Abstract
Background
To identify potential molecular prognostic markers in core binding factor (CBF) AML, we analyzed incidences and prognostic impacts of mutations in c-KIT, WT1, CEBPA, CBL, and a number of epigenetic genes in CBF AML.
Methods
Seventy one and 21 AML patients with t(8;21) and inv(16) were enrolled in this study, respectively. NPM1, CEBPA, c-KIT, IDH1/2, DNMT3A, EZH2, WT1, and CBL mutations were analyzed by direct sequencing. Patients were categorized with respect to c-KIT and WT1 mutation status, and both clinical features and prognoses were compared.
Results
The incidences of FLT3 internal tandem duplication (ITD), NPM1, CEBPA, IDH1/2, DNMT3A, EZH2, and CBL mutations were low (≤5%) in CBF AML patients. However, c-KIT and WT1 mutations occurred frequently (10.9% and 13.8%, respectively). t(8;21) patients with c-KIT mutations showed significantly shorter overall survival (OS) and disease free survival (DFS) periods than those without mutations (P<0.001, for both); however, although the limited number of t(8;21) patients were analyzed, WT1 mutation status did not affect prognosis significantly. Relapse or death during follow-up occurred more frequently in t(8;21) patients carrying c-KIT mutations than in those without the mutation, although the difference was significant only in a specific patient subgroup with no WT1 mutations (P=0.014).
Conclusions
The incidences of mutations in epigenetic genes are very low in CBF AML; however, c-KIT and WT1 mutations occur more frequently than others. The poor prognostic impact of c-KIT mutation in t(8;21) AML patients only applies in a specific patient subgroup without WT1 mutations. The prognostic impact of WT1 mutation in CBF AML is not evident and further investigation is required.
Keywords: Acute myeloid leukemia, Core binding factor, c-KIT, Epigenetic modification, Incidence, Prognosis, WT1
INTRODUCTION
Mutations related to the development of myeloid malignancy are categorized into five classes, namely those in genes related to signaling pathways (e.g. fms-related tyrosine kinase 3 [FLT3], c-KIT, and casitas b-lineage lymphoma [CBL]; class I), genes encoding transcription factors (e.g. CCAAT/enhancer-binding protein alpha [CEBPA] and nucleophosmin1 [NPM1]; class II), genes related to epigenetic modification (e.g. enhancer of zeste homolog 2 [EZH2], DNA-methyltransferase 3 alpha [DNMT3A], and isocitrate dehydrogenase 1/2 [IDH1/2]; class III), tumor suppressor genes (e.g. Wilms' tumor 1 [WT1]; class IV), and genes associated with RNA maturation (e.g. subunit 1 of splicing factor 3b protein complex [SF3B1], serine/arginine-rich splicing factor 2 [SRSF2], and U2 small nuclear RNA auxiliary factor 1 [U2AF1]; class V) [1, 2].
Of these mutations, FLT3 internal tandem duplication (ITD) is a commonly observed aberration associated with poor prognosis in AML [3]. NPM1 and CEBPA mutations, associated with a favorable prognosis in normal karyotype (NK) AML, are also used for risk classification [4]. In addition, both DNMT3A and IDH1/2 mutations were recently introduced as potential adverse and favorable prognosis indicators in NK AML, respectively [5, 6, 7, 8]. However, both EZH2 and CBL mutations were reported to occur very rarely and offer no prognostic impact for AML [9, 10, 11]; in addition, WT1 mutation was reported to occur only rarely in AML [12]. Despite the low incidence of WT1 mutation, its prognostic impact in AML cases still needs to be investigated since several studies have shown that WT1 overexpression, possibly due to WT1 mutation, may constitute an unfavorable [13, 14, 15] or favorable [16] prognostic indicator.
Although a recent study reported that CBL mutation occurs in core binding factor (CBF) AML at a frequency of 6% and was associated with a favorable prognosis in CBF AML patients with a CBL mutant level of 25% or higher, there have been fewer studies on the identification of prognostic markers for AML when compared with the number of studies concerning NK AML, and a reliable marker other than c-KIT mutation has not been identified [17, 18, 19, 20, 21]. Given that the five-year survival rate for CBF AML is only 50% and the incidence of FLT3 ITD mutation in this disease is low, occurring in 5 to 6% of patients [19, 21], an investigation into the identification of potential molecular prognostic markers in CBF AML needs to be performed.
Therefore, we aimed to compare clinical features between patients with t(8;21) and those with inv(16), and to evaluate the incidence and prognostic impact of genetic mutations associated with epigenetic modifications, such as IDH1/2, DNMT3A, and EZH2 mutations, as well as tumor suppressor WT1 mutation, and CBL mutation in CBF AML patients, while including FLT3 ITD, NPM1, CEBPA, and c-KIT mutations in a multicenter study on the Korean population.
METHODS
1. Patient selection and treatment
A total of 92 patients diagnosed with CBF AML at four tertiary hospitals in Korea from January 2002 to December 2010 were retrospectively enrolled in this study, including 71 patients with t(8;21)(q22;q22) and 21 patients with inv(16)(p31.1;q22)/t(16;16)(p13.1;q22). All patients received induction chemotherapy with cytarabine at 100 mg/m2 per day for seven days plus daunorubicin at 45 mg/m2 per day for three days (the AD regimen), or cytarabine at 100 mg/m2 per day for seven days plus idarubicin 12 mg/m2 per day for three days (the AI regimen). Complete remission (CR) was defined as the presence of<5% blasts on bone marrow (BM) aspirates and ≥20% cellularity in BM biopsy after induction chemotherapy. In total, 67 (72.8%) patients reached CR. Relapse was defined as the presence of ≥5% leukemic blasts on BM aspirates for patients who had previously achieved CR. In total, 33 (35.9%) patients experienced relapse or died during follow-up periods (median: 18.5 months, range: 0-150 months). Patients underwent stem cell transplantation (SCT) depending on the patient's age and the availability of a suitable donor. In total, 35 (38.0%) patients received SCT during follow-up period. This study was approved by the institutional review board of each institution.
