Abstract
A yet-undescribed bacterial species, tentatively named “Porphyromonas katsikii,” was isolated from individuals of a small goat herd with pyogranulomatous pneumonia during an outbreak of acute respiratory disease. The isolated bacteria grew in the form of black-pigmented colonies after 14 days of incubation under anaerobic conditions at 37°C on a tryptic soy blood agar medium. The bacteria were identified as a yet-undescribed Porphyromonas species by determination of the nucleotide sequence of the rrs 16S rRNA gene, and this species was tentatively named Porphyromonas katsikii. PCR amplification with specific primers for this yet-undescribed species revealed the presence of P. katsikii in the lung tissue of all affected animals, while no PCR signals were evidenced from the lungs of healthy goats or from goats with pasteurellosis caused by Mannheimia haemolytica. These data indicate P. katsikii as the causative agent of acute respiratory distress. P. katsikii is phylogenetically related to Porphyromonas somerae and Porphyromonas levii, which cause pathologies in humans and animals, respectively. P. katsikii was not detected by PCR from samples of the gingival pockets or of the faces of healthy goats.
INTRODUCTION
Porphyromonas species belonging to the phylum Bacteroidetes are nonmotile, Gram-negative, rod-shaped, anaerobic bacteria that are considered emerging pathogens in humans and animals (1, 2). In general, Porphyromonas species, and in particular Porphyromonas gingivalis, the best-studied representative of the genus, are known as oral pathogens causing gingivitis, periodontitis, endodontic diseases, and apical abscesses in human and animals (3–5). However, several Porphyromonas species are involved in other human and animal pathologies, such as metritis, peritonitis, interdigital necrobacillosis, and necrotic vulvovaginitis (2, 6–8). Given that phenotypic identification and discrimination of Porphyromonas and Prevotella species had been inconsistent (9), genotypic species identification based on 16S rRNA gene sequences was developed and was used successfully thereafter (10).
Porphyromonas somerae and Porphyromonas levii, two closely related species, are regarded as pathogenic species of humans and cattle, respectively. Clinical manifestations of P. somerae (a P. levii-like species) include soft tissue and bone infections, brain abscesses, and otitis media with mastoiditis (6, 7, 11). In ruminants, P. levii has been isolated from cows with necrobacillosis (8), papillomatous digital dermatitis (12), and acute interdigital phlegmon (13) and from cows during an outbreak of bovine necrotic vulvovaginitis (6). However, P. levii was also reported as an opportunistic pathogen in the rumen of cattle (2, 14) and was found in healthy cattle herds (15).
Herein, we report data on the bacteriological isolation and genotypic identification of, and the clinical signs, laboratory results, and pathology that are indicative of an outbreak of pyogranulomatous pneumonia associated with acute respiratory disease in goats (Capra aegagrus hircus) caused by, a yet-unknown Porphyromonas species, which we tentatively named “Porphyromonas katsikii.”
MATERIALS AND METHODS
Description of the goat herd.
The herd that was affected by pyogranulomatous pneumonia consisted of 25 domestic goats (Capra aegagrus hircus) between 1 and 6 years of age. The animals were in the high milking period and not pregnant. They were kept in a closed building with good natural ventilation (permanently open windows), were fed with silage, and were allowed grazing for 4 to 5 h daily during months with a moderate climate. Animal traffic was strongly controlled in the area. Twice per year, all adult goats were vaccinated against clostridial diseases (Bravoxin 10; MSD Animal Health, United Kingdom). As a preventive measure against pneumonic and intestinal endoparasites, all adult animals were treated with netobimin (Hapadex 5%; MSD Animal Health) once per year. A brief study of the clinical records showed that, in 2011, 11 milking goats in this herd showed clinical symptoms of contagious agalactia and were successfully treated with tylosin (Tylan; Elanco).
Gross pathology and histology.
A field necropsy was performed 1 h postmortem on all 6 affected animals of the goat herd after an outbreak of acute respiratory distress. The lung specimens were fixed in 10% neutral buffered formalin, embedded in paraffin wax, and cut into 5-μm-thick sections for subsequent hematoxylin-eosin and May-Grünwald Giemsa staining.
