SUMMARY
In metazoan adherens junctions, β-catenin links the cytoplasmic tail of classical cadherins to the F-actin-binding protein α-catenin. Phosphorylation of a Ser/Thr rich region in the cadherin tail dramatically enhances affinity for β-catenin and promotes cell-cell adhesion in cell culture systems, but its importance has not been demonstrated in vivo. Here, we identify a critical phosphorylated serine in the C. elegans cadherin HMR-1 required for strong binding to the β-catenin homolog HMP-2. Ablation of this phospho-serine interaction produces developmental defects that resemble full loss-of-function (Hammerhead and Humpback) phenotypes. Most metazoans possess a single gene for β-catenin, which is also a transcriptional coactivator in Wnt signaling. Nematodes and planaria, however, have a set of paralogous β-catenins; for example, C. elegans HMP-2 functions only in cell-cell adhesion, whereas SYS-1 mediates transcriptional activation through interactions with POP-1/Tcf. Our structural data define critical sequence differences responsible for the unique ligand specificities of these two proteins.
INTRODUCTION
Cell-cell adhesion is fundamental to the generation and structure of multicellular tissues. The classical cadherin cell adhesion molecules mediate homophilic adhesion, and are linked to the actin cytoskeleton through the catenins (Pokutta and Weis, 2007). β-Catenin binds to the cytoplasmic tail of classical cadherins, and to α-catenin, an F-actin binding protein. β-Catenin is a highly conserved metazoan protein that functions in both cell-cell adhesion and as a transcriptional coactivator in the Wnt/β-catenin pathway. β-Catenin has an N-terminal region of roughly 150 amino acids, followed by 12 armadillo (arm) repeats each comprised of three α helices, and a C-terminal acidic tail that mediates interactions with components of the general transcription apparatus. Structural studies have shown that the different partners of β-catenin involved in adhesion (the classical cadherin cytoplasmic domain) and in Wnt signaling (TCF transcription factors, ICAT, the Adenomatous Polyposis Coli protein, and Axin) interact with the arm domain almost identically (Pokutta and Weis, 2007).
In most metazoans, a single gene encodes a β-catenin that functions in both cadherin-based adhesion and transcriptional coactivation via Wnt signaling. An exception is the nematode C. elegans, which expresses four distinct β-catenin paralogs, only one of which, HMP-2, functions in cell adhesion. It is therefore of interest to understand how these different paralogs have evolved to mediate distinct functions by binding to partners involved in adhesion vs. signaling. C. elegans also expresses the cadherin HMR-1, whose cytoplasmic domain bears significant sequence homology to that of mammalian classical cadherins, and the α-catenin homolog HMP-1 (Costa et al., 1998; Cox and Hardin, 2004).
Studies in cell culture have revealed that interactions among adherens junction components can be regulated by phosphorylation. A serine-rich region of the classical cadherin tail is phosphorylated (Stappert and Kemler, 1994), and in vitro phosphorylation of purified cadherin tail by GSK-3β and CKII strengthens its affinity for β-catenin about 800 fold by creating an additional interaction surface (Choi et al., 2006; Huber and Weis, 2001; Lickert et al., 2000). Mutation of the phosphorylated serines to alanine reduced cell-cell adhesion when these constructs were transfected into NIH 3T3 cells (Lickert et al., 2000). Gottardi and colleagues recently narrowed these phosphorylation sites to three residues that are required for high-affinity β-catenin binding, cell adhesion, inhibition of cell migration and surface stability of cadherin in cultured cells (McEwen et al., 2014). In contrast, Src-mediated phosphorylation of β-catenin Tyr654 reduces affinity for cadherin (Roura et al., 1999), and CKII phosphorylation of β-catenin regulates its interaction with α-catenin (Bek and Kemler, 2002).
Although there are strong correlations between dysregulation of AJ assembly and disease such as metastatic cancers (Benjamin and Nelson, 2008; van Roy, 2014), the functional effects of phosphorylation of AJ components have not been examined in a true in vivo setting. C. elegans offers a system with which to test effects of phosphorylative regulation in vivo. Here, we use biochemical and structural methods to identify a critical phosphorylated serine in the tail of the C. elegans cadherin HMR-1 required for binding to the β-catenin homolog HMP-2. Mutation of this serine, or a set of arginine residues in HMP-2 that coordinate the phosphate group, produces severe developmental defects, demonstrating the physiological importance of a posttranslational modification identified from biochemical and structural approaches. Likewise, a phosphomimetic mutation of a conserved tyrosine in β-catenin/HMP-2 produces significant developmental phenotypes that correlate with the effect of phosphorylation of this residue in higher organisms. The structural data also allow us to define critical sequence differences responsible for the unique ligand specificities of HMP-2 and SYS-1.
RESULTS
Similarities between HMP-2 and β-catenin
HMP-2 is homologous to mammalian β-catenin, with a central 12 arm repeat domain (33% identity and 52% similarity to the mouse β-catenin arm domain) flanked by N- and C-terminal tails that are shorter than those of mammalian β-catenin (Figs. 1A, S1). In particular, HMP-2 lacks the N-terminal phosphorylation-dependent degron present in mammalian β-catenins that is critical for regulation of β-catenin in Wnt signaling (Stamos and Weis, 2013). Nonetheless, the N-terminal region contains sequences homologous to the α-catenin binding site of β-catenin that mediate similar interactions with α-catenin (Kwiatkowski et al., 2010). The C-terminal tail of mammalian β-catenins is the interaction site for general transcription factors needed for β-catenin function as a transcriptional coactivator (Barker et al., 2001; Hecht et al., 1999; Hecht et al., 2000; Miyagishi et al., 2000; Takemaru and Moon, 2000; Tutter et al., 2001); the HMP-2 tail is shorter and is not homologous in this region.
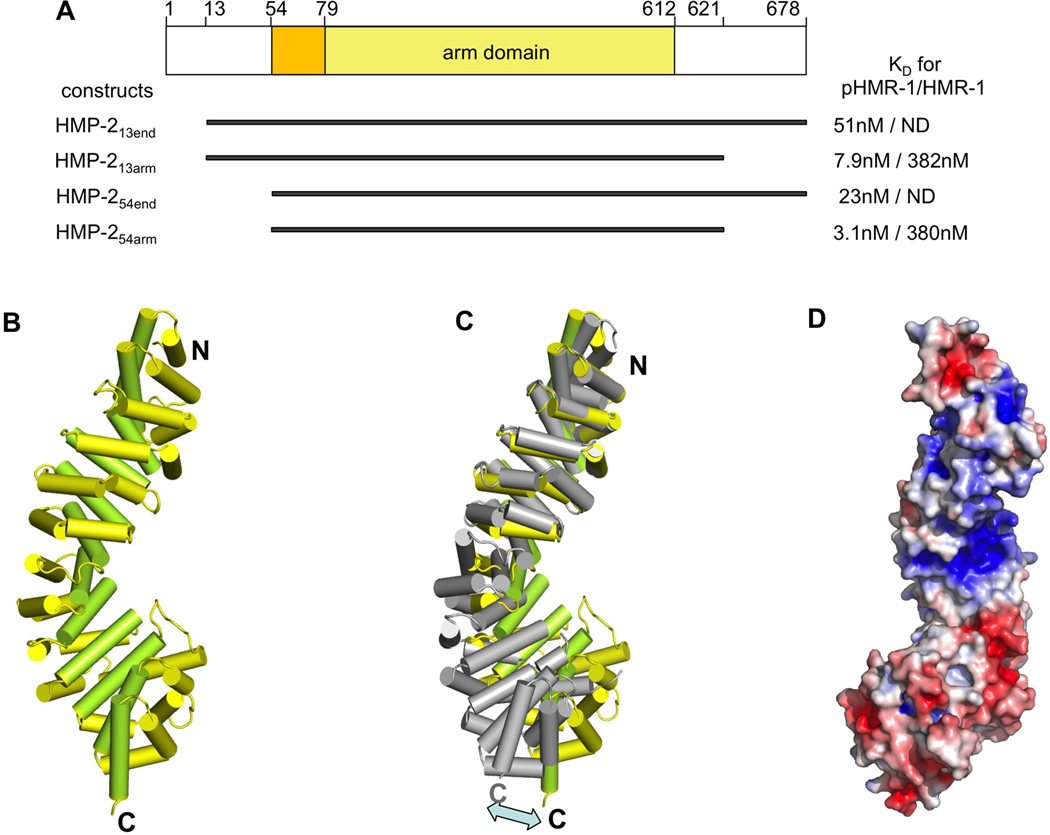
Figure 1. HMP-2 structure and dependence of binding on the phosphorylation state of HMR-1.
(A) Four constructs of HMP-2, which were used in our study, are shown and their residue boundaries indicated. On the right side of each construct, KD values for pHMR-1cyto80 and HMR-1cyto80, measured by ITC experiments are shown. HMP-213end and HMP-254end did not give measurable ITC signals for the interaction with HMR-1cyto80 and are labeled as ND.
(B) Structure of HMP-254arm (residues 77–613). Helices 1 and 2 of each arm repeat are colored yellow, and helix 3 is colored light green. The structures of HMP-254arm and HMP-254end are very similar except for the N-terminal region. Residues 56–79 are seen only in the HMP-254end structure, and form an extra N-terminal helix whose position appears to be a consequence of crystal packing.
(C) Superposition of arm repeats 1 to 4 of HMP-2 and mouse β-catenin (PDB ID 1I7W). HMP-2 helices are colored as in Figure 2A and β-catenin is colored grey.
(D) Electrostatic surface of the HMP-2 arm domain, with negative and positive regions colored red and blue, respectively. Contoured at ± 5 kBT/e.
We determined the structure of a HMP-2 construct consisting of residues 54–678, designated HMP-254end, which comprises a portion of the HMP-1/α-catenin binding site, the arm domain and C-terminal region (Fig. 1A) at 2.0Å resolution (Table S1; Fig. 1B). The visible structure contains the twelve predicted arm repeats, and an extra helix (residues 54–71) at the N-terminus preceding the arm repeats, which corresponds to the α-catenin binding helix formed by mouse β-catenin 121–149. This helix is engaged in lattice packing interactions, so it is unclear if its position is functionally relevant.
