Abstract
gyrB is used to improve the identification of the Nocardia species N. brasiliensis, N. higoensis, N. ignorata, N. otitidiscaviarum, N. paucivorans, N. pneumoniae, N. puris, N. takedensis, N. veterana, and N. vinacea, but it does not improve the identification of another 12 Nocardia studied species. gyrB provides typing and phylogenetic markers for N. carnea, N. transvalensis, N. brasiliensis, and N. otitidiscaviarum.
INTRODUCTION
Nocardia species are soilborne aerobic Gram-positive bacilli that can cause severe cutaneous, pulmonary, and central nervous system infections (1). 16S rRNA analysis has been the gold standard in the identification of these species, but their low mutation rates make closely related species difficult to distinguish.
gyrB has been used on its own in phylogenetic studies of Nocardia (2, 3) and in multilocus sequence typing pattern strategies (4) and has allowed new species to be described (5, 6). The phylogenetic relationships between very similar species can be difficult to establish when using 16S rRNA-based trees (7). However, knowledge of gyrB sequences may help in this respect. Certainly, gyrB shows remarkable variation across Nocardia species and has been used in the identification, typing, and phylogenetic examination of populations of the more common Nocardia species in Spain (i.e., those involved in 67% of clinical cases: N. abscessus, N. cyriacigeorgica, N. farcinica, and N. nova) (3).
Here, we report an extension of the latter study (3), taking into account a collection of 75 strains belonging to 22 species and clustered into two groups: a commonly reported group (responsible for 13.0% of clinical cases of nocardiosis: N. brasiliensis [n = 10 strains], N. carnea [n = 10], N. otitidiscaviarum [n = 10], and N. transvalensis [n = 10]) and an unusually reported group, including 35 strains belonging to 18 species (responsible for <4.0% of clinical cases) (Table 1). The identifying and typing capacities of gyrB, a 16S target, and a 606-bp 16S fragment were compared.
TABLE 1.
Characteristics of the 75 selected strains belonging to 22 Nocardia speciesa
| Nocardia sp. (no. of strains; no. of provinces) | Clinical origin(s) | Strain agreement between 16S and gyrB (no. of strains/total no. of strains) |
|---|---|---|
| Commonly reported group (40 isolates) | ||
| N. brasiliensis (10; 8) | 4 sputum, 2 cutaneous abscess, 4 wound | 10/10 |
| N. carnea (10; 5) | 7 sputum, 1 gastric juice, 1 BAL fluid,b 1 perianal exudate | 3/10 |
| N. otitidiscaviarum (10; 8) | 7 sputum, 2 pleural liquid, 1 BAL fluid | 10/10 |
| N. transvalensis (10; 8) | 6 sputum, 1 cornea, 1 wound, 1 BAL fluid, 1 cutaneous abscess | 0/10 |
| Unusually reported species (35 isolates) | ||
| N. arthritidis (1; 1) | 1 BAL fluid | 1/1 |
| N. asteroides (2; 2) | 2 sputum | 1/2 |
| N. beijingensis (4; 3) | 3 sputum, 1 BAL fluid | 2/4 |
| N. elegans (1; 1) | 1 sputum | 0/1 |
| N. exalbida (1; 1) | 1 pulmonary puncture | 0/1 |
| N. flavorosea (1; 1) | 1 sputum | 0/1 |
| N. higoensis (1; 1) | 1 sputum | 0/1 |
| N. ignorata (4; 2) | 2 sputum, 1 wound, 1 gastric juice | 4/4 |
| N. jiangxiensis (1; 1) | 1 sputum | 0/1 |
| N. paucivorans (2; 2) | 1 sputum, 1 BASc | 2/2 |
| N. pneumoniae (1; 1) | 1 BAS | 1/1 |
| N. puris (2; 2) | 2 sputum | 2/2 |
| N. rhamnosiphila cluster (4; 3) | 3 sputum, 1 BAS | 1/4 |
| N. takedensis (3; 2) | 3 sputum | 3/3 |
| N. testacea (3; 1) | 3 sputum | 2/3 |
| N. veterana (1; 1) | 1 sputum | 1/1 |
| N. vinacea (1; 1) | 1 nodule | 1/1 |
| N. wallacei (2; 2) | 2 sputum | 0/2 |
Nocardia species were classified according to their full 16S sequences.
