Abstract
Folate is a water-soluble vitamin that is critical for nucleotide synthesis and can modulate methylation of DNA by altering one-carbon metabolism. Previous studies have shown that folate status during pregnancy is associated with various congenital defects including the risk of aberrant neural tube closure. Maternal exposure to a methyl supplemented diet also can alter DNA methylation and gene expression, which may influence the phenotype of offspring. We investigated if higher gestational folic acid (FA) in the diet dysregulates the expression of genes in the cerebellum of offspring in C57BL/6 J mice. One week before gestation and throughout the pregnancy, groups of dams were supplemented with FA either at 2 mg/kg or 20 mg/kg of diet. Microarray analysis was used to investigate the genome wide gene expression profile in the cerebellum from day old pups. Our results revealed that exposure to the higher dose FA diet during gestation dysregulated expression of several genes in the cerebellum of both male and female pups. Several transcription factors, imprinted genes, neuro-developmental genes and genes associated with autism spectrum disorder exhibited altered expression levels. These findings suggest that higher gestational FA potentially dysregulates gene expression in the offspring brain and such changes may adversely alter fetal programming and overall brain development.
Keywords: folic acid, DNA methylation, microarray, autism, gene expression, epigenetics, genomic imprinting, prenatal nutrition, brain
1. Introduction
The changes in the epigenetic patterning occurring in utero may result in alterations in the gene expression and susceptibility to various diseases, which may have a lifelong impact [1,2]. Such changes in DNA methylation and gene expression can be induced by maternal lifestyle and environmental factors including nutrition during the pregnancy. The critical role of folic acid (FA) in the complex of cellular activities was suggested as early as 1944 by the observation that folate deficiency results in an increased incidence of prematurity [3]. Later in 1966 the seminal work by Smithells and Hibbard suggested that folate status during pregnancy is an important contributing factor in the origin of neural tube defects (NTDs) [4,5]. Considering the benefits of FA in reducing the incidence of NTDs, various countries have recommended mandatory fortification of grain and flour since the late 1990’s, and women of childbearing age are recommended to take 400–800 μg/day FA [6,7]. For women with a history of complicated pregnancy, such as a prior NTD-affected baby, the recommended daily FA intake is ten times higher, 4000 μg/day [8]. However, in addition to the recommended FA intake through fortified foods and prescription multivitamin tablets, intake of FA through other sources, such as over the counter vitamins and energy drinks can lead to even higher levels FA exposure. Concern has been raised regarding adverse potential health outcomes of these high levels of FA [5,9]. Recent studies in animal models have shown that exposure to higher FA levels during gestation can induce alterations in methylation and gene expression in the cerebral hemispheres in offspring brain. Some of the genes altered are associated with neural development [10]. Since a dysfunctional folate-methionine pathway has been implicated in many individuals with autism [11], dysregulation in gene expression induced by higher gestational FA may have a profound impact on neural functioning in the offspring.
In addition to cerebral circuits, defects in cerebellar circuits have also been proposed as being involved with autism [12], and such changes in cerebellar circuitry may impair cognition and emotion resulting in autism spectrum disorders (ASDs) [13]. Furthermore, a recent study reported that individuals with autism have reduced number and density of Purkinje cells in the cerebellum compared to controls, suggesting a pattern of developmental alteration in the cerebellum may contribute to the etiology of autism [14]. In order to reveal the clinical significance of the higher FA intake in developing brain, in this pilot study, we conducted a genome wide microarray analysis to evaluate the effect of increased gestational FA on gene expression in the cerebellum of offspring.
2. Results
2.1. Higher Maternal FA during Gestation Alters Expression of Several Genes in the Cerebellum of Offspring
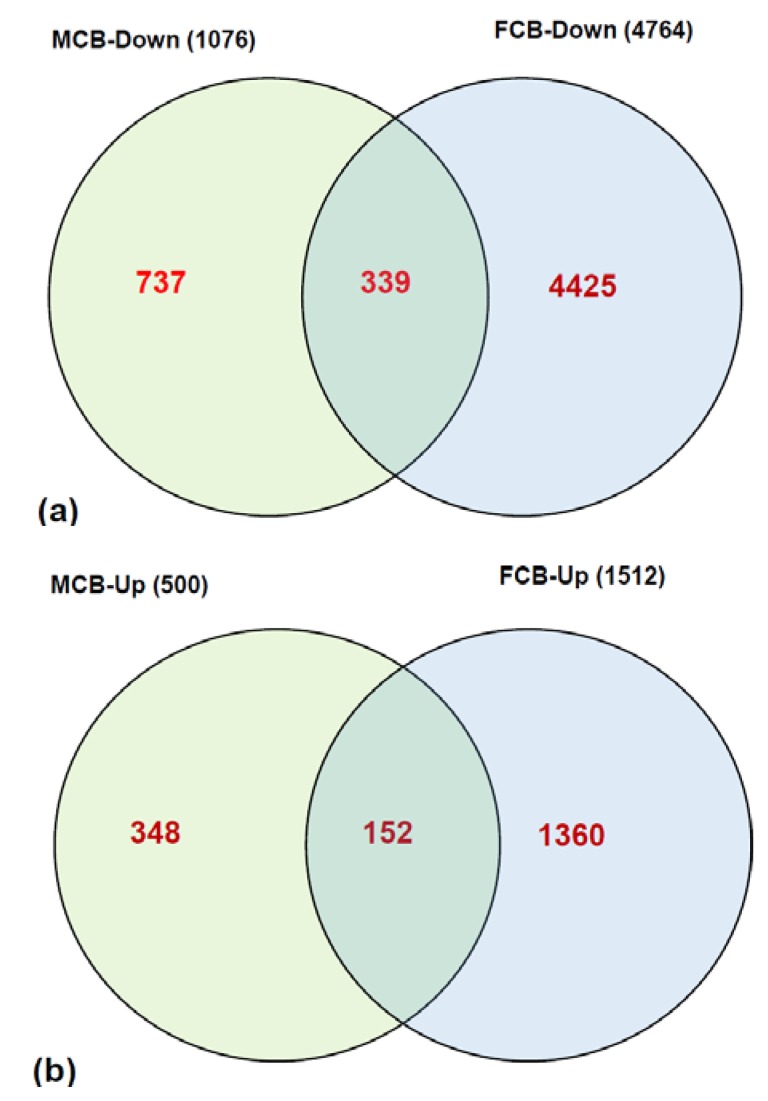
In the microarray analysis of transcripts from 2 mg FA group vs. 20 mg FA group of postnatal day one (P1) pups, the expression pattern of a significant number of genes were found to be altered by ≥2.5 fold at a significance of p > 0.05. Overall, in male pups the expression patterns of 1076 transcripts were down-regulated (Supplementary Table S1) and 499 transcripts were up-regulated (Supplementary Table S2) in the cerebellum from the 20 mg maternal FA group. In female pups, the expressions of 4764 transcripts (Supplementary Table S3) were down-regulated and 1511 transcripts (Supplementary Table S4) were up-regulated. The expression of 339 transcripts that were down-regulated and 152 transcripts that were up-regulated by ≥2.5 fold in pups’ cerebellum were common for both the genders in the 20 mg maternal FA group (Figure 1). These results of microarray data illustrate that higher maternal FA during gestation can induce significant alterations in the expression of transcripts in offspring brain cerebellum. The data further suggests that in addition to the FA dose, there is an interaction between maternal FA dose and its role in modulating the gene expression levels depending on the sex of the offspring.
Figure 1.
Overview of differential gene expression in the cerebellum of P1 pups. The distribution of (a) down-regulated genes, as well as (b) up-regulated genes, between male (MCB) and female (FCB) pups from higher gestational FA.
