Abstract
Mycoplasma bovis is a major bovine pathogen associated with bovine respiratory disease complex and is responsible for substantial economic losses worldwide. M. bovis is also associated with other clinical presentations in cattle, including mastitis, otitis, arthritis, and reproductive disorders. To gain a better understanding of the genetic diversity of this pathogen, a multilocus sequence typing (MLST) scheme was developed and applied to the characterization of 137 M. bovis isolates from diverse geographical origins, obtained from healthy or clinically infected cattle. After in silico analysis, a final set of 7 housekeeping genes was selected (dnaA, metS, recA, tufA, atpA, rpoD, and tkt). MLST analysis demonstrated the presence of 35 different sequence types (STs) distributed in two main clonal complexes (CCs), defined at the double-locus variant level, namely, CC1, which included most of the British and German isolates, and CC2, which was a more heterogeneous and geographically distant group of isolates, including European, Asian, and Australian samples. Index of association analysis confirmed the clonal nature of the investigated M. bovis population, based on MLST data. This scheme has demonstrated high discriminatory power, with the analysis showing the presence of genetically distant and divergent clusters of isolates predominantly associated with geographical origins.
INTRODUCTION
Mycoplasma diseases cause substantial economic losses, particularly in intensively farmed cattle production systems worldwide, as a result of poor growth, morbidity, and deaths, as well as the costs associated with increased control and prophylactic measures. Mycoplasma bovis has increasingly been recognized as one of the main pathogens involved in the bovine respiratory disease complex, on its own or in association with other respiratory pathogens (1). M. bovis can also be found in association with mastitis, in which outbreaks can affect more than 20% of the cows in a herd, regardless of the stage of lactation, and infections are usually refractory to treatment. Arthritis and otitis have also been associated with M. bovis, usually appearing once pneumonia or mastitis is already established in the herd (1, 2). The control of M. bovis infections relies strongly on antimicrobial therapy, which has variable success rates in the field (3). Vaccination has been used in the early stages of cattle development and is mostly based on autogenous vaccines, which limits their use and the potential for widespread control of M. bovis infections (3).
Taking into consideration the limited tools available for M. bovis disease management, the development of a dependable molecular typing scheme able to offer robust and reproducible epidemiological information would provide a valuable addition to control measures targeting this pathogen. M. bovis isolates have been characterized previously using multiple molecular typing methods, including amplified fragment length polymorphism (AFLP) analysis (4), random amplified polymorphic DNA (RAPD) analysis (5), pulsed-field gel electrophoresis (PFGE) analysis (6), insertion sequence analysis (7, 8), and, more recently, multilocus variable-number tandem repeat (VNTR) analysis (8–10). Although some of these techniques have demonstrated their usefulness as reference molecular typing schemes for other important microbial pathogens, including PFGE for Escherichia coli (11) and VNTR analysis for Mycobacterium bovis (12), the lack of universal comparability, transferability of data, and unambiguous interpretation has hampered their use as routine typing techniques for the characterization of M. bovis.
Multilocus sequence typing (MLST), a technique initially described for the analysis of the pathogenic bacteria Neisseria meningitidis (13), is a sequence-based typing technique that focuses on characterizing single mutations in sets of housekeeping genes in order to establish relationships between populations of microorganisms. The availability of rapid affordable DNA-sequencing services in recent years has enabled the use of this technique for molecular epidemiological studies, which are increasingly recognized as the gold standard for typing analysis of microbial populations (14). The high accuracy and repeatability of current sequencing technology, the universal comparability of DNA sequences, and the easy transfer of data between laboratories make this technique the most complete, robust, and reliable typing method described to date (15). MLST has been successfully applied to the analysis of Mycoplasma agalactiae (16), a Mycoplasma species closely related to M. bovis, and also Mycoplasma hyopneumoniae (17), a major porcine respiratory pathogen. This study describes the development of an MLST system for the characterization of M. bovis isolates and its use for the analysis of the population structure of a selection of 137 isolates.
MATERIALS AND METHODS
Bacterial isolates.