Strains and culture conditions.
Microbiologic examination of the lung samples, including aerobic and anaerobic cultures for bacteria and mycoplasma, were performed according to standard methods (16). Clinical samples were cultured on a tryptic soy blood agar base medium (CM0271; Oxoid Ltd., Basingstoke, Hampshire, England) containing 5% sheep erythrocytes at 37°C under aerobic and anaerobic atmospheres and on a mycoplasma Hayflick medium (17) under an atmosphere of 5% CO2 for 14 days with intermittent inspection of the medium plates. To determine growth on antibiotics, the tryptic soy blood agar base medium was supplemented with 200 μg/ml kanamycin (Roche Pharma, Reinach, Switzerland), 5 μg/ml vancomycin (Sigma-Aldrich, St. Louis, MO, USA), or 100 μg/ml ampicillin (Sigma-Aldrich).
DNA extraction, PCR amplification, and sequence analysis of rrs gene.
DNA from the isolates was extracted using the guanidinium thiocyanate extraction method (18) with two subsequent phenol extractions and two ethanol precipitations.
The 16S rRNA genes were amplified from bacterial DNA using FIREPol master mix (Solis BioDyne, Tartu, Estonia) and the universal 16S rRNA gene primers 16SUNI-L (AGAGTTTGATCATGGCTCAG) and 16SUNI-R (GTGTGACGGGCGGTGTGTAC), with an annealing temperature of 54°C as described previously (10). These primers amplify a fragment of the 16S rRNA gene corresponding to nucleotide positions 8 to 1410 of the reference 16S rRNA gene of Escherichia coli K-12 strain MG1655 (GenBank accession number J01859). To check the quality of the PCR products, they were run on a 1.5% agarose gel. Ten microliters of each amplicon was purified in order to remove residual deoxynucleotides and primers by adding 2.0 μl of 1-U/μl rAPid alkaline phosphatase (Roche Diagnostics, Rotkreuz, Switzerland), 0.4 μl of the corresponding buffer, and 0.1 μl of exonuclease I (New England BioLabs, Ipswich, MA, USA) and by incubation at 37°C for 20 min and then at 80°C for 20 min to inactivate the enzymes. Primers used for the PCRs were also applied for sequencing. Internal primers for 16S rRNA were universal primers, as described previously (10). Five picomoles of the appropriate primer was added to about 20 ng (1.0 μl) of the purified PCR product and sequenced with the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems) in a thermocycler for 25 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 1 min. After ethanol precipitation of the sequencing products, the samples were run on an ABI Prism 3130 XL genetic analyzer (Applied Biosystems). The sequences were edited using the Sequencher software version 5.0 (Gene Code Corp., Ann Arbor, MI, USA).
In order to directly detect the newly discovered Porphyromonas katsikii in clinical samples, we designed specific primers based on the rrs gene sequence of this novel species and a comparison with rrs gene sequences of related Porphyromonas species, resulting in the primers P._katsikii 16S_L (ATAGAGTCGGGCACGTGTG) and P._katsikii 16S_R (CAGCACCTACATACAGACCC). The PCR conditions were the same as those described for the universal primers.
Total DNA from 125-mm3 pieces of lung tissue, 1 mg of feces, or swabs taken from subgingival pockets of goats was extracted using a guanidinium-thiocyanate magnetic bead method, as described earlier (19). The samples were suspended in 1 ml SV lysis buffer (4 M guanidinium thiocyanate, 0.01 M Tris-HCl [pH 7.5], 1% β-mercaptoethanol) and were incubated at room temperature for 60 min with gentle shaking, followed by centrifugation at 4,500 × g for 10 min to remove debris. Then, 500 μl of the supernatant was mixed with 30 μl MagneSil Red magnetic bead suspension (Promega, Madison, WI, USA) in a 1.5-ml disposable Eppendorf tube and mixed for 10 min at room temperature with gentle shaking. With the aid of a magnetic separator (scil Magnetic separator 24; Promega), the supernatant was removed from the magnetic beads. The liquid phase was removed again, as described above. The magnetic beads were then washed twice with 500 μl of absolute ethanol, and the beads were air dried for approximately 40 min. Then, the DNA retained on the beads was eluted with 50 μl of pyrogen-free water by pipetting the bead pellet up and down. The DNA eluate (40 to 42 μl) was removed after 5 min of incubation at room temperature using the magnetic rack to retain the magnetic beads, and it was stored at −20°C until further use.