2.2. Higher Maternal FA Alters Expression of Several Transcription Factors and Imprinted Genes in the Cerebellum of Offspring
Analyses of our data have revealed that a higher intake of maternal FA altered the expression levels of several transcription factors and imprinted genes in the offspring cerebellum (Supplementary Tables S1–S4). Transcriptional regulators such as forkhead box or homeobox genes Foxa1, Foxa2, Hoxc11, Lhx6 in male pups and Foxc2, Foxf1a, Hoxa3, Hoxa4, Hoxa5 in female pups were altered by ≥2.5 fold. Though the direction of expression changes varied between male and female pups, some transcription factors (Esr1, Sp3, Sqstm1 and Zfp93) altered were common in both the genders from the higher maternal FA group.
Further analysis of our data revealed dysregulation in the expression of several imprinted genes in both male and female pups from the higher maternal FA group. In male pups, the expression of imprinted genes Kcnq1, Tsix were down-regulated, while the expression of Snrpn was up-regulated. In contrast, in female pups while the expression of imprinted genes Mirg and Rhox5 were up-regulated, several genes including H19, H12, Peg12, Peg3 and Xist were down regulated by ≥2.5 fold. Since transcription factors and imprinted genes play a key role in developmental outcomes, such dysregulation in gene expression as a result of higher gestational FA may have adverse outcomes in overall development of the offspring cerebellum.
2.3. Higher Maternal FA Alters the Expression Profile of Several Neural Genes in the Cerebellum of Offspring
In order to study the effect of higher gestational FA on gene expression at a functional level, we analyzed the differential expression of neural genes and grouped them based on their functional role in specific neural pathways (Table 1 and Table 2). We found higher amounts of gestational FA altered expression of several neural genes in both male and female pups. The extent of variation was between 6-fold down-regulation in males and 13-fold down-regulation in females. In male pups from the higher maternal FA group, the expression of genes relating to circadian rhythm (Cry1), dopaminergic and serotonergic pathways (Slc18a2, Pde4c, Slc6a4, Pde4b), gap junctions (Hras1, Tubb3, Tubb2b), neurogenesis (Hes1, Rac1), including genes related to neuronal-ion channel, neurotrophin-receptor and synaptic plasticity exhibited dysregulation in expression. In female pups from the higher gestational FA group, the expression of several genes relating to circadian rhythm (Prkar2b, Smad4, Bhlhe41, Mtnr1b, Ncoa3), dopaminergic and serotonergic pathways (Dusp1, Pde10a, Fos), GABA-glutamate pathway (Avp, Cacna1a, Gabrb1, Grin1, Slc1a6), gap junctions (Kras, Map2k1, Gja5, Cav1, Gja3), neurogenesis (Vegfa, Ntn1, Dvl3, Robo1, Pax5, Stat3), including neurotransmitter, neuronal-ion channel, neurotrophin-receptor and synaptic plasticity were dysregulated significantly. Further analysis of the data revealed several neural genes (Cdk1, Csnk1a1, Frs3, Nsf and Tuba3a) were down-regulated, and genes related to the dopaminergic and serotonergic pathway (Adcy3) were up-regulated in both male and female pups from the higher maternal FA group. Thus our data suggests that higher maternal FA significantly alters expression of several neural genes in the cerebellum of offspring, and some of these changes varied in a gender specific manner.
Table 1.
Neural genes altered ≥2.5 fold in the cerebellum of male pups from mothers having FA supplementation during gestation at 20 mg/kg in comparison to mothers at 2 mg/kg diet.
| Accession | Symbol | Fold Change | Direction of Change | Pathway |
|---|---|---|---|---|
| ref|NM_146087 | Csnk1a1 | −3.90 | ↓ | Circadian |
| ref|NM_007771 | Cry1 | −2.70 | ↓ | Circadian |
| ref|NM_172523 | Slc18a2 | −3.81 | ↓ | Dopa-Serotonin |
| ref|NM_201607 | Pde4c | −3.11 | ↓ | Dopa-Serotonin |
| ref|NM_010484 | Slc6a4 | −3.06 | ↓ | Dopa-Serotonin |
| ref|NM_008740 | Nsf | −3.40 | ↓ | GABA-glutamate |
| ref|NM_001130444 | Hras1 | −4.63 | ↓ | Gap-Junctions |
| ref|NM_009446 | Tuba3a | −3.17 | ↓ | Gap-Junctions |
| ref|NM_023279 | Tubb3 | −2.76 | ↓ | Gap-Junctions |
| ref|NM_023716 | Tubb2b | −2.71 | ↓ | Gap-Junctions |
| ref|NM_007659 | Cdk1 | −2.53 | ↓ | Gap-Junctions |
| ref|NM_008235 | Hes1 | −3.17 | ↓ | Neurogenesis |
| ref|NM_001039104 | Trpm1 | −3.36 | ↓ | Neuronal-Ion channel |
| ens|ENSMUST00000105918 | Kcnq1 | −3.29 | ↓ | Neuronal-Ion channel |
| ref|NM_144939 | Frs3 | −6.24 | ↓ | Neurotrophin-receptor |
| ref|NM_053075 | Rheb | −3.13 | ↓ | Synaptic Plasticity |
| ref|NM_138305 | Adcy3 | 2.71 | ↑ | Dopa-Serotonin |
| ref|NM_019840 | Pde4b | 2.57 | ↑ | Dopa-Serotonin |
| ref|NM_009007 | Rac1 | 2.73 | ↑ | Neurogenesis |
| ref|NM_011913 | Best1 | 2.62 | ↑ | Neuronal-Ion channel |
| ref|NM_009314 | Tacr2 | 2.73 | ↑ | Neurotransmitter |
| ref|NM_009750 | Ngfrap1 | 2.92 | ↑ | Neurotrophin-receptor |
↓ = Down regulated; ↑ = Up regulated.
Table 2.
Neural genes altered ≥2.5 fold in the cerebellum of female pups from mothers having FA supplementation during gestation at 20 mg/kg in comparison to mothers at 2 mg/kg diet.