A total of 137 M. bovis isolates from cattle, including the type strain PG45, were analyzed in this study. Field isolates from 12 different countries, including representatives from Europe, Asia, and Australia in addition to the type strain PG45, which was isolated in North America, were analyzed. The isolate selection covered the maximum number of variables possible, including isolates from outbreaks and routine surveillance, samples from healthy animals, historical and recent samples, and isolates obtained from different clinical presentations and specimen types (see Table S1 in the supplemental material).
Culture and DNA extraction.
M. bovis isolates were propagated at 37°C in 5% CO2 for 24 h, in 3-ml volumes of Eaton's broth (18). Ten microliters of culture was then plated on Eaton's agar and incubated for 72 h. After this incubation, single colonies were selected and grown in 3 ml of Eaton's broth for 48 h. DNA was then extracted from 400 μl of each culture by using an automated nucleic acid extraction system (Maxwell 16 cell DNA purification kit; Promega), following the manufacturer's instructions. The species and purity of the extracted DNA were verified by universal and Mycoplasma-specific PCR-denaturing gradient gel electrophoresis (DGGE) assays (19, 20) prior to use in the MLST PCRs.
MLST.
The most common targets used in published MLST schemes were found at the PubMLST website (http://pubmlst.org) (21), and their presence in M. bovis was assessed. The final selection of MLST targets was performed after comparison of the numbers of nucleotide polymorphisms in selected genes obtained from the complete genomes of two geographically distant M. bovis isolates, namely, PG45 (GenBank accession number NC_014760) isolated in the United States and Hubei-1 (GenBank accession number NC_015725) isolated in China. A final set of 7 genes showing the greatest number of nucleotide polymorphisms among the isolates described previously was selected for this scheme; the genes selected were atpA, dnaA, metS, recA, rpoD, tkt, and tufA). The primer sequences and characteristics of each selected locus can be found in Table 1.
TABLE 1.
Primer sequences and main characteristics of selected loci in M. bovis MLST typing scheme
| Gene | Primer name | Primer sequence (5′ to 3′) | Product size (bp) | Analyzed sequence size (bp) | Nucleotide positions within genea | Hb | No. of alleles | No. of polymorphic sites | Z test resultc |
|---|---|---|---|---|---|---|---|---|---|
| dnaA | dnaA-F | TCAAATCGTGAAGTCGACAA | 924 | 498 | 532–1029 | 0.51 | 11 | 28 | dN < dS |
| dnaA-R | CTTCCCAATTTGTTCCAGTG | ||||||||
| metS | metS-F | TCGGACAACTGATCCTAAGC | 867 | 596 | 439–1009 | 0.79 | 12 | 29 | NS |
| metS-R | AATAGGCCTTCTGGGAAAGA | ||||||||
| recA | recA-F | TCCTAAAGGCAGAATCGTTG | 711 | 463 | 248–710 | 0.47 | 6 | 12 | NS |
| recA-R | TAAGCATATCAAGCGCCTTT | ||||||||
| tufA | tufA-F | TGATTACTGGTGCTGCTCAA | 798 | 499 | 447–945 | 0.54 | 6 | 17 | dN > dS |
| tufA-R | TTTCACCAGGTTGAACGAAT | ||||||||
| atpA | atpA-F | TGTTGCTATATTTGGTAACGC | 787 | 589 | 322–910 | 0.70 | 7 | 23 | dN < dS |
| atpA-R | AACATTAGTAGGAATATAAGCTG | ||||||||
| rpoD | rpoD-F | ATATTATGGATGAAGTTGATGC | 861 | 600 | 381–980 | 0.74 | 11 | 21 | dN > dS |
| rpoD-R | CGCCATTTTCTTGTTGTAGC | ||||||||
| tkt | tkt-F | CATGTGAATATAAATGCACTGC | 677 | 394 | 743–1136 | 0.76 | 9 | 30 | dN < dS |
| tkt-R | TGATAAGGCAATTACTGAAGC |
Based on the complete genome of M. bovis strain PG45.
H, genetic diversity.
dN < dS, test for purifying selection statistically significant; dN > dS, test for positive selection statistically significant; NS, no statistical significance found.