2. Analyses of FLT3 ITD, NPM1, c-KIT, and CEBPA mutations
FLT3 ITD, NPM1, c-KIT, and CEBPA mutations were analyzed on DNA samples obtained from each patient at initial diagnosis. FLT3 ITD mutation was analyzed by multiplex PCR using a Seeplex FLT3 Genotyping Kit (Seegene, Seoul, Korea). NPM1 mutation was analyzed using a primer set designed in-house and PCR conditions as described previously [22]. The size of PCR products was determined by capillary electrophoresis using ABI 3130 genetic analyzer and GeneScan Analysis software (Applied Biosystems Inc., Foster City, CA, USA). For samples with an additional peak in their profile, direct sequencing was performed to confirm the mutation. For analysis of mutations in exons 8 and 17 of c-KIT, PCR and direct sequencing were also performed with a primer set designed in-house, using the PCR conditions and analysis strategy described previously [18]. For CEBPA mutation analysis, four primer sets were used for PCR and direct sequencing, using a detection strategy and PCR conditions identical to those applied in a previous study [23]. All information regarding the sequences and melting temperatures of primers used to amplify c-KIT, NPM1, and CEBPA genes, and the size of PCR products, is provided in Supplemental Data Table S1.
3. Analyses of IDH1, IDH2, DNMT3A, EZH2, WT1, and CBL mutations
WT1, CBL, and four genes associated with epigenetic modification (IDH1, IDH2, DNMT3A, and EZH2) were analyzed by PCR and direct sequencing. Since the quantity of DNA in samples from 11 patients with t(8;21) and one patient with inv(16) was not sufficient for analysis, a total of 80 CBF AML patients were finally included in these analyses. Mutation hotspots in IDH1 (codon 123 and 132 in exon 4), IDH2 (codon 140 and 172 in exon 4), DNMT3A (codon 882 in exon 23), EZH2 (exons 17 to 19), WT1 (exons 7 and 9), and CBL (exons 8 and 9) genes were determined as amplification targets to be analyzed. The following PCR conditions were used to amplify a total of 10 exons from six genes: 5 min at 95℃, followed by 35 cycles of 30 sec at 95℃ (denaturation), 45 sec at 55℃ (annealing) and 30 sec at 72℃ (extension), and a final 10 min extension at 72℃. All information regarding the sequences and melting temperatures of primers used to amplify each gene, and the size of PCR products, is provided in Supplemental Data Table S1.
4. Comparison of clinical features and incidences of genetic mutations between AML patients with t(8;21) and those with inv(16)
Both clinical features and incidences of genetic mutations were compared between patients with t(8;21) and those with inv(16). The clinical features compared included gender, age, percentage of patients with additional chromosomal abnormalities, SCT performance, relapse and death rates, length of follow-up periods, complete blood cell count data at diagnosis, and peripheral blood (PB) and BM blast counts as percentages at diagnosis. In addition, all genetic mutations detected were aligned against reference sequences (NM_004119.2 for FLT3 ITD, NM_002520.6 for NPM1, NM_000222.2 for c-KIT, NM_004364.3 for CEBPA, NM_005896.2 for IDH1, NM_002168.2 for IDH2, NM_175629.2 for DNMT3A, NM_001203247.1 for EZH2, NM_000378.4 for WT1 and NM_005188.3 for CBL) and any resulting protein changes were analyzed. These results are represented in Tables 1 and 2.
Table 1. Comparison of clinical features and incidences of genetic mutations between acute myeloid leukemia patients with t(8;21) and acute myeloid leukemia patients with inv(16).
| Variables | AML with t(8;21), 71 patients (% of total patients) | AML with inv(16), 21 patients (% of total patients) | Total, 92 patients | P |
|---|---|---|---|---|
| Sex (M:F)* | 38 : 33 | 13 : 8 | 51 : 41 | 0.497 |
| Age, yr, median (range)† | 41.0 (5.0-78.0) | 47.0 (16.0-82.0) | 43.0 (5.0-82.0) | 0.126 |
| Additional chromosomal abnormalities* | 17/71 (23.9%) | 5/21 (23.8%) | 22/92 (23.9%) | 0.990 |
| SCT during follow-up* | 24/71 (33.8%) | 11/21 (52.4%) | 35/92 (38.0%) | 0.123 |
| Relapse or death during follow-up* | 22/71 (31.0%) | 11/21 (52.4%) | 33/92 (35.9%) | 0.073 |
| Follow-up period, months, median (range)† | 22.0 (0.0-150.0) | 17.0 (0.0-119.0) | 18.5 (0.0-150.0) | 0.955 |
| Laboratory findings at diagnosis† | ||||
| WBC ( × 109/L), median (range) | 8.34 (1.2-192.9) | 54.4 (2.7-277.6) | 10.3 (1.2-277.6) | < 0.001 |
| Hemoglobin (g/dL), median (range) | 7.8 (1.5-13.2) | 8.1 (4.6-12.4) | 7.9 (1.5-13.2) | 0.488 |
| Platelets ( × 109/L), median (range) | 31.0 (3.0-155.0) | 33.0 (6.0-307.0) | 31.0 (3.0-307.0) | 0.551 |
| PB blasts (%), median (range) | 27.0 (0.0-92.0) | 63.0 (7.0-87.0) | 37.5 (0.0 -92.0) | 0.002 |
| BM blasts (%), median (range) | 48.0 (21.0-90.0) | 65.0 (21.0-91.0) | 57.0 (21.0-91.0) | 0.003 |
| Mutation analysis results* | ||||
| FLT3 ITD | 1/71 (1.4%) | 2/21 (9.5%) | 3/92 (3.3%) | 0.129 |
| NPM1 | 1/71 (1.4%) | 0/21 (0.0%) | 1/92 (1.1%) | 0.584 |
| c-KIT | 7/71 (9.9%) | 3/21 (14.3%) | 10/92 (10.9%) | 0.690 |
| CEBPA | 0/71 (0.0%) | 0/21 (0.0%) | 0/92 (0.0%) | NC |
| IDH1 | 0/60 (0.0%) | 0/20 (0.0%) | 0/80 (0.0%) | NC |
| IDH2 | 2/60 (3.3%) | 0/20 (0.0%) | 2/80 (2.5%) | 0.408 |
| DNMT3A | 1/60 (1.7%) | 3/20 (15.0%) | 4/80 (5.0%) | 0.046 |
| EZH2 | 2/60 (3.3%) | 2/20 (10.0%) | 4/80 (5.0%) | 0.259 |
| WT1 | 7/60 (11.7%) | 4/20 (20.0%) | 11/80 (13.8%) | 0.454 |
| CBL | 1/60 (1.7%) | 0/20 (0.0%) | 1/80 (1.3%) | 0.561 |
P values were obtained using Chi-square or Fisher's exact tests (for numbers less than five in each group)* and Mann-Whitney U test†.