| Accession | Symbol | Fold Change | Direction of Change | Pathway |
|---|---|---|---|---|
| ref|NM_146087 | Csnk1a1 | −7.80 | ↓ | Circadian |
| ref|NM_011158 | Prkar2b | −6.84 | ↓ | Circadian |
| ref|NM_013672 | Sp1 | −5.80 | ↓ | Circadian |
| ref|NM_008540 | Smad4 | −5.02 | ↓ | Circadian |
| ref|NM_172563 | Hlf | −4.27 | ↓ | Circadian |
| ref|NM_009602 | Chrnb2 | −4.10 | ↓ | Circadian |
| ref|NM_001174053 | Camk2b | −3.37 | ↓ | Circadian |
| ref|NM_015822 | Fbxl3 | −3.27 | ↓ | Circadian |
| ref|NM_009516 | Wee1 | −3.05 | ↓ | Circadian |
| ref|NM_010098 | Opn3 | −3.04 | ↓ | Circadian |
| ref|NM_008904 | Ppargc1a | −2.90 | ↓ | Circadian |
| ref|NM_011144 | Ppara | −2.56 | ↓ | Circadian |
| ref|NM_013642 | Dusp1 | −4.63 | ↓ | Dopa-Serotonin |
| ref|NM_011866 | Pde10a | −4.55 | ↓ | Dopa-Serotonin |
| ref|NM_010234 | Fos | −4.47 | ↓ | Dopa-Serotonin |
| ref|NM_001131020 | Gfap | −4.29 | ↓ | Dopa-Serotonin |
| ref|NM_001012765 | Adcy5 | −3.83 | ↓ | Dopa-Serotonin |
| ref|NM_172778 | Maob | −3.67 | ↓ | Dopa-Serotonin |
| ref|NM_001111015 | Syn2 | −3.11 | ↓ | Dopa-Serotonin |
| gb|AK032648 | Pde4d | −2.63 | ↓ | Dopa-Serotonin |
| ref|NM_019827 | Gsk3b | −2.55 | ↓ | Dopa-Serotonin |
| ref|NM_009732 | Avp | −13.07 | ↓ | GABA-glutamate |
| ref|NM_007578 | Cacna1a | −8.80 | ↓ | GABA-glutamate |
| ref|NM_008069 | Gabrb1 | −7.02 | ↓ | GABA-glutamate |
| ref|NM_146072 | Grik1 | −5.57 | ↓ | GABA-glutamate |
| ref|NM_001037724 | Adcy7 | −5.50 | ↓ | GABA-glutamate |
| ref|NM_008174 | Grm8 | −5.34 | ↓ | GABA-glutamate |
| ref|NM_008171 | Grin2b | −5.07 | ↓ | GABA-glutamate |
| ref|NM_001039195 | Gria2 | −5.07 | ↓ | GABA-glutamate |
| ref|NM_001146311 | Cln3 | −5.01 | ↓ | GABA-glutamate |
| ref|NM_001013385 | Grm4 | −4.57 | ↓ | GABA-glutamate |
| ref|NM_176942 | Gabra5 | −4.20 | ↓ | GABA-glutamate |
| ref|NM_010251 | Gabra4 | −4.12 | ↓ | GABA-glutamate |
| ref|NM_011393 | Slc1a2 | −3.99 | ↓ | GABA-glutamate |
| ref|NM_001113383 | Gls | −3.95 | ↓ | GABA-glutamate |
| ref|NM_182959 | Slc17a8 | −3.91 | ↓ | GABA-glutamate |
| ref|NM_016886 | Gria3 | −3.89 | ↓ | GABA-glutamate |
| ref|NM_008075 | Gabrr1 | −3.83 | ↓ | GABA-glutamate |
| ref|NM_080853 | Slc17a6 | −3.49 | ↓ | GABA-glutamate |
| ref|NM_175328 | Slc6a15 | −3.15 | ↓ | GABA-glutamate |
| ref|NM_001143834 | Grm5 | −3.10 | ↓ | GABA-glutamate |
| ref|NM_008740 | Nsf | −3.09 | ↓ | GABA-glutamate |
| ref|NM_019691 | Gria4 | −3.03 | ↓ | GABA-glutamate |
| ref|NM_001042451 | Snca | −2.80 | ↓ | GABA-glutamate |
| ref|NM_147176 | Homer1 | −2.69 | ↓ | GABA-glutamate |
| ref|NM_021284 | Kras | −6.85 | ↓ | Gap-Junctions |
| ref|NM_009446 | Tuba3a | −6.70 | ↓ | Gap-Junctions |
| ref|NM_008927 | Map2k1 | −4.92 | ↓ | Gap-Junctions |
| ref|NM_010937 | Nras | −4.37 | ↓ | Gap-Junctions |
| ref|NM_011101 | Prkca | −3.98 | ↓ | Gap-Junctions |
| ref|NM_007659 | Cdk1 | −3.84 | ↓ | Gap-Junctions |
| ref|NM_001080971 | Tubb1 | −3.67 | ↓ | Gap-Junctions |
| ref|NM_011100 | Prkacb | −3.64 | ↓ | Gap-Junctions |
| ref|NM_009447 | Tuba4a | −3.62 | ↓ | Gap-Junctions |
| ref|NM_010288 | Gja1 | −3.52 | ↓ | Gap-Junctions |
| ref|NM_175452 | Gjc2 | −2.91 | ↓ | Gap-Junctions |
| ref|NM_010930 | Nov | −2.91 | ↓ | Gap-Junctions |
| ref|NM_007912 | Egfr | −2.71 | ↓ | Gap-Junctions |
| ref|NM_009231 | Sos1 | −2.70 | ↓ | Gap-Junctions |
| ref|NM_028751 | Tjap1 | −2.53 | ↓ | Gap-Junctions |
| ref|NM_001025250 | Vegfa | −6.08 | ↓ | Neurogenesis |
| ref|NM_008744 | Ntn1 | −5.16 | ↓ | Neurogenesis |
| ref|NM_007889 | Dvl3 | −4.83 | ↓ | Neurogenesis |
| ref|NM_010894 | Neurod1 | −4.13 | ↓ | Neurogenesis |
| ref|NM_008781 | Pax3 | −4.07 | ↓ | Neurogenesis |
| ref|NM_177821 | Ep300 | −3.87 | ↓ | Neurogenesis |
| ref|NM_011443 | Sox2 | −3.54 | ↓ | Neurogenesis |
| ref|NM_009599 | Ache | −3.50 | ↓ | Neurogenesis |
| ref|NM_033620 | Pard3 | −3.34 | ↓ | Neurogenesis |
| ref|NM_010928 | Notch2 | −3.25 | ↓ | Neurogenesis |
| ref|NM_008737 | Nrp1 | −3.07 | ↓ | Neurogenesis |
| ref|NM_007553 | Bmp2 | −2.86 | ↓ | Neurogenesis |
| ref|NM_022312 | Tnr | −2.85 | ↓ | Neurogenesis |
| ref|NM_010883 | Ndp | −2.62 | ↓ | Neurogenesis |
| ref|NM_001039934 | Mtap2 | −2.60 | ↓ | Neurogenesis |
| ref|NM_007865 | Dll1 | −2.59 | ↓ | Neurogenesis |
| ref|NM_008973 | Ptn | −2.54 | ↓ | Neurogenesis |
| ref|NM_008421 | Kcnc1 | −10.41 | ↓ | Neuronal-Ion channel |
| ref|NM_010597 | Kcnab1 | −5.07 | ↓ | Neuronal-Ion channel |
| ref|NM_009900 | Clcn2 | −3.81 | ↓ | Neuronal-Ion channel |
| ref|NM_001025581 | Kcnc2 | −3.45 | ↓ | Neuronal-Ion channel |
| ref|NM_001083616 | Cacna1d | −3.26 | ↓ | Neuronal-Ion channel |
| ref|NM_008226 | Hcn2 | −3.23 | ↓ | Neuronal-Ion channel |
| ref|NM_011930 | Clcn7 | −3.04 | ↓ | Neuronal-Ion channel |
| ref|NM_010408 | Hcn1 | −2.85 | ↓ | Neuronal-Ion channel |
| ref|NM_145983 | Kcna5 | −2.67 | ↓ | Neuronal-Ion channel |
| ref|NM_001099298 | Scn2a1 | −2.60 | ↓ | Neuronal-Ion channel |
| ref|NM_001044308 | Cacna1i | −2.59 | ↓ | Neuronal-Ion channel |
| ref|NM_018732 | Scn3a | −2.51 | ↓ | Neuronal-Ion channel |
| ref|NM_010610 | Kcnma1 | −2.50 | ↓ | Neuronal-Ion channel |
| ref|NM_153087 | Hrh4 | −4.01 | ↓ | Neurotransmitter |
| ref|NM_001081147 | Oxtr | −3.68 | ↓ | Neurotransmitter |
| ref|NM_008285 | Hrh1 | −2.90 | ↓ | Neurotransmitter |
| ref|NM_021382 | Tacr3 | −2.71 | ↓ | Neurotransmitter |
| ref|NM_009313 | Tacr1 | −2.70 | ↓ | Neurotransmitter |
| ref|NM_008747 | Ntsr2 | −2.61 | ↓ | Neurotransmitter |
| ref|NM_009219 | Sstr4 | −2.61 | ↓ | Neurotransmitter |