All PCRs were performed in an iCycler thermal cycler (Bio-Rad), with an initial step of denaturation at 95°C for 5 min, 30 cycles of denaturation at 95°C for 1 min, annealing at 56°C for 1 min, and extension at 72°C for 1 min, and a final step of extension at 72°C for 10 min. Each PCR was performed in a final volume of 50 μl, which included 10 μl of 5× Green GoTaq Flexi buffer (Promega), 1.5 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate, 1.25 units of GoTaq DNA polymerase (Promega), 1 μl of each forward and reverse primer (Table 1) at a concentration of 25 pmol/μl, and 1 μl of DNA. PCR amplification was confirmed after electrophoresis of the amplified products in 2% agarose gels, with detection under UV illumination after staining with GelRed (Biotium).
PCR products in the reaction mixture were purified using a QIAquick PCR purification kit (Qiagen), following the manufacturer's instructions, and were quantified using a NanoDrop 1000 spectrophotometer (Thermo Scientific). Sequencing reactions were performed using a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems), using the primers described in Table 1, and were run in an ABI 3130xl genetic analyzer (Applied Biosystems). Both strands from each PCR amplicon were sequenced and analyzed.
Sequence data were aligned using SeqMan v8.1.5 (DNASTAR), and each individual sequence was trimmed according to the nucleotide positions selected for each gene (Table 1). Trimmed DNA sequences were analyzed using the nonredundant database (NRDB) comparison tool (http://pubmlst.org/analysis). Each distinct allele present in an individual locus was identified by the assignment of an arbitrary number. The combination of the seven allele identifiers formed an allelic profile, which was used to determine a unique identifier or sequence type (ST).
The possible evolutionary relationship between isolates and M. bovis population structure was determined using PHYLOViZ (22) and was evaluated by analysis of a minimum spanning tree (MST) created using the global optimal eBURST (goeBURST) algorithm (23). The null hypothesis of linkage equilibrium for multilocus data was tested using the standardized index of association (IA) in order to determine the role of recombination in population evolution, using the LIAN 3.5 application (24) (http://pubmlst.org/analysis). LIAN 3.5 was also used to calculate the genetic diversity (H) (25) of each locus, analyzed as a measure of the expected heterozygosity or genetic variability of each gene. This value ranges from 0 (no heterozygosity) to 1. The discriminatory power of this typing scheme was determined after calculation of Simpson's index of diversity (SID), as described previously (26). This index, which ranges from 0 (no discriminatory power) to 1 (highest discriminatory power), facilitates comparisons of the discriminatory powers of typing techniques. It is recommended that SID be greater than 0.9 to allow typing results to be interpreted with confidence. The number of polymorphic sites was obtained using START v2 (27). The number of nonsynonymous substitutions per nonsynonymous site (dN) and the number of synonymous substitutions per synonymous site (dS) were calculated, and the null hypothesis of strict neutrality (dN = dS) versus the alternative hypotheses of purifying (dN < dS) or positive (dN > dS) selection was tested for each locus using the Z test in MEGA5 software (28). MEGA5 was also used to align and to create a phylogenetic reconstruction based on the minimum evolution method described previously (29), using concatenated amino acid sequences of the allelic profiles described for each isolate.
Nucleotide sequence accession numbers.
The sequences of all alleles found for each gene can be found in GenBank (accession numbers KP229456 to KP229517).
RESULTS
Discriminatory power and locus characteristics.
The discriminatory power of this typing scheme, as calculated using the SID, was 0.91 (95% confidence interval, 0.88 to 0.93). This value indicates that, if two M. bovis isolates were selected at random from the population, they would fall in different MLST types on 91% of the occasions. Genetic diversity values obtained for each locus ranged from 0.47 for recA (less genetically diverse locus) to 0.79 for metS (more genetically diverse locus). The numbers of alleles found ranged from 6 (recA and tufA) to 12 (dnaA and metS). The numbers of polymorphic sites varied from 12 in recA to 30 in tkt. The numbers of polymorphic sites and genetic diversity (H) values for each locus can be seen in Table 1.
Based on the Z test of selection, tufA and rpoD were found to be under positive selection in M. bovis (P < 0.05), showing a higher rate of nonsynonymous substitutions. The atpA, dnaA, and tkt genes were found to be under purifying or neutral selection (P < 0.05), and no statistical differences were found for the metS and recA genes (Table 1).
Evolutionary relationships and population structure.