Although the C-terminal tail (residues 613–678) is part of the crystallized construct, it was not visible in the structure; in particular, the extra α helix (“helix C”) found just after the arm repeats in crystals of full-length zebrafish β-catenin (Xing et al., 2008) and in plakoglobin (Choi et al., 2009) is absent. The sequence of helix C is conserved in human, mouse, zebrafish and Drosophila β-catenins, whereas the corresponding portion of the HMP-2 sequence lacks these residues and is predicted to have no secondary structure. Helix C contributes to the interaction with the Wnt transcription antagonists ICAT and Chibby (Xing et al., 2008). The absence of helix C in HMP-2 is consistent with its specialized function as a cell adhesion protein.
Superposition of the arm domains of HMP-2 and β-catenin shows that although the individual arm repeats are similar, the curvatures of the superhelix are different; the C-terminal halves of the domain are displaced by about 20Å when the N-terminal halves are aligned, and vice versa (Fig. 1C). The arm domains of HMP-2, β-catenin, p120, plakoglobin, plakophilin, and SYS-1 have different curvatures, likely due to small differences in the packing of arm repeats. However, all of these proteins feature a positively charged groove formed by the superhelix, and their overall pIs are about 8 or above. In β-catenin, this groove is the common binding site for many different ligands, including cadherins, TCFs, APC and ICAT (Pokutta and Weis, 2007). Curiously, the overall pI of the arm domain of HMP-2 is about 5.5, but it contains a positively charged groove similar to these other arm domain structures (Fig. 1D).
HMR-1 must be phosphorylated in order to bind strongly to HMP-2
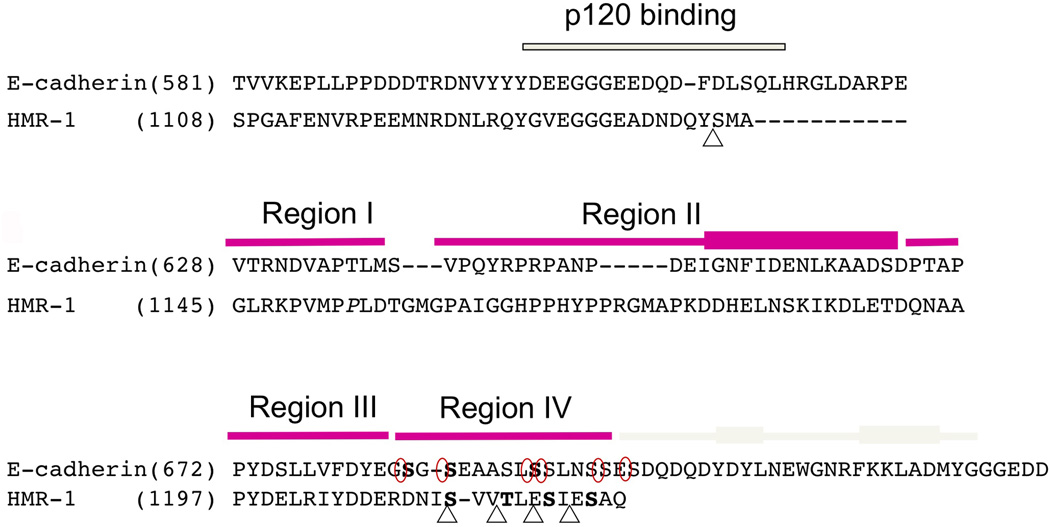
The cytoplasmic domains of HMR-1 and murine E-cadherin bear significant sequence homology (25% identity, 34% similarity) (Fig. 2). The crystal structure of the 151 amino acid E-cadherin cytoplasmic domain (Ecyto) bound to the β-catenin arm domain revealed that the last 100 amino acids of Ecyto constitute the β-catenin binding site. Ecyto features five regions of interaction with β-catenin (Huber and Weis, 2001). Region I, the N-terminal portion of the binding site, packs against arm repeats 8 and 9 and also interacts with region III. Region II includes an amphipathic α helix that forms a hydrophobic interface with β-catenin arm repeats 11 and 12. Region III is an extended peptide that binds in the groove formed by arm repeats 5–10, and includes the Dxθθxϕx2–7E (θ=non-polar aliphatic interaction, ϕ=aromatic) motif that defines the interactions of β-catenin with Wnt pathway ligands including TCFs, APC and ICAT (Pokutta and Weis, 2007). Region IV is a serine-rich motif that binds to β-catenin only when phosphorylated, a modification that increases the affinity of Ecyto for β-catenin from 46 nM to 52 pM. The C-terminal region V, also called the hydrophobic cap, contains two short α helices that pack against what would otherwise be exposed hydrophobic residues at the start of the arm domain. Sequence alignment (Fig. 2) indicates that the amphipathic helix in region II and the region III Dxθθxϕx2–7E motif are present in HMR-1, and a serine-rich sequence is present in region IV. However, HMR-1 is truncated relative to E-cadherin and lacks the region V cap.
Figure 2. Sequence alignment of the cytoplasmic tails of mouse E-cadherin and HMR-1 reveals key conserved residues.
Juxtamembrane region including p120 (JAC-1 in C. elegans) binding site was aligned base on the sequence homology and β-catenin (HMP-2) binding region was aligned based on the crystal structures of pEcyto and pHMR-1cyto80. HMP-2 binding domain is divided into four regions and each region is indicated with magenta bar on top of the sequences. The C-terminal cap region is only present in Ecyto and is shown with grey colored bar. The thickened rectangles represent α-helices. Five CKI-phosphorylated sites in HMR-1cyto are marked with triangle and six phosphorylation sites by GSK-3β and CKII in Ecyto are circled. Phosphorylated residues observed in the structures are written in bold (3 residues in pEcyto and 4 in pHMRcyto80). The starting residue numbers are indicated.
Despite the similarities between the C. elegans and mammalian proteins, there was no detectable interaction between bacterially expressed HMR-1 cytoplasmic domain (HMR-1cyto) and HMP-254end, as assessed by isothermal titration calorimetry (ITC) (Figs. 1A, S2, Table S2) and co-migration on a gel filtration column (Kwiatkowski et al., 2010). This is consistent with the absence of the region V cap in HMR-1, as deletion of this region significantly weakens the interaction of E-cadherin with β-catenin (Finnemann et al., 1997; Stappert and Kemler, 1994). Curiously, HMP-2 lacking the C-terminal tail (HMP-254arm) bound weakly, but detectably, to HMR-1cyto (KD= M) in ITC (Figs. 1A, S2, Table S2). This behavior is reminiscent of mammalian β-catenin binding to its weaker ligands Axin and APC, where deletion of its C-terminal tail increases the affinity for axin and APC-R3 by 7-fold and 15-fold (Choi et al., 2006). However, the C-terminal tail does not affect binding of β-catenin to tightly binding ligands including E-cadherin and LEF-1 (Choi et al., 2006). We also tested the effect of the N-terminal tail of HMP-2. Full-length HMP-2 could not be expressed as a soluble protein in E. coli, so constructs in which the N-terminal 12 amino acids were deleted (HMP-213end and HMP-213arm; Figs. 1A, S2) were used for these experiments. The data showed that residues 13–53 of HMP-2 had no significant effect on the affinity for HMR-1 (Table S2). This is consistent with ITC data of β-catenin indicating that the N-terminal domain has no or very minor effect on ligand binding (Choi et al., 2006).
Phosphorylation of Ecyto by GSK-3β and casein kinase II (CKII) increases the affinity for β-catenin about 800-fold, due to the interaction of the phosphorylated region IV with arm repeats 3–5 (Choi et al., 2006; Huber and Weis, 2001). The HMR-1 cytoplasmic domain contains four consensus casein kinase I (CKI) sites in region IV, as well as a 5th CKI site in the juxtamembrane region (Fig. 2). Specifically, CKI prefers substrates with an acidic side chain or a phosphorylated Ser/Thr three residues N-terminal to the target phosphorylation site: in region IV, Ser1212 is preceded by Asp1209, and is followed by Thr1215, Ser1218 and Ser1221 (Fig. 2). Anion exchange chromatography following in vitro phosphorylation of HMR-1cyto with CKI gave two peaks, both of which eluted at a higher salt concentration relative to non-phosphorylated HMR-1cyto (data not shown). Mass spectrometric analysis confirmed that the two peaks have 4 and 5 phosphorylated residues, respectively. It is likely that the four serines in region IV are sequentially phosphorylated by CKI, with the 5th site the juxtamembrane Ser1142.
CKI-mediated phosphorylation of purified HMR-1cyto produced a dramatic increase in affinity for HMP-2: both phosphorylated HMR-1cyto (pHMR-1cyto) fractions, with either 4 or 5 phosphorylated residues, were found to co-elute with HMP-2 on a size exclusion column (Kwiatkowski et al., 2010). The β-catenin binding region of E-cadherin is homologous to the last 80 amino acids of HMR-1 (Fig. 2), so a construct corresponding to this region, designated HMR-1cyto80, was tested for HMP-2 binding by ITC. The 380 nM dissociation constant of HMP-254arm for HMR-1cyto80 was decreased to 3.1 nM for pHMR-1cyto80 (Figs. 1A, S2, Table S2). To test whether the tails of HMP-2 affect the affinity for pHMR-1cyto80, ITC experiments were performed using HMP-213end and HMP-254end. The presence of the C-terminal tail of HMP-2 weakens the affinity approximately 7-fold (23 nM vs. 3.1 nM), whereas the presence of the N-terminal tail did not significantly affect binding to phosphorylated HMR-1 (Figs. 1A, S2, Table S2).
The increase in affinity for HMP-2 associated with phosphorylation of HMR-1 parallels the change in murine E-cadherin affinity for β-catenin; in both cases, phosphorylation increases affinity more than two orders of magnitude. However, the absolute affinities differ dramatically: without phosphorylation, E-cadherin binds to β-catenin with an affinity of 46 nM, whereas comparable affinity is achieved for HMR-1 only when it is phosphorylated.