| ref|NM_008177 | Grpr | −2.53 | ↓ | Neurotransmitter |
| ref|NM_001025074 | Ntrk2 | −7.28 | ↓ | Neurotrophin-receptor |
| ref|NM_009365 | Tgfb1i1 | −6.21 | ↓ | Neurotrophin-receptor |
| ref|NM_144939 | Frs3 | −5.29 | ↓ | Neurotrophin-receptor |
| ref|NM_011640 | Trp53 | −4.40 | ↓ | Neurotrophin-receptor |
| ref|NM_010560 | Il6st | −4.00 | ↓ | Neurotrophin-receptor |
| ref|NM_019791 | Maged1 | −3.69 | ↓ | Neurotrophin-receptor |
| ens|ENSMUST00000052164 | Ppyr1 | −3.47 | ↓ | Neurotrophin-receptor |
| ref|NM_009911 | Cxcr4 | −3.39 | ↓ | Neurotrophin-receptor |
| ref|NM_022024 | Gmfg | −3.10 | ↓ | Neurotrophin-receptor |
| ref|NM_010849 | Myc | −3.06 | ↓ | Neurotrophin-receptor |
| ref|NM_139149 | Fus | −2.84 | ↓ | Neurotrophin-receptor |
| ref|NM_177410 | Bcl2 | −2.64 | ↓ | Neurotrophin-receptor |
| ref|NM_001025074 | Ntrk2 | −7.28 | ↓ | Synaptic Plasticity |
| ref|NM_028736 | Grip1 | −6.77 | ↓ | Synaptic Plasticity |
| ref|NM_020493 | Srf | −3.25 | ↓ | Synaptic Plasticity |
| ens|ENSMUST00000111939 | Nos1 | −2.97 | ↓ | Synaptic Plasticity |
| ref|NM_013498 | Crem | −2.88 | ↓ | Synaptic Plasticity |
| ref|NM_009952 | Creb1 | −2.83 | ↓ | Synaptic Plasticity |
| ref|NM_024469 | Bhlhe41 | 5.46 | ↑ | Circadian |
| ref|NM_145712 | Mtnr1b | 4.18 | ↑ | Circadian |
| ref|NM_008679 | Ncoa3 | 3.70 | ↑ | Circadian |
| ref|NM_017376 | Tef | 3.48 | ↑ | Circadian |
| ref|NM_011281 | Rorc | 3.19 | ↑ | Circadian |
| ref|NM_138305 | Adcy3 | 4.41 | ↑ | Dopa-Serotonin |
| ref|NM_008169 | Grin1 | 5.56 | ↑ | GABA-glutamate |
| ref|NM_009200 | Slc1a6 | 2.80 | ↑ | GABA-glutamate |
| ref|NM_008121 | Gja5 | 4.27 | ↑ | Gap-Junctions |
| ref|NM_007616 | Cav1 | 3.57 | ↑ | Gap-Junctions |
| ref|NM_016975 | Gja3 | 2.98 | ↑ | Gap-Junctions |
| ref|NM_011840 | Map2k5 | 2.72 | ↑ | Gap-Junctions |
| ref|NM_008361 | Il1b | 2.67 | ↑ | GABA-glutamate |
| ens|ENSMUST00000114274 | Robo1 | 4.31 | ↑ | Neurogenesis |
| ref|NM_008782 | Pax5 | 3.39 | ↑ | Neurogenesis |
| ref|NM_011486 | Stat3 | 2.92 | ↑ | Neurogenesis |
| ref|NM_001077403 | Nrp2 | 2.52 | ↑ | Neurogenesis |
| ref|NM_183000 | Accn3 | 4.44 | ↑ | Neuronal-Ion channel |
| ref|NM_007699 | Chrm4 | 3.82 | ↑ | Neurotransmitter |
| ref|NM_013462 | Adrb3 | 2.61 | ↑ | Neurotransmitter |
| ref|NM_008006 | Fgf2 | 2.65 | ↑ | Neurotrophin-receptor |
| ref|NM_008416 | Junb | 2.86 | ↑ | Synaptic Plasticity |
| ref|NM_007664 | Cdh2 | 2.82 | ↑ | Synaptic Plasticity |
↓ = Down regulated; ↑ = Up regulated.
3. Discussion
In this study, microarray analysis of gene expression was utilized as a tool to investigate the effect of gestational FA on the transcriptome of cerebellum from P1 pups. Our data has shown that higher maternal FA during gestation dysregulated the expression of several genes that may play a causal role in developmental anomalies. In particular, the expression of several genes that code for genomic imprinting, transcriptional regulation and neural plasticity were significantly altered as a result of higher amounts of maternal FA. Genomic imprinting is a specialized epigenetic phenomenon resulting in monoallelic parent-specific expression of genes due to differential DNA methylation and histone modifications of specific gene regions [15]. Perturbations in dosage compensation can cause developmental issues including changes in growth and behavior [15]. Since the dynamics of genomic imprinting take place during gametogenesis and early embryogenesis; FA, by virtue of being a methyl group modulator during gestation, may play a critical role in affecting DNA methylation in the offspring epigenome. Our results have shown that higher gestational FA dysregulated expression of several imprinted genes in the cerebellum of P1 offspring. For example, in male pups the expression of the gene Kcnq1, a voltage-gated potassium channel, which has been associated with Jervell and Lange-Nielsen syndrome and hereditary long QT syndrome 1 [16,17], and the gene Tsix that expresses the non-coding antisense transcript required for imprinted X inactivation [18], were down-regulated. In contrast, the expressions of the gene Snrpn that plays a role in Angelman syndrome or Prader-Willi syndrome [19] was up-regulated. Similarly, in female pups, the miRNA containing gene (Mirg), and reproductive homeobox-5 gene (Rhox5), were up-regulated, while the expression of several imprinted genes, including H13, H19, Igf2 and Xist were down-regulated. Since, a methylation defect at the Igf2/H19 locus can result in imprinting disorders, and has been associated with Beckwith-Wiedemann syndrome [20,21], such gene dysregulation as a result of higher maternal FA may modulate epigenetic abnormalities in the pathogenesis of growth and development.
Next, our analysis further revealed dysregulation in the expression of several transcription factors in the cerebellum of P1 pups from higher maternal FA compared to pups from dams having FA at 2 mg/kg diet. The dysregulation was observed in many genes including the evolutionarily conserved class of homeobox genes in both male and female pups. The expression of several transcription factors such as Esr1, Sp3, Zfp93, Sqstm1 were dysregulated in pups of both genders from dams having higher gestational FA, though the direction of changes were not always same. Since transcription factors orchestrate gene expression of complex cellular machineries, such gene dysregulation may modulate the development of many organ systems, including the brain.