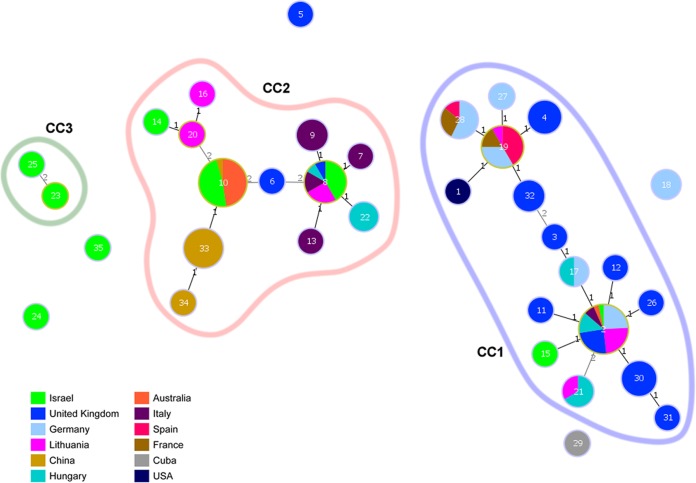
The 137 isolates analyzed gave a total of 35 different STs (see Table S1 in the supplemental material), divided into three clonal complexes (CCs) found after goeBURST and MST analysis (Fig. 1). In our study, a clonal complex was defined as a group of similar isolates in which every isolate shared at least five of seven identical alleles with at least one other ST in the group (30). The main clonal complex, CC1, included 73 isolates (53.28% of the total). Two common ancestors were identified in CC1, i.e., ST2, which was the most common ST found in the characterized M. bovis population and included isolates from multiple geographical origins (United Kingdom, Lithuania, Hungary, Germany, Italy, Israel, and Australia), and ST19, which was represented by isolates originating from mainland Europe only (Spain, Germany, France, and Lithuania). The latter was shown to have the greatest similarity to the type strain PG45, which was isolated in the United States, with differences being observed only at the single-locus variant level. The majority of the German and United Kingdom isolates clustered in this clonal complex, with approximately 90% of the characterized isolates from these countries being found in this cluster.
FIG 1.
Minimum spanning tree (MST) representing the evolutionary relationships between M. bovis sequence types (STs). Numbers inside circles, STs. Numbers over connecting lines, locus variant levels (1, single-locus variant; 2, double-locus variant). CC, clonal complex.
CC2 included 56 of the 137 analyzed isolates (40.88% of the total). This group presented a high level of diversity in the geographical origins of the isolates analyzed, including all Chinese isolates and the majority of the Israeli and Australian isolates.
A minor clonal complex, CC3, which included only two Israeli isolates (1.46% of the total), was identified in addition to the two main complexes described. Furthermore, five singleton STs that did not cluster in any of the main CCs were also observed. The list of singletons included ST5, ST18, ST24, ST29, and ST35, with a total of 6 isolates (4.38% of the total) (see Table S1 in the supplemental material).
No clear pattern of distribution of isolates according to clinical presentation was observed. Approximately 60% of the analyzed samples were obtained from cases of respiratory infection. Although this set of samples included isolates from two well-differentiated pathological presentations, i.e., cranioventral consolidation of the lung and caseous necrosis, no clear differential clustering was detected.
Some of the most diverse isolates analyzed, which included the majority of the singleton STs and CC3, were isolated from mastitis cases. The remaining singleton isolates, 144B08 (ST5) and 422/88 (ST29), were obtained from cases of pneumonia. Isolate 144B08, a highly pathogenic M. bovis isolate, was obtained from an experimental respiratory infection in cattle in the United Kingdom that was induced with a U.S. isolate. Isolate 422/88, which was isolated in 1988 in Cuba, is one of the oldest and more geographically and genetically atypical samples analyzed in this study. Other historical isolates were also characterized; these were isolated between 1978 and 1993 in various European countries and included ST19, ST27, and ST28, which were similar to the type strain PG45, with maximal differences of two loci.
