert J, Mangeol A, Branger C, Petit JC. 2005. Clinical features of Clostridium difficile-associated diarrhea due to binary toxin (actin-specific ADP-ribosyltransferase)-producing strains. J Med Microbiol 54:181–185. doi: 10.1099/jmm.0.45804-0. [DOI] [PubMed] [Google Scholar]
- 12.Eckert C, Emirian A, Le Monnier A, Cathala L, De Montclos H, Goret J, Berger P, Petit A, De Chevigny A, Jean-Pierre H, Nebbad B, Camiade S, Meckenstock R, Lalande V, Marchandin H, Barbut F. 2015. Prevalence and pathogenicity of binary toxin-positive Clostridium difficile strains that do not produce toxins A and B. New Microbes New Infect 3:12–17. doi: 10.1016/j.nmni.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Couturier B, Schlaberg R, Konzk C, Nicholes J, Law C, She RC. 2013. tcdA as a diagnostic target in a loop-mediated amplification assay for detecting toxigenic Clostridium difficile. J Clin Lab Anal 27:171–176. doi: 10.1002/jcla.21577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott B, Squire MM, Thean S, Chang BJ, Brazier JS, Rupnik M, Riley TV. 2011. New types of toxin A-negative, toxin B-positive strains among clinical isolates of Clostridium difficile in Australia. J Med Microbiol 60:1108–1111. doi: 10.1099/jmm.0.031062-0. [DOI] [PubMed] [Google Scholar]
- 15.Squire MM, Carter GP, Mackin KE, Chakravorty A, Norén T, Elliott B, Lyras D, Riley TV. 2013. Novel molecular type of Clostridium difficile in neonatal pigs, Western Australia. Emerg Infect Dis 19:790–792. doi: 10.3201/eid1905.121062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneeberg A, Neubauer H, Schmoock G, Grossmann E, Seyboldt C. 2013. Presence of Clostridium difficile PCR ribotype clusters related to 033, 078 and 045 in diarrhoeic calves in Germany. J Med Microbiol 62:1190–1198. doi: 10.1099/jmm.0.056473-0. [DOI] [PubMed] [Google Scholar]