Higher maternal FA altered expression of several genes involved in dopamine-serotonin, GABA-glutamate, gap-junctions, neuronal-ion channel, neurogenesis, synaptic plasticity and the circadian pathway (Table 1 and Table 2). Higher gestational FA also altered expression of several genes that were earlier shown to be associated with ASDs as well as cognitive development. For example, in male pups from higher gestational FA, the serotonin transporter gene Slc6a4 that encodes an integral membrane protein, which plays a crucial role in serotonergic neurotransmission [22], was down-regulated by three fold; Fezf2, a gene belonging to zinc finger family [23], and Mbd3 a gene belonging to a family of nuclear proteins having methyl-CpG binding domain [24], were significantly up-regulated more than three fold. Of note, it has been shown that the alterations in the expression of Slc6a4 can be mediated due to epigenetic modulation in the promoter associated CpG island by environmental factors [25]. Such changes in Slc6a4 expression have been associated with alterations in brain structure and various neuropsychiatric illnesses including depression [26,27,28]. In female pups, the expression of several genes associated with ASDs (Table 1 and Table 2, Supplementary Tables S3 and S4) e.g., Fmr1, Grid1, Nrxn1, Grip1 were down-regulated, and Nrp2, a gene belonging to the neuropilin family of receptor proteins was up-regulated from dams having higher gestational FA. Fmr1 has long been associated with developmental cognitive impairment such as fragile X syndrome (FXS) and, found to be accompanied by epigenetic modification along with genetic anomalies [29,30,31]. Earlier, by using lymphoblastoid cells, we have shown that FA supplementation down-regulates the expression of the FMR1 gene, both at mRNA and protein levels [32]. Moreover, a recent study has suggested strong evidence for multiple medical, cognitive and psychiatric difficulties among women with the FMR1 premutation [33]. Similarly, when the ionotropic glutamate receptor Grid1 is deleted, it results in aberrant emotional and social behavior in mice [34]. Here Grid1 was found to be down-regulated in female pups from dams with higher FA. In contrast, Nrp2, a neuropilin family gene that controls neuronal migration and axon guidance, and has been associated with autism, wasup-regulated in female pups from dams having higher gestational FA [35]. Previous studies have shown that pups exposed to higher gestational and post-weaning FA exhibited increased ultrasonic vocalizations, greater anxiety-like behavior and hyperactivity [36]. Indeed, higher gestational FA during gestation altered methylation in the cerebral hemispheres of the offspring in both intergenic and intragenic regions, and thus modulated the expression of several genes [10]. In the current study, we found higher FA dysregulates the expression of several genes including those associated with neural development in the cerebellum of the offspring. Mechanistically, such dysregulations may have been induced by the precise changes in the DNA methylation in utero, modulated by higher gestational FA. The dysregulation of gene expression may be due to the direct impact of site or gene specific changes in the promoter or gene body region of specific genes or it may result from distant effects from changes in signaling pathways.
Metabolically, FA (pteroylmonoglutamate) consumed through supplementation or fortified foods is reduced by the enzyme dihydrofolate reductase primarily to dihydrofolate, and chemically reduced to tetrahydrofolate in the liver, where it collects a formyl group which is reduced to methyl to form 5-methyl-tetrahydrofolate [5]. However with the supplementation of high doses of FA this process is swamped by saturation of dihydrofolate reductase activity, and these may result in a considerable increase of unmetabolized FA in plasma [37]. Such evidence was further confirmed by the presence of unmetabolized FA in cord blood [38]. Though the exact mechanism by which this excess maternal FA is modulating the fetal brain development is not known, we speculate it may be a combined effect of pteroylmonoglutamate and methyl-tetrahydrofolate. One possibility is, with higher supplementation of maternal FA, a proportion of high dose FA that enters tissue becomes a substrate for polyglutamate synthase but is inert and thus excess FA in blood may compete with methyl-tetrahydrofolate to bind with the folate receptor [39] in the choroid plexus and inhibit methyl-tetrahydrofolate transport in the developing brain and thus interfere with regulatory functions. Alternatively an increased methyl-folate via maternal metabolism may modulate the methylation profile of imprinted genes, which have key roles in maintaining brain homeostasis and development.
Since, early life experience may alter the brain structure and function including behavioral and neural development, higher gestational FA may have untoward side effects in overall development of the offspring resulting in lifelong effects. The causal link between higher FA supplementation and ASD incidence rates was hypothesized by previous studies [40,41], and a recent study from the Rochester Epidemiological Project in Rochester, MN (1976–1997), found a strong correlation (0.87) of ASD incidence in offspring for maternal consumption of prenatal vitamins containing >1 mg of folic acid [42,43]. Such evidence suggests that while too little FA results in damage to nervous tissue as evident from several studies, too much FA may disrupt normal function of nervous tissue associated with autism. Indeed concern has been raised with the role of excess maternal FA with several developmental outcomes including congenital heart defects, oral clefts, asthma, cancer and ASD [5,44].
While analysis of the microarray data revealed that the expression of a number of genes exhibited the same direction of change in both male and female pups from the higher gestational FA group (Figure 2), the expressions of several other genes were sex biased. Indeed, we found that the expression of genes in the cerebellum of female pups were more vulnerable to the changes induced by higher gestational FA. It shows that the in utero exposure of gestational FA may have a different impact depending upon the gender of the progeny, and it may have distinct mechanisms of alterations and consequences. Though the precise mechanism of such changes cannot be established from the present study, it confirms our previous data showing that the gestational FA induces differential alterations in methylation and gene expression in the brain of offspring depending upon gender of the progeny [10]. Of note, several previous studies have shown strong evidence that the susceptibility to neuropsychiatric and neurodegenerative diseases varies between men and women [45,46,47]; and maternal diet can significantly influence fetal outcomes including biasing offspring sex ratio [48]. Moreover, a recent study has shown widespread differences in the splicing and expression of genes in human brain between male and female [49]. Another study has shown that the maturation of GABAA signaling follows sex-specific patterns, which correlate with brain developmental expression profiles [50]. The switch from GABA being excitatory to inhibitory occurs earlier in females than in males.
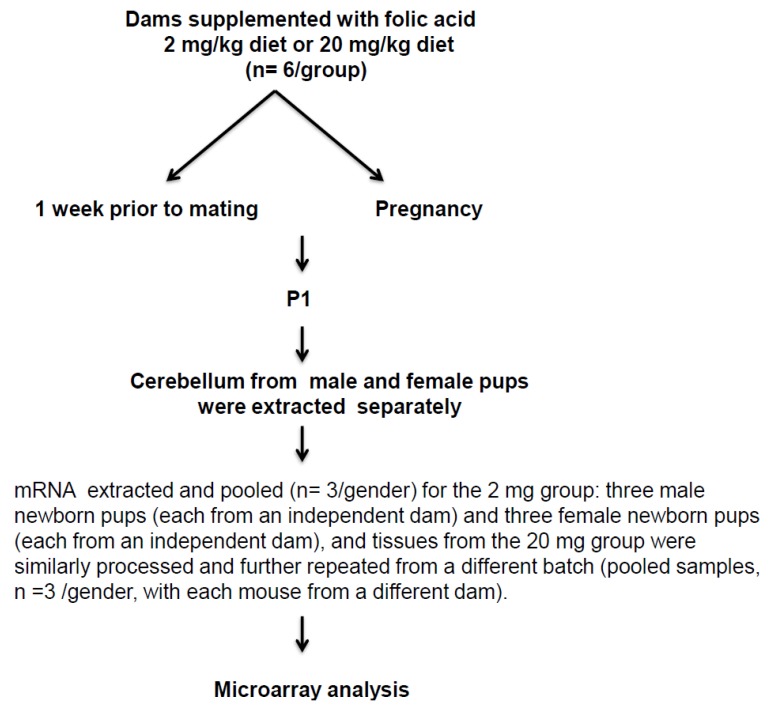
Figure 2.
Schematic diagram of the study design illustrating main experimental approach.