BAL, bronchoalveolar lavage.
BAS, bronchoaspirate.
Isolates were grown, their DNA was extracted, sequencing was performed, and phylogenetic trees were constructed as previously described (3). Isolates that were ≥99.0% (for the 606-bp 16S fragment and full 16S gene sequence [8]) and ≥93.5% (for gyrB [2]) similar to those with sequences in the GenBank database were deemed to be of the same species.
Partial and full 16S sequence analyses matched most of the time, but 5.5% and 8.5% of the strains from commonly reported groups and unusually reported species, respectively, showed discrepancies between 606-bp 16S fragment and full 16S gene sequence analyses because discrimination grows when the length increases. In any case, full 16S gene sequences were selected for classification.
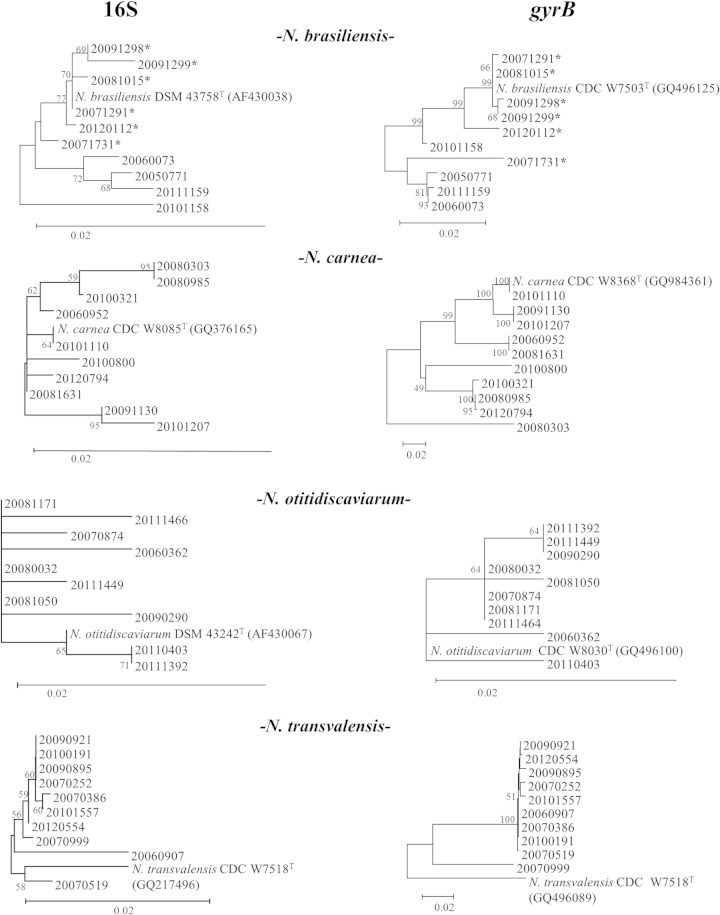
Within the commonly reported group, the identification of all N. brasiliensis and N. otitidiscaviarum strains by all three methods was fully concordant. However, discrepancies were seen in the identification of 70% of the N. carnea strains and all N. transvalensis strains. Their relationships, based on the 16S and gyrB results, are shown in their corresponding phylogenetic trees (Fig. 1 and Table 2).
FIG 1.
16S- and gyrB-based phylogenetic trees for N. brasiliensis, N. carnea, N. transvalensis, and N. otitidiscaviarum (neighbor-joining method). Each bootstrap value is expressed as a percentage of 1,000 replications. Bar, 0.02 substitutions per nucleotide position; *, strain from a cutaneous sample.
TABLE 2.