Phylogenetic analysis.
DNA sequences were analyzed for comparison with known sequences by BLAST (20). For the generation of a phylogenetic tree, the 16S rRNA sequence of the novel Porphyromonas species was analyzed by MEGA 6 software (21) with the following parameters: gap opening penalty, 15; gap extension penalty, 6.6; DNA weight matrix, International Union of Biochemistry (IUB); and transition weight, 0.5 using the 16S rRNA gene sequences of the type strains of the most closely related Porphyromonas species (P. levii [GenBank accession number L16493], P. somerae [NR_043312.1], P. cangingivalis [NR_113080.1], P. cansulci [NR_026137.1], P. gingivalis [NR_119038.1], P. gulae [NR_113088.1], and P. endodontalis [NR_042803.1]) and related species of other genera (Bacteroides merdae [X83954], Bacteroides fragilis [AB510701], and Prevotella oralis [L16480]). The 16S rRNA reference sequence from Escherichia coli K-12 strain MG1655 (J01859) was used as the outgroup for construction of the phylogenetic tree.
Nucleotide sequence accession number.
The sequence determined in this study has been deposited in GenBank under accession number KM360064.
RESULTS
An outbreak of acute respiratory disease affecting 6 out of 25 goats occurred in a small farm in northwest Greece. The animals that were presented to a private veterinarian showed loss of appetite for 3 days. The clinical examination revealed high fever (41.5°C), lethargy, an empty-appearing abdomen, hyperactive rumen motility, purulent nasal discharge, open-mouth breathing, and tachypnea. On auscultation, wheezing sounds were detected. A biochemical blood analysis of a characteristic diseased animal, a 2-year-old female goat, indicated that globulin (2.9 g/dl [reference range, 2.7 to 4.1 g/dl]), creatinine (1.4 mg/dl [reference range, 1.0 to 1.8 mg/dl]), glucose (68 mg/dl [reference range, 50 to 75 mg/dl]), and total bilirubin (1.1 mg/dl [reference range, 0.1 to 1.7 mg/dl]) concentrations were within their respective reference ranges. In addition, hepatic enzyme activities (sorbitol dehydrogenase, 17 IU/liter [reference range, 14 to 23.6 IU/liter]; gamma-glutamyl transferase, 26 IU/liter [reference range, 20 to 56 IU/liter]) were also within their respective reference ranges. A dipstick screening for proteinuria, hematuria, and ketone bodies in the urine was negative. Additionally, microscopic examination of Giemsa-stained thin- and thick-blood films did not reveal the presence of any blood parasites (Anaplasma ovis, Babesia ovis, or Babesia motasi). The animals were treated intramuscularly with 20 mg/kg of body weight of tylosin (Tylan 50; Elanco, Greensfield, IN, USA) twice per day. However, no clinical improvement was achieved, and all 6 affected animals died on the 1st to 3rd day of therapy. The remaining animals showed no symptoms. There were no risk factors, such as stress, crowding, or viral infections, recorded for the herd prior to the outbreak.
