Abstract
The integration of matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) in clinical microbiology has revolutionized species identification of bacteria, yeasts, and molds. However, beyond straightforward identification, the method has also been suggested to have the potential for subspecies-level or even type-level epidemiological analyses. This minireview explores MALDI-TOF MS-based typing, which has already been performed on many clinically relevant species. We discuss the limits of the method's resolution and we suggest interpretative criteria allowing valid comparison of strain-specific data. We conclude that guidelines for MALDI-TOF MS-based typing can be developed along the same lines as those used for the interpretation of data from pulsed-field gel electrophoresis (PFGE).
INTRODUCTION
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has recently been integrated into the routine diagnostic workflow of many industrial, pharmaceutical, and medical microbiology laboratories. It has already been thoroughly evaluated for the identification of clinically relevant bacterial species, including anaerobes, Gram-positive rods and cocci, Enterobacteriaceae, and miscellaneous Gram-negative (including nonfermentative) rods, with adequate to excellent results (1, 2).
Species identification, however, is only a first step in the diagnostic workflow. The ability to quickly and reliably distinguish or “type” related bacterial isolates is essential for bacterial transmission studies and larger-scale epidemiological surveillance projects. “Conventional” phenotyping methods, such as multiple-susceptibility testing, phage typing, serotyping, biochemical typing, and several others, have been important contributors to our understanding of the epidemiology of community- and health care-associated infections. However, these methods all have practical limitations which render them largely unsuitable for comprehensive bacterial population analyses as well as for scientifically less ambitious but critical infection surveillance. Furthermore, most phenotypic methods have been developed for individual bacterial taxa and are not transferable to other taxa without considerable adaptation.
Hence, over the past 2 decades, phenotyping has been largely replaced by “molecular” genotyping. Clonal reproduction by binary fission imprints the evolutionary history of the organism in genotypic markers amenable to analysis by nucleic acid-mediated methods. However, in practice, the ease with which recombination, transfection, and transformation can take place in bacteria necessitates that data from multiple genetic markers are analyzed in defining a “precise” genotype and even then there is no guarantee that an adequate natural taxonomy will be derived. Polyphasic taxonomy currently uses combinations of different phenotypic and/or genotypic data sets to define genera, species, and entities at or below the subspecies level. Still, all these approaches remain time-consuming and relatively costly and are often restricted to specialized laboratories. Of these, pulsed-field gel electrophoresis (PFGE) is generally considered to be the “gold standard” for the epidemiological typing of microorganisms (3) while waiting for the advent of easy whole-genome sequencing.
In this context, a key issue is whether MALDI-TOF MS could be employed in the epidemiological typing of clinically relevant bacterial species. This approach is attractive since whole-cell mass spectrometry is commonly used for species identification with potentially exploitable data already available from routine analyses. Hence, the goal would be that MALDI-TOF MS data used for isolate identification might be secondarily exploited for epidemiological typing either directly or in an additional data interpretation step at no added costs. A number of examples of the potential usefulness of this relatively cheap, easy-to-perform proteomic technology for epidemiological typing have been published recently for a variety of problem pathogens. However, at this point it is not clear whether the typing of microorganisms by MALDI-TOF MS (MALDI typing) will be as successful as MALDI identification. One reason for a delay in the use of MALDI typing likely relates to a lack of guidelines for data interpretation. In this review, possible reasons and putative solutions for this missing link are discussed.
Current experience with MALDI typing suggests its usefulness in identifying strains of clinical significance, which in essence is a simplified form of typing, and for the detailed epidemiological typing of a variety of human pathogens, examples of which are reviewed below.
(i) Streptococcus agalactiae.
The use of an Ultraflex III machine and ClinProTools software 2.0 (Bruker Daltonics, Bremen, Germany) to analyze 197 sequence-typed (ST) strains from various patient samples revealed that ST-17 strains could be identified by a peak shift from m/z 7,650 to m/z 7,625 (4). This peak shift, however, is also found in the closely related ST-201, ST-315, and ST-405 strains, which are single-locus variants of ST-17. Thus, this rapid method could be of value in evaluating the risk of acquiring ST-17-mediated neonatal disease. It was also possible to tentatively identify isolates of the emerging ST-1 strain, responsible for infections in immunocompromised adults and neonates, by a single peak shift from m/z 6,888 to m/z 6,250. However, this peak shift was also found in unrelated STs and no consistent “clonal” biomarker was identified for other major sequence types (ST-8, ST-19, and ST-23).