Crystal structure of pHMR-1 bound to HMP-2
To define the mechanism of phosphorylation-dependent binding of HMR-1 to HMP-2, we obtained crystal structures of the pHMR-1/HMP-2 complex. The complex of individually purified HMP-254arm and in vitro CKI-phosphorylated HMR-1cyto80 (pHMR-1cyto80) was prepared by gel filtration chromatography and crystallized in two different conditions (Table S1). The two structures were determined at 2.3Å and 2.8Å resolution, and provided four independent views of the complex, one in the higher-resolution P43 crystal form and three in the P21 form (Fig. 3A). The four copies of HMR-1cyto80 align well in regions II and III, whereas region I is visible only in three copies. The phosphorylated region IV adopts two different conformations; three copies (the single P43 and two of the P21 copies) adopt the same conformation, designated conformation A, whereas the other is distinct (conformation B).
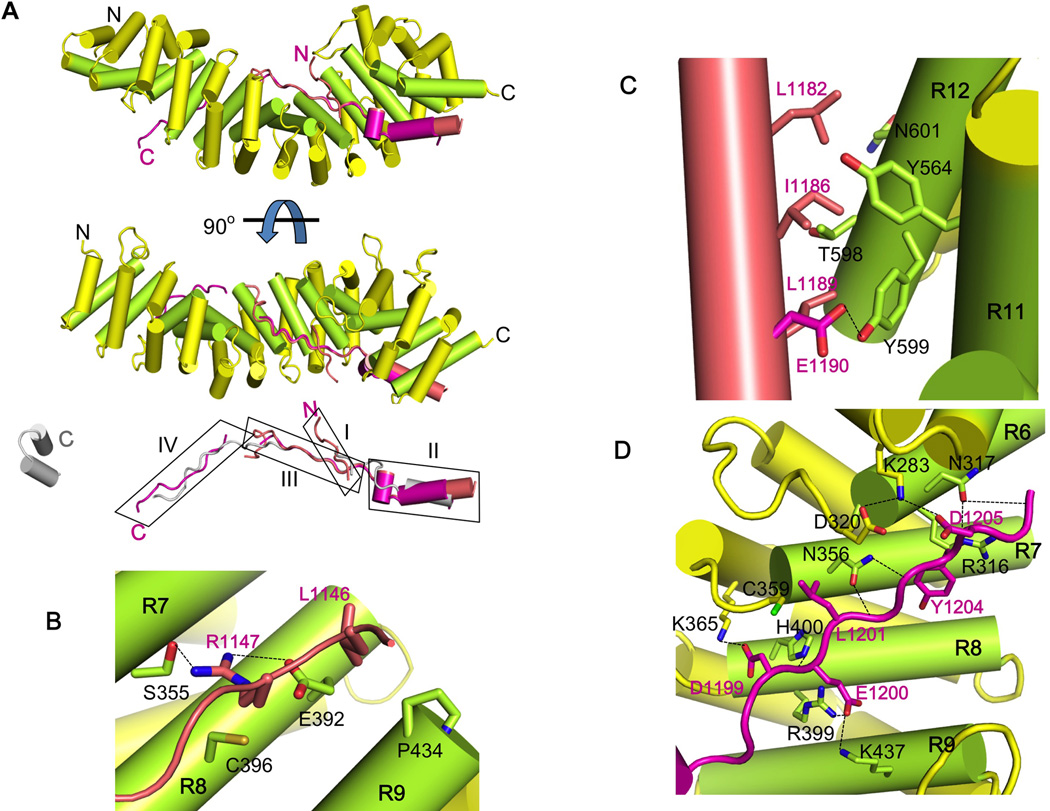
Figure 3. Crystal structure of HMP-254arm/pHMR-1cyto80 complex identifies key interaction regions.
In each panel, the arm domain colored as in Figure 1B and the structures of pHMR-1cyto80 are shown in salmon (conformation A from the high-resolution P43 crystal form) and magenta (conformation B).
(A) Ribbon diagram of the complex. The N and C terminus of each protein is labeled. Phosphorylated Ecyto is aligned to pHMR-1cyto80 and shown in grey. The four regions of pHMR-1 are boxed and labeled as I, II, III and IV.
(B) Interaction region I. Arm repeats 7, 8 and 9 are labeled as R7, R8 and R9. Side chains of amino acids that make direct contacts between HMP-2 and pHMR-1cyto are shown as sticks, and HMP-2 and pHMR-1 residues are labeled in black and magenta, respectively. Hydrogen bonds are shown as dotted lines.
(C) Interaction region II.
(D) Interaction region III. Most of the contacts between HMP-2 and pHMR-1cyto are formed by side chains, except for N317, N356, and H400 of HMP-2, which form hydrogen bonds with amide and carbonyl groups of a polypeptide backbone of pHMR-1cyto.
Apart from the absence of region V, the overall binding mode of HMR-1cyto80 for HMP-2 is very similar to that of E-cadherin binding to β-catenin (Fig. 3A). Region I interacts with side chains from arm repeats 7, 8 and 9 (Fig. 3B). Residues 1153–1178, which connect regions I and II, are disordered. Hydrophobic residues on the amphipathic helix in region II pack against HMP-2 residues located on arm repeats 11 and 12, including Tyr599 (see below) (Fig. 3C). The polypeptide then makes a sharp turn into region III, which adopts an extended conformation that interacts with the basic groove of the arm domain in essentially the same way as observed between murine E-cadherin and β-catenin (Fig. 3D). The polypeptide backbone of region III forms hydrogen bonds with polar residues on arm repeats 6, 7, and 8 of HMP-2. Asp1199, the first residue of the Dxθθxϕx2–7E motif, forms a salt bridge with HMP-2 Lys365. Leu1201 packs against Cys359, and Tyr1204 packs against HMP-2 Arg316. In two of the four independent copies of the complex, Glu1207, the last residue of the consensus region III motif, forms a salt bridge with Lys240 of HMP-2. In addition to the interactions in common with the E-cadherin/β-catenin complex, Glu1200 of HMR-1 forms salt bridges with Arg399 and Lys437 of HMP-2.
The mass spectrometry analysis of in vitro phosphorylated HMR-1 suggested that four residues in region IV are sequentially phosphorylated by CKI. In pHMR-1 conformation A, only the first phosphorylated residue, Ser1212, is observed; all of the HMR-1 residues C-terminal to this position are disordered (Fig. 4A,B). In conformation B, region IV is disconnected from region III by three missing residues, 1208–1210, but it continues until A1222, the penultimate HMR-1 residue. Notably, all four phosphorylated residues (S1212, T1215, S1218, S1221) are visible (Fig. 4C) in conformation B, which superimposes closely with that of phosphorylated E-cadherin bound to β-catenin (Fig. 4C,D). In contrast, conformation A appears to arise from displacement by neighboring molecules in the crystal. We conclude that conformation B represents the solution conformation of the bound phosphoHMR-1.
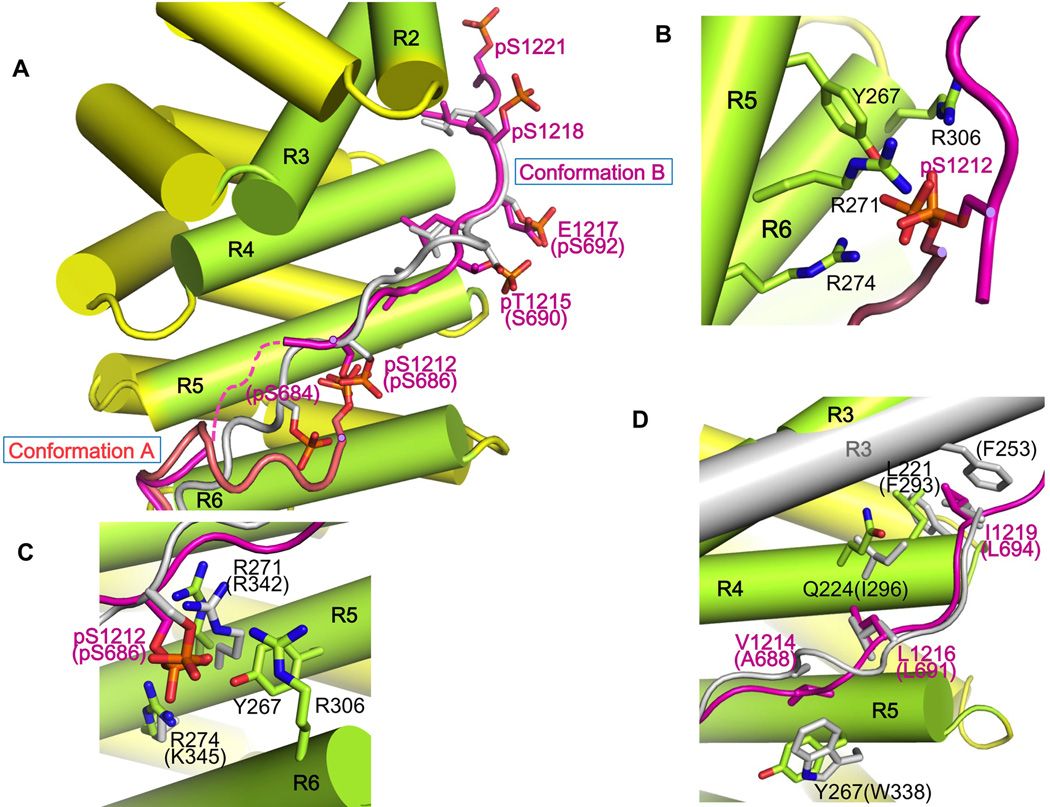
Figure 4. Phosphorylation generates specific interactions of HMR-1 region IV with HMP-2.
(A) Structures of HMP-2 bound to the two different conformations of pHMR-1cyto are aligned, with conformations A and B shown in salmon and magenta. Also shown is a superposition of pEcyto (grey) from the β-cateninarm/pEcyto complex structure (PDB ID 1I7W) with conformation B of pHMR-1cyto; E-cadherin residues are indicated in parentheses. For clarity, the superimposed arm domain β-catenin is not shown. All phosphorylated residues observed in the structures of pEcyto and pHMR-1cyto are represented.
(B) Close-up of the interactions between HMP-2 and pS1212 of pHMR-1cyto from conformations A and B.
(C) Comparison of HMR-1 pS1212 interactions with HMP-2 and E-cadherin pS686 with β-catenin.
(D) Comparison of β-catenin–pEcyto interactions with those of HMP-2–pHMR-1cyto residues 1214–1219.