In the affected animals, gross lesions were confined to the lungs and consisted of generalized interstitial edema with multiple light-tan to light-yellow branching and anastomosing tracts. The edema moderately elevated the surface of the whole lung parenchyma. These pathologies were consistent with severely ectatic airways frequently distended to form a prominent nodular collection of exudate ranging from 1 mm to 4 cm in diameter. Morphologically, these lesions were consistent with a subacute, severe, and multifocal to coalescing bronchopneumonia with severe bronchiectasis, presumably of an infectious origin (Fig. 1). Furthermore, microscopic examination of the histologic section of the lung of the affected 2-year-old female goat confirmed the macroscopic morphological diagnosis by revealing multifocal to coalescing severe active suppurate bronchopneumonia characterized by bronchial and bronchiolar lumen filled with amorphous basophilic mucoid material admixed with viable and degenerating neutrophils, sloughed degenerated epithelial cells, macrophages, and cellular debris. Furthermore, the alveolar walls were multifocally expanded by infiltrating lymphocytes, macrophages, plasma cells, and neutrophils. Additionally, there was moderate hyperplasia of the bronchus-associated lymphoid tissue (BALT) with mild to moderate multifocal hyperplasia of the smooth muscles of the airways, diffuse edema, and thickening of the pleura. May-Grünwald Giemsa-stained lung tissue revealed the presence of 5-μm-long rods, indicating the presence of bacteria in the affected lung foci.
FIG 1.
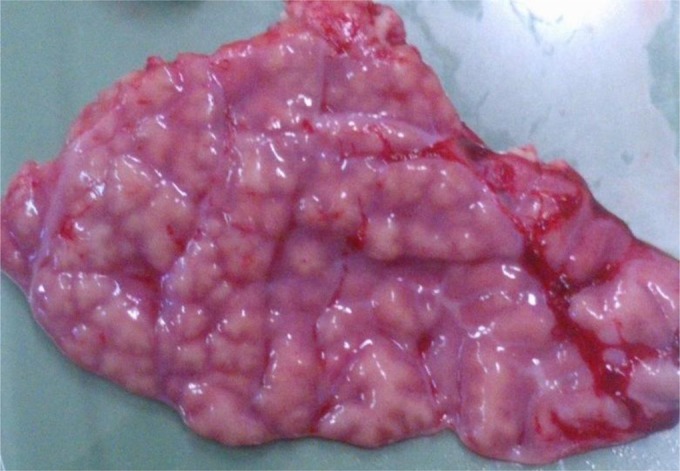
Diaphragmatic lobe of the goat. Generalized interstitial edema and multiple ectatic airways elevated the lung surface and formed branching and anastomosing tracts. Morphologically, these lesions are consistent with a subacute, severe, multifocal to coalescing bronchopneumonia with bronchiectasis.
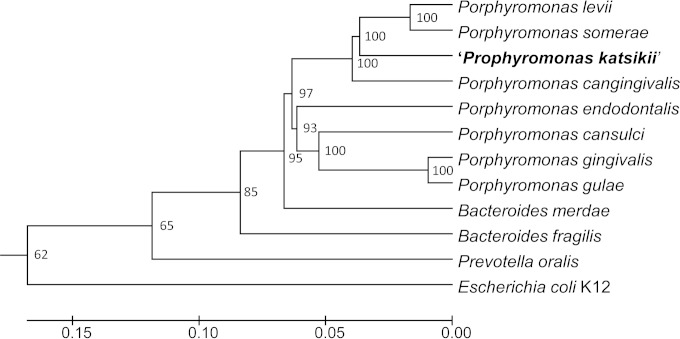
Microbiological examination of lung samples taken from multiple ectatic airway lumina revealed no growth under any of the conditions tested after 96 h. However, after 10 to 14 days under anaerobic conditions, growth of small black-pigmented colonies of rod-shaped Gram-negative bacteria were found on the blood agar medium from samples of 3 affected animals. These bacteria also grew on the blood agar medium supplemented with kanamycin or ampicillin but not with vancomycin. DNA amplification from the black-pigmented colonies and subsequent sequence analysis of the amplified rrs 16S rRNA gene showed the same 1,408-bp sequence for isolates of all 3 samples (GenBank accession number KM360064). A comparison of this DNA sequence by BLAST revealed 92% sequence similarity (identical nucleotides) with the rrs genes of Porphyromonas levii (GenBank accession number L16493) and Porphyromonas somerae (GenBank accession number L16493) as the closest known bacterial species. P. levii and P. somerae are pathogenic Porphyromonas species of cattle and humans, respectively. The interspecies variability of the 16S rRNA gene sequence of this yet-undescribed Porphyromonas sp. from goats compared to that of the 16S rRNA gene sequence of P. levii or P. somerae is 8%. This is largely above the 3% difference that is generally recommended to distinguish a new species (22). We therefore conclude that the Porphyromonas strains isolated from the diseased goats represent a novel species that we tentatively named Porphyromonas katsikii (κατσικα [katsika], Greek for goat). Figure 2 shows the phylogenetic position of Porphyromonas katsikii in relation to the closest relatives of Porphyromonas and selected species of Bacteroides and Prevotella.