4. Material and Methods
4.1. Mice Strain and Feeding
Adult, 8- to 10-week-old C57BL/6J mice were used in all the experiments, and handled according to the protocol reviewed and approved by the Institute for Basic Research Institutional Animal Care and Use Committee. One week prior to mating and throughout the pregnancy, female mice were fed with a custom AIN-93G amino acid-based diet (Research Diet, Inc. New-Brunswick, NJ, USA), having FA at 2 mg/kg (n = 6) and 20 mg/kg (n = 6) diet. While the diet containing 2 mg of FA/kg diet is the general recommended basal dietary requirements for rodents [51]; the level of 20 mg FA/kg diet is tenfold higher than the basal dietary requirements. These amounts were chosen in this study, considering women with previous history of NTD affected pregnancy are recommended ten-fold higher FA (4 mg FA/day) in comparison to normal pregnant women (400–800 μg/day). Studies have shown in human Tolerable Upper Intake Level (1 mg/day adults) becomes limiting and intake of higher than 1mg/day escalates circulating unmetabolized FA [37]. The supplemented dietary FA level used in this study may not accurately reflect the precise level of FA in human, considering the inherent physiological differences in folate metabolism. Indeed, dihydrofolate reductase activity of rodent liver is perhaps higher than in human liver and to determine the impact of systemic exposure to FA and to elicit the same circulating plasma concentration of FA as human, perhaps rodents have to ingest much greater than pro-rata amounts of FA as human [37,52].
At postnatal day one (P1), for 2 mg group: male pups n = 3 and female pups n = 3 were sacrificed, and the cerebellum tissues were collected. For 20 mg group: male pups n = 6 and female pups n = 6 were similarly processed. All tissues were rapidly frozen after dissection and stored at −80 °C until downstream analysis was performed. The schematic diagram of the experimental design for this study is shown in Figure 2.
4.2. RNA Preparation
Cerebellum tissues of P1 pups were pooled (n = 3/gender) for the 2 mg group with three male pups (each from an independent dam) and three female pups (each from an independent dam), for a total of six pups (n = 6). Tissues from 20 mg group were similarly processed. RNA extractions for 20 mg group were further repeated from a different batch (pooled samples, n = 3/gender, with each mouse from a different dam), for a total of twelve pups (n = 12). Total RNA was extracted with Trizol reagent (Life Technologies, Carlsbad, CA, USA) and further purified by Qiagen RNeasy kit (Qiagen, Valencia, CA, USA), according to the manufacturer’s instructions and processed as described earlier [10,36]. All RNA samples were stored at −80 °C until downstream analysis was performed.
4.3. Microarrays
To minimize the differences of individual variability and increase the statistical power, microarray analysis was performed using purified RNA from pooled samples segregated by gender for each group as previously described [36]. Total RNA was reverse-transcribed, and cyanine-3-labelled cRNA was generated by following the manufacturer’s instructions using the one-color, low-input QuickAmp labeling kit (Agilent Technologies, Santa Clara, CA, USA). Following purification of cRNA with RNeasy kit (Qiagen, Valencia, CA, USA), the incorporation of cyanine-3 was assessed by a Nano Drop spectrophotometer. For each sample, a total of 600 ng of cRNA was fragmented and hybridized to the SurePrint G3 mouse gene expression 8 × 60 k arrays (G4852A, Agilent Technologies, Santa Clara, CA, USA). Using Feature Extraction software v10.7.3.1, the data were processed for image analysis and initial quality control. Our Minimum Information about a Microarray Experiment (MIAME)—compliant data has been deposited in the Gene Expression Omnibus database of the National Center for Bioinformatics, with the accession number GSE60531.
4.4. Data Analysis
Further analyses of microarray data were conducted with the publicly available BRB ArrayTools 4.3.1-stable release as previously described [36,53]. Quantile-Normalization was used to normalize the expression data and log-transformed. The differential expression of transcripts were assessed by following a series of filtering steps based on intensity filter, p value of log intensity variation and fold changes. For each spot if the intensity was below the minimum, a threshold to minimum 10 was used. To further filter the data, a cut-off set at p > 0.05 and percent missing exceeds 50% was used to exclude the corresponding spots for an entire gene from all arrays having variation in log intensity. Following filtering, a transcript was considered as differentially expressed if the difference in the mean expression in between two groups was ≥2.5 fold.
5. Conclusions
The role of FA in the prevention of NTDs has been well established for several years. However, considering the key role of FA in one-carbon metabolism and modulating the methylation profile, excess FA during gestation may alter the DNA methylation and gene expression of offspring. In this pilot study, we employed genome-wide profiling and found that higher gestational FA altered gene expression of several imprinted genes, transcription factors and genes associated with various neural pathways, including those reported to impact autism etiology, in the cerebellum of the offspring. In addition, several genes were dysregulated differentially in a gender-biased manner in pups from higher gestational FA. Since the cerebellum plays an important role to maintain abundant connections with non-motor brain regions [12], such dysregulations in the expression of genes associated with autism and neurodevelopment may influence overall cognitive development. In light of a recent report that higher FA intake can moderately modify the behavior of offspring [36,54], we suggest moderation of FA supplementation be exercised during gestation. Indeed, considering the shared metabolism between vitamin B12 and folate, high FA supplementation may mask vitamin B12 deficiency in pregnant woman and thus can have adverse consequences in the health of their children [55,56]. Since, vitamin B12 plays an important role in myelination and brain myelination is concentrated from mid-gestation, such imbalance in metabolism may result in impairment in early brain development [57].
This study was conducted as part of a pilot study to elucidate the overall difference in the transcriptional profile as a result of higher gestational FA in the cerebellum of offspring. One of the limitations of our current study is the limited sample size. Moreover as autism is a wider spectrum of disorders not a single entity and now diagnosis is more refined, caution has to be taken in relating findings in other species to humans as biochemical pathways may concur but have different rate limiting determinants. In the future, additional biochemical studies including behavioral analysis with larger numbers of samples will help to more precisely determine the influence of gestational FA in neural disease and would make it a fertile ground for exploration of ASDs.
Acknowledgements
Financial support from the March of Dimes Research Foundation (12-FY12-170) and the New York State Office for People with Developmental Disabilities is gratefully acknowledged. We thank Michael Fenko for assistance with the microarray hybridization and data collection.
Supplementary Information
Table S1. Genes down-regulated ≥2.5 fold in the cerebellum of male pups from mothers having FA supplementation during gestation at 20 mg/kg in comparison to mothers at 2 mg/kg diet.
Table S2. Genes up-regulated ≥2.5 fold in the cerebellum of male pups from mothers having FA supplementation during gestation at 20 mg/kg in comparison to mothers at 2 mg/kg diet.
Table S3. Genes down-regulated ≥2.5 fold in the cerebellum of female pups from mothers having FA supplementation during gestation at 20 mg/kg in comparison to mothers at 2 mg/kg diet.
Table S4. Genes up-regulated ≥2.5 fold in the cerebellum of female pups from mothers having FA supplementation during gestation at 20 mg/kg in comparison to mothers at 2 mg/kg diet.