(ii) Streptococcus pyogenes.
A study using an AB 4700 Proteomics Analyzer (Applied Biosystems, Foster City, USA) combined with the Tag-ident proteomics tool and the ExPASy sequence retrieval system (http://us.expasy.org) has shown that S. pyogenes isolates from necrotizing fasciitis clustered together and could be separated from noninvasive isolates, despite being the same emm types (5). However, the number of strains in the study was small (n = 9) and, even using a larger collection, different emm types were not all separated by MS. The potentially improved discrimination of MS compared to emm typing is not unexpected since the former provides a more global proteomic analysis. Nevertheless, further studies comparing MALDI typing with emm and other typing methods using better-documented isolate collections are necessary.
(iii) Streptococcus pneumoniae.
The study of 25 S. pneumoniae strains on an AB 4700 Proteomics Analyzer showed that MALDI-TOF MS can differentiate conjunctival S. pneumoniae (cPnc) from nonconjunctival controls (6). Eleven signals with high intensity were observed in the spectra of all S. pneumoniae isolates, while a peak at m/z 2,944 was common and uniquely recorded among all cPnc isolates. The cPnc proteomic signatures could ultimately be important in the diagnosis of this infection.
(iv) Staphylococcus aureus.
A first study performed on a Microflex LT benchtop (Bruker Daltonics) and involving 60 spa-typed methicillin-resistant S. aureus (MRSA) isolates suggested that discrimination of strains belonging to the five major multilocus sequence typing (MLST) clonal complexes (CC5, CC8, CC22, CC30, and CC45) was possible based on 13 distinctive peaks whose presence or absence differed for individual CCs. The discriminatory indices of MALDI typing and spa typing were found to be comparable (7). However, the set of strains was limited and missing important clonal complexes.
These findings were extended by a later study using a Biflex III Biotyper (Bruker Daltonics) and more than 400 MRSA strains, significantly increasing CC coverage (8). The sets of discriminating peaks identified in the two studies overlapped by eight CC-specific peaks. This result suggests that the MALDI typing data for S. aureus are equivalent and transferable from equipment of different brands, which is an important requirement for the broad applicability of a novel typing method.
Another promising study using an UltraFlex III machine proposed three protein peaks as specific for 197 of 224 (88%) USA300 isolates (9). However, a previous study (7) identified one of the peaks, m/z 6,423, as species specific, while a second peak, m/z 6,592, was more generally associated with CC8, to which USA300 belongs. The study suggested that the markers and the mathematical model for spectra analysis were suitable for rapid identification of USA300. But this finding requires confirmation since the analyzed strains were all derived from a single hospital, potentially implying clonal bias, and two of the three MS peaks are potentially nonspecific for USA300. The need for further clarification and optimization of MALDI typing for S. aureus is also evidenced in these and other studies by the inability to differentiate methicillin-susceptible from methicillin-resistant isolates (i.e., similar to MLST). For example, a recent study demonstrated no differences of mass spectra for a pair of isogenic MRSA and methicillin-susceptible S. aureus (MSSA) strains (10).
(v) Escherichia coli.
E. coli is one of the most frequently isolated clinically relevant species. Thus, the rapid recognition of pathogenic or potentially pathogenic types is of interest. In a study by Clark et al., 136 unrelated strains of E. coli, encompassing six pathotypes, could be grouped by MALDI-TOF MS using an Autoflex III machine (11). One interesting result was the association of E. coli O157:H7 with two specific MS peaks (12).
The differentiation of E. coli and Shigella species was targeted by Khot et al. using a set of strains originating from a wide geographical region (13). The spectral data acquired on a Microflex instrument were analyzed with ClinProTools to establish group-identifying models using fully automated or manual methods or a hybrid approach. The proposed models were challenged with independent strains and resulted in correct identification for 90% of Shigella and E. coli isolates.
(vi) Salmonella spp.
A BioTyper 3.0 system (Bruker Daltonics) was used to successfully differentiate Salmonella enterica serovar Typhi and non-S. Typhi isolates on the basis of the presence or absence of six peaks (14). The strains originated from sub-Saharan Africa and represented 12 serovars, of which S. Typhi, S. Enteritidis, and S. Typhimurium were the predominant strains. A similar study was performed on 913 strains of 89 non-Typhi Salmonella serovars originating from different regions and outbreaks in Germany (15). MS spectra allowed for decision tree classification for non-Typhi Salmonella serovars, although the study was limited by a lack of some clinically relevant serovars.
(vii) Clostridium difficile.
