Abstract
Enterovirus D68 (EV-D68) has been recognized as an important cause of acute respiratory infections. Here we report the molecular epidemiology of EV-D68 in Philippines from 2013 to 2014; we found cases in areas affected by Typhoon Haiyan and found new strains in the country.
TEXT
Enterovirus D68 (EV-D68) is a member of the Picornaviridae family and is primarily associated with respiratory illness (1). Since its discovery in 1962 (2), EV-D68 has been detected only sporadically among patients with respiratory illness until recently (3). Since 2004, reports of EV-D68 infections have increased worldwide in Philippines, Japan, Thailand, the Netherlands, the United States, New Zealand, and Kenya (4–8). Besides, the United States and Canada have continued to experience a significant EV-D68 outbreak since August 2014, which has resulted in many severe infections, particularly among children (9). However, there have been few reports of the virus in other countries in 2013 and 2014.
Nasopharyngeal swab samples were collected between September 2012 and February 2014 from pediatric patients hospitalized with severe acute respiratory infection (sARI) at three hospitals in Philippines: Eastern Visayas Regional Medical Center (EVRMC; Tacloban City, Lyete); Biliran Provincial Hospital (BPH; Naval, Biliran); and Ospital ng Palawan (ONP; Puerto Princesa City, Palawan). Nucleic acids were extracted from all samples and tested for respiratory viruses, including EV-D68, adenovirus, cytomegalovirus, human metapneumovirus, influenza virus, parainfluenza virus, respiratory syncytial virus, rhinovirus, and other enteroviruses (including coxsackievirus, echovirus, and poliovirus), by reverse transcription-PCR using previously described methods with slight modifications (10). We targeted the 5′ untranslated region of the EV-D68 genome for detection and the VP1 gene of the virus for sequencing analysis. A phylogenetic tree based on the genetic sequence of a partial VP1 region (542 nucleotides) was inferred using the maximum-likelihood method based on the T92 model with gamma distribution using MEGA6 software (11). The present study was approved by the Institutional Review Board of each institute, and informed consent was obtained from guardians of all enrolled pediatric patients.
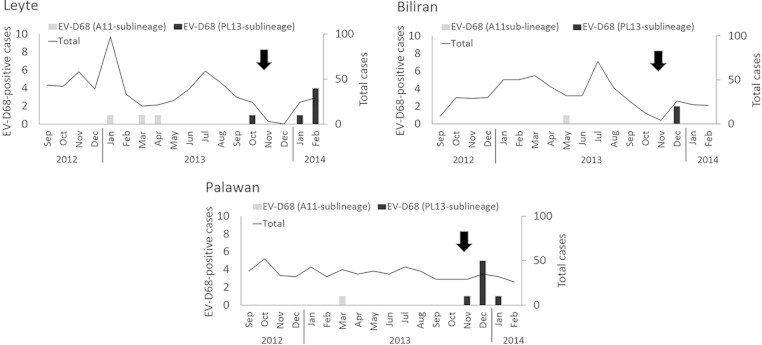
We identified 20 EV-D68-positive cases among 1,854 hospitalized patients with sARI. The most common viral pathogen identified among sARI cases was respiratory syncytial virus (29.3%), followed by rhinovirus (14.0%) and influenza virus (5.1%). The detection rate of EV-D68 in this study was 1.0% (20/1,854). The ages of EV-D68-positive patients ranged from 1 month to 4 years (median, 14 months). During the study period, Typhoon Haiyan (Yolanda), which hit Philippines on 8 November 2013 (12), caused extensive damage to large areas of the country, including some of our study sites. Our study sites included EVRMC in Tacloban City, which is located on Leyte Island, one of the municipalities most severely damaged by the typhoon. It should be noted that our research projects were temporarily discontinued after the typhoon; thus, we were unable to collect continuous samples in this region (Fig. 1). Interestingly, we found 13 positive cases after the typhoon hit. It is, however, unclear whether the typhoon had any impact on EV-D68 circulation.
FIG 1.