FIG 2.
Phylogenetic comparison of the newly discovered species Porphyromonas katsikii in relation to the most closely related Porphyromonas species and to selected species of Bacteroides and Prevotella, based on the 16S rRNA gene sequences using unweighted-pair group method using average linkages. Escherichia coli K-12 was used as the outgroup. Bootstrap values of 1,000 simulations are shown at the branches. The scale represents sequence divergence.
In order to determine whether Porphyromonas katsikii was present as the major bacterial species in the lungs of all the diseased animals, total DNA extracted from lung tissues of the affected animals was analyzed by PCR with the universal 16S rRNA gene primers 16SUNI-L and 16SUNI-R and by sequencing of the PCR-amplified fragments. Sequence analysis of the PCR fragments we obtained resulted in the same sequence as that found for the strains (GenBank accession number KM360064). As no double or multiple peaks were found in the chromatograms of Sanger sequencing of the rrs gene amplicons from the lung samples, we concluded that Porphyromonas katsikii was the predominant or sole bacterial species found in the lungs of the diseased animals.
Direct PCR analysis of lung tissues using specific primers designed for the amplification of a specific part of the rrs gene of Porphyromonas katsikii revealed a strong positive PCR signal of 800 bp for all lung samples of the diseased animals. Lung tissue samples from 6 healthy goats taken from slaughter were used as negative controls, and samples from 7 goats with pasteurellosis pneumonia caused by Mannheimia haemolytica were used as specificity controls. The controls showed no signal upon PCR amplification with the specific Porphyromonas katsikii primers. Furthermore, subgingival samples taken from the 6 healthy goats and fecal samples from healthy goats, where Porphyromonas species are expected, showed no amplicons when tested with the P. katsikii-specific PCR, in contrast to PCR with the universal 16S rRNA primers, which was positive in these samples.
DISCUSSION
To our knowledge, this is the first report of a Porphyromonas lung infection in goats. As bacteriological diagnostics using culture methods often result in overgrowth by secondary and much faster-growing bacteria resulting from contaminations of the dead carcasses, the impact of Porphyromonas species in specimens of diseased animals is not easy to pinpoint, and the primary pathogen quite often remains undetected due to its slow growth properties. In the current case, Porphyromonas grew only after 10 days or more and was identified in only 3 animals. However, we were able to detect Porphyromonas katsikii in the lung samples of all affected animals by PCR with universal 16S rRNA gene primers followed by DNA sequencing, where it was revealed to be the only species recognized. It has to be noted that universal bacterial 16S rRNA gene primers used for PCR resulted uniquely in detectable 16S rRNA gene fragments corresponding to Porphyromonas katsikii, hence indicating that other bacteria were comparatively at very low abundance or absent in the affected lung tissue. Furthermore, P. katsikii was also detected in the affected lung tissues by the specific PCR primers. In contrast, tissues from the lungs of healthy goats or from goats with other pneumonic infections were devoid of the specific PCR signal. Hence, we postulate that the new Porphyromonas species, tentatively named Porphyromonas katsikii, was the causative agent of lung disease in the goats affected in this outbreak. The closest relatives to P. katsikii are P. levii and P. somerae, which cause various infections in animals and humans, respectively, such as vulvovaginitis, chronic skin and soft tissue infections, and bone infections (6, 7, 15).