Abbreviations: SCT, stem cell transplantation; WBC, white blood cell; PB, peripheral blood; BM, bone marrow; FLT3, fms-related tyrosine kinase 3; NPM, nucleophosmin; CEBPA, CCAAT/enhancer binding protein alpha; WT, Wilms' tumor; IDH, isocitrate dehydrogenase; DNMT3A, DNA (cytosine-5-)-methyltransferase 3 alpha; EZH, enhancer of zeste homolog; CBL, casitas b-lineage lymphoma; NC, not calculated.
Table 2. Summary of detected mutations in patients with core binding factor acute myeloid leukemia.
| Genes | AML with t(8;21) | AML with inv(16) | Alignment reference sequence | N of patients with mutation/under mutation analysis | ||
|---|---|---|---|---|---|---|
| Mutation results (protein change) | N of patients | Mutation results (protein change) | N of patients | |||
| c-KIT mutation (Exon 8) | c.1250_1255delCTTACG (p.Thr417_Asp419delinsAsn) | 1 | c. 1256_1257insTTTTCGA | 1 | NM_000222.2 | 10/92 |
| c-KIT mutation (Exon 17) | c.2447A>T (p.Asp816Val) | 1 | c.2447A>T (p.Asp816Val) | 1 | ||
| c.2446G>T (p.Asp816Tyr) | 2 | c.2446G>T (p.Asp816Tyr) | 1 | |||
| c.2446G>C (p.Asp816His) | 3 | |||||
| IDH2 mutation | c.419G>A (p.Arg140Gln) | 2 | NM_002168.2 | 2/80 | ||
| DNMT3A mutation | c.2638A>C (p.Met880Val) | 1 | c.2638A>C (p.Met880Val) | 2 | NM_175629.2 | 4/80 |
| c.2644C>T (p. Arg882Cys) | 1 | |||||
| EZH2 mutation (Exon 17) | c.1978G>A (p.Gly660Arg) | 1 | c.1996T>C (p.Tyr666Asn) | 1 | NM_001203247.1 | 4/80 |
| EZH2 mutation (Exon 18) | c.2068C>T (p.Arg690Cys) | 1 | c.2068C>T (p.Arg690Cys) | 1 | ||
| WT1 mutation (Exon 7) | c.1102G > A (p.Val367Met) | 1 | NM_000378.4 | 11/80 | ||
| c.1105C>G (p.Arg369Gly) | 1 | |||||
| c.1112C > T (p.Val371Ala) | 1 | |||||
| c.1131_1132insT | 1 | |||||
| c.1141T > A (p.Ser381Thr) | 2 | |||||
| c.1147T > A (p.Ser383Thr) | 1 | c.1147T>A (p.Ser383Thr) | 2 | |||
| WT1 mutation (Exon 9) | c.1372C>T (p.Arg458X) | 1 | ||||
| c.1379T > A (p.Phe460Tyr) | 1 | |||||
| CBL mutation | c.1196T > C (p.Leu399Pro) | 1 | NM_005188.3 | 1/80 | ||
Abbreviations: WT, Wilms' tumor; IDH, isocitrate dehydrogenase; DNMT3A, DNA (cytosine-5-)-methyltransferase 3 alpha; EZH, enhancer of zeste homolog; CBL, casitas b-lineage lymphoma; del, deletion; ins, insertion; X, stop codon.
5. Comparison of clinical features and prognoses between patients with or without c-KIT or WT1 mutations
Patients with t(8;21) for whom c-KIT and WT1 mutation data were available (71 and 60 patients, respectively) were categorized into two subgroups based on their c-KIT and WT1 mutation status, and their clinical features were compared. c-KIT and WT1 were selected because they showed higher mutation frequencies than other genes. To evaluate the prognostic impact of both c-KIT and WT1 mutations, both overall survival (OS) and disease free survival (DFS) were compared between the two patient subgroups. Since two patients showed FLT3 ITD or NPM1 mutations, they were excluded from the survival analysis to exclude the prognostic impact of these mutations. OS was defined as the time from diagnosis to death or last follow-up. DFS was defined as the time from CR to relapse (for patients who experienced relapse), death (for non-relapsed patients who did not survive), or last follow-up (for non-relapsed patients who survived). Patients who underwent SCT were censored at the time of transplantation. In addition, identical comparisons were performed for 21 patients with inv(16). These results are summarized in Table 3 and Fig. 1.
Table 3. Comparison of clinical features between patients with c-KIT or WT1 mutations and those without c-KIT or WT1 mutations.
| 71 patients with t(8;21) | Patient subgroups | |||
|---|---|---|---|---|
| c-KIT(-), N=64 | c-KIT(+), N=7 | WT1(-), N=53 | WT1(+), N=7 | |
| Sex (M:F)* | 35:29 | 3:4 | 30:23 | 4:3 |
| Age, yr, median (range)† | 41.5 (5.0-78.0) | 37 (18.0-51.0) | 41 (5.0-78.0) | 36 (18.0-64.0) |
| Additional chromosomal abnormalities* | 17/64 (26.6%) | 0/7 (0.0%) | 12/53 (22.6%) | 1/7 (14.3%) |
| SCT during follow-up* | 20/64 (31.3%) | 4/7 (57.1%) | 21/53 (39.6%) | 1/7 (14.3%) |
| Relapse or death during follow-up* | 16/64 (25.0%) | 6/7‡ (85.7%) | 14/53 (26.4%) | 3/7 (42.9%) |
| Follow-up period, months, median (range)† | 29 (0.0-150.0) | 10 (4.0-35.0) | 29 (0.0-109.0) | 15 (0.0-82.0) |
| Laboratory findings at diagnosis† | ||||
| WBC ( × 109/L), median (range) | 8.62 (1.20-192.90) | 5.91 (2.90-25.33) | 8.34 (1.20-102.37) | 10.7 (3.20-192.90) |
| Hemoglobin (g/dL), median (range) | 8 (1.5-13.2) | 6.3 (5.4-8.7) | 8.3 (2.3-13.2) | 8.4 (5.4-12.3) |
| Platelets ( × 109/L), median (range) | 32 (3.0-155.0) | 24 (9.0-59.0) | 34 (3.0-155.0) | 32 (8.0-59.0) |
| PB blasts (%), median (range) | 24 (0.0-92.0) | 45 (13.0-54.0) | 26 (0.0-86.0) | 47 (15.0-92.0) |
| BM blasts (%), median (range) | 47 (21.0-90.0) | 58 (30.0-81.0) | 44 (21.0-90.0) | 60 (21.0-89.0) |
P values were obtained using Chi-square or Fisher's exact tests (for numbers less than five in each group)* and Mann-Whitney U test†. In t(8;21), the patients with c-KIT mutation experienced relapse or died during follow-up more frequently than those without c-KIT mutation (P=0.003). In inv(16), the patients with c-KIT mutation showed lower platelet counts (P=0.024) and higher BM blasts (P=0.017) than those without c-KIT mutation, and the patients with WT1 mutation were older than those without WT1 mutation (P=0.029). Comparison items which showed statistically significant differences with respect to c-KIT and WT1 mutation status were indicated with superscripts (‡) and (§), respectively.