Within-farm variations were observed for some of the isolates analyzed. For example, isolates 63982 (ST13) and 105880 (ST8), which were obtained at the same time on the same farm from different animals presenting different clinical signs (pneumonia and mastitis), had diverse STs, which differed only in a single mutation in the dnaA gene. Other samples obtained from the same farms at different time points also presented different STs. For example, isolates 223B09 (ST31) and 263B09 (ST12), which were obtained from pneumonia cases on the same farm with a 10-day gap between sampling times, presented differences in two alleles (dnaA and rpoD). Within-farm differences were also observed in mastitis cases; various isolates obtained from a mastitis outbreak (isolates 343B09, 345B09, 346B09, and 393B09 [ST2]) differed in one allele (rpoD) from isolate 392B09 (ST30), which was obtained 20 days later (Fig. 1; also see Table S1 in the supplemental material).
A phylogenetic tree showing the evolutionary history of isolates based on the analysis of concatenated amino acid sequences is shown in Fig. S1 in the supplemental material. All positions containing gaps and missing data were eliminated. There were a total of 1,165 positions in the final data set. A clear split into two main M. bovis populations, similar to that observed after goeBURST analysis, was found after the phylogenetic analysis of the amino acid sequences. However, the presence of synonymous substitutions reduced the number of unique M. bovis types. Based on this analysis, ST11, ST12, and ST17 were found to be identical to ST2, ST32 was found to be identical to ST28, and ST33 and ST34 were found to be identical to ST10. In spite of the reduced discriminatory power found in analyses of amino acid sequences, an apparent clustering of isolates based on geographical origins can be seen with this approach. For example, ST8, found in CC2 after goeBURST analysis and found mainly in mainland European isolates, clustered in the main branch of the concatenated amino acid phylogenetic tree that included ST2 and ST19, the two main European STs found in mainland European and British isolates. Also, geographically diverse isolates such as Chinese and Australian isolates found in ST10, ST33, and ST34 clustered together in the same branch of the phylogenetic tree, in close relationships with Israeli isolates obtained from animals in direct contact with imported Australian cattle.
DISCUSSION
This study describes a standardized MLST scheme that was successfully applied to the analysis of a diverse population of M. bovis isolates. The newly developed MLST scheme, based on seven housekeeping genes (dnaA, metS, recA, tufA, atpA, rpoD, and tkt) selected after in silico analysis, enabled clear differentiation of M. bovis isolates mainly on the basis of their geographical origins, with good discriminatory power (SID = 0.91). An attempt to develop an MLST scheme for the analysis of M. bovis isolates was described previously (31). Although the scheme was useful for differentiation between closely related Mycoplasma species, such as M. agalactiae, the small number of strains tested (n = 8) did not provide enough evidence to support its selection as a MLST scheme of choice for M. bovis analysis, based on the guidelines for the validation and application of typing methods for use in bacterial epidemiology (15).
LIAN analysis based on MLST data from the described set of M. bovis housekeeping genes suggests that the investigated bacterial population may be in linkage disequilibrium, meaning that its evolution is not associated with recombination. Therefore, natural genetic drift may explain the differential evolution of this population. This scenario is similar to that seen following MLST analysis of M. agalactiae (16), a closely related Mycoplasma species that shares up to 99% of its 16S rRNA gene sequence with M. bovis (32). However, other authors have demonstrated, using complete genome analysis, a possible role for sexual reproduction in the evolution of M. agalactiae (33). Moreover, recent studies have shown how recombination is directly involved in the evolution of members of the Mycoplasma mycoides cluster (34), as well as the porcine mycoplasmas M. hyopneumoniae and Mycoplasma flocculare (35). Given the extreme relatedness to M. agalactiae and the evidence of the effects of recombination in the evolution of other mycoplasmas, it is possible that M. bovis has followed a similar evolutionary pathway and therefore the lack of recombination observed after LIAN analysis can be explained by the choice of the housekeeping genes analyzed. Based on our results and the previous evidence described above for other mycoplasmas, we suggest that the analysis of recombination for M. bovis should be based on whole-genome sequence data, in order to get a better understanding of the recombinatorial events involved in the evolution of this pathogen.