Monthly distribution of EV-D68-positive cases at each study site. The total number and number of EV-D68-positive cases are shown. Arrows indicate the occurrence of Typhoon Haiyan. The genetic sublineages of EV-D68 are shown in Fig. 2. Jan, January; Feb, February; Mar, March; Apr, April; Jun, June; Jul, July; Aug, August; Sep, September; Oct, October; Nov, November; Dec, December.
In terms of coinfection, we found 3 cases positive for both EV-D68 and respiratory syncytial virus. One patient (2 months of age) coinfected with the viruses died during the study period. All the other patients who were infected with EV-D68 recovered. Moreover, patients with asthma tend to develop severe symptoms from EV-D68 infection (9, 13). During this study period, 35% (7/20) of EV-D68-positive sARI patients exhibited wheezing.
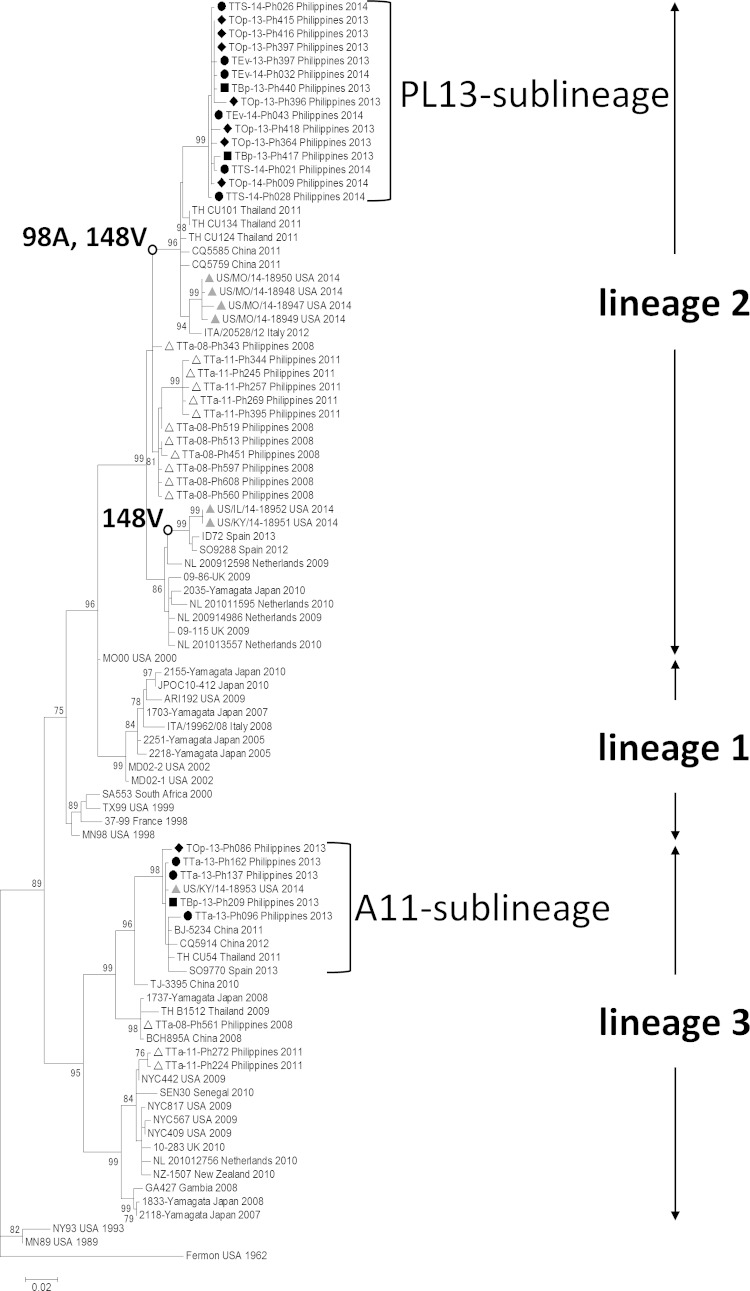
Next, we constructed a phylogenetic tree to compare the VP1 gene sequences of the EV-D68 strains, which showed that the samples were divided into two distinct sublineages: A11 (in lineage 3) and PL13 (in lineage 2) (Fig. 2; the numbering of the lineages is described in reference 14). All EV-D68-positive samples collected before May 2013 were classified into the A11 sublineage, which also consisted of strains from Thailand and China collected in 2011 and 2012. However, EV-D68-positive samples after October 2013 formed a distinct sublineage, PL13. Although the strains in the PL13 sublineage were closely related to the strains collected in Thailand and China in 2011 and 2012, this sublineage was clearly distinct. Among our samples, we found no obvious clustering by study site. As mentioned above, the PL13 sublineage emerged in October 2013 and all strains detected thereafter were classified as PL13. Notably, we found amino acid substitutions in the predicted antigenic sites of the VP1 gene in the PL13 sublineage: alanine at position 98 (98A) in the BC loop and valine at position 148 (148V) in the DE loop. Furthermore, sequence data from seven strains from the current 2014 outbreak in North America became available as of October 2014 (15). One of them was classified into the A11 sublineage (in lineage 3), whereas the 6 others were in lineage 2 (Fig. 2). Four of the 6 strains in