In conformation B, the side chains of V1214, L1216 and I1219 pack against residues from HMP-2 arm repeats 4 and 5, analogous to the hydrophobic interactions mediated by phosphorylated E-cadherin A688, L691 and L694 with β-catenin (Fig. 4D). Moreover, HMR-1 pT1215 forms a salt bridge with HMP-2 K264; the equivalent residue in E-cadherin, S690, is not phosphorylated and points away from the surface of β-catenin. Given the disorder of region IV in the non-phosphorylated mammalian E-cadherin/β-catenin complex, we expect that region IV of non-phosphorylated HMR-1 is disordered. Thus, as in the mammalian case, phosphorylation stabilizes an extensive set of electrostatic and hydrophobic packing interactions that accounts for the large increase in affinity.
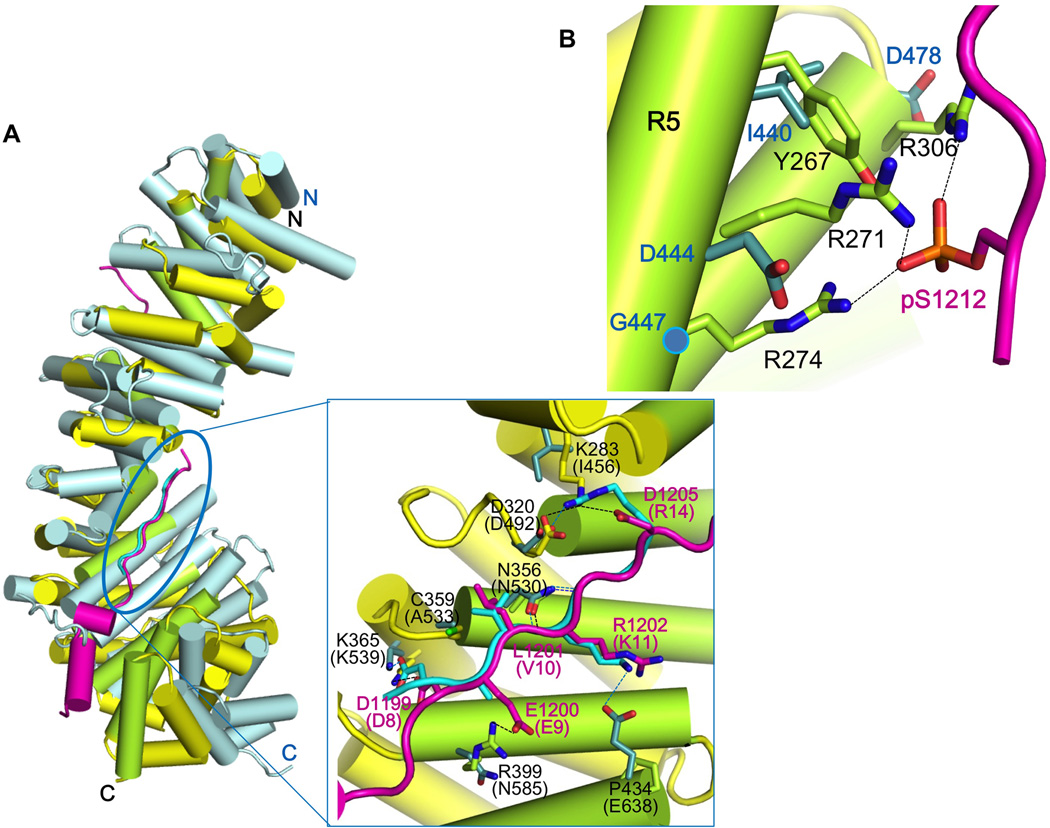
Although the backbone at residues 1211 and 1212 is positioned differently in conformations A and B (Fig. 4A), the phosphate group of pS1212 is in almost the same position relative to HMP-2, and interacts with Tyr267, Arg271, Arg274, and Arg306 of HMP-2 (Fig. 4B). Moreover, Ser1212 is the only phosphorylated residue that can be superimposed with the corresponding phospho-Ser (Ser686) among three visible phospho-serines (Ser684, Ser686, and Ser692) seen in the complex of phosphorylated Ecyto and β-catenin (Fig. 4C).
To assess the contributions of S1212 and the other phosphosites to the affinity of HMR-1 for HMP-2, we produced and purified two different HMR-1 cytoplasmic tail mutants, S1212A and T1215A, and phosphorylated them in vitro with CKI. As expected for the consensus CKI sites processively phosphorylated starting at S1212 (Fig. 2), mass spectrometry confirmed that S1212A was not phosphorylated in region IV, whereas T1215A retained phosphorylation at S1212 but lacked phosphorylation at S1218 and S1221 (data not shown). ITC measurements showed that HMP-254-arm bound to HMR-1(S1212A)cyto80 with essentially the same affinity as wild-type, non-phosphorylated HMR-1cyto80, whereas it bound strongly to pHMR-1(T1215A)cyto80 with KD = 26 nM, 8x weaker than that of the wild-type phosphorylated HMR-1cyto80 (Table S2, Figure S2). Non-phosphorylated HMR-1(T1215A)cyto80 bound with KD = 610 nM, 24x weaker than the phosphorylated protein (Table S2, Figure S2). Like HMR-1(S1212A), there was no significant difference in affinity of HMP-2 for the non-phosphorylated HMR-1(T1215A) vs. wild-type HMR-1 (Table S2, Figure S2).
HMR-1 S1212 and S1218 are equivalent to two of the three residues recently identified as bearing the majority of vertebrate E-cadherin phosphorylation (Ser686 and Ser693, numbered from the mature sequence) (McEwen et al., 2014); the third phosphosite in the vertebrate cadherin, Ser692, is equivalent to HMR-1 E1217, a potential phosphomimetic. It is not known which of these phosphorylated residues contribute to the enhanced adhesion observed by McEwan et al. In the crystal structure of phosphorylated E-cadherin bound to β-catenin, pSer692 interacts extensively with β-catenin, whereas HMR-1 E1217 forms no contacts with HMP-2. Ser693 was not phosphorylated in the study of Huber and Weis (2001), and the equivalent HMR-1 pS1218 does not form direct contacts with HMP-2.
Collectively, the thermodynamic and structural data indicate that phosphorylation of the conserved S1212 contributes the majority of the affinity enhancement when the HMR-1 tail is phosphorylated by CK1, serving as a critical anchor point for the binding of region IV to HMP-2, enabling the large enhancement in affinity relative to the unphosphorylated HMR-1 (Fig. 1A).
Phosphorylation of HMR-1 Ser1212 is essential for cell adhesion in vivo
To test the functional significance of the HMR-1 pSer1212 interaction with HMP-2, in vivo rescue experiments were performed. The hmr-1 allele zu389 contains a nonsense mutation very early in the transcript and behaves as a genetic null, with homozygous mutant offspring dying embryonically with the Hammerhead phenotype. A full length hmr-1::gfp transgene driven by the endogenous promoter (courtesy J. Nance) localizes to epidermal adherens junctions and was able to rescue hmr-1(zu389) homozygotes to viability (Fig. 5AB). Site-directed mutagenesis was used to create a nonphosphorylatable HMR-1::GFP S1212A construct, which was then expressed in hmr-1(zu389)/+ unbalanced heterozygotes to assay for rescue. Although HMR-1(S1212A)::GFP localizes to junctions and is expressed at levels comparable to wildtype HMR-1 (Figs. 5D, S3A), presumably due to having an intact extracellular adhesive domain, it fails to rescue hmr-1(zu389) homozygotes to viability. Most of these embryos die as Hammerhead (Fig. 5C), indicative of full hmr-1 loss of function.
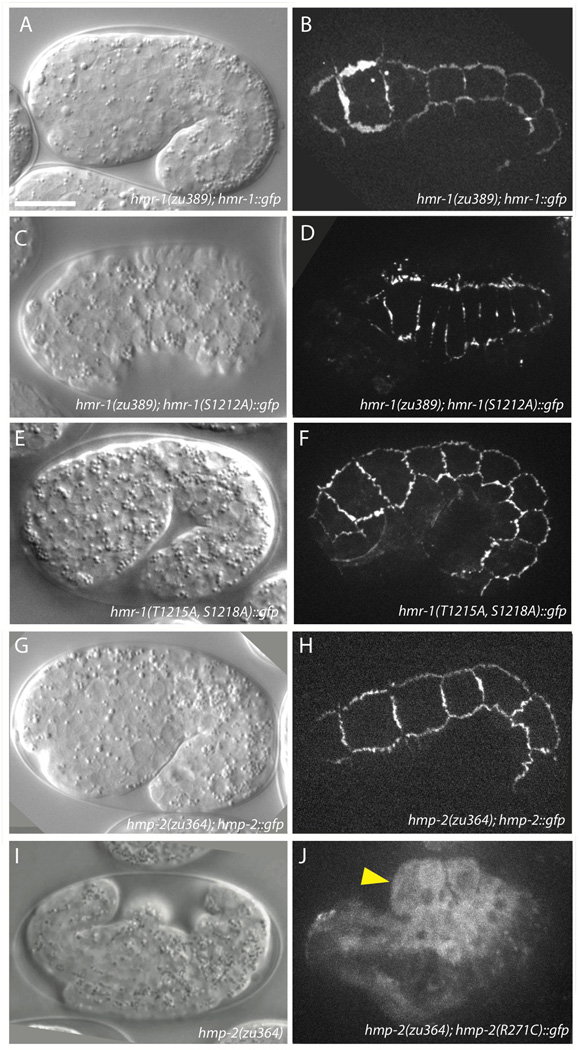
Figure 5. HMR-1 pS1212 is required for interaction between HMP-2 and HMR-1.
(A, B) DIC (A) and confocal (B) images of 1.5-fold elongating hmr-1(zu389) homozygotes rescued to viability by HMR-1::GFP. Lethality in offspring of hmr-1(zu389); hmr-1::gfp mothers is 85.2% (n = 209). Signal localizes to epidermal adherens junctions similarly to HMR-1 immunostaining (not shown). Scale bar is 10 µm.
(C, D) The phospho-null construct HMR-1(S1212A)::GFP localizes to junctions but is unable to rescue hmr-1(zu389) homozygotes to viability. Lethality in offspring of hmr-1(zu389)/+; hmr-1(S1212A)::gfp mothers is 27.4% (n = 1072). Cells in (D) appear compressed due to retraction of the hypodermis subsequent to failure of epiboly.