Authors Contributions
Mohammed Junaid and W. Ted Brown conceived and designed the experiments; Subit Barua, Salomon Kuizon, Kathryn Chadman and Mohammed Junaid performed the experiments; Subit Barua and Mohammed Junaid analyzed the data; Subit Barua, Salomon Kuizon, Kathryn Chadman, W. Ted Brown and Mohammed Junaid contributed reagents/materials/analysis tools; Subit Barua wrote the paper, Kathryn Chadman, W. Ted Brown and Mohammed Junaid critically revised the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Heijmans B.T., Tobi E.W., Stein A.D., Putter H., Blauw G.J., Susser E.S., Slagboom P.E., Lumey L.H. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl. Acad. Sci. USA. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vanhees K., Vonhogen I.G., van Schooten F.J., Godschalk R.W. You are what you eat, and so are your children: The impact of micronutrients on the epigenetic programming of offspring. Cell Mol. Life Sci. 2014;71:271–285. doi: 10.1007/s00018-013-1427-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Callender S. A critical review of pernicious anaemia of pregnancy. Q. J. Med. 1944;13:75–105. [Google Scholar]
- 4.Hibbard E.D., Smithells R.W. Folic acid metabolism and human embryopathy. Lancet. 1965;285 doi: 10.1016/S0140-6736(65)91895-7. [DOI] [Google Scholar]
- 5.Barua S., Kuizon S., Junaid M.A. Folic acid supplementation in pregnancy and implications in health and disease. J. Biomed. Sci. 2014;21 doi: 10.1186/s12929-014-0077-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Honein M.A., Paulozzi L.J., Mathews T.J., Erickson J.D., Wong L.Y. Impact of folic acid fortification of the US food supply on the occurrence of neural tube defects. JAMA. 2001;285:2981–2986. doi: 10.1001/jama.285.23.2981. [DOI] [PubMed] [Google Scholar]
- 7.De Wals P., Tairou F., van Allen M.I., Uh S.H., Lowry R.B., Sibbald B., Evans J.A., van den Hof M.C., Zimmer P., Crowley M., et al. Reduction in neural-tube defects after folic acid fortification in Canada. N. Engl. J. Med. 2007;357:135–142. doi: 10.1056/NEJMoa067103. [DOI] [PubMed] [Google Scholar]
- 8.U.S. Preventive Services Task Force Folic acid for the prevention of neural tube defects: U.S. Preventive Services Task Force recommendation statement. Ann. Intern. Med. 2009;150:626–631. doi: 10.7326/0003-4819-150-9-200905050-00009. [DOI] [PubMed] [Google Scholar]
- 9.Choumenkovitch S.F., Selhub J., Wilson P.W., Rader J.I., Rosenberg I.H., Jacques P.F. Folic acid intake from fortification in United States exceeds predictions. J. Nutr. 2002;132:2792–2798. doi: 10.1093/jn/132.9.2792. [DOI] [PubMed] [Google Scholar]
- 10.Barua S., Kuizon S., Chadman K.K., Flory M.J., Brown W.T., Junaid M.A. Single-base resolution of mouse offspring brain methylome reveals epigenome modifications caused by gestational folic acid. Epigenetics Chromatin. 2014;7 doi: 10.1186/1756-8935-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Main P.A., Angley M.T., Thomas P., O’Doherty C.E., Fenech M. Folate and methionine metabolism in autism: A systematic review. Am. J. Clin. Nutr. 2010;91:1598–1620. doi: 10.3945/ajcn.2009.29002. [DOI] [PubMed] [Google Scholar]
- 12.Wang S.S., Kloth A.D., Badura A. The Cerebellum, Sensitive Periods, and Autism. Neuron. 2014;83:518–532. doi: 10.1016/j.neuron.2014.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reeber S.L., Otis T.S., Sillitoe R.V. New roles for the cerebellum in health and disease. Front. Syst. Neurosci. 2013;7 doi: 10.3389/fnsys.2013.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wegiel J., Flory M., Kuchna I., Nowicki K., Ma S., Imaki H., Wegiel J., Cohen I.L., London E., Wisniewski T., Brown W. Stereological study of the neuronal number and volume of 38 brain subdivisions of subjects diagnosed with autism reveals significant alterations restricted to the striatum, amygdala and cerebellum. Acta Neuropathol. Commun. 2014;2 doi: 10.1186/s40478-014-0141-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horsthemke B. In brief: Genomic imprinting and imprinting diseases. J. Pathol. 2014;232:485–487. doi: 10.1002/path.4326. [DOI] [PubMed] [Google Scholar]
- 16.Tranebjaerg L., Bathen J., Tyson J., Bitner-Glindzicz M. Jervell and Lange-Nielsen syndrome: A Norwegian perspective. Am. J. Med. Genet. 1999;89:137–146. doi: 10.1002/(SICI)1096-8628(19990924)89:3<137::AID-AJMG4>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 17.Crotti L., Celano G., Dagradi F., Schwartz P.J. Congenital long QT syndrome. Orphanet J. Rare Dis. 2008;3 doi: 10.1186/1750-1172-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Senner C.E., Brockdorff N. Xist gene regulation at the onset of X inactivation. Curr. Opin. Genet. Dev. 2009;19:122–126. doi: 10.1016/j.gde.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 19.Plagge A. Non-Coding RNAs at the Gnas and Snrpn-Ube3a Imprinted Gene Loci and Their Involvement in Hereditary Disorders. Front. Genet. 2012;3 doi: 10.3389/fgene.2012.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riccio A., Sparago A., Verde G., de Crescenzo A., Citro V., Cubellis M.V., Ferrero G.B., Silengo M.C., Russo S., Larizza L., Cerrato F. Inherited and Sporadic Epimutations at the IGF2-H19 locus in Beckwith-Wiedemann syndrome and Wilms’ tumor. Endocr. Dev. 2009;14:1–9. doi: 10.1159/000207461. [DOI] [PubMed] [Google Scholar]
- 21.Soejima H., Higashimoto K. Epigenetic and genetic alterations of the imprinting disorder Beckwith-Wiedemann syndrome and related disorders. J. Hum. Genet. 2013;58:402–409. doi: 10.1038/jhg.2013.51. [DOI] [PubMed] [Google Scholar]
- 22.Antypa N., Serretti A., Rujescu D. Serotonergic genes and suicide: A systematic review. Eur. Neuropsychopharmacol. 2013;23:1125–1142. doi: 10.1016/j.euroneuro.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 23.Kwan K.Y. Transcriptional dysregulation of neocortical circuit assembly in ASD. Int. Rev. Neurobiol. 2013;113:167–205. doi: 10.1016/B978-0-12-418700-9.00006-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cukier H.N., Rabionet R., Konidari I., Rayner-Evans M.Y., Baltos M.L., Wright H.H., Abramson R.K., Martin E.R., Cuccaro M.L., Pericak-Vance M.A., Gilbert J.R. Novel variants identified in methyl-CpG-binding domain genes in autistic individuals. Neurogenetics. 2010;11:291–303. doi: 10.1007/s10048-009-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vijayendran M., Beach S.R., Plume J.M., Brody G.H., Philibert R.A. Effects of genotype and child abuse on DNA methylation and gene expression at the serotonin transporter. Front. Psychiatry. 2012;3 doi: 10.3389/fpsyt.2012.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang C.H., Santangelo S.L. Autism and serotonin transporter gene polymorphisms: A systematic review and meta-analysis. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2008;147B:903–913. doi: 10.1002/ajmg.b.30720. [DOI] [PubMed] [Google Scholar]