MALDI typing successfully identified ribotypes 001, 027, and 078/126 despite the fact that some MS peaks were shared between these ribotypes (16). All ribotype 001 isolates were recognized by the presence of 9 to 18 masses, while for ribotype 027 isolates, only three specific masses were found. Ribotype 126 and hypervirulent ribotype 078 could not be discriminated by MALDI typing and presented as a separate group (16). For other ribotypes, the number of strains was considered too small to establish specific profiles. Toxin detection and identification were not assessed since these proteins are far beyond the mass range usually used for identification and typing, although MALDI-TOF MS technically allows the detection of such large molecules.
(viii) Acinetobacter baumannii.
A study performed using a Vitek MS RUO system showed that MS was able to separate outbreak-associated A. baumannii isolates (n = 14) from control isolates (n = 10) and non-outbreak-associated isolates (n = 4) (17). These results were in general agreement with MLST, but more work is needed to better define interstrain relationships. These results were confirmed on a Microflex system analyzing 35 multidrug-resistant strains isolated from colonized or infected patients. The nosocomial strains could be segregated into three clusters, showing a good correlation with repetitive-PCR technology (rep-PCR) results (18).
(ix) Legionella pneumophila.
A study of 23 L. pneumophila reference and environmental strains performed using an Autoflex II machine (Bruker Daltonics) showed the potential for rapid epidemiological typing of L. pneumophila (19). Analyses using full-mass fingerprints showed resolving power and clustering similar to those of PFGE. In the study, serotyping was not considered and 13 of the strains originated from two locations in Japan possibly representing single clones.
TYPING BY MALDI-TOF MS: WHAT CAN WE LEARN FROM PFGE?
Current literature shows positive results for microbial whole-cell MALDI-TOF MS typing for a number of species, although a number of studies have described a failure to achieve satisfying resolution. For example, Lasch et al. (2014) reported insufficient discriminatory power for typing of Enterococcus faecium and Staphylococcus aureus isolates (20) and Schirmeister et al. (2013) reported the same for Vibrio cholerae isolates (21) and such studies may represent the tip of the difficult-to-publish iceberg. After examination of successful reports on MALDI-TOF MS typing, it becomes evident that for some bacterial taxa, the available data are not consistent. Discrepancies between individual studies and failure to perform may be partly explained by the following considerations related to the methodology.
(i) Technological and biological MS issues.
MALDI-TOF MS fingerprints are clearly different from genomic data. While the latter have an essentially digital nature, MS peaks are waveform data that are more analog in nature. Presence versus absence, m/z, and (relative) intensity levels are subject to analytical error, biological and technical variation (including complex, sometimes low-level protein expression and posttranslational modification and its regulation), and analyte incorporation in matrix crystals. Thus, some typing schemes, such as the one proposed by Clark et al. (11), are largely based on absence of peaks and may thus be more sensitive to technical issues, such as noise in spectra, compared to approaches based on presence of more stable and specific biomarkers.
(ii) Clonality versus correlation.
MALDI-TOF MS typing is a tool for phylotyping and records all intracellular and cell-wall-bound proteins. However, acquisition of virulence or resistance factors which are not directly detectable because of their high molecular mass (m/z of ≫20 kDa) may not be reflected by changes in whole-cell mass spectra. Hence, apparent correlations of mass fingerprints to virulence and resistance need to be thoroughly evaluated, taking into account possible effects of clonality, as some previous erroneous results have already been published (22).
(iii) Sensitivity, specificity, and strain sets.
Any MALDI typing scheme is a priori only valid for the set of strains tested (this applies, in principle, also to MALDI identification, although in this case the size and quality of the underlying database often allow broader application). When strains from a small number of clonal complexes, sequence types, or serotypes can be distinguished, this cannot always be extrapolated to a larger number of phylotypes. The same applies to strains from geographically close entities.
(iv) Definition and limitation of specific peaks.