(E, F) HMR-1(T1215A, S1218A)::GFP localizes to junctions and rescues hmr-1(zu389) embryonic enclosure and elongation. The hmr-1 transgene reduces embryonic lethality of hmr-1(zu389)/+ offspring from 24.9% (n=1101) to 16.9% (n=1055, 3 independent lines). Pictured is a representative offspring from a hmr-1(zu389)/+; hmr-1(T1215A, S1218A)::gfp mother.
(G, H) Wildtype HMP-2::GFP localizes to junctions (H) and rescues hmp-2(zu364) to viability (G).
(I) hmp-2(zu364) homozygotes fail to elongate and die with the Humpback phenotype. The zu364 lesion was identified as a point mutation resulting in HMP-2 R271C.
(J) HMP-2(R271C)::GFP localizes exclusively to the cytoplasm, and embryos die with the Hmp phenotype, consistent with a model in which this residue is crucial for interacting with HMR-1 pS1212. Arrowhead indicates dorsal humps.
HMR-1 pS1212 may also be necessary to prime additional phosphorylations at C-terminal CKI targets; as noted above, phosphorylation of these C-terminal sites contributes modestly to the affinity for HMP-2. In order to distinguish between the priming and direct HMP-2 interaction functions of HMR-1 S1212, we created a hmr-1(T1215A, S1218A)::gfp construct in which S1212 remains phosphorylatable, but the C-terminal phosphorylation cascade is blocked. HMR-1(T1215A, S1218A)::GFP localizes junctionally in wildtype and hmr-1(zu389)/+ backgrounds and is able to partially rescue hmr-1(zu389) embryonic lethality (Fig. 5E–F), but larval lethality precluded generation of a rescued hmr-1(zu389) homozygous line.
We also examined multiple conserved residues in HMP-2 that coordinate with pS1212 in HMR-1. Significantly, sequencing of a canonical strong hmp-2 allele (zu364) revealed that one of the coordinating residues, Arg271, is mutated to a cysteine. A full length hmp-2::gfp construct driven by 2 kb of endogenous promoter was expressed at epidermal adherens junctions and was able to rescue hmp-2(zu364) homozygotes to viability (Fig. 5G–H). Re-creating the zu364 allele in a hmp-2::gfp construct revealed that the protein is no longer recruited to adherens junctions and instead localizes diffusely to the cytoplasm (Fig. 5I–J). As expected from its location on the exterior surface of the protein, this mutation does not alter protein folding or stability, as the recombinant protein could be prepared as a stable protein and was expressed at similar levels in animals (Fig. S3B). ITC measurements revealed that HMP-2 R271C bound to pHMR-1 and HMR-1 with affinities that were the same as wild-type HMP-2 binding to non-phosphorylated HMR-1, as expected for ablating the interaction of the R271 side chain with the phosphorylated S1212 (Table S2). Mutation of the other HMP-2 arginines in Region IV to glutamic acids is predicted to ablate their ability to electrostatically bind HMR-1 pS1212. Consistent with this prediction, HMP-2::GFP R274E, R306E fails to rescue hmp-2(zu364) and localizes cytoplasmically, similarly to the R271C construct (data not shown).
Phosphorylation of HMP-2 Y599 may regulate junction formation
Previous in vitro work has shown that Src phosphorylation of vertebrate β-catenin at Tyr654 in Arm repeat 12 reduces its affinity for E-cadherin (Roura 1999, and dephosphorylation of this residue is maintained in cell culture by the phosphatase PTP1B (Xu 2004). The crystal structure of mouse β-catenin bound to E-cadherin revealed that Tyr654 forms a hydrogen bond with Asp665 of E-cadherin, so phosphorylation would disrupt this interaction (Huber and Weis, 2001). In two of the four crystallographically independent copies of the HMR-1/HMP-2 structure presented here, the equivalent tyrosine, Tyr599, forms a hydrogen bond with Glu1190 of HMR-1 (Fig. 3C).
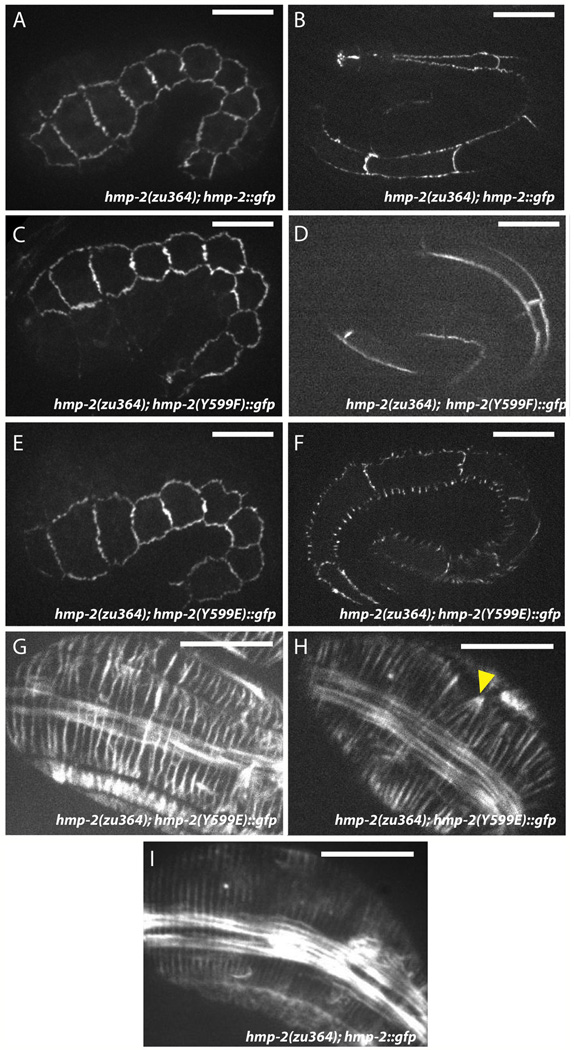
Although it is not known whether Tyr599 is phosphorylated in vivo, these data led us to examine the potential effect of this modification. We created phosphomimetic and phospho-null mutations at amino acid 599 in hmp-2::gfp. Surprisingly, both Y599E and Y599F constructs were able to rescue hmp-2(zu364) to a similar extent (45.9% n=283 and 50.2% n=305, respectively). However, although HMP-2::GFP Y599E initially localizes to adherens junctions similarly to wildtype HMP-2::GFP (Figs. 6A,E, S3C), the junctional distribution becomes punctate during late elongation, and fluorescent signal forms excursions orthogonal to the junctions between lateral seam cells and their dorsal and ventral neighbors (Fig. 6F). Morphological defects observed in cadherin-catenin mutants often result from aberrant circumferential F-actin filament (CFB) organization in C. elegans embryos (Costa et al., 1998; Cox-Paulson et al., 2012; Maiden et al., 2013), as embryonic elongation is driven in large part by radially directed actomyosin-mediated tension transmitted to adherens junctions via CFBs (Costa et al., 1998; Piekny et al., 2003). Phalloidin staining of hmp-2(zu364); hmp-2::gfp Y599E embryos revealed modest defects in CFB organization, including wavy, irregularly spaced filaments, which could indicate a lack of maintenance of tension (Fig. 6 D,E), as well as occasional co-termination of multiple CFBs at a single point along the junction (Fig. 6H). Surprisingly, HMP-2 Y599E and HMR-1 appear to colocalize in the excursions (Fig. S4). This result suggests that loss of affinity between HMR-1 and HMP-2 leads to loss of resistance to lateral tension at junctions without complete abrogation of their interaction.
Fig 6. HMP-2 Y599E localizes aberrantly and produces mild circumferential F-actin bundle (CFB) defects.
(A,B) Wildtype HMP-2::GFP signal is well established by early elongation (A) and is maintained at junctions through late elongation (B). Rescued zu364 homozygotes display 53.9% lethality (n = 178), approximately commensurate with the transmission rate of the extrachromosomal array carrying the transgene. Scale bar is 10 µm.
(C,D) HMP-2(Y599F)::GFP rescues hmp-2(zu364) homozygotes and localizes to epidermal adherens junctions identically to wildtype HMP-2::GFP both in early (C) and late (D) elongation. Rescued zu364 homozygotes display 50.2% lethality (n = 305).
(E,F) HMP-2::GFP Y599E rescues hmp-2(zu364). Signal initially localizes to adherens junctions (E), but it becomes punctate during late elongation, and fluorescent excursions form orthogonally to junctions between lateral (seam) cells and their dorsal and ventral neighbors (F). Rescued zu364 homozygotes display 45.9% lethality (n = 283).
(G–I) CFBs visualized by phalloidin staining in hmp-2(zu364) homozygotes rescued by wildtype (I) or Y599E (G, H) HMP-2::GFP constructs. Wildtype HMP-2::GFP embryos display evenly-distributed radial F-actin bundles (I). CFBs in Y599E embryos become irregularly spaced (G, H), and occasionally multiple bundles aggregate to a single locus on the junction (H, arrowhead). Scale bar is 10 µm.
To understand the molecular mechanism of the Y599E phenotype, Glu was modeled at the position of Tyr599. The side chain of Glu can be accommodated in the interface with HMR-1 without steric collision or charge repulsion. However, Glu cannot form the extensive hydrophobic contacts with Ile1186 and Leu1189 of HMR-1, which could decrease HMP-2 affinity for HMR-1. Conversely, retention of these contacts may also explain the lack of obvious effects of the Y599F mutation. Furthermore, modeling suggests that phosphorylation at Tyr599 cannot be accommodated, as the phosphate group would be too close to the backbone carbonyl group of HMR-1 Leu1189 and the side chain of Glu1190. Thus, phosphorylation may reduce the interaction with the region II of HMR-1.