Abbreviations: SCT, stem cell transplantation; WBC, white blood cell; PB, peripheral blood; BM, bone marrow; WT, Wilms' tumor.
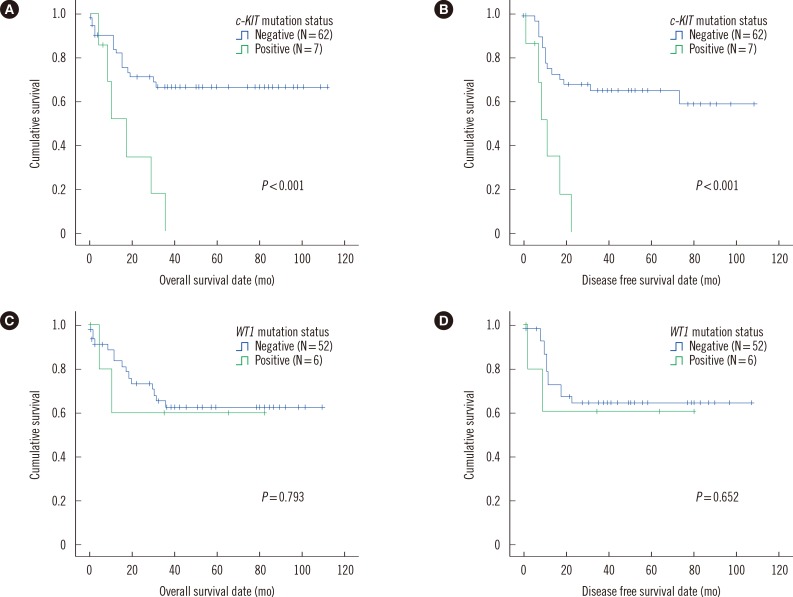
Fig. 1. Comparisons of overall survival and disease free survival lengths in core binding factor acute leukemia patients with t(8;21) and no FLT3 ITD or NPM1 mutations, between patients with c-KIT mutations and those without c-KIT mutations (N=69, A, overall survival; B, disease free survival). Identical comparisons between patients with WT1 mutations and those without WT1 mutations (N=58, C, overall survival; D, disease free survival) are also given in this figure.
Abbreviation: WT, Wilms' tumor.
6. Comparison of clinical features and prognoses in patients with t(8;21) among 4 patient subgroups categorized by c-KIT and WT1 mutation status
The 60 patients with t(8;21) for whom both c-KIT and WT1 mutation data were available were categorized into four patient subgroups: 1) c-KIT(-)/WT1(-), N=48; 2) c-KIT(+)/WT1(-), N=5; 3) c-KIT(-)/WT1(+), N=5; and 4) c-KIT(+)/WT1(+), N=2. Both clinical features and survival rates were compared between patient subgroups. As mentioned above, two patients with FLT3 ITD or NPM1 mutations were excluded from the survival analysis. These results are summarized in Table 4 and Fig. 2.
Table 4. Comparison of clinical features in 60 patients with t(8;21) according to c-KIT and WT1 mutation status.
| Variables | Patient subgroups | |||
|---|---|---|---|---|
| c-KIT(-)/WT1(-), N = 48 | c-KIT(+)/WT1(-), N = 5 | c-KIT(-)/WT1(+), N = 5 | c-KIT(+)/WT1(+), N = 2 | |
| Sex (M:F)* | 27:21 | 3:2 | 4:1 | 0:2 |
| Age, yr, median (range)† | 41.0 (5.0-78.0) | 42.0 (37.0-51.0) | 53.0 (18.0-64.0) | 18.5 (18.0-19.0) |
| Additional chromosomal abnormalities* | 12/48 (25.0%) | 0/5 (0.0%) | 1/5 (20.0%) | 0/2 (0.0%) |
| SCT during follow-up* | 18/48 (37.5%) | 3/5 (60.0%) | 0/5 (0.0%) | 1/2 (50.0%) |
| Relapse or death during follow-up* | 10/48 (20.8%) | 4/5‡ (80.0%) | 1/5 (20.0%) | 2/2 (100.0%) |
| Follow-up period, months, median (range)† | 30.5 (0.0-109.0) | 17.0 (6.0-35.0) | 35.0 (0.0-82.0) | 7.0 (4.0-10.0) |
| Laboratory findings at diagnosis† | ||||
| WBC ( × 109/L), median (range) | 8.62 (1.20-102.37) | 5.91 (2.90-8.40) | 10.70 (3.20-192.90) | 14.27 (3.20-25.33) |
| Hemoglobin (g/dL), median (range) | 8.5 (2.3-13.2) | 6.3 (5.6-8.4) | 8.4 (6.5-12.3) | 7.1 (5.4-8.7) |
| Platelets ( × 109/L), median (range) | 34.5 (3.0-155.0) | 24.0 (9.0-59.0) | 35.0 (8.0-59.0) | 21.5 (11.0-32.0) |
| PB blasts (%), median (range) | 23.0 (0.0-86.0) | 45.0 (13.0-54.0) | 52.0 (15.0-92.0) | 37.0 (27.0-47.0) |
| BM blasts (%), median (range) | 44.0 (21.0-90.0) | 58.0 (30.0-81.0) | 60.0 (21.0-89.0) | 62.0 (44.0-80.0) |
P values were obtained using Chi-square or Fisher's exact tests (for numbers less than five in each group)* and Mann-Whitney U test†. In patients without WT1 mutation, those with c-KIT mutation experienced relapse or died during follow-up more frequently than those without c-KIT mutation (P=0.014). Comparison items which showed statistically significant differences between two patient subgroups were indicated with superscript (‡).