lineage 2 clustered near the PL13 sublineage, and 2 formed a distinct cluster. None of the strains from the United States seems to be a direct descendant of PL13 sublineage. Therefore, the phylogenetic analysis did not support the idea that PL13 sublineage viruses were transmitted in 2013 and 2014 from Philippines to the United States, leading to the ongoing widespread outbreak. Interestingly, strains in the PL13 sublineage and some American strains in 2014 share common mutations in the antigenic sites mentioned above (Fig. 2).
FIG 2.
A phylogenetic tree of EV-D68 strains in Philippines and reference strains. Significant mutations (98A and 148V) mentioned in the body are shown next to nodes. Bootstrap values of >70% (in 1,000 tests) are shown next to the branches. A black circle, square, and rhombus indicate strains included in this study collected from Leyte, Biliran, and Palawan, respectively. The white triangles indicate strains previously reported from Philippines in 2001 and 2008. The gray triangles indicate strains reported in the United States in 2014.
In this study, we performed detection and molecular analysis of EV-D68 in Philippines from 2013 to 2014. Our analyses revealed the current situation with respect to EV-D68 infection in the country. EV-D68 has been circulating in the population, and we found a large number of infections in the postdisaster setting. We also detected the emergence of new strains (PL13 sublineage) in the latter part of 2013. To better understand infectious disease outbreaks in postdisaster settings and associations between local circulation and global circulation of viral pathogens, continuous global surveillance is necessary.
Nucleotide sequence accession numbers.
Sequence data have been deposited in the GenBank database under accession numbers AB992413 to AB992443.
ACKNOWLEDGMENTS
We thank the staff of EVRMC, BPH, and ONP.
This work was supported by Grants-in-Aid from the Japan Initiative for Global Research Network on Infectious Diseases (J-GRID); the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan; Science and Technology Research Partnership for Sustainable Development (SATREPS); the Japan Science and Technology Agency (JST); and Japan International Cooperation Agency (JICA).