Comparison with the SYS-1/POP-1 complex
Previous yeast two-hybrid analyses suggested that HMR-1 cannot bind the other β-catenin paralogs in C. elegans, i.e., BAR-1, WRM-1 and SYS-1 (Kidd, 2005; Korswagen et al., 2000). Among the other β-catenin paralogs in C. elegans, only the structure of SYS-1 bound to POP-1, the C. elegans TCF homolog, is available (Liu et al., 2008). Comparison with the structure of the HMR-1/HMP-2 complex provides insights into why HMR-1 does not bind to these other β-catenin paralogs. Even though a POP-1 construct spanning residues 1–200 was present in the crystals of Liu et al (2008), only residues 7–14 were visible. The polypeptide backbone of POP-1 residues 8–14 aligns well to pHMR-1cyto80 region III, residues 1199–1205, when arm repeats 7–9 of HMP-2 and SYS-1 are superimposed (Fig. 7A). The critical salt bridge made by Asp8 of POP-1 and Lys539 of SYS-1 is preserved in HMP-2/pHMR-1 structure (HMR-1 Asp1199 – HMP-2 Lys365), as are hydrogen bonds between Asn530 of SYS-1 and the polypeptide backbone of POP-1 (Fig. 7A). Key hydrophobic packing interactions mediated by the Dxθθxϕx2–7E motif are also present. Nonetheless, we found that purified POP-1 (1–45), which is known to be sufficient for the interaction with SYS-1 and BAR-1 (Liu et al., 2008), does not bind to purified HMP-2 (data not shown).
Figure 7. Structural comparison of HMP-2/pHMR-1cyto80 complex with SYS-1/POP-1 complex suggests why only HMP-2 can bind pHMR-1.
(A) Superposition of SYS-1/POP-1 (PDB ID 3C2G) into HMP-2/pHMR-1cyto80 complex. SYS-1 and POP-1 are colored light blue and cyan, respectively. Repeats 7 to 9 of SYS-1 and POP-1 are manually aligned to corresponding regions of HMP-2 and pHMR-1 in Coot. POP-1 (amino acids 8–14) aligns well with pHMR-1 region III, as shown in the close-up (boxed) view. SYS-1 and POP-1 residue numbers are in parentheses, and only those SYS-1 side chains that interact with POP-1 interaction are shown. Side chain of Glu9 of POP-1 is not modeled in the original structure. Hydrogen bonds in HMP-2/pHMR-1cyto80 and SYS-1/POP-1 complexes are shown as black and blue dotted lines, respectively.
(B) Repeats 5 and 6 of SYS-1 are aligned to corresponding region of HMP-2. Three arginine residues of HMP-2 interacting with pS1212 are not conserved in SYS-1.
The lack of binding between POP-1 and HMP-2 can be rationalized by aligning POP-1 with pHMR-1cyto80 bound to HMP-2. Most POP-1 residues are accommodated without clashes with HMP-2. However, Arg14 is close to the side chain of Lys283 of HMP-2, which corresponds to Ile456 in SYS-1 (Fig. 7A). POP-1 Arg14 forms a salt bridge with Asp492 of SYS-1, but in the HMP-2arm/pHMR-1cyto80 structure, Asp320 of HMP-2, which is equivalent to Asp492 of SYS-1, makes an intramolecular salt bridge with Lys283 and intermolecular interaction with Asp1205 of HMR-1 (Fig. 3D). Although a different rotamer of Arg14 might avoid clashes, it would not be able to contribute to the interaction. Moreover, POP-1 Lys11 forms a salt bridge with SYS-1 Glu638, but this glutamate is replaced with Pro434 in HMP-2.
Although modeling suggests that HMR-1 regions II and III might be able to interact with SYS-1, the requirement for phosphorylation of region IV indicates that SYS-1 cannot bind to HMR-1 (pHMR-1). The HMP-2 residues that interact with the phosphate group of pSer1212, including Arg271, Arg274 and Arg306, are replaced with Asp444, Gly447 and Asp478 in SYS-1, which would prevent binding to the phosphorylated region IV (Fig. 7B) by acting similarly to the charge reversal mutations in HMR-1 described above. Moreover, whereas HMP-2 Asn218 and Lys220 present on helix 3 of arm repeat 4 form hydrogen bonds with the polypeptide backbone of pHMR-1, this helix is shorter in SYS-1 and there are no residues in the same position as Asn218 and Lys220.
DISCUSSION
In this study, detailed biochemical and structural analysis of the interaction between HMR-1 and HMP-2, as well as comparison to mammalian E-cadherin and β-catenin, revealed conserved cadherin-catenin regulatory mechanisms that could be examined in a genetically tractable organism. Crucial among the interaction motifs we identified was HMR-1 S1212, homologous to murine E-cadherin S686, which functions as part of a phosphorylation switch for HMR-1/HMP-2 binding. While this work was in progress, a phosphoproteomics analysis of C. elegans embryos revealed that HMR-1 S1212 is in fact phosphorylated in vivo (J. Audhya, personal communication). Our results thus provide direct in vivo demonstration of how phosphorylation regulates cell-cell adhesion during tissue development.
HMR-1 S1212 is the first in a series of four consensus CKI sites which, when phosphorylated, increase affinity for HMP-2 120 fold in vitro (Fig. 1, Table S2). This is consistent with previous in vitro studies of vertebrate homologs, in which phosphorylation of E-cadherin increased affinity for β-catenin by up to 800-fold (Choi et al., 2006). Vertebrate E-cadherin/β-catenin binding not only enables linkage between the cadherin-catenin complex and the actin cytoskeleton, but also it imposes structure on the E-cadherin cytoplasmic tail. This structure renders the conserved ubiquitination and proteasomal degradation motif L-S-S-L within the PEST sequence of E-cadherin's cytoplasmic tail inaccessible (Huber et al., 2001; Huber and Weis, 2001). The purified HMR-1 cytoplasmic tail runs about 3x larger than its predicted size on a gel filtration column, similar to the behavior of the unstructured E-cadherin cytoplasmic domain (Huber et al., 2001), consistent with its being an intrinsically unstructured protein. Thus it is likely that the region around HMR-1 S1212 is disordered unless it is phosphorylated and bound to HMP-2. Whether this interaction protects a true PEST sequence in this region remains undetermined, though it is notable that HMR-1 contains the amino acid sequence L-E-S-I in a position homologous to the L-S-S-L motif in E-cadherin.
Phosphorylation of HMR-1 S1212 appears to be critical for stabilizing the HMR-1/HMP-2 association. In C. elegans embryos, nonphosphorylatable HMR-1 S1212A protein is unable to functionally substitute for wildtype HMR-1. That HMR-1(S1212A)::GFP and HMR-1(T1215A, S1218A)::GFP both target to adherens junctions, but only the latter rescues hmr-1(zu389) embryonic lethality, suggests that mechanical uncoupling from HMP-2 due to the drastic loss in affinity is predominantly responsible for failure to rescue. Nevertheless, the inability of hmr-1(T1215A, S1218A)::gfp to fully rescue the hmr-1(zu389) mutation suggests that while pS1212 is essential for strong binding to HMP-2, the C-terminal phosphorylations primed by pS1212, in particular T1215, are required for full HMR-1 function, consistent with its modest, but significant, contribution to binding of phosphorylated HMR-1 to HMP-2 (Table S2). Although we cannot rule out a minor contribution due to proteasomal degradation of HMR-1, we did not detect appreciable quantitative reduction of HMR-1(S1212A)::GFP at junctions compared to wild-type HMR-1::GFP (Fig. S3).
The HMR-1/HMP-2 co-crystal also identified the conserved HMP-2 residues R271, R274, and R306 as electrostatic partners with HMR-1 pS1212. Mutation of HMP-2 at these sites localizes the protein cytoplasmically and renders it unable to rescue hmp-2(zu364) embryos, confirming the importance of these residues in vivo. Further confirmation of this interaction comes from sequencing of the hmp-2(zu364) allele, which revealed that the mutation R271C is responsible for the genetic null phenotype, and ablates the affinity enhancement provided by phosphorylation of the HMR-1 tail in vitro (Table S2). The importance of Arg271 to phosphoHMR-1 binding is highlighted by its replacement by Asp444 in SYS-1, Gln324 of BAR-1 and Gln403 of WRM-1, none of which bind HMR-1. Analysis of HMP-2 also helps to explain phylogenetic diversification of β-catenins in C. elegans. Comparison of the HMR-1/HMP-2 and POP-1/SYS-1 structures, as well as the mutational data, indicates that SYS-1 lacks residues required for the high-affinity interaction with HMR-1. It may be that the arginine cluster needed for interaction of HMP-2 with phosphorylated HMR-1 Ser1212 is incompatible with high-affinity binding to Wnt pathway partners.
HMR-1 lacks the C-terminal hydrophobic cap present in vertebrate cadherins, which contributes significantly to binding to β-catenin: even when not phosphorylated, E-cadherin binds to β-catenin with a Kd of 46 nM, and phosphorylation increases the affinity to 52 pM (Choi et al., 2006). It is interesting that phosphorylated HMR-1 binds to HMP-2 with about the same affinity as non-phosphorylated E-cadherin binding to β-catenin, and that phosphorylation also provides 2–3 orders of magnitude enhancement of affinity (Fig. 1A). We speculate that the differences in absolute affinity reflect differences in the mechanical load experienced by the adherens junctions in these two systems. Alternatively, it may be that C. elegans development requires a binary on/off switch for cell-cell adhesion that is provided by kinases and phosphatases acting on Ser1212.
HMP-2 Y599 is homologous to human and mouse β-catenin Y654, a phosphorylation site targeted by Src, FLT3, Abl, and PTP1B (Kajiguchi et al., 2012; Roura et al., 1999; Tamada et al., 2012; Xu et al., 2004). Previous in vitro work suggests that phosphorylation at β-catenin Y654 modestly decreases affinity for E-cadherin (Roura et al., 1999; van Veelen et al., 2011; Xu et al., 2004), suggesting a possible method of fine-tuning binding affinity. Our modeling indicates that phosphorylation of HMP-2 Y599 would not be accommodated in the region II interface (see above), but that HMP-2 Y599E could be accommodated into HMP-2/HMR-1 complex, albeit with a loss of some packing interactions. In Drosophila melanogaster, a putative phosphomimetic mutation at the homologous residue in Armadillo (Y667E) did not perturb binding to E-cadherin (Tamada et al., 2012); similarly, HMP-2 Y599E remains colocalized with HMR-1 in junctional excursions. These observations suggest that the Tyr→Glu mutation may not fully disrupt binding at the region II interface.