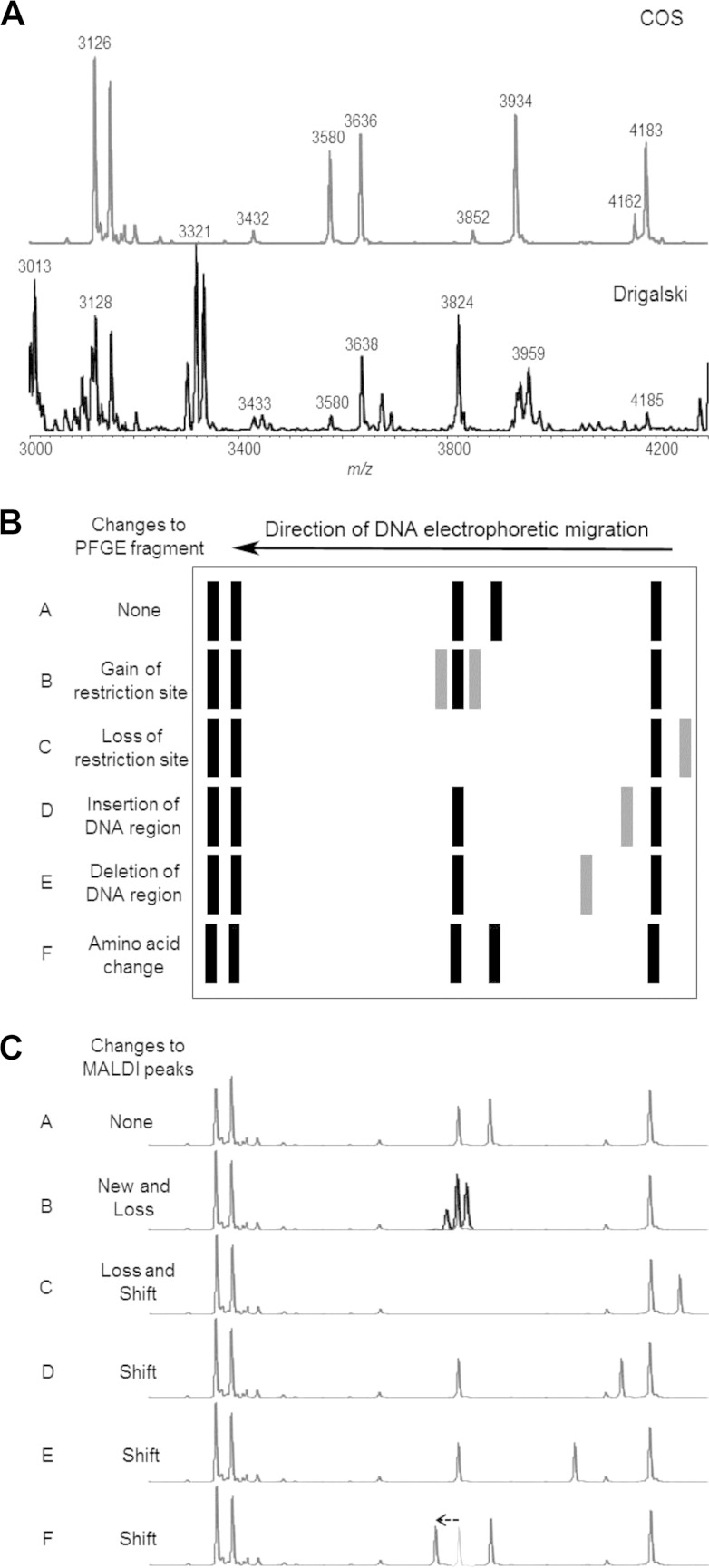
Peaks that are considered type-specific biomarkers should be recorded consistently with reasonable signal intensities to avoid false interpretation based on analytical variability. Further, proteins corresponding to biomarker peaks may not be expressed under different cultivation conditions (see Fig. 1A). Thus, some potentially specific peaks only expressed in a specific medium cannot be used for the purpose of typing. Consequently, typing schemes should be standardized as much as possible or the robustness needs to be tested prior to a transfer to other systems. This includes analyzing a sufficient number of isolates (e.g., 50) to ensure the validity of specific biomarker-strain associations (23).
FIG 1.
(A) MALDI-TOF MS spectra obtained for a single strain of E. coli grown on different media. The top spectrum shows results obtained after culture on Columbia–5% sheep blood agar, whereas the data shown below were from cells grown on Drigalski agar. (B) Theoretical representation of a PFGE gel after staining. Restriction fragments are visible, and differences between the individual lanes can be explained on the basis of presumed genetic events which are indicated on the right. Based on theoretical exercises such as the one presented, adapted from Tenover et al. (3), rules suggesting that up to three fragment differences can be ignored in order to still define clear epidemiological relatedness were developed. (C) The MALDI-TOF MS analogs for the PFGE data displayed in panel B. Fragments have been translated into peaks, and the events potentially leading to changes from the reference spectrum in row A are proposed (left). This relates to the emergence and disappearance of protein peaks and distinct shifts in protein peaks for various (epi)genetic reasons.