Local infections due to Porphyromonas species are generally caused by bacterial strains present in tissues prior to the development of the clinical infection (1). In a previous study (6), cases of necrotic vulvovaginitis were associated with P. levii in cows. In that study, lesions of the genital tract and immunosuppressive stressors, such as calving, age, and primiparity of the cows, were considered risk factors of that outbreak. In our current report, the primary site of infection was not determined. P. katsikii was not detected by specific PCR from the gingival pockets of healthy goats, where the habitat of Porphyromonas species is expected, or from the feces of healthy goats. Furthermore, there was no evidence of immune suppression in the goat. It could be speculated that lungworm, which is very common among goats, especially in goats reared in backyard farms, could be the primary site of infection prior to dissemination of the bacterium to the lung. However, no ova or larvae of nematodes were evidenced in the affected animals. Porphyromonas katsikii is an anaerobic bacterium, although slight growth of this bacterium in a 5% CO2 atmosphere on mycoplasma medium was observed. Hence, one can speculate that airways filled with exudate might have served as anaerobic conditions in order to initiate the infection. The absence of P. katsikii in healthy goats, as revealed by analyzing subgingival samples, where Porphyromonas species most likely are expected to have their ecological niche, and feces suggests that the infectious agent may not be a commensal organism in these animals.
Porphyromonas infections are most probably underdiagnosed in veterinary and also human medicine, because appropriate media and growth conditions and particularly long incubation times of up to 2 weeks are not implemented in the procedures of routine diagnostic laboratories. However, in the case of a disease outbreak with an undetermined etiology, we suggest implementation of procedures with extended incubation times and the use of various growth conditions in order to isolate and identify rare bacterial pathogens, such as Porphyromonas species and other slow-growing Bacteroidetes, as potential infectious agents. Furthermore, the amplification of 16S rRNA genes using universal primes and subsequent DNA sequence analysis was shown to be a valuable rapid diagnostic tool. The current report indicates that the newly identified species Porphyromonas katsikii should be considered a possible etiological agent of lung infections in goats and related animal species.
ACKNOWLEDGMENTS
We are grateful to Paola Pilo and Francesco Origgi, Institute of Veterinary Bacteriology, Bern, Switzerland, for their expert help in editing the manuscript and Amandine Ruffieux for excellent technical support.
This project was financed by the internal research fund of the Institute of Veterinary Bacteriology, University of Bern, Bern, Switzerland.
We declare no conflict of interest.
REFERENCES
- 1.Falagas ME, Siakavellas E. 2000. Bacteroides, Prevotella, and Porphyromonas species: a review of antibiotic resistance and therapeutic options. Int J Antimicrob Agents 15:1–9. doi: 10.1016/S0924-8579(99)00164-8. [DOI] [PubMed] [Google Scholar]
- 2.Finegold SM, Jousimies-Somer H. 1997. Recently described clinically important anaerobic bacteria: medical aspects. Clin Infect Dis 25(Suppl 2):S88–S93. doi: 10.1086/516237. [DOI] [PubMed] [Google Scholar]
- 3.Klarstrom Engstrom K, Khalaf H, Kalvegren H, Bengtsson T. 8 July 2014. The role of Porphyromonas gingivalis gingipains in platelet activation and innate immune modulation. Mol Oral Microbiol doi: 10.1111/omi.12067. [DOI] [PubMed] [Google Scholar]