Interestingly, Drosophila Armadillo Y667E rescues β-catenin function during germband extension better than Y667F, in contrast to our work, in which the HMP-2 Y599E construct causes morphogenetic defects not seen with wild-type or Y599F rescue constructs. The different outcomes in the two systems may nevertheless reflect differing biological requirements for junction dynamics. In the elongating D. melanogaster germ band, adherens junctions must be quickly remodeled in order to form multicellular rosettes that resolve into an elongated tissue. This could be facilitated through phosphorylation of β-catenin at Y667. In contrast, in the elongating C. elegans embryo, the organism as a whole is radially compressed via tensile force applied to CFBs, which are anchored at epidermal adherens junctions. In this case, stabilizing β-catenin by dephosphorylation at Y599 could be advantageous for withstanding the force applied during embryonic elongation. Testing this idea awaits a means of unambiguously identifying individual posttranslational modifications of β-catenin in vivo at a subcellular level.
In summary, we have identified key cadherin/β-catenin interactions at atomic resolution and tested their functional significance in a living organism. Our data connect detailed atomic properties with specific developmental outcomes, providing structural insights into conserved binding interfaces through which cadherin-catenin binding affinity, and consequently intercellular adhesion, can be modulated according to the demands of a particular tissue or process. Additionally, the functionally specialized β-catenin paralogs of C. elegans have enabled us to distinguish which features are critical for the association of E-cadherin to β-catenin and to discern adhesion-specific consequences of post-translational modifications at these sites.
METHODS
Construct design and protein purification
Expression of full length HMP-2 in E. coli did not yield soluble protein, whereas removing the first 4 amino acids produced a soluble protein. This construct lost another 8 residues during purification, so the construct HMP-213end, was produced for subsequent studies. Shorter constructs, based on the N-terminal boundary of the proteolytically defined arm domain of mouse β-catenin (Huber 1997), start at residue 54. The C-terminal boundary of the arm domain was determined by the crystal structure of HMP-254end and two constructs, HMP-213arm and HMP-254arm, comprising residues 13–621 and 54–621, respectively, were designed. The cytoplasmic domain of HMR-1, consisting of residues 1108–1223 and the minimal HMP-2 binding region, containing residues 1144–1223, were designed and labeled as HMR-1cyto and HMR-1cyto80, respectively.
Each protein was overexpressed with a tobacco etch virus (TEV) protease-cleavable GST tag at its N-terminus. Proteins were purified by glutathione agarose affinity chromatography, and GST removed by overnight treatment with TEV protease (20:1 substrate:TEV w/w) at 4°C. Each protein was subsequently purified by anion exchange (Mono Q; 20 mM Tris pH 8.0, 0.5 mM EDTA, 2mM DTT, run with a 100–350 mM NaCl gradient), followed by size exclusion chromatography on Superdex 200 (H buffer: 20mM HEPES pH 8.0, 150mM NaCl, 1mM DTT).
To generate phosphorylated HMR-1, in vitro phosphorylation reaction with CKI was performed as described (Kwiatkowski et al., 2010). Briefly, phosphorylation reaction was carried out for 12 hours at 30°C and phosphorylated protein was separated by Mono Q chromatography. Mass spectrometric analysis confirmed 5 phosphorylation sites in phospho-HMR-1cyto and 4 sites in phospho-HMR-1cyto80.
The HMP-2/phosphoHMR-1 complex was purified by size exclusion chromatography after incubating the mixture of HMP-254arm and phospho HMR-1cyto80 in a 1:1.5 molar ratio for an hour at room temperature. Purified protein complex was concentrated to 20 mg ml−1 and used for crystallization.
Crystallization and data collection
Crystals of HMP-254end were grown by hanging drop vapor diffusion at 21°C by mixing protein solution with mother liquor of 1.3M sodium formate and 50mM HEPES (pH 7.5). The crystals were cryoprotected in perfluoropolyether oil (PFO). These crystals belong to space group C2, and there is one molecule in the asymmetric unit.
The complex of HMP-254arm and phospho-HMR-1cyto80 were crystallized as two different forms, needle shaped and thin plate shaped ones in a similar condition. Needle shaped crystals, space group P21 with 3 complexes in the asymmetric unit, were grown in 17% PEG3350, 0.1M Bis-Tris-Propane (pH 8.0) and 0.2M sodium sulfate. Crystals with plate-like morphology, space group P43 with one complex molecule in the asymmetric unit, were obtained with the same condition except that 0.2M sodium iodide was used instead of sodium sulfate. In both cases, the crystals were cryoprotected in the same mother liquor except that the PEG3350 concentration was raised to 20%, and 20% glycerol was present. Diffraction data were measured at 100K on beamline 11-1 at Stanford Synchrotron Radiation laboratory (SSRL), and processed with HKL2000 (Supplemental Table 1).
Structure determination & refinement
The structure of HMP-254end was solved by molecular replacement (MR) with Phaser, using the mouse β-catenin arm domain structure as a search model (PDB ID =1I7W). The structure was refined with PHENIX and Coot. The final refined model consists of HMP-2 residues 56–612.
The structure of the HMP-2/phosphoHMR-1cyto80 complex was solved by MR using the refined structure of HMP-254end as a search model. The three copies of the complex and a single copy of the complex in the asymmetric unit were refined with PHENIX. Initial maps calculated using phases from the model of MR solution showed extra density corresponding to HMR-1. Iterative cycles of manual rebuilding with Coot and refinement with Phenix were performed to produce the final model (Supplemental Table 1). The final model in P43 contains HMP-2 residues 77–613, and HMR-1 residues 1145–1152 and 1179–1212. In the P21 crystal, Copy A consists of HMP-2 residues 81–88 and 91–614, and HMR-1 1145–1152 and 1179–1213; copy B comprises HMP-2 78–613 and HMR-1 1145–1152, and 1179–1213; copy C comprises HMP-2 82–107, 120–522, and 531–607 and HMR-1 1182–1207 and 1211–1222.
Isothermal titration calorimetry
ITC experiments were carried out in a VP-ITC calorimeter (Microcal, GE Healthcare) at 25°C. The cell contained 8–12 µM of HMP-2 protein, and 7–10µl of 100–150µM HMR-1 (nonphospho- and phospho-) was injected 20–36 times, following two initial 2µl injections. Data were analyzed using Microcal Origin software package.
C. elegans constructs and strains used
pJN455[Phmr-1::hmr-1::gfp] (Achilleos et al., 2010) was mutagenized using circle PCR to generate pTDL51[Phmr-1::hmr-1(S1212A)::gfp] and pTDL128[Phmr-1::hmr-1(T1215A, S1218A)::gfp]. Forward S1212A PCR primer: 5′ -GCT GTC GTC ACG TTG GAG AGT ATC-3′. Reverse PCR primer: 5′ -AAT ATT GTC CCG CTC ATC G-3′ Forward T1215A, S1218A primer: 5′ -GCT ATC GAA AGT GCC CAA GG-3′. Reverse primer: 5′ -CTC CAA CGC GAC GAC AGA AAT ATT-3′. Underlined bases indicate mutation sites.
2kb of endogenous hmp-2 promoter was amplified from genomic DNA and cloned into pPD95.75 to generate pTDL33, into which hmp-2 cDNA yk1047c6 was cloned to generate pTDL34[Phmp-2::hmp-2::gfp]. R271 and Y599 mutations were generated using circle PCR with the pTDL34 template.
The hmr-1 null allele zu389 was maintained as an unbalanced heterozygote. Presence of the zu389 allele was determined by dead embryo counts and by restriction digest and gel electrophoresis of PCR amplicons from worm lysates. Hermaphrodites were allowed to lay embryos for 24h before being lysed as described previously (Williams et al., 1992). Forward primer flanking the zu389 locus: 5′ -TTT CAT GCT TTT TCC CCA AA-3′. Reverse flanking primer: 5′ -CCA CAG CCT ACA ACC CTT TT-3′. PCR produces a 966 nt amplicon which is cleaved into 650 nt and 316 nt fragments by DdeI when the zu389 mutation is present.
hmr-1(zu389)/+ I; jcEx198 was generated by injecting pTDL51 into N2 at a concentration of 2ng/µL and crossing into hmr-1(zu389) heterozygotes. hmr-1(zu389) I; jcEx208 and hmr-1(zu389)/+ I; jcEx247 were generated by injecting pJN455 and pTDL128, respectively, into hmr-1(zu389) heterozygotes.
hmp-2(zu364)/hIn1[unc-54(h1040)] I. To genotype the hmp-2(zu364) allele, Humpback zu364 homozygous embryos were isolated, lysed, and sequenced as described previously (Maiden et al., 2013). All hmp-2 transgenic lines were generated by injecting pTDL34 or one of its mutant derivatives at 20 ng/µL into hmp-2(zu364) heterozygotes.
Bristol N2 was used as the wildtype C. elegans strain. All worms were fed OP50 nematode growth media and maintained at 20°C (Brenner, 1974).
Phalloidin Staining
Embryos were collected by dissolving gravid adults in a solution of 250 mM KOH and 0.5% NaOCl for 5 minutes, then washed twice with distilled deionized water and mounted on poly-L-lysine-coated ring slides. Embryos were fixed in a solution of 4% paraformaldehyde, 10 mM EGTA pH 8, 25 mM HEPES pH 6.8, 48 mM PIPES pH 6.8, 2 mM MgCl2, and 0.2% Triton X-100 for 20 minutes at room temperature in a humid chamber. Fixed embryos were washed twice with PBS for 5 minutes, and then incubated with TexasRed-conjugated phalloidin for 2 hours at room temperature in a humid chamber, protected from light. Slides were then washed twice with PBS for 5 minutes. Antifade reagent was added to each sample before coverslips were applied and sealed with nail polish.
Microscopy and Image Analysis
Rabbit HMR-1 and HMP-2 antibodies were used at 1 µg/mL. Staining was performed as described previously (Maiden et al., 2013). Mouse monoclonal GFP antibody was used at 0.2 µg/mL (Molecular Probes).
Microscopy of live embryos was performed using a Nikon Eclipse E600 microscope with a Yokogawa CSU10 spinning disk scanhead. Images were obtained using a Hamamatsu ORCA-ER CCD camera and Micromanager software. 4D movies were processed and analyzed using ImageJ (Abramoff, 2004). Images are Z-projections of 3–4 focal planes spaced 1.0 µm apart. FRAP images were acquired with an Olympus Fluoview FV1000 microscope.