Diversities of the N. brasiliensis, N. carnea, N. transvalensis, and N. otitidiscaviarum strains as determined by 16S, 606-bp 16S fragment, and gyrB analyses
| Species (no. of strains) | Gene or protein (bp or aa)a | Haplotype no. (HGDI, S2, SD)b | No. of SNPs or amino acid changes (divergence rate)c | No. of SNPs per strain (range [mean, mode]) |
|---|---|---|---|---|
| N. brasiliensis (10) | 606-bp 16S rRNA (571) | 9 (0.978, 0.00292, 0.054) | 15 (0.0–1.8)d | 1–10 (4.5, 5)d |
| 16S (1,182) | 10 (1.000, 0.00200, 0.045) | 25 (0.2–1.5)d | 0–1 (0.9, 1)d | |
| gyrB (756) | 10 (1.000, 0.00200, 0.045) | 51 (0.1–5.8)e | 0–33 (14.4, 1)e | |
| GyrB (252) | 5 (0.500) | 19e | 0–18 (5.7, 6)e | |
| N. carnea (10) | 606-bp 16S rRNA (560) | 10 (1.000, 0.00200, 0.0459) | 7 (0.2–1.5)d | 0- 6 (3.3, 6)d |
| 16S (1,202) | 9 (0.978, 0.00292, 0.054) | 14 (0.1–0.8)d | 0–6 (3.2, 2)d | |
| gyrB (735) | 8 (0.956, 0.00353, 0.059) | 211 (0.0–22.3)e | 0–77 (37.0, 39)e | |
| GyrB (245) | 7 (0.7) | 61e | 0–50 (24.6, 24)e | |
| N. otitidiscaviarum (10) | 606-bp 16S rRNA (556) | 1 (0,0,0) | 0d | 0 (0, 0)d |
| 16S (1,208) | 7 (0.911, 0.00598, 0.077) | 10 (0.1–0.3)d | 1–3 (1.8, 1)d | |
| gyrB (765) | 5 (0.800, 0.01003, 0.100) | 7 (0.0–0.5)e | 1–2 (1.6, 2)e | |
| GyrB (255) | 2 (0.2) | 1e | 0–1 (0.1, 0)e | |
| N. transvalensis (10) | 606-bp 16S rRNA (551) | 7 (0.911, 0.00598, 0.077) | 18 (0.2–6.0)d | 5–8 (6.1, 5)d |
| 16S (1,214) | 7 (0.867, 0.01149, 0.107) | 37 (0.0–1.6)d | 16–26 (18.7, 18)d | |
| gyrB (771) | 8 (0.933, 0.00597, 0.077) | 150 (0.0–16.0)e | 103–112 (108.3, 108)e | |
| GyrB (257) | 6 (0.5) | 55e | 62–69 (51.9, 51)e |
aa, amino acid.
HGDI, Hunter and Gaston discrimination index; S2, variance. For GyrB, only HGDI was calculated.
The divergence rate is expressed as a percentage for each group.
Three levels of diversity were seen for the gyrB gene and GyrB protein in the commonly reported group: a high level for N. carnea (211 single nucleotide polymorphisms [SNPs], 61 amino acid changes) and N. transvalensis (150 SNPs, 55 amino acid changes), a middle level for N. brasiliensis (51 SNPs, 19 amino acid changes), and a low level for N. otitidiscaviarum (7 SNPs, 1 amino acid changes).
The N. carnea strains showed the largest numbers of SNPs in gyrB and a similarity range of 80.9% to 100% compared to that of N. carnea W8368T gyrB (GenBank accession no. GQ984361), followed by 79.2% to 94.0% compared to that of N. flavorosea CDC<USA-GA>:W9741T gyrB (GQ496113). Three GyrB protein motives were found; five strains produced a protein with positions 121Hys(CAC) to 122Asp(GAC) as seen in the GyrB protein of N. carnea ATCC 6847T (ACX70140), four with a 122Asp deletion, and one with the 121Leu(CTC) to 122Asn(AAC) motif. These findings established three clonal lineages within the present N. carnea population (Fig. 1). Only three strains were confirmed by their gyrB genes as belonging to N. carnea. The remaining strains were identified as N. flavorosea (n = 2, both with the 122Asp deletion), N. rhamnosiphila (n = 1), N. blacklockiae and N. wallacei (n = 1), and N. testacea, N. jinanensis, and N. sienata (n = 3). In previous taxonomic studies, N. carnea and N. flavorosea were clustered together by gyrB (2, 4).
The gyrB sequences of the N. transvalensis strains returned low similarity scores (below the cutoff value) with respect to N. transvalensis CDC<USA-GA>:W7518T (GenBank accession no. GQ496089) (85.3% to 86.1%), while greater similarity was seen with N. blacklockiae CDC<USA-GA>:W8088T (GQ496126) (89% to 95.5%) and N. wallacei CDC<USA-GA>:W7672T (GQ496086) (88.6% to 98.4%). None of the present strains seemed to belong to N. transvalensis sensu stricto (9), as was reflected by the considerable number of SNPs accumulated per strain (103 to 112, with 68 common SNPs present in every strain).