MLST has been successfully used to determine the population structure of various geographically diverse microorganisms (36, 37). In this study, 137 historical and recent isolates of M. bovis from four different continents and from 12 different countries were analyzed. Interestingly, two of the United Kingdom isolates that clustered out of CC1, namely, isolates 144B08 (ST5, singleton) and 197B08 (ST6, CC2), were originally obtained from the United States and were subsequently isolated in the United Kingdom after experimental infection of cattle performed in that country (R. A. J. Nicholas and R. D. Ayling, unpublished data), which explains the genetic divergence of those isolates from the rest of the British isolates. Five singleton STs were found in our analysis (ST5, ST18, ST24, ST29, and ST35). Together with CC3 (ST23 and ST25), these STs are the most genetically diverse. Strikingly, the majority of these diverse isolates were obtained from mastitis cases. Although most of the mastitis isolates characterized in this study clustered largely with pneumonia isolates, there was greater diversity in the small group of isolates described above, possibly due to adaptation to the specific ecological niche of the bovine mammary gland.
Analysis of the genetic variability of M. bovis within herds demonstrated the presence of single or multiple profiles, depending on the farms studied (see Table S1 in the supplemental material). Previous studies demonstrated that husbandry conditions influence the genetic variation of M. bovis, with isolates obtained from closed herds being less variable than those obtained from multiple-source or open herds (5). The possibility of finding multiple genotypes on the same farm in both pneumonia and mastitis outbreaks may need to be taken into consideration when designing prophylactic strategies.
The clear clustering of isolates after the analysis of concatenated amino acid sequences further supports the hypothesis of population evolution primarily based on geographical isolation. This can be clearly seen in Australian, Chinese, and some Israeli isolates (ST10, ST33, and ST34), which grouped apart from the majority of the European isolates. A similar distribution of geographically distant isolates was observed previously (9) after VNTR analysis of a cohort of M. bovis isolates that included some Australian, Israeli, Lithuanian, and Hungarian isolates, most of which were characterized in this study (see Table S1 in the supplemental material). VNTR analysis demonstrated the presence of two main groups, groups A and B, in which isolates clustered predominantly on the basis of geographical origins. Our data also support a clear linkage between the type strain PG45, which was obtained from the United States, and the main European group of isolates. Similar findings supporting the European linkage of PG45 were described previously (4) after the finding of great homogeneity between Danish isolates and PG45 by AFLP analysis. Although not representative, the similarity between PG45 and most of the European isolates highlights a possible genetic linkage between these two distant geographical origins that could be explained by the extensive livestock trade between Europe and United States in the past.
Furthermore, the range of different STs and scattered clustering observed for isolates obtained from Israel (Fig. 1; also see Fig. S1 in the supplemental material) can also be explained by the role of cattle import trade in that country, where calves from diverse geographical origins, including Europe and Australia, are sourced for the supply of feedlots with a high turnover of livestock (9), supporting the theory of geographically independent evolution of M. bovis isolates and the role of trade in the introduction and spread of new genotypes of this microorganism in farms and countries. The ability of MLST to discriminate between these geographical sources demonstrates the usefulness of this typing scheme for the analysis of the epidemiology of M. bovis and its potential use as a universal typing technique for disease traceability and control of this pathogen.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded as part of Coordination of European Research on Emerging and Major Infectious Diseases of Livestock project FP73-ERA NET.
We thank Miroslav Hlusek (Animal Health and Veterinary Laboratories Agency, United Kingdom) for technical support, as well as all colleagues involved in providing samples for this study. We are grateful to the Department for Environment, Food, and Rural Affairs (United Kingdom) for continued support.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01910-14.