- 27.Oberlander T.F. Fetal serotonin signaling: Setting pathways for early childhood development and behavior. J. Adolesc. Health. 2012;51(Suppl. S2):9–16. doi: 10.1016/j.jadohealth.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 28.Dannlowski U., Kugel H., Redlich R., Halik A., Schneider I., Opel N., Grotegerd D., Schwarte K., Schettler C., Ambree O., et al. Serotonin transporter gene methylation is associated with hippocampal gray matter volume. Hum. Brain Map. 2014;35:5356–5367. doi: 10.1002/hbm.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tabolacci E., Chiurazzi P. Epigenetics, fragile X syndrome and transcriptional therapy. Am. J. Med. Genet. A. 2013;161A:2797–2808. doi: 10.1002/ajmg.a.36264. [DOI] [PubMed] [Google Scholar]
- 30.Brown W.T. Clinical aspects of the fragile X syndrome. Results Probl. Cell Differ. 2012;54:273–279. doi: 10.1007/978-3-642-21649-7_15. [DOI] [PubMed] [Google Scholar]
- 31.Tsiouris J.A., Brown W.T. Neuropsychiatric symptoms of fragile X syndrome: Pathophysiology and pharmacotherapy. CNS Drugs. 2004;18:687–703. doi: 10.2165/00023210-200418110-00001. [DOI] [PubMed] [Google Scholar]
- 32.Junaid M.A., Kuizon S., Cardona J., Azher T., Murakami N., Pullarkat R.K., Brown W.T. Folic acid supplementation dysregulates gene expression in lymphoblastoid cells—Implications in nutrition. Biochem. Biophys. Res. Commun. 2011;412:688–692. doi: 10.1016/j.bbrc.2011.08.027. [DOI] [PubMed] [Google Scholar]
- 33.Wheeler A.C., Bailey D.B., Jr., Berry-Kravis E., Greenberg J., Losh M., Mailick M., Mila M., Olichney J.M., Rodriguez-Revenga L., Sherman S., et al. Associated features in females with an FMR1 premutation. J. Neurodev. Disord. 2014;6 doi: 10.1186/1866-1955-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yadav R., Gupta S.C., Hillman B.G., Bhatt J.M., Stairs D.J., Dravid S.M. Deletion of glutamate delta-1 receptor in mouse leads to aberrant emotional and social behaviors. PLoS One. 2012;7:e32969. doi: 10.1371/journal.pone.0032969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu S., Yue W., Jia M., Ruan Y., Lu T., Gong X., Shuang M., Liu J., Yang X., Zhang D. Association of the neuropilin-2 (NRP2) gene polymorphisms with autism in Chinese Han population. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007;144B:492–495. doi: 10.1002/ajmg.b.30495. [DOI] [PubMed] [Google Scholar]
- 36.Barua S., Chadman K.K., Kuizon S., Buenaventura D., Stapley N.W., Ruocco F., Begum U., Guariglia S.R., Brown W.T., Junaid M.A. Increasing Maternal or Post-Weaning Folic Acid Alters Gene Expression and Moderately Changes Behavior in the Offspring. PLoS One. 2014;9:e101674. doi: 10.1371/journal.pone.0101674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bailey S.W., Ayling J.E. The extremely slow and variable activity of dihydrofolate reductase in human liver and its implications for high folic acid intake. Proc. Natl. Acad. Sci. USA. 2009;106:15424–15429. doi: 10.1073/pnas.0902072106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sweeney M.R., McPartlin J., Weir D.G., Daly S., Pentieva K., Daly L., Scott J.M. Evidence of unmetabolised folic acid in cord blood of newborn and serum of 4-day-old infants. Br. J. Nutr. 2005;94:727–730. doi: 10.1079/BJN20051572. [DOI] [PubMed] [Google Scholar]
- 39.Smith A.D., Kim Y.I., Refsum H. Is folic acid good for everyone? Am. J. Clin. Nutr. 2008;87:517–533. doi: 10.1093/ajcn/87.3.517. [DOI] [PubMed] [Google Scholar]
- 40.Rogers E.J. Has enhanced folate status during pregnancy altered natural selection and possibly Autism prevalence? A closer look at a possible link. Med. Hypotheses. 2008;71:406–410. doi: 10.1016/j.mehy.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 41.Leeming R.J., Lucock M. Autism: Is there a folate connection? J. Inherit. Metab. Dis. 2009;32:400–402. doi: 10.1007/s10545-009-1093-0. [DOI] [PubMed] [Google Scholar]
- 42.Beard C.M., Panser L.A., Katusic S.K. Is excess folic acid supplementation a risk factor for autism? Med. Hypotheses. 2011;77:15–17. doi: 10.1016/j.mehy.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 43.O’Neill R.J., Vrana P.B., Rosenfeld C.S. Maternal methyl supplemented diets and effects on offspring health. Front. Genet. 2014;5:289. doi: 10.3389/fgene.2014.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neggers Y.H. Increasing prevalence, changes in diagnostic criteria, and nutritional risk factors for autism spectrum disorders. ISRN Nutr. 2014;2014 doi: 10.1155/2014/514026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cahill L. Why sex matters for neuroscience. Nat. Rev. Neurosci. 2006;7:477–484. doi: 10.1038/nrn1909. [DOI] [PubMed] [Google Scholar]
- 46.Cosgrove K.P., Mazure C.M., Staley J.K. Evolving knowledge of sex differences in brain structure, function, and chemistry. Biol. Psychiatry. 2007;62:847–855. doi: 10.1016/j.biopsych.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jazin E., Cahill L. Sex differences in molecular neuroscience: From fruit flies to humans. Nat. Rev. Neurosci. 2010;11:9–17. doi: 10.1038/nrn2754. [DOI] [PubMed] [Google Scholar]
- 48.Rosenfeld C.S. Periconceptional influences on offspring sex ratio and placental responses. Reprod. Fertil. Dev. 2011;24:45–58. doi: 10.1071/RD11906. [DOI] [PubMed] [Google Scholar]
- 49.Trabzuni D., Ramasamy A., Imran S., Walker R., Smith C., Weale M.E., Hardy J., Ryten M. Widespread sex differences in gene expression and splicing in the adult human brain. Nat. Commun. 2013;4 doi: 10.1038/ncomms3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Galanopoulou A.S. Sexually dimorphic expression of KCC2 and GABA function. Epilepsy Res. 2008;80:99–113. doi: 10.1016/j.eplepsyres.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reeves P.G., Nielsen F.H., Fahey G.C., Jr. AIN-93 purified diets for laboratory rodents: Final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J. Nutr. 1993;123:1939–1951. doi: 10.1093/jn/123.11.1939. [DOI] [PubMed] [Google Scholar]
- 52.Deghan M.S., Ishiguro L., Sohn K.J., Medline A., Renlund R., Croxford R., Kim Y.I. Folic acid supplementation promotes mammary tumor progression in a rat model. PLoS One. 2014;9:e84635. doi: 10.1371/journal.pone.0084635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simon R., Lam A., Li M.C., Ngan M., Menenzes S., Zhao Y. Analysis of gene expression data using BRB-Array Tools. Cancer Inform. 2007;3:11–17. [PMC free article] [PubMed] [Google Scholar]
- 54.Shorter K.R., Anderson V., Cakora P., Owen A., Lo K., Crossland J., South A.C., Felder M.R., Vrana P.B. Pleiotropic effects of a methyl donor diet in a novel animal model. PLoS One. 2014;9:e104942. doi: 10.1371/journal.pone.0104942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dwarkanath P., Barzilay J.R., Thomas T., Thomas A., Bhat S., Kurpad A.V. High folate and low vitamin B-12 intakes during pregnancy are associated with small-for-gestational age infants in South Indian women: A prospective observational cohort study. Am. J. Clin. Nutr. 2013;98:1450–1458. doi: 10.3945/ajcn.112.056382. [DOI] [PubMed] [Google Scholar]
- 56.Smith A.D. Folic acid fortification: The good, the bad, and the puzzle of vitamin B-12. Am. J. Clin. Nutr. 2007;85:3–5. doi: 10.1093/ajcn/85.1.3. [DOI] [PubMed] [Google Scholar]
- 57.Black M.M. Effects of vitamin B12 and folate deficiency on brain development in children. Food Nutr. Bull. 2008;29(Suppl. S2):126–131. doi: 10.1177/15648265080292S117. [DOI] [PMC free article] [PubMed] [Google Scholar]