For N. brasiliensis, the main cause of tissue infections (10), clustering by sample origin (6 cutaneous versus 4 respiratory strains) was observed in the 16S and gyrB trees (Fig. 1). The N. brasiliensis strains showed 7 out of the 19 amino acid changes to lie between positions 123 and 136 with respect to N. brasiliensis CDC<USA-GA>:W7503T (GenBank accession no. GQ496125). In every other species, amino acid changes were distributed along the length of GyrB.
The N. otitidiscaviarum strains were the most homogeneous in terms of their 606-bp 16S fragment, 16S, and gyrB and GyrB sequences, with smaller haplotype numbers for the two genes.
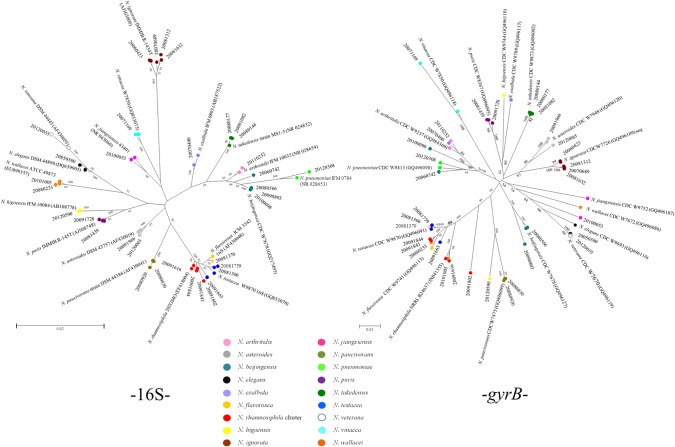
In the unusually reported species, agreement between the 16S and gyrB results was observed for 21 strains (60.0%) of the species N. higoensis, N. ignorata, N. paucivorans, N. pneumoniae, N. puris, N. takedensis, N. veterana, and N. vinacea and one-half the N. asteroides strains, two-fourths of the N. beijingensis strains, one-fourth of the N. rhamnosiphila cluster, and one-third of the N. testacea strains (see Table S1 in the supplemental material). No agreement was seen for 14 strains (40.0%), i.e., the strains of N. arthritidis, N. elegans, N. exalbida, N. flavorosea, N. jiangxiensis, and N. wallacei.
SNP numbers were analyzed as markers of variation in the unusually reported species. The greatest diversity was seen for the N. asteroides 16S sequence (37 SNPs) and the N. wallacei gyrB and GyrB sequence (144 SNPs, 45 amino acid changes compared to those of the reference sequences) (see Table S1 in the supplemental material).
When the phylogenetic trees based on 16S and gyrB were compared, the strains classified as N. paucivorans, N. veterana, and N. ignorata remained together in both representations. However, strains belonging to N. beijingensis or N. wallacei grouped together in the 16S-based tree by species and separately in the gyrB-based tree. To a lesser extent, this happens in strains proceeding from N. elegans, N. flavorosea, N. higoensis, N. rhamnosiphila, and N. testacea. This shows the greater discriminating capacity of gyrB (Fig. 2). Changes in GyrB showed that >70% of the SNPs were silent, above all in the strains of N. arthritidis and N. veterana. Nonsynonym substitution gathered predominantly between positions 778 and 787 with respect to the N. farcinica IFM10152 gyrB gene partial sequence (GenBank accession no. NC_006361), which corresponds to positions 260 to 264 of the N. farcinica IFM10152 GyrB protein sequence (YP_116212).
FIG 2.
16S- and gyrB-based phylogenetic trees for the unusually reported species of Nocardia (neighbor-joining method). Each bootstrap value is expressed as a percentage of 1,000 replications. Bar, 0.02 substitutions per nucleotide position.
In the studied Nocardia strains, gyrB analysis improves identification based on full 16S gene sequences of infrequently isolated Nocardia spp. by confirming the classifications of strains which need further investigation for identification in 57.5% and 60% of the commonly reported group and unusually reported species, respectively.