Abbreviations: SCT, stem cell transplantation; WBC, white blood cell; PB, peripheral blood; BM, bone marrow; WT, Wilms' tumor.
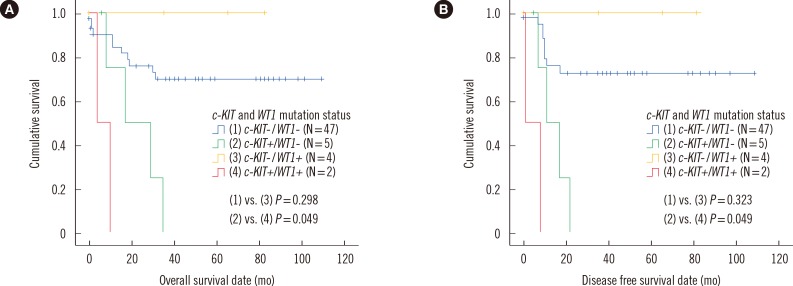
Fig. 2. Comparisons of overall survival and disease free survival in core binding factor acute leukemia patients with t(8;21) and no FLT3 ITD or NPM1 mutations, among four patient subgroups categorized by c-KIT and WT1 mutation status (N=58, A, overall survival; B, disease free survival).
Abbreviation: WT, Wilms' tumor.
7. Statistical analysis
Pearson chi-square or Fisher's exact tests were performed to compare categorical variables. The Mann-Whitney U test was used to compare continuous variables. The log-rank test was applied for the comparison of OS and DFS, and Kaplan-Meier survival curves were generated. All tests were two-tailed, and P values <0.05 were considered statistically significant. SPSS 13.0.1 for Windows (SPSS Inc, Chicago, IL, USA) was used for statistical analysis.
RESULTS
1. Comparison of clinical features and incidences of genetic mutations between AML patients with t(8;21) and those with inv(16)
Patients with inv(16) showed significantly higher white blood cell (WBC) counts (median 54.4×109/L vs. 8.34×109/L, P<0.001), and levels of PB blasts (median 63.0% vs. 27.0%, P=0.002) and BM blasts (median 65.0% vs. 48.0%, P=0.003) than those with t(8;21). Other clinical features were not significantly different between the two patient subgroups.
Among the total of 92 patients, incidences of FLT3 ITD, NPM1, CEBPA, CBL, IDH1, IDH2, EZH2, and DNMT3A mutations were 3.3%, 1.1%, 0%, 1.3%, 0%, 2.5%, 5.0%, and 5.0%, respectively. The incidences of both c-KIT mutations (10.9%) and WT1 mutations (13.8%) were relatively higher than those of the other gene mutations analyzed. Mutation frequencies in patients with t(8;21) and inv(16) were not significantly different, except for that of DNMT3A, which showed significantly higher incidence in patients with inv(16) than in those with t(8;21) (15.0% vs. 1.7%, P=0.046; Table 1).
2. Summary of detected mutations in 92 CBF AML patients
In total, two and eight patients showed c-KIT mutations in exons 8 and 17, respectively. The mutations detected included four previously reported (c.1250_1255delCTTACG, c.2447A>T, c.2446G>T, and c.2446G>C), and one novel mutation (c.1256_ 1257insTTTTCGA). In the CEBPA gene, no mutations were detected but a known polymorphism (c.584_589dup ACCCGC) [23] was observed in 26 (28.3%) patients. In the case of IDH2, only two patients harbored a mutation, which was previously reported (c.419G>A), and for DNMT3A, four patients showed mutations, both of which were reported previously (c.2638A>C and c.2644C>T). Regarding EZH2, three previously reported mutations (c.1978G>A, c.1996T>C, and c.2068C>T) were found in four patients, and one previously reported intronic variant (c.2110 +6T>G), predicted as being benign in the ClinVar database (http://www.ncbi.nlm.nih.gov/clinvar/variation/137273/, last reviewed on 14 Mar 2014), was detected in one patient. One previously reported CBL mutation (c.1196T>C) was detected in one patient. Analysis of WT1 revealed six previously reported mutations (c.1102G>A, c.1105C>G, c.1141T>A, c.1147T>A, c.1372C>T, and c.1379T>A) and two novel mutations (c.1112C>T and c.1131_1132insT) in 11 patients.
In summary, the present study identified 20 mutations (17 known and three novel mutations) in 32 patients, one intronic variation of the EZH2 gene from one patient, and one polymorphism of the CEBPA gene in 26 patients (Table 2).
3. Comparison of clinical features and prognoses between patients with or without c-KIT or WT1 mutations
With regard to patients with t(8;21), clinical features did not differ significantly between patients with c-KIT mutations and those without. However, those with such mutations experienced relapse or died during follow-up more frequently than those without (85.7% vs. 25.0%, P=0.003), and both OS and DFS lengths were significantly shorter in patients with c-KIT mutations than in those without (P<0.001 for both). Neither clinical features nor prognoses differed significantly with respect to WT1 mutation status.
As for patients with inv(16), neither c-KIT nor WT1 mutation status significantly affected clinical features and prognoses, with the exception of significantly lower platelet counts in patients with c-KIT mutations than in those without (median 15.0×109/L vs. 34.5×109/L, P=0.024; Table 3 and Fig. 1).
4. Comparison of clinical features and prognoses in patients with t(8;21) among four patient subgroups categorized by c-KIT and WT1 mutation status
Within the subgroup showing no WT1 mutation, the patients harboring c-KIT mutations were more likely to experience relapse or die during follow-up than those without c-KIT mutations (80.8% vs. 20.8%, P=0.014). However, this difference was not statistically significant (100.0% vs. 20.0%, P=0.143) for the patient subgroup with WT1 mutations, which suggests that the poor prognostic impact of c-KIT mutation in AML patients with t(8;21) may apply only in patients with wild type WT1.
Regarding the subgroup showing c-KIT mutations, patients with WT1 mutations did not show significant differences in clinical outcomes, including relapse or death during follow-up (100.0% vs. 80.0%, P=0.495), compared with those without WT1 mutation. In the patient subgroup harboring c-KIT mutations, although the survival analysis showed significantly shorter OS and DFS in patients with WT1 mutations than in those without WT1 mutations (P=0.049 in both), the statistical power of this analysis was seriously limited due to the very low number of patients in each group. In the subgroup without c-KIT mutations, the patients with WT1 mutations also showed no differences in clinical outcomes, including rate of relapse or death during follow-up (20.0% vs. 20.8%, P=0.965), and prognosis (P=0.298 for OS and P=0.323 for DFS), compared with those without WT1 mutation (Table 4 and Fig. 2).