The recording of a peak in a mass spectrum cannot be taken as representing unambiguous detection of a particular protein without further analysis. The apparent m/z value is only one (simple) characteristic of an individual protein and could be considered the “shadow” of a protein. As described before, a large number of proteins with entirely different functions and sequences can produce peaks with similar m/z values (24). In addition, the m/z value for a given protein can change depending on, for instance, the degree of posttranslational modification or complexing of cofactors. This induces peak position variability (Fig. 1C, row F) that should be taken into account. However, it is possible that such unstable peaks may serve as stable epidemiological markers.
(v) Statistical analysis.
Statistical analysis of peak patterns is a crucial step in the discovery and evaluation of type-specific biomarkers and peak patterns, respectively. The selection of the algorithms, models, etc., should be guided by biomathematical reasoning combined with microbiological expertise. Despite the apparent persuasiveness of complex statistical procedures, punctual visual inspection of original spectra and consideration of the biology of microorganisms are good control measures to avoid overinterpretation of mass spectral data.
(vi) Primary MALDI-TOF identification limitations.
Some specific groups of organisms such as E. coli and Shigella sp. or S. pneumoniae have proven to be potentially difficult to identify in the past. These difficulties have probably slowed their potential evaluation by MALDI typing. However, primary identification of S. pneumoniae using specific instruments (25) and recent success with clone-specific MALDI typing (6, 12) have demonstrated the potential of MALDI typing for species that may be difficult to identify with this technology.
As noted earlier, PFGE has a long history as the gold standard of epidemiological strain typing, due in large part to the guidelines available to assist in data interpretation (3). However, based on the points presented above, a consideration of guidelines for the interpretation of MALDI typing data must note that PFGE typing and MALDI typing measure completely different cellular properties. Thus, it would be simplistic to attempt to superimpose PFGE interpretative guidelines on data generated by whole-cell MS. Nevertheless, from a purely numerical standpoint, PFGE and MS data sets are quite similar, although, on average, the number of peaks in a MS spectrum outnumbers the sum of restriction fragments visible on a PFGE gel (Table 1). Where changes in PFGE patterns can be explained on the basis of insertions, deletions, inversions, and nucleotide mutations, there may be additional, more complicated reasons for differences in mass spectra. Obviously, the same set of genetic events relevant for PFGE will give rise to changes in the mobility of proteins during MS: additions, mass shifts, and lack of protein are direct consequences of the genomic changes and protein expression regulation (Fig. 1B and C).
TABLE 1.
Comparison of the average number of peaks in MALDI-TOF MS spectra to the average number of restriction fragments observed upon PFGEa
| Species | No. of MS database strains | Avg no. of peaks per spectrum | No. of variable peaks |
Avg no. of PFGE RF | Ratio of variable peak no./PFGE RF | |
|---|---|---|---|---|---|---|
| 80% | 70% | |||||
| Acinetobacter baumannii | 440 | 66 | 50 | 33 | 30 | 2.2 |
| Acinetobacter calcoaceticus | 4 | 66 | 44 | 20 | 23 | 2.9 |
| Bacteroides fragilis | 17 | 103 | 69 | 27 | 9 | 11.4 |
| Bordetella pertussis | 4 | 84 | 40 | 19 | 25 | 3.4 |
| Burkholderia cepacia | 6 | 102 | 83 | 47 | 23 | 4.5 |
| Campylobacter fetus subsp. fetus | 8 | 110 | 39 | 17 | 13 | 8.8 |
| Campylobacter jejuni | 14 | 77 | 46 | 30 | 9 | 8.6 |
| Clostridium difficile | 253 | 69 | 42 | 21 | 13 | 5.5 |
| Clostridium perfringens | 6 | 90 | 41 | 8 | 11 | 8.2 |
| Enterobacter cloacae | 14 | 84 | 70 | 44 | 20 | 4.2 |
| Enterococcus faecalis | 97 | 65 | 45 | 32 | 18 | 3.7 |
| Escherichia coli | 121 | 98 | 57 | 24 | 16 | 6.1 |
| Haemophilus influenzae | 60 | 87 | 66 | 46 | 11 | 7.9 |
| Klebsiella pneumoniae | 168 | 70 | 48 | 26 | 20 | 3.5 |
| Legionella pneumophila | 22 | 148 | 62 | 25 | 10 | 14.8 |
| Mycobacterium tuberculosis | 11 | 95 | 13 | 0 | 16 | 5.9 |
| Neisseria gonorrhoeae | 7 | 102 | 53 | 26 | 15 | 7.0 |
| Neisseria meningitidis | 10 | 111 | 60 | 27 | 25 | 4.4 |
| Proteus mirabilis | 166 | 80 | 49 | 24 | 8 | 10.0 |
| Pseudomonas aeruginosa | 149 | 88 | 72 | 48 | 35 | 2.5 |
| Salmonella serovar Typhi | 6 | 118 | 63 | 27 | 45 | 2.6 |