REFERENCES
- 1.Oberste MS, Maher K, Schnurr D, Flemister MR, Lovchik JC, Peters H, Sessions W, Kirk C, Chatterjee N, Fuller S, Hanauer JM, Pallansch MA. 2004. Enterovirus 68 is associated with respiratory illness and shares biological features with both the enteroviruses and the rhinoviruses. J Gen Virol 85:2577–2584. doi: 10.1099/vir.0.79925-0. [DOI] [PubMed] [Google Scholar]
- 2.Schieble JH, Fox VL, Lennette EH. 1967. A probable new human picornavirus associated with respiratory diseases. Am J Epidemiol 85:297–310. [DOI] [PubMed] [Google Scholar]
- 3.Khetsuriani N, Lamonte-Fowlkes A, Oberst S, Pallansch MA; Centers for Disease Control and Prevention. 2006. Enterovirus surveillance–United States, 1970-2005. MMWR Surveill Summ 55:1–20. http://www.cdc.gov/MMWR/preview/mmwrhtml/ss5508a1.htm. [PubMed] [Google Scholar]
- 4.Wang Z, Malanoski AP, Lin B, Long NC, Leski TA, Blaney KM, Hansen CJ, Brown J, Broderick M, Stenger DA, Tibbetts C, Russell KL, Metzgar D. 2010. Broad spectrum respiratory pathogen analysis of throat swabs from military recruits reveals interference between rhinoviruses and adenoviruses. Microb Ecol 59:623–634. doi: 10.1007/s00248-010-9636-3. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2011. Clusters of acute respiratory illness associated with human enterovirus 68—Asia, Europe, and United States, 2008-2010. MMWR Morb Mortal Wkly Rep 60:1301–1304. [PubMed] [Google Scholar]
- 6.Todd AK, Hall RJ, Wang J, Peacey M, McTavish S, Rand CJ, Stanton JA, Taylor S, Huang QS. 2013. Detection and whole genome sequence analysis of an enterovirus 68 cluster. Virol J 10:103. doi: 10.1186/1743-422X-10-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Opanda SM, Wamunyokoli F, Khamadi S, Coldren R, Bulimo WD. 2014. Genetic diversity of human enterovirus 68 strains isolated in Kenya using the hypervariable 3′-end of VP1 gene. PLoS One 9:e102866. doi: 10.1371/journal.pone.0102866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linsuwanon P, Puenpa J, Suwannakarn K, Auksornkitti V, Vichiwattana P, Korkong S, Theamboonlers A, Poovorawan Y. 2012. Molecular epidemiology and evolution of human enterovirus serotype 68 in Thailand, 2006-2011. PLoS One 7:e35190. doi: 10.1371/journal.pone.0035190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Midgley CM, Jackson MA, Selvarangan R, Turabelidze G, Obringer E, Johnson D, Giles BL, Patel A, Echols F, Oberste MS, Nix WA, Watson JT, Gerber SI. 2014. Severe respiratory illness associated with enterovirus d68—Missouri and Illinois, 2014. MMWR Morb Mortal Wkly Rep 63:798–799. [PMC free article] [PubMed] [Google Scholar]
- 10.Imamura T, Fuji N, Suzuki A, Tamaki R, Saito M, Aniceto R, Galang H, Sombrero L, Lupisan S, Oshitani H. 2011. Enterovirus 68 among children with severe acute respiratory infection, the Philippines. Emerg Infect Dis 17:1430–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan EY, Liu S, Hung KK. 2013. Typhoon Haiyan and beyond. Lancet 382:1873. doi: 10.1016/S0140-6736(13)62415-0. [DOI] [PubMed] [Google Scholar]
- 13.Hasegawa S, Hirano R, Okamoto-Nakagawa R, Ichiyama T, Shirabe K. 2011. Enterovirus 68 infection in children with asthma attacks: virus-induced asthma in Japanese children. Allergy 66:1618–1620. doi: 10.1111/j.1398-9995.2011.02725.x. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda T, Mizuta K, Abiko C, Aoki Y, Itagaki T, Katsushima F, Katsushima Y, Matsuzaki Y, Fuji N, Imamura T, Oshitani H, Noda M, Kimura H, Ahiko T. 2012. Acute respiratory infections due to enterovirus 68 in Yamagata, Japan between 2005 and 2010. Microbiol Immunol 56:139–143. doi: 10.1111/j.1348-0421.2012.00411.x. [DOI] [PubMed] [Google Scholar]
- 15.Brown BA, Nix WA, Sheth M, Frace M, Oberste MS. 2014. Seven strains of enterovirus D68 detected in the United States during the 2014 severe respiratory disease outbreak. Genome Announc 2:e01201-14. doi: 10.1128/genomeA.01201-14. [DOI] [PMC free article] [PubMed] [Google Scholar]