DISCUSSION
The present study found that the incidences of FLT3 ITD and NPM1 mutations were extremely low, representing 3.3% and 1.1% respectively of the CBF AML patients evaluated, and these results are consistent with previous studies, which reported similarly low frequencies of FLT3 ITD mutation (5 to 6%) and NPM1 mutation (0%) in cases of CBF AML [19, 21, 24]. Our study also demonstrated that patients with this disease are unlikely to harbor CEBPA mutations. These results indicate that FLT3 ITD, NPM1, and CEBPA mutations are not involved in leukemogenesis and imply no prognostic impact for CBF AML. Our data also support previous studies that have implicated mutations in both RAS and c-KIT as major leukemogenic factors in CBF AML [21, 25].
In addition, our study suggests that the frequencies of mutation in CBL and in genes associated with epigenetic modification, such as IDH1, IDH2, DNMT3A, and EZH2, are low in this disease, present in 1.3%, 0%, 2.5%, 5.0%, and 5.0% of the total number of patients, respectively. These results may support the idea that mutations involved in epigenetic modification do not contribute to leukemogenesis and have no significant prognostic value in CBF AML, since IDH2, DNMT3A, EZH2, and CBL mutation status did not significantly affect prognosis in our study (data not shown). In contrast, the present study showed that incidences of c-KIT and WT1 mutation in CBF AML were relatively higher than the mutations in other genes, being found in 10.9% and 13.8% of patients, respectively. Our results confirm the consistency of c-KIT mutation frequency in CBF AML between previous publications (6 to 48%) and Korean population [17, 18, 19, 20, 21].
The present work demonstrated that the poor prognostic impact of c-KIT mutation in patients carrying t(8;21) was evident only in the subgroup lacking WT1 mutations. Although this trend was also evident in the patient subgroup with WT1 mutations, the difference was not statistically significant. We also found that WT1 mutation status does not affect clinical features and prognosis, regardless of c-KIT mutation status, in patients with t(8;21). Within the subgroup harboring c-KIT mutations, a worse prognosis was identified for patients with WT1 mutations compared with those without WT1 mutations, but the statistical power of this result is seriously limited owing to the fact that only seven patients were included in the survival analysis.
On the basis of these results, we can speculate that both c-KIT and WT1 mutations constitute important genetic aberrations involved in leukemogenesis of CBF AML and show relatively high incidences in these patients. In addition, c-KIT mutation is a significant indicator of poor prognosis in CBF AML patients carrying t(8;21), but this prognostic value may exist only for a specific patient subgroup without WT1 mutations. It may be suggested that the presence of WT1 mutations does not affect clinical features and prognosis significantly, but further analysis involving a larger number of patients is required to address this point more conclusively.
The present study has two major limitations. First, since only 21 patients with inv(16) were included, comparisons of clinical features and prognoses for inv(16) patients with respect to c-KIT and WT1 mutation status could not be performed with sufficient statistical power. Given that there were also only 71 patients carrying t(8;21) in our study, suggestions and speculations based on this work should be interpreted with much caution. Second, we were unable to evaluate the effect of WT1 mutation on the expression of the corresponding WT1 protein, which is necessary to interpret our results at the protein expression level. Since previous studies involved evaluation of protein expression, stratification of patients based only on WT1 mutation status, as performed in the present study, our results need to be interpreted carefully. More focused analysis on this point is required in future studies.
In conclusion, our study demonstrates that the incidences of genetic mutations associated with epigenetic modification are very low and that both c-KIT and WT1 mutations occur more frequently than other mutations in CBF AML. In addition, our results suggest that the poor prognostic impact of c-KIT mutation in t(8;21)-positive CBF AML patients may apply only to a specific patient subgroup without WT1 mutations. The poor prognostic impact of WT1 mutation was not evident in t(8;21)-positive CBF AML patients owing to the small number of patients in this study; further study will be required to confirm this speculation in a large number of patients.
Acknowledgments
This study was supported by a Biomedical Research Institute Grant (2011-00), Pusan National University Hospital.
Footnotes
Authors' Disclosures of Potential Conflicts of Interest: No potential conflicts of interest relevant to this article were reported.
Supplementary Material
Supplementary table 1. Summary of all informations about primer sequences, melting temperatures of each primer used in the amplification of nine genes and size of PCR product.
| Genes | Forward primers | Reverse primers | Size of PCR product (bp) | ||
|---|---|---|---|---|---|
| Sequences | Tm (℃) | Sequences | Tm (℃) | ||
| c-KIT exon 8 | GCTGAGGTTTTCCAGCACTC | 61.7 | AATTGCAGTCCTTCCCCTCT | 61.3 | 219 |
| c-KIT exon 17 | TGGTTTTCTTTTCTCCTCCAA | 57.4 | TGCAGGACTGTCAAGCAGAG | 62.6 | 185 |
| NPM1 exon 12 | GTTTCTTTTTTTTTTTTTCCAGGCTATTCAAG | 59.9 | CACGGTAGGGAAAGTTCTCACTCTGC | 65.6 | 170 |
| CEBPA exon 1 A | CGCCATGCCGGGAGAACTCT | 67.3 | CTTCTCCTGCTGCCGGCTGT | 67.5 | 299 |
| CEBPA exon 1 B | GCCGCCTTCAACGACGAGTT | 66.2 | CTTGGCTTCATCCTCCTCGC | 63.4 | 303 |
| CEBPA exon 1 C | CCGCTGGTGATCAAGCAGGA | 65.1 | CCGGTACTCGTTGCTGTTCT | 62.6 | 390 |
| CEBPA exon 1 D | CAAGGCCAAGAAGTCGGTGGACA | 66.5 | CACGGTCTGGGCAAGCCTCGAGAT | 69.7 | 356 |
| WT1 exon 7 | CAGTGCTCACTCTCCCTCAAG | 62.2 | AGTGTGAGAGCCTGGAAAGG | 61.9 | 300 |
| WT1 exon 9 | GTGAGGCAGATGCAGACATTG | 61.7 | CCTCTCATCACAATTTCATTCCA | 58.3 | 297 |
| IDH1 exon 4 | GCCATCACTGCAGTTGTAGGTT | 62.9 | CACATACAAGTTGGAAATTTCTGG | 57.8 | 439 |
| IDH2 exon 4 | GGGGTTCAAATTCTGGTTGAA | 58.8 | CTGTGGCCTTGTACTGCAGAG | 63.1 | 323 |
| DNMT3A exon 23 | ACTAAGCAGGCGTCAGAGGAG | 63.7 | TCCATCCTCATGTTCTTGGTG | 59.8 | 393 |
| EZH2 exon 17 | CCTTTTTGTTGCGTTTTCTCC | 58.9 | ATCCTCCTTCTGGTCACCTCA | 62.2 | 310 |
| EZH2 exon 18 | AGGCAAACCCTGAAGAACTGT | 62.0 | GATGGCTCTCTTGGCAAAAAT | 59.3 | 396 |
| EZH2 exon 19 | CGTTTTGCAAATCATTCGGTA | 57.9 | ATTCCCCACTAATGCTCATGG | 60 | 413 |
| CBL exon 8 | ACCCAGACTAGATGCTTTCTG | 59.3 | AGGCCACCCCTTGTATCAGT | 63.3 | 385 |
| CBL exon 9 | CCTGGCTTTTGGGGTTAGGTTT | 62.9 | GACAACTCACAATGGATTTTGCC | 60.4 | 375 |
Abbreviations: NPM, nucleophosmin; CEBPA, CCAAT/enhancer binding protein alpha; WT, Wilms tumor; IDH, isocitrate dehydrogenase; DNMT3A, DNA (cytosine-5-)-methyltransferase 3 alpha; EZH, enhancer of zeste homolog; CBL, casitas b-lineage lymphoma; Tm, melting temperature of primer; bp, base pairs.