| Shigella dysenteriae | 5 | 108 | 67 | 24 | 19 | 5.7 |
| Staphylococcus aureus | 171 | 67 | 48 | 27 | 15 | 4.5 |
| Staphylococcus epidermidis | 204 | 56 | 44 | 31 | 18 | 3.2 |
| Stenotrophomonas maltophilia | 83 | 97 | 77 | 40 | 15 | 6.5 |
| Streptococcus mitis | 19 | 84 | 62 | 36 | 18 | 4.8 |
| Streptococcus pneumoniae | 201 | 87 | 64 | 34 | 15 | 6.0 |
| Vibrio cholerae | 9 | 113 | 59 | 37 | 25 | 4.5 |
| Yersinia pestis | 2 | 111 | 42 | 10 | 20 | 5.5 |
Adapted from Tenover et al. (22). As shown in columns 4 and 5, a peak was considered constant when found in either 80% or 70% of every spectrum for the same species. RF, restriction fragments.
Up to three band differences for 20 to 30 DNA fragments are tolerated in PFGE in order to still define an epidemiological link between two strains (3). As an average of 100 peaks (three to five times more than PFGE) is usually seen in a microbial mass spectrum, we similarly propose that an average difference of 15 peaks could be tolerated (i.e., could define related types) in MALDI-TOF MS typing. However, it is evident from the published data that the discriminatory potential for MALDI typing (i.e., corresponding to what constitutes a “significant” difference) is likely not the same for all bacterial species since some show a substantially lesser share of variable peaks than others. For example, strongly clonal species such as Mycobacterium tuberculosis for which the number of variable peaks is limited demonstrated only 13 variable peaks among 95 average peaks when a peak was considered constant if present in 80% of every spectra for the same species (Table 1). However, the number of peaks and their variability necessary for robust typing could potentially be calibrated for individual species using data as shown in Table 1. To help build these specific guidelines, ratios of the number of variable peaks to the number of PFGE restriction fragments are shown in Table 1 for a number of species. For instance, Burkholderia cepacia has a variable-peak number/PFGE restriction fragment number ratio of 4.5; for this species, then, a three-band PFGE difference would equate to 14 MALDI peak differences and could be tolerated within an average peak number of 102 to define related types (Table 1).
In conclusion, MALDI-TOF MS appears to hold promise as a strain identification/epidemiological typing tool, especially since the data are available in most cases at no additional cost: the typing information, or at least part of it, is contained within the primary spectra that are used for species identification. Even in cases in which a second spectrum acquisition may be necessary, for example, after a more elaborate extraction procedure, application of another matrix, or a change of the covered mass range, MALDI-TOF MS remains a rapid and relatively inexpensive tool for potential identification of health care-associated outbreaks caused by various surveillance organisms. Given the rapidity of the method, MALDI typing could for the first time allow broad and prospective typing of a majority of all clinical isolates detected in clinical settings without too much of an add-on with regard to costs and the requirement for specific laboratory expertise. In addition, interpretation of the data is supposedly simple, as shown by the overlap in methodology and guidelines as developed for the current gold standard method, PFGE. However, the reproducibility of the technology, portability of results, effects of storage and culture conditions of the strains, careful comparison with widely accepted typing technologies, and many more features remain to be carefully assessed, preferably by multicenter validation studies on MALDI typing.
Biography

Sébastien Spinali received his medical education at Paris Descartes University in Paris. He did his internship at Centre Hospitalier Universitaire (Pr. R. Courcol) in Lille until he moved to Paris and joined the microbiology team at Cochin Port Royal (Pr. C. Poyart) for 2 years. He then joined the Clinical Unit of bioMérieux in Lyon (R&D Microbiology) in November 2013. He has a major interest in mass spectrometry as a huge breakthrough for bacteria and yeast identification and more to come. He has been working in this field for 5 years now since one of the first mass spectrometers was installed in Lille hospital. He is also interested in general microbiology and especially in new diagnosis technologies. He also contributed to a recently published article concerning evaluation of MALDI-TOF mass spectrometry for the identification of medically important yeasts (26).