REFERENCES
- 1.Pfutzner H, Sachse K. 1996. Mycoplasma bovis as an agent of mastitis, pneumonia, arthritis and genital disorders in cattle. Rev Sci Tech 15:1477–1494. [DOI] [PubMed] [Google Scholar]
- 2.Ball HJ, Nicholas RA. 2010. Mycoplasma bovis-associated disease: here, there and everywhere. Vet J 186:280–281. doi: 10.1016/j.tvjl.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Nicholas RA, Ayling RD, McAuliffe L. 2008. Bovine respiratory disease, p 132–168. In Nicholas RA, Ayling RD, McAuliffe L (ed), Mycoplasma diseases of ruminants. CAB International, Norfolk, United Kingdom. [Google Scholar]
- 4.Kusiluka LJ, Kokotovic B, Ojeniyi B, Friis NF, Ahrens P. 2000. Genetic variations among Mycoplasma bovis strains isolated from Danish cattle. FEMS Microbiol Lett 192:113–118. doi: 10.1111/j.1574-6968.2000.tb09368.x. [DOI] [PubMed] [Google Scholar]
- 5.Butler JA, Pinnow CC, Thomson JU, Levisohn S, Rosenbusch RF. 2001. Use of arbitrarily primed polymerase chain reaction to investigate Mycoplasma bovis outbreaks. Vet Microbiol 78:175–181. doi: 10.1016/S0378-1135(00)00286-8. [DOI] [PubMed] [Google Scholar]
- 6.McAuliffe L, Kokotovic B, Ayling RD, Nicholas RA. 2004. Molecular epidemiological analysis of Mycoplasma bovis isolates from the United Kingdom shows two genetically distinct clusters. J Clin Microbiol 42:4556–4565. doi: 10.1128/JCM.42.10.4556-4565.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miles K, McAuliffe L, Persson A, Ayling RD, Nicholas RA. 2005. Insertion sequence profiling of UK Mycoplasma bovis field isolates. Vet Microbiol 107:301–306. doi: 10.1016/j.vetmic.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Pinho L, Thompson G, Rosenbusch R, Carvalheira J. 2012. Genotyping of Mycoplasma bovis isolates using multiple-locus variable-number tandem-repeat analysis. J Microbiol Methods 88:377–385. doi: 10.1016/j.mimet.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Amram E, Freed M, Khateb N, Mikula I, Blum S, Spergser J, Sharir B, Ozeri R, Harrus S, Lysnyansky I. 2013. Multiple locus variable number tandem repeat analysis of Mycoplasma bovis isolated from local and imported cattle. Vet J 197:286–290. doi: 10.1016/j.tvjl.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 10.Spergser J, Macher K, Kargl M, Lysnyansky I, Rosengarten R. 2013. Emergence, re-emergence, spread and host species crossing of Mycoplasma bovis in the Austrian Alps caused by a single endemic strain. Vet Microbiol 164:299–306. doi: 10.1016/j.vetmic.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 11.Gerner-Smidt P, Kincaid J, Kubota K, Hise K, Hunter SB, Fair MA, Norton D, Woo-Ming A, Kurzynski T, Sotir MJ, Head M, Holt K, Swaminathan B. 2005. Molecular surveillance of Shiga toxigenic Escherichia coli O157 by PulseNet USA. J Food Prot 68:1926–1931. [DOI] [PubMed] [Google Scholar]
- 12.Allix-Beguec C, Harmsen D, Weniger T, Supply P, Niemann S. 2008. Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J Clin Microbiol 46:2692–2699. doi: 10.1128/JCM.00540-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A 95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urwin R, Maiden MC. 2003. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol 11:479–487. doi: 10.1016/j.tim.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 15.van Belkum A, Tassios PT, Dijkshoorn L, Haeggman S, Cookson B, Fry NK, Fussing V, Green J, Feil E, Gerner-Smidt P, Brisse S, Struelens M, European Society of Clinical Microbiology and Infectious Diseases Study Group on Epidemiological Markers . 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin Microbiol Infect 13(Suppl 3):S1–S46. doi: 10.1111/j.1469-0691.2007.01786.x. [DOI] [PubMed] [Google Scholar]
- 16.McAuliffe L, Gosney F, Hlusek M, de Garnica ML, Spergser J, Kargl M, Rosengarten R, Ayling RD, Nicholas RA, Ellis RJ. 2011. Multilocus sequence typing of Mycoplasma agalactiae. J Med Microbiol 60:803–811. doi: 10.1099/jmm.0.028159-0. [DOI] [PubMed] [Google Scholar]
- 17.Mayor D, Jores J, Korczak BM, Kuhnert P. 2008. Multilocus sequence typing (MLST) of Mycoplasma hyopneumoniae: a diverse pathogen with limited clonality. Vet Microbiol 127:63–72. doi: 10.1016/j.vetmic.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 18.Nicholas R, Baker S. 1998. Recovery of mycoplasmas from animals. Methods Mol Biol 104:37–43. [DOI] [PubMed] [Google Scholar]