References
- 1.Conway O'Brien E, Prideaux S, Chevassut T. The epigenetic landscape of acute myeloid leukemia. Adv Hematol. 2014;2014:103175. doi: 10.1155/2014/103175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murati A, Brecqueville M, Devillier R, Mozziconacci MJ, Gelsi-Boyer V, Birnbaum D. Myeloid malignancies: mutations, models and management. BMC Cancer. 2012;12:304. doi: 10.1186/1471-2407-12-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kiyoi H, Naoe T, Nakano Y, Yokota S, Minami S, Miyawaki S, et al. Prognostic implication of FLT3 and N-RAS gene mutations in acute myeloid leukemia. Blood. 1999;93:3074–3080. [PubMed] [Google Scholar]
- 4.Port M, Böttcher M, Thol F, Ganser A, Schlenk R, Wasem J, et al. Prognostic significance of FLT3 internal tandem duplication, nucleophosmin 1, and CEBPA gene mutations for acute myeloid leukemia patients with normal karyotype and younger than 60 yr: a systematic review and meta-analysis. Ann Hematol. 2014;93:1279–1286. doi: 10.1007/s00277-014-2072-6. [DOI] [PubMed] [Google Scholar]
- 5.Thol F, Damm F, Lüdeking A, Winschel C, Wagner K, Morgan M, et al. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J Clin Oncol. 2011;29:2889–2896. doi: 10.1200/JCO.2011.35.4894. [DOI] [PubMed] [Google Scholar]
- 6.Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43:309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 7.Fried I, Bodner C, Pichler MM, Lind K, Beham-Schmid C, Quehenberger F, et al. Frequency, onset and clinical impact of somatic DNMT3A mutations in therapy-related and secondary acute myeloid leukemia. Haematologica. 2012;97:246–250. doi: 10.3324/haematol.2011.051581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcucci G, Maharry K, Wu YZ, Radmacher MD, Mrózek K, Margeson D, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:2348–2355. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Dai H, Wang Q, Wang Q, Xu Y, Wang Y, et al. EZH2 mutations are related to low blast percentage in bone marrow and -7/del(7q) in de novo acute myeloid leukemia. PLoS One. 2013;8:e61341. doi: 10.1371/journal.pone.0061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naoe T, Kiyoi H. Gene mutations of acute myeloid leukemia in the genome era. Int J Hematol. 2013;97:165–174. doi: 10.1007/s12185-013-1257-4. [DOI] [PubMed] [Google Scholar]
- 11.Abdel-Wahab O, Pardanani A, Patel J, Wadleigh M, Lasho T, Heguy A, et al. Concomitant analysis of EZH2 and ASXL1 mutations in myelofibrosis, chronic myelomonocytic leukemia and blast-phase myeloproliferative neoplasms. Leukemia. 2011;25:1200–1202. doi: 10.1038/leu.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rocquain J, Carbuccia N, Trouplin V, Raynaud S, Murati A, Nezri M, et al. Combined mutations of ASXL1, CBL, FLT3, IDH1, IDH2, JAK2, KRAS, NPM1, NRAS, RUNX1, TET2 and WT1 genes in myelodysplastic syndromes and acute myeloid leukemias. BMC Cancer. 2010;10:401. doi: 10.1186/1471-2407-10-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inoue K, Sugiyama H, Ogawa H, Nakagawa M, Yamagami T, Miwa H, et al. WT1 as a new prognostic factor and a new marker for the detection of minimal residual disease in acute leukemia. Blood. 1994;84:3071–3079. [PubMed] [Google Scholar]
- 14.Schmid D, Heinze G, Linnerth B, Tisljar K, Kusec R, Geissler K, et al. Prognostic significance of WT1 gene expression at diagnosis in adult de novo acute myeloid leukemia. Leukemia. 1997;11:639–643. doi: 10.1038/sj.leu.2400620. [DOI] [PubMed] [Google Scholar]
- 15.Bergmann L, Maurer U, Weidmann E. Wilms tumor gene expression in acute myeloid leukemias. Leuk Lymphoma. 1997;25:435–443. doi: 10.3109/10428199709039030. [DOI] [PubMed] [Google Scholar]
- 16.Miglino M, Colombo N, Pica G, Grasso R, Clavio M, Bergamaschi M, et al. WT1 overexpression at diagnosis may predict favorable outcome in patients with de novo non-M3 acute myeloid leukemia. Leuk Lymphoma. 2011;52:1961–1969. doi: 10.3109/10428194.2011.585673. [DOI] [PubMed] [Google Scholar]
- 17.Kim HJ, Ahn HK, Jung CW, Moon JH, Park CH, Lee KO, et al. KIT D816 mutation associates with adverse outcomes in core binding factor acute myeloid leukemia, especially in the subgroup with RUNX1/RUNX1T1 rearrangement. Ann Hematol. 2013;92:163–171. doi: 10.1007/s00277-012-1580-5. [DOI] [PubMed] [Google Scholar]