REFERENCES
- 1.Dubois D, Segonds C, Prere M-F, Marty N, Oswald E. 2013. Identification of clinical Streptococcus pneumoniae isolates among other alpha and nonhemolytic streptococci by use of the Vitek MS matrix-assisted laser desorption ionization–time of flight mass spectrometry system. J Clin Microbiol 51:1861–1867. doi: 10.1128/JCM.03069-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eigner U, Holfelder M, Oberdorfer K, Betz-Wild U, Bertsch D, Fahr A-M. 2009. Performance of a matrix-assisted laser desorption ionization-time-of-flight mass spectrometry system for the identification of bacterial isolates in the clinical routine laboratory. Clin Lab 55:289–296. [PubMed] [Google Scholar]
- 3.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lartigue M-F, Kostrzewa M, Salloum M, Haguenoer E, Héry-Arnaud G, Domelier A-S, Stumpf S, Quentin R. 2011. Rapid detection of “highly virulent” group B Streptococcus ST-17 and emerging ST-1 clones by MALDI-TOF mass spectrometry. J Microbiol Methods 86:262–265. doi: 10.1016/j.mimet.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 5.Moura H, Woolfitt AR, Carvalho MG, Pavlopoulos A, Teixeira LM, Satten GA, Barr JR. 2008. MALDI-TOF mass spectrometry as a tool for differentiation of invasive and noninvasive Streptococcus pyogenes isolates. FEMS Immunol Med Microbiol 53:333–342. doi: 10.1111/j.1574-695X.2008.00428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williamson YM, Moura H, Woolfitt AR, Pirkle JL, Barr JR, Carvalho MDG, Ades EP, Carlone GM, Sampson JS. 2008. Differentiation of Streptococcus pneumoniae conjunctivitis outbreak isolates by matrix-assisted laser desorption ionization–time of flight mass spectrometry. Appl Environ Microbiol 74:5891–5897. doi: 10.1128/AEM.00791-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolters M, Rohde H, Maier T, Belmar-Campos C, Franke G, Scherpe S, Aepfelbacher M, Christner M. 2011. MALDI-TOF MS fingerprinting allows for discrimination of major methicillin-resistant Staphylococcus aureus lineages. Int J Med Microbiol 301:64–68. doi: 10.1016/j.ijmm.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Josten M, Reif M, Szekat C, Al-Sabti N, Roemer T, Sparbier K, Kostrzewa M, Rohde H, Sahl H-G, Bierbaum G. 2013. Analysis of the matrix-assisted laser desorption ionization-time of flight mass spectrum of Staphylococcus aureus identifies mutations that allow differentiation of the main clonal lineages. J Clin Microbiol 51:1809–1817. doi: 10.1128/JCM.00518-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boggs SR, Cazares LH, Drake R. 2012. Characterization of a Staphylococcus aureus USA300 protein signature using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Med Microbiol 61:640–644. doi: 10.1099/jmm.0.037978-0. [DOI] [PubMed] [Google Scholar]
- 10.Szabados F, Kaase M, Anders A, Gatermann SG. 2012. Identical MALDI TOF MS-derived peak profiles in a pair of isogenic SCCmec-harboring and SCCmec-lacking strains of Staphylococcus aureus. J Infect 65:400–405. doi: 10.1016/j.jinf.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 11.Clark CG, Kruczkiewicz P, Guan C, McCorrister SJ, Chong P, Wylie J, van Caeseele P, Tabor HA, Snarr P, Gilmour MW, Taboada EN, Westmacott GR. 2013. Evaluation of MALDI-TOF mass spectroscopy methods for determination of Escherichia coli pathotypes. J Microbiol Methods 94:180–191. doi: 10.1016/j.mimet.2013.06.020. [DOI] [PubMed] [Google Scholar]
- 12.Fagerquist CK, Zaragoza WJ, Sultan O, Woo N, Quiñones B, Cooley MB, Mandrell RE. 28 February 2014. Top-down proteomic identification of Shiga toxin 2 subtypes from Shiga toxin-producing Escherichia coli by matrix-assisted laser desorption ionization-tandem time of flight mass spectrometry. Appl Environ Microbiol doi: 10.1128/AEM.04058-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khot PD, Fisher MA. 2013. Novel approach for differentiating Shigella species and Escherichia coli by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 51:3711–3716. doi: 10.1128/JCM.01526-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhns M, Zautner AE, Rabsch W, Zimmermann O, Weig M, Bader O, Groß U. 2012. Rapid discrimination of Salmonella enterica serovar Typhi from other serovars by MALDI-TOF mass spectrometry. PLoS One 7:e40004. doi: 10.1371/journal.pone.