Abstract
Signal transduction in bacteria is complex, ranging across scales from molecular signal detectors and effectors to cellular and community responses to stimuli. The unicellular, photosynthetic cyanobacterium Synechocystis sp. PCC6803 transduces a light stimulus into directional movement known as phototaxis. This response occurs via a biased random walk toward or away from a directional light source, which is sensed by intracellular photoreceptors and mediated by Type IV pili. It is unknown how quickly cells can respond to changes in the presence or directionality of light, or how photoreceptors affect single-cell motility behavior. In this study, we use time-lapse microscopy coupled with quantitative single-cell tracking to investigate the timescale of the cellular response to various light conditions and to characterize the contribution of the photoreceptor TaxD1 (PixJ1) to phototaxis. We first demonstrate that a community of cells exhibits both spatial and population heterogeneity in its phototactic response. We then show that individual cells respond within minutes to changes in light conditions, and that movement directionality is conferred only by the current light directionality, rather than by a long-term memory of previous conditions. Our measurements indicate that motility bias likely results from the polarization of pilus activity, yielding variable levels of movement in different directions. Experiments with a photoreceptor (taxD1) mutant suggest a supplementary role of TaxD1 in enhancing movement directionality, in addition to its previously identified role in promoting positive phototaxis. Motivated by the behavior of the taxD1 mutant, we demonstrate using a reaction-diffusion model that diffusion anisotropy is sufficient to produce the observed changes in the pattern of collective motility. Taken together, our results establish that single-cell tracking can be used to determine the factors that affect motility bias, which can then be coupled with biophysical simulations to connect changes in motility behaviors at the cellular scale with group dynamics.
Introduction
The movement of cells toward a favorable environment results from a complex regulatory network of many components that are responsible for detecting input signals and transducing them into a motility response (1–3). Motility in response to light has been extensively studied in the freshwater, coccoid photoautotrophic cyanobacterium, Synechocystis sp. PCC6803 (hereafter Synechocystis) (4–6). Synechocystis exhibits biased random walk behavior during positive phototaxis toward a light source (1,7,8) or negative phototaxis away from ultraviolet radiation that can damage DNA and other cellular components (1,8,9). On a soft, moist agarose surface cells typically exhibit positive phototaxis that yields group behavior such that subcommunities resembling finger-like projections emerge from an initially homogeneous population (10,11). This surface-dependent phototaxis is mediated by Type IV pili (12–14). In addition, several components of the signal transduction pathway including photoreceptors have been identified using mutant screens and biochemical analyses (5,15–19). It has been suggested that cells sense light direction rather than flux (7), but we have limited understanding of how this information is sensed and transduced to Type IV pili to cause directional motility. Biased movement can be generated by asymmetric localization and/or activity of the pili, as has been observed in the rod-shaped bacterium Pseudomonas aeruginosa during chemotaxis (20–22). Such mechanisms can be explored using single-cell imaging under conditions in which the presence of light is rapidly altered, and through quantification of movement features such as speed, orientation, and bias.
Several photoreceptors have been identified to play a role in phototaxis; they absorb light at specific wavelengths of the visible and ultraviolet spectrum (5,17,18,23–27) that overlap to a large extent with the absorption spectrum of pigmented proteins in the photosynthetic apparatus (28). Some of the photoreceptors also have overlapping absorption spectra such that they absorb at approximately the same wavelengths as each other (e.g., TaxD1 and PixD at 435 nm, and TaxD1 and UirS at 535 nm) (18,24,25). Previous studies of the behavior of mutants have established the role of each photoreceptor in either positive or negative phototaxis (7,8,18,29,30). The first photoreceptor identified to play a role in phototaxis was TaxD1, a cyanobacteriochrome that exists in two photo-reversible states, a blue light-absorbing form and a green light-absorbing form (24,31). taxD1 mutants lose positive phototaxis and exhibit negative phototaxis instead (5,8).