- 18.Park SH, Chi HS, Min SK, Park BG, Jang S, Park CJ. Prognostic impact of c-KIT mutations in core binding factor acute myeloid leukemia. Leuk Res. 2011;35:1376–1383. doi: 10.1016/j.leukres.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Boissel N, Leroy H, Brethon B, Philippe N, de Botton S, Auvrignon A, et al. Incidence and prognostic impact of c-Kit, FLT3, and Ras gene mutations in core binding factor acute myeloid leukemia (CBF-AML) Leukemia. 2006;20:965–970. doi: 10.1038/sj.leu.2404188. [DOI] [PubMed] [Google Scholar]
- 20.Paschka P, Marcucci G, Ruppert AS, Mrózek K, Chen H, Kittles RA, et al. Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t(8;21): a Cancer and Leukemia Group B Study. J Clin Oncol. 2006;24:3904–3911. doi: 10.1200/JCO.2006.06.9500. [DOI] [PubMed] [Google Scholar]
- 21.Allen C, Hills RK, Lamb K, Evans C, Tinsley S, Sellar R, et al. The importance of relative mutant level for evaluating impact on outcome of KIT, FLT3 and CBL mutations in core-binding factor acute myeloid leukemia. Leukemia. 2013;27:1891–1901. doi: 10.1038/leu.2013.186. [DOI] [PubMed] [Google Scholar]
- 22.Park SH, Min SK, Park BG, Jang S, Park CJ, Chi HS. Clinical usefulness of plasma specimens for detection of nucleophosmin 1 gene mutations in patients with normal karyotype acute myeloid leukemia. Leuk Res. 2011;35:e159–e160. doi: 10.1016/j.leukres.2011.04.023. [DOI] [PubMed] [Google Scholar]
- 23.Park SH, Chi HS, Cho YU, Jang S, Park CJ. CEBPA single mutation can be a possible favorable prognostic indicator in NPM1 and FLT3-ITD wild-type acute myeloid leukemia patients with intermediate cytogenetic risk. Leuk Res. 2013;37:1488–1494. doi: 10.1016/j.leukres.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 24.Verhaak RG, Goudswaard CS, van Putten W, Bijl MA, Sanders MA, Hugens W, et al. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood. 2005;106:3747–3754. doi: 10.1182/blood-2005-05-2168. [DOI] [PubMed] [Google Scholar]
- 25.Mrózek K, Marcucci G, Paschka P, Bloomfield CD. Advances in molecular genetics and treatment of core-binding factor acute myeloid leukemia. Curr Opin Oncol. 2008;20:711–718. doi: 10.1097/CCO.0b013e32831369df. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table 1. Summary of all informations about primer sequences, melting temperatures of each primer used in the amplification of nine genes and size of PCR product.
| Genes | Forward primers | Reverse primers | Size of PCR product (bp) | ||
|---|---|---|---|---|---|
| Sequences | Tm (℃) | Sequences | Tm (℃) | ||
| c-KIT exon 8 | GCTGAGGTTTTCCAGCACTC | 61.7 | AATTGCAGTCCTTCCCCTCT | 61.3 | 219 |
| c-KIT exon 17 | TGGTTTTCTTTTCTCCTCCAA | 57.4 | TGCAGGACTGTCAAGCAGAG | 62.6 | 185 |
| NPM1 exon 12 | GTTTCTTTTTTTTTTTTTCCAGGCTATTCAAG | 59.9 | CACGGTAGGGAAAGTTCTCACTCTGC | 65.6 | 170 |
| CEBPA exon 1 A | CGCCATGCCGGGAGAACTCT | 67.3 | CTTCTCCTGCTGCCGGCTGT | 67.5 | 299 |
| CEBPA exon 1 B | GCCGCCTTCAACGACGAGTT | 66.2 | CTTGGCTTCATCCTCCTCGC | 63.4 | 303 |
| CEBPA exon 1 C | CCGCTGGTGATCAAGCAGGA | 65.1 | CCGGTACTCGTTGCTGTTCT | 62.6 | 390 |
| CEBPA exon 1 D | CAAGGCCAAGAAGTCGGTGGACA | 66.5 | CACGGTCTGGGCAAGCCTCGAGAT | 69.7 | 356 |
| WT1 exon 7 | CAGTGCTCACTCTCCCTCAAG | 62.2 | AGTGTGAGAGCCTGGAAAGG | 61.9 | 300 |
| WT1 exon 9 | GTGAGGCAGATGCAGACATTG | 61.7 | CCTCTCATCACAATTTCATTCCA | 58.3 | 297 |
| IDH1 exon 4 | GCCATCACTGCAGTTGTAGGTT | 62.9 | CACATACAAGTTGGAAATTTCTGG | 57.8 | 439 |
| IDH2 exon 4 | GGGGTTCAAATTCTGGTTGAA | 58.8 | CTGTGGCCTTGTACTGCAGAG | 63.1 | 323 |
| DNMT3A exon 23 | ACTAAGCAGGCGTCAGAGGAG | 63.7 | TCCATCCTCATGTTCTTGGTG | 59.8 | 393 |
| EZH2 exon 17 | CCTTTTTGTTGCGTTTTCTCC | 58.9 | ATCCTCCTTCTGGTCACCTCA | 62.2 | 310 |
| EZH2 exon 18 | AGGCAAACCCTGAAGAACTGT | 62.0 | GATGGCTCTCTTGGCAAAAAT | 59.3 | 396 |
| EZH2 exon 19 | CGTTTTGCAAATCATTCGGTA | 57.9 | ATTCCCCACTAATGCTCATGG | 60 | 413 |
| CBL exon 8 | ACCCAGACTAGATGCTTTCTG | 59.3 | AGGCCACCCCTTGTATCAGT | 63.3 | 385 |
| CBL exon 9 | CCTGGCTTTTGGGGTTAGGTTT | 62.9 | GACAACTCACAATGGATTTTGCC | 60.4 | 375 |
Abbreviations: NPM, nucleophosmin; CEBPA, CCAAT/enhancer binding protein alpha; WT, Wilms tumor; IDH, isocitrate dehydrogenase; DNMT3A, DNA (cytosine-5-)-methyltransferase 3 alpha; EZH, enhancer of zeste homolog; CBL, casitas b-lineage lymphoma; Tm, melting temperature of primer; bp, base pairs.