- 19.McAuliffe L, Ellis RJ, Ayling RD, Nicholas RA. 2003. Differentiation of Mycoplasma species by 16S ribosomal DNA PCR and denaturing gradient gel electrophoresis fingerprinting. J Clin Microbiol 41:4844–4847. doi: 10.1128/JCM.41.10.4844-4847.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McAuliffe L, Ellis RJ, Lawes JR, Ayling RD, Nicholas RA. 2005. 16S rDNA PCR and denaturing gradient gel electrophoresis: a single generic test for detecting and differentiating Mycoplasma species. J Med Microbiol 54:731–739. doi: 10.1099/jmm.0.46058-0. [DOI] [PubMed] [Google Scholar]
- 21.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Francisco AP, Vaz C, Monteiro PT, Melo-Cristino J, Ramirez M, Carrico JA. 2012. PHYLOViZ: phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinformatics 13:87. doi: 10.1186/1471-2105-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francisco AP, Bugalho M, Ramirez M, Carrico JA. 2009. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics 10:152. doi: 10.1186/1471-2105-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haubold B, Hudson RR. 2000. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Bioinformatics 16:847–848. doi: 10.1093/bioinformatics/16.9.847. [DOI] [PubMed] [Google Scholar]
- 25.Nei M. 1973. Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci U S A 70:3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol 26:2465–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jolley KA, Feil EJ, Chan MS, Maiden MC. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230–1231. doi: 10.1093/bioinformatics/17.12.1230. [DOI] [PubMed] [Google Scholar]
- 28.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rzhetsky A, Nei M. 1992. A simple method for estimating and testing minimum-evolution trees. Mol Biol Evol 9:945–967. [Google Scholar]
- 30.Feil EJ, Holmes EC, Bessen DE, Chan MS, Day NP, Enright MC, Goldstein R, Hood DW, Kalia A, Moore CE, Zhou J, Spratt BG. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc Natl Acad Sci U S A 98:182–187. doi: 10.1073/pnas.98.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manso-Silvan L, Dupuy V, Lysnyansky I, Ozdemir U, Thiaucourt F. 2012. Phylogeny and molecular typing of Mycoplasma agalactiae and Mycoplasma bovis by multilocus sequencing. Vet Microbiol 161:104–112. doi: 10.1016/j.vetmic.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 32.Pettersson B, Uhlen M, Johansson KE. 1996. Phylogeny of some mycoplasmas from ruminants based on 16S rRNA sequences and definition of a new cluster within the hominis group. Int J Syst Bacteriol 46:1093–1098. doi: 10.1099/00207713-46-4-1093. [DOI] [PubMed] [Google Scholar]
- 33.Sirand-Pugnet P, Lartigue C, Marenda M, Jacob D, Barre A, Barbe V, Schenowitz C, Mangenot S, Couloux A, Segurens B, de Daruvar A, Blanchard A, Citti C. 2007. Being pathogenic, plastic, and sexual while living with a nearly minimal bacterial genome. PLoS Genet 3:e75. doi: 10.1371/journal.pgen.0030075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fischer A, Shapiro B, Muriuki C, Heller M, Schnee C, Bongcam-Rudloff E, Vilei EM, Frey J, Jores J. 2012. The origin of the ‘Mycoplasma mycoides cluster’ coincides with domestication of ruminants. PLoS One 7:e36150. doi: 10.1371/journal.pone.0036150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siqueira FM, Thompson CE, Virginio VG, Gonchoroski T, Reolon L, Almeida LG, da Fonseca MM, de Souza R, Prosdocimi F, Schrank IS, Ferreira HB, de Vasconcelos AT, Zaha A. 2013. New insights on the biology of swine respiratory tract mycoplasmas from a comparative genome analysis. BMC Genomics 14:175. doi: 10.1186/1471-2164-14-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonzalez-Escalona N, Martinez-Urtaza J, Romero J, Espejo RT, Jaykus LA, DePaola A. 2008. Determination of molecular phylogenetics of Vibrio parahaemolyticus strains by multilocus sequence typing. J Bacteriol 190:2831–2840. doi: 10.1128/JB.01808-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arvand M, Raoult D, Feil EJ. 2010. Multi-locus sequence typing of a geographically and temporally diverse sample of the highly clonal human pathogen Bartonella quintana. PLoS One 5:e9765. doi: 10.1371/journal.pone.0009765. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.