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dieckmann R, Malorny B. 2011. Rapid screening of epidemiologically important Salmonella enterica subsp. enterica serovars by whole-cell matrix-assisted laser desorption ionization–time of flight mass spectrometry. Appl Environ Microbiol 77:4136–4146. doi: 10.1128/AEM.02418-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reil M, Erhard M, Kuijper EJ, Kist M, Zaiss H, Witte W, Gruber H, Borgmann S. 2011. Recognition of Clostridium difficile PCR-ribotypes 001, 027 and 126/078 using an extended MALDI-TOF MS system. Eur J Clin Microbiol Infect Dis 30:1431–1436. doi: 10.1007/s10096-011-1238-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Houck HJ, Santiago I, Delano JP, Archibald L, Rand KH. Multi-drug resistant Acinetobacter burn unit outbreak: application of matrix-assisted laser desorption ionization time-of-flight mass spectrometry in determining strain differentiation, poster C-1370 Abstr 113th Gen Meet Am Soc Microbiol, Denver, CO. American Society for Microbiology, Washington, DC. [Google Scholar]
- 18.Mencacci A, Monari C, Leli C, Merlini L, De Carolis E, Vella A, Cacioni M, Buzi S, Nardelli E, Bistoni F, Sanguinetti M, Vecchiarelli A. 2013. Typing of nosocomial outbreaks of Acinetobacter baumannii by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 51:603–606. doi: 10.1128/JCM.01811-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujinami Y, Kikkawa HS, Kurosaki Y, Sakurada K, Yoshino M, Yasuda J. 2011. Rapid discrimination of Legionella by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Microbiol Res 166:77–86. doi: 10.1016/j.micres.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 20.Lasch P, Fleige C, Stämmler M, Layer F, Nübel U, Witte W, Werner G. 2014. Insufficient discriminatory power of MALDI-TOF mass spectrometry for typing of Enterococcus faecium and Staphylococcus aureus isolates. J Microbiol Methods 100:58–69. doi: 10.1016/j.mimet.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 21.Schirmeister F, Dieckmann R, Bechlars S, Bier N, Faruque SM, Strauch E. 10 November 2013, posting date Genetic and phenotypic analysis of Vibrio cholerae non-O1, non-O139 isolated from German and Austrian patients. Eur J Clin Microbiol Infect Dis 33:767–778. doi: 10.1007/s10096-013-2011-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szabados F, Becker K, von Eiff C, Kaase M, Gatermann S. 2011. The matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF MS)-based protein peaks of 4448 and 5302 Da are not associated with the presence of Panton-Valentine leukocidin. Int J Med Microbiol 301:58–63. doi: 10.1016/j.ijmm.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 23.Van Belkum A, Tassios PT, Dijkshoorn L, Haeggman S, Cookson B, Fry NK, Fussing V, Green J, Feil E, Gerner-Smidt P, Brisse S, Struelens M, European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Study Group on Epidemiological Markers (ESGEM) . 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin Microbiol Infect 13(Suppl 3):1–46. doi: 10.1111/j.1469-0691.2007.01786.x. [DOI] [PubMed] [Google Scholar]
- 24.Fagerquist CK, Bates AH, Heath S, King BC, Garbus BR, Harden LA, Miller WG. 2006. Sub-speciating Campylobacter jejuni by proteomic analysis of its protein biomarkers and their post-translational modifications. J Proteome Res 5:2527–2538. doi: 10.1021/pr050485w. [DOI] [PubMed] [Google Scholar]
- 25.Branda JA, Markham RP, Garner CD, Rychert JA, Ferraro MJ. 2013. Performance of the Vitek MS v2.0 system in distinguishing Streptococcus pneumoniae from nonpneumococcal species of the Streptococcus mitis group. J Clin Microbiol 51:3079–3082. doi: 10.1128/JCM.00824-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sendid B, Ducoroy P, François N, Lucchi G, Spinali S, Vagner O, Damiens S, Bonnin A, Poulain D, Dalle F. 2013. Evaluation of MALDI-TOF mass spectrometry for the identification of medically-important yeasts in the clinical laboratories of Dijon and Lille hospitals. Med Mycol 51:25–32. doi: 10.3109/13693786.2012.693631. [DOI] [PubMed] [Google Scholar]