Fluorescence quantitation was performed in ImageJ. Junctions were isolated by thresholding Z projections of 2 focal planes to 99% and selected using the wand tool. Mean fluorescence per pixel for identical seam-dorsal and seam-ventral junctions were compared between embryos. To compare junctional HMP-2::GFP expression to cytoplasmic HMP-2(R271C)::GFP, 20 focal planes of each embryo were sum projected, and mean fluorescence per pixel was calculated for each embryo after subtraction of background signal.
Supplementary Material
Acknowledgements
We thank Dr. Jon Audhya for sharing his unpublished mass spectrometry data on HMR-1 and Bethany Lucas for aid with DNA microinjections. This work was supported by the Research Settlement Fund for the new faculty of SNU and Basic Science Research Program through the NRF funded by the Ministry of Education (NRF-2013R1A1A2061541) (HJC), NIH grants R21 HD072769 and R01 GM58038 (JH), and NIH grants R01 GM56169 and U01 GM09463 (WIW). Portions of this work were performed at the Stanford Synchrotron Radiation Lightsource, supported by the U.S. Department of Energy and the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barker N, Hurlstone A, Musisi H, Miles A, Bienz M, Clevers H. The chromatin remodelling factor Brg-1 interacts with b-catenin to promote target gene activation. EMBO J. 2001;20:4935–4943. doi: 10.1093/emboj/20.17.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bek S, Kemler R. Protein kinase II regulates the interaction of β-catenin with α-catenin and its protein stability. J Cell Sci. 2002;115:4743–4753. doi: 10.1242/jcs.00154. [DOI] [PubMed] [Google Scholar]
- Benjamin JM, Nelson WJ. Bench to bedside and back again: Molecular mechanisms of α-catenin function and roles in tumorigenesis. Semin Cancer Biol. 2008;18:53–64. doi: 10.1016/j.semcancer.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H-J, Huber AH, Weis WI. Thermodynamics of β-catenin–ligand interactions. The roles of the N- and C-terminal tails in modulating binding affinity. J Biol Chem. 2006;281:1027–1038. doi: 10.1074/jbc.M511338200. [DOI] [PubMed] [Google Scholar]
- Choi HJ, Gross JC, Pokutta S, Weis WI. Interactions of plakoglobin and β-catenin with desmosomal cadherins: basis of selective exclusion of α- and β-catenin from desmosomes. J Biol Chem. 2009;284:31776–31788. doi: 10.1074/jbc.M109.047928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Raich W, Agbunag C, Leung B, Hardin J, Priess JR. A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J Cell Biol. 1998;141:297–308. doi: 10.1083/jcb.141.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox EA, Hardin J. Sticky worms: adhesion complexes in C. elegans. J Cell Sci. 2004;117:1885–1897. doi: 10.1242/jcs.01176. [DOI] [PubMed] [Google Scholar]
- Cox-Paulson EA, Walck-Shannon E, Lynch AM, Yamashiro S, Zaidel-Bar R, Eno CC, Ono S, Hardin J. Tropomodulin protects α-catenin-dependent junctionalactin networks under stress during epithelial morphogenesis. Curr Biol. 2012;22:1500–1505. doi: 10.1016/j.cub.2012.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnemann S, Mitrik I, Hess M, Otto G, Wedlich D. Uncoupling of XB/Ucadherin- catenin complex formation from its function in cell-cell adhesion. J Biol Chem. 1997;272:11856–11862. doi: 10.1074/jbc.272.18.11856. [DOI] [PubMed] [Google Scholar]
- Hecht A, Litterst CM, Huber O, Kemler R. Functional characterization of multiple transactivating elements in b-catenin, some of which interact with the TATA-binding protein in vitro. J Biol Chem. 1999;274:18017–18025. doi: 10.1074/jbc.274.25.18017. [DOI] [PubMed] [Google Scholar]
- Hecht A, Vleminckx K, Stemmler MP, van Roy F, Kemler R. The p300/CBP acetyltransferases function as transcriptional coactivators of b-catenin in vertebrates. EMBO J. 2000;19:1839–1850. doi: 10.1093/emboj/19.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber AH, Stewart DB, Laurents DV, Nelson WJ, Weis WI. The cadherin cytoplasmic domain is unstructured in the absence of β-catenin: a possible mechanism for regulating cadherin turnover. J Biol Chem. 2001;276:12301–12309. doi: 10.1074/jbc.M010377200. [DOI] [PubMed] [Google Scholar]
- Huber AH, Weis WI. The structure of the β-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by β-catenin. Cell. 2001;105:391–402. doi: 10.1016/s0092-8674(01)00330-0. [DOI] [PubMed] [Google Scholar]
- Kajiguchi T, Katsumi A, Tanizaki R, Kiyoi H, Naoe T. Y654 of β-catenin is essential for FLT3/ITD-related tyrosine phosphorylation and nuclear localization of β-catenin. Eur J Haematol. 2012;88:314–320. doi: 10.1111/j.1600-0609.2011.01738.x. [DOI] [PubMed] [Google Scholar]
- Kidd AR. In Cellular and Molecular Biology. Madison, WI: University of Wisconsin; 2005. SYS-1, a β-catenin identified by functional critiera is required for asymetric cell divisions in the nemadoe Caenorhabditis elegans. [Google Scholar]
- Korswagen HC, Herman MA, Clevers HC. Distinct β-catenins mediate adhesion and signalling functions in C. elegans. Nature. 2000;406:527–532. doi: 10.1038/35020099. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski AV, Maiden SL, Pokutta S, Choi HJ, Benjamin JM, Lynch AM, Nelson WJ, Weis WI, Hardin J. In vitro and in vivo reconstitution of the cadherin-catenin-actin complex from Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2010;107:14591–14596. doi: 10.1073/pnas.1007349107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickert H, Bauer A, Kemler R, Stappert J. Casein Kinase II phosphorylation of E-cadherin Increases E-cadherin/β-catenin interaction and strengthens cell-cell adhesion. J Biol Chem. 2000;275:5090–5095. doi: 10.1074/jbc.275.7.5090. [DOI] [PubMed] [Google Scholar]
- Liu J, Phillips BT, Amaya MF, Kimble J, Xu W. The C. elegans SYS-1 protein is a bona fide β-catenin. Dev Cell. 2008;14:751–761. doi: 10.1016/j.devcel.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden SL, Harrison N, Keegan J, Cain B, Lynch AM, Pettitt J, Hardin J. Specific conserved C-terminal amino acids of Caenorhabditis elegans HMP-1/α- catenin modulate F-actin bindign independently of vinculin. J Biol Chem. 2013;288:5694–5706. doi: 10.1074/jbc.M112.438093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen AE, Maher MT, Mo R, Gottardi CJ. E-cadherin phosphorylation occurs during its biosynthesis to promote its cell surface stability and adhesion. Mol Biol Cell. 2014 doi: 10.1091/mbc.E14-01-0690. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishi M, Fujii R, Hatta M, Yoshida E, Araya N, Nagafuchi A, Ishihara S, Nakajima T, Fukamizu A. Regulation of Lef-mediated transcription and p53- dependent pathway by associating b-catenin with CPB/p300. J Biol Chem. 2000;275:35170–35175. doi: 10.1074/jbc.C000258200. [DOI] [PubMed] [Google Scholar]
- Piekny AJ, Johnson J-LF, Cham GD, Mains PE. The Caenorhabditis elegans nonmuscle myosin genes nmy-1 and nmy-2 funtion as redundant components of the let-502/Rho-binding kinase and mel-11/myosin phosphatase pathway during embryonic morphogeneisis. Development. 2003;130:5695–5704. doi: 10.1242/dev.00807. [DOI] [PubMed] [Google Scholar]
- Pokutta S, Weis WI. Structure and mechanism of cadherins and catenins in cell-cell contacts. Annu Rev Cell Dev Biol. 2007;23:237–261. doi: 10.1146/annurev.cellbio.22.010305.104241. [DOI] [PubMed] [Google Scholar]
- Roura S, Miravet S, Piedra J, García de Herreros A, Dunach M. Regulation of E-cadherin/catenin association by tyrosine phosphorylation. J Biol Chem. 1999;274:36734–36740. doi: 10.1074/jbc.274.51.36734. [DOI] [PubMed] [Google Scholar]
- Stamos JL, Weis WI. The β-catenin destruction complex. Cold Spring Harb Perspect Biol. 2013;5:a007898. doi: 10.1101/cshperspect.a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappert J, Kemler R. A short core region of E-cadherin is essential for catenin binding and is highly phosphorylated. Cell Adhes Commun. 1994;2:319–327. doi: 10.3109/15419069409014207. [DOI] [PubMed] [Google Scholar]
- Takemaru K, Moon RT. The transcriptional coactivator CBP interacts with b-catenin to activate gene expression. J Cell Biol. 2000;149:249–254. doi: 10.1083/jcb.149.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamada M, Farrell DL, Zallen JA. Abl regulates planar polarized junctional dynamics through β-catenin tyrosine phosphorylation. Dev Cell. 2012;22:309–319. doi: 10.1016/j.devcel.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutter AV, Fryer CJ, Jones KA. Chromatin-specific regulation of LEF-1-β- catenin transcription activation and inhibition in vitro. Genes Devel. 2001;15:3342–3354. doi: 10.1101/gad.946501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roy F. Beyond E-cadherin: roles of other cadherin superfamily members in cancer. Nat Rev Cancer. 2014;14:121–134. doi: 10.1038/nrc3647. [DOI] [PubMed] [Google Scholar]
- van Veelen W, Le NH, Helvensteijn W, Blonden L, Theeuwes M, Bakker ERM, Franken PF, van Gurp L, Meijlink F, van der Valk MA, et al. β-catenin tyrosine 654 phosphorylation increases Wnt signalling and intestinal tumorigenesis. Gut. 2011;60:1204–1212. doi: 10.1136/gut.2010.233460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Takemaru K, Liu J, Berndt JD, Zheng JJ, Moon RT, Xu W. Crystal structure of a full-length β-catenin. Structure. 2008;16:478–487. doi: 10.1016/j.str.2007.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Craig AWB, Greer P, Miller M, Anastasiadis PZ, Lilien J, Balsamo J. Continuous association of cadherin with β-catenin requires the non-receptor tyrosine-kinase Fer. J Cell Sci. 2004;117:3207–3219. doi: 10.1242/jcs.01174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.