To summarize, in the studied Nocardia strains, the combination of 16S and gyrB analysis improves the identification of commonly reported species, such as N. brasiliensis and N. otitidiscaviarum, and unusually reported strains, such as N. higoensis, N. ignorata, N. paucivorans, N. pneumoniae, N. puris, N. takedensis, N. veterana, and N. vinacea, but it does not improve the identification of N. carnea or N transvalensis of the commonly reported group or N. arthritidis, N. asteroides, N. beijingensis, N. elegans, N. exalbida, N. flavorosea, N. jiangxiensis, N. rhamnosiphila, N. testacea, or N. wallacei of the unusually reported species.
As the number of gyrB sequences in databases increases, gyrB sequencing should play an increasingly important role in the discrimination and typing of Nocardia spp.
Nucleotide sequence accession numbers.
The new 16S and gyrB sequences were deposited in GenBank under the accession numbers KP010715 through KP010826.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by a grant to N.G. from the Instituto de Salud Carlos III (MPY-1446/11).
We are grateful to the CNM Biopolymers Unit for assistance in sequencing and Adrian Burton for language assistance. We are very grateful to the sample providers.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03072-14.
REFERENCES
- 1.Brown-Elliott BA, Brown JM, Conville PS, Wallace RJ Jr. 2006. Clinical and laboratory features of the Nocardia spp. based on current molecular taxonomy. Clin Microbiol Rev 19:259–282. doi: 10.1128/CMR.19.2.259-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takeda K, Kang Y, Yazawa K, Gonoi T, Mikami Y. 2010. Phylogenetic studies of Nocardia species based on gyrB gene analyses. J Med Microbiol 59:165–171. doi: 10.1099/jmm.0.011346-0. [DOI] [PubMed] [Google Scholar]
- 3.Carrasco G, Valdezate S, Garrido N, Villalón P, Medina-Pascual MJ, Sáez-Nieto JA. 2013. Identification, typing, and phylogenetic relationships of the main clinical Nocardia species in Spain according to their gyrB and rpoB genes. J Clin Microbiol 51:3602–3608. doi: 10.1128/JCM.00515-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McTaggart LR, Richardson SE, Witkowska M, Zhang SX. 2010. Phylogeny and identification of Nocardia species on the basis of multilocus sequence analysis. J Clin Microbiol 48:4525–4533. doi: 10.1128/JCM.00883-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Everest GJ, Cook AE, le Roes-Hill M, Meyers PR. 2011. Nocardia rhamnosiphila sp. nov., isolated from soil. Syst Appl Microbiol 34:508–512. doi: 10.1016/j.syapm.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Moser BD, Klenk HP, Schumann P, Pötter G, Lasker BA, Steigerwalt AG, Hinrikson HP, Brown JM. 2011. Nocardia niwae sp. nov., isolated from human pulmonary sources. Int J Syst Evol Microbiol 61:438–442. doi: 10.1099/ijs.0.020370-0. [DOI] [PubMed] [Google Scholar]
- 7.Tamura T, Matsuzawa T, Oji S, Ichikawa N, Hosoyama A, Katsumata H, Yamazoe A, Hamada M, Suzuki K, Gonoi T, Fujita N. 2012. A genome sequence-based approach to taxonomy of the genus Nocardia. Antonie Van Leeuwenhoek 102:481–491. doi: 10.1007/s10482-012-9780-5. [DOI] [PubMed] [Google Scholar]
- 8.Kong F, Chen SC, Chen X, Sintchenko V, Halliday C, Cai L, Tong Z, Lee OC, Sorrell TC. 2009. Assignment of reference 5′-end 16S rDNA sequences and species-specific sequence polymorphisms improves species identification of Nocardia. Open Microbiol J 3:97–105. doi: 10.2174/1874285800903010097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conville PS, Brown JM, Steigerwalt AG, Brown-Elliott BA, Witebsky FG. 2008. Nocardia wallacei sp. nov. and Nocardia blacklockiae sp. nov., human pathogens and members of the “Nocardia transvalensis complex.” J Clin Microbiol 46:1178–1184. doi: 10.1128/JCM.02011-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen KW, Lu CW, Huang TC, Lu CF, Liau YL, Lin JF, Li SY. 2013. Cutaneous manifestations of Nocardia brasiliensis infection in Taiwan during 2002–2012—clinical studies and molecular typing of pathogen by gyrB and 16S gene sequencing. Diagn Microbiol Infect Dis 77:74–78. doi: 10.1016/j.diagmicrobio.2013.05.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.