- 4.Mysak J, Podzimek S, Sommerova P, Lyuya-Mi Y, Bartova J, Janatova T, Prochazkova J, Duskova J. 2014. Porphyromonas gingivalis: major periodontopathic pathogen overview. J Immunol Res 2014:476068. doi: 10.1155/2014/476068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riggio MP, Lennon A, Taylor DJ, Bennett D. 2011. Molecular identification of bacteria associated with canine periodontal disease. Vet Microbiol 150:394–400. doi: 10.1016/j.vetmic.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Elad D, Friedgut O, Alpert N, Stram Y, Lahav D, Tiomkin D, Avramson M, Grinberg K, Bernstein M. 2004. Bovine necrotic vulvovaginitis associated with Porphyromonas levii. Emerg Infect Dis 10:505–507. doi: 10.3201/eid1003.020592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Summanen PH, Durmaz B, Vaisanen ML, Liu C, Molitoris D, Eerola E, Helander IM, Finegold SM. 2005. Porphyromonas somerae sp. nov., a pathogen isolated from humans and distinct from Porphyromonas levii. J Clin Microbiol 43:4455–4459. doi: 10.1128/JCM.43.9.4455-4459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sweeney M, Watts J, Portis E, Lucas M, Nutsch R, Meeuwse D, Bade D, Oliver V, Morck DW, Shinabarger D, Poppe S, Peterson M, Sweeney D, Knechtel M, Zurenko G. 2009. Identification of Porphyromonas levii isolated from clinical cases of bovine interdigital necrobacillosis by 16S rRNA sequencing. Vet Ther 10:E1–E10. [PubMed] [Google Scholar]
- 9.Frandsen EV, Poulsen K, Kilian M. 1995. Confirmation of the species Prevotella intermedia and Prevotella nigrescens. Int J Syst Bacteriol 45:429–435. doi: 10.1099/00207713-45-3-429. [DOI] [PubMed] [Google Scholar]
- 10.Kuhnert P, Frey J, Lang NP, Mayfield L. 2002. Phylogenetic analysis of Prevotella nigrescens, Prevotella intermedia and Porphyromonas gingivalis clinical strains reveals a clear species clustering. Int J Syst Evol Microbiol 52:1391–1395. doi: 10.1099/ijs.0.02021-0. [DOI] [PubMed] [Google Scholar]
- 11.Bekana M, Jonsson P, Ekman T, Kindahl H. 1994. Intrauterine bacterial findings in postpartum cows with retained fetal membranes. Zentralbl Veterinarmed A. doi: 10.1111/j.1439-0442.1994.tb00134.x. [DOI] [PubMed] [Google Scholar]
- 12.Moe KK, Yano T, Misumi K, Kubota C, Nibe K, Yamazaki W, Muguruma M, Misawa N. 2010. Detection of antibodies against Fusobacterium necrophorum and Porphyromonas levii-like species in dairy cattle with papillomatous digital dermatitis. Microbiol Immunol 54:338–346. doi: 10.1111/j.1348-0421.2010.00220.x. [DOI] [PubMed] [Google Scholar]
- 13.Lobb DA, Loeman HJ, Sparrow DG, Morck DW. 1999. Bovine polymorphonuclear neutrophil-mediated phagocytosis and an immunoglobulin G2 protease produced by Porphyromonas levii. Can J Vet Res 63:113–118. [PMC free article] [PubMed] [Google Scholar]
- 14.Jousimies-Somer H, Summanen P. 1999. Microbiology terminology update: clinically significant anaerobic Gram-positive and Gram-negative bacteria (excluding spirochetes). Clin Infect Dis 29:724–727. doi: 10.1086/520422. [DOI] [PubMed] [Google Scholar]
- 15.Blum S, Brenner J, Friedgut O, Stram Y, Koren O, Dagoni I, Munbaz A, Elad D. 2008. Isolation of Porphyromonas levii from vaginal samples from cows in herds negative for bovine necrotic vulvovaginitis. Vet Rec 163:745–747. [PubMed] [Google Scholar]
- 16.Quinn PJ, Carter ME, Markey B, Carter GR. 1994. Clinical veterinary microbiology. Mosby, London, United Kingdom. [Google Scholar]
- 17.Hayflick L. 1965. Tissue cultures and mycoplasmas. Tex Rep Biol Med 23(Suppl 1):285–303. [PubMed] [Google Scholar]
- 18.Pitcher DG, Saunders NA, Owen RJ. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol 8:151–156. doi: 10.1111/j.1472-765X.1989.tb00262.x. [DOI] [Google Scholar]
- 19.Stäuble A, Steiner A, Normand L, Kuhnert P, Frey J. 2014. Molecular genetic analysis of Dichelobacter nodosus proteases AprV2/B2, AprV5/B5 and BprV/B in clinical material from European sheep flocks. Vet Microbiol 168:177–284. doi: 10.1016/j.vetmic.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 20.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol. Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, Feil EJ, Stackebrandt E, Van de PY, Vandamme P, Thompson FL, Swings J. 2005. Opinion: re-evaluating prokaryotic species. Nat Rev Microbiol 3:733–739. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]