Collective behavior requires some form of communication between cells, such as the chemical signaling that takes place during bacterial quorum sensing (32). Secreted extracellular polymeric substances (EPS) have been demonstrated to enhance surface-dependent social motility in the soil-dwelling bacterium Myxococcus xanthus and in the pathogen Pseudomonas aeruginosa (33,34). Although the role of chemical signals and EPS is less well established in Synechocystis, we have shown in previous work that if a finger-like projection was redirected into the trail of another finger by changing the direction of incident light, cells rapidly increased their speed (10,11). To assess the role of EPS in influencing community spatial structure, we developed a biophysical model and used simulations to demonstrate that the combination of a biased random walk with mobility-enhancing EPS secretion was sufficient to generate communities with similar spatial patterns and on similar time and length scales to our experimental data (11).
To determine how cells respond to changes in light, we employed time-lapse video microscopy to quantitatively analyze the response of single cells to input signals and to probe heterogeneities across the population. This method also permits analysis of key mutants in terms of changes in the motility of single cells, thereby expanding on previous studies that relied on qualitative characteristics of community behavior (8,12,13,18,23,25,29). Here, we demonstrate that heterogeneous behavior exists across spatial locations in the community; cells in finger-like projections displayed higher motility bias and a decreased fraction of nonmotile cells. When the incident light direction was changed, cells rapidly altered their motility bias within minutes to align with the change in light direction. Removal of light quickly resulted in the elimination of forward bias, indicating that cells do not maintain a long-term memory of previous light conditions. Tracking taxD1 photoreceptor mutant single cells revealed that they exhibited increased movement perpendicular to the light and a lower (negative) bias, indicating that TaxD1 both promotes positive phototaxis and enhances movement directionality. Finally, we provide further evidence that changes in single-cell motility can predictably alter community behavior through a combination of experimental observations and biophysical simulations, demonstrating that increased movement of taxD1 cells perpendicular to the light abolishes the formation of finger-like projections seen in wild-type communities.
Materials and Methods
Strains and growth conditions
Synechocystis sp. PCC6803 cells derived from an original single colony of phototaxis-positive cells were grown in BG-11 medium (35) at 30°C with continuous shaking at 100 rpm under overhead warm white fluorescent light (Super Saver Warm White F40WW/SS, 34 W, Osram Sylvania, Inc., Danvers, MA). The incident flux was 10 μmol photons m-2s−1. All imaging experiments were performed using exponentially growing cells with optical density (OD) at 730 nm of 0.6 to 1.3 (25,000 to 55,000 cells/μL) as measured with an Ultrospec 3100 pro spectrophotometer (Amersham Biosciences, Piscataway, NJ). The taxD1 strain was grown in the presence of spectinomycin at a final concentration of 25 μg/mL (4).
Motility assays
Motility assays were carried out on 0.4% (w/v) agarose in BG-11 in 50 mm plastic petri dishes (BD, Franklin Lakes, NJ) at 30°C. One microliter of cells from an exponentially growing culture (OD730 = 0.8) was placed in the center of a plate, and then the plate was inverted to minimize evaporation. A warm white LED (5 mm, 7000 mcd, 35° spread; Fig. S1 in the Supporting Material; Super Bright LEDs, St. Louis, MO) was used to illuminate each plate. To induce directed phototaxis, the LED was placed 50 mm from the center of the initial droplet (diameter of ∼2.5 mm) at the level of the agarose. The incident light flux was ∼20 μmol photons m-2s−1, as measured with an LI-189 light meter (LI-COR Biosciences, Lincoln, NE).
Time-lapse video microscopy
Entire droplets of cells were imaged using a Canon 60D DSLR camera (Canon U.S.A., Inc., Melville, NY) attached to a Leica MZ12 stereoscope (Leica Microsystems, Buffalo Grove, IL). In the light manipulation experiments (see Figs. 3 and 4), cells were placed under directional light for at least 24 h, until finger-like projections formed. In the change-of-light-direction experiment (Fig. 3 A), the plate was rotated 90° relative to the LED. In the light on/off experiment (Fig. 3 B), the power supply to the LED was turned off or on, depending on the desired light condition. Time-lapse imaging at single-cell resolution was conducted at 20× magnification and 1 frame/s using a Coolsnap-Pro Monochrome camera (Photometrics, Tucson, AZ) attached to a Nikon TE-300 inverted microscope (Nikon Instruments, Inc., Melville, NY). The temperature was maintained at 30°C in all time-lapse imaging experiments using an environmental chamber (HaisonTech, Taipei, Taiwan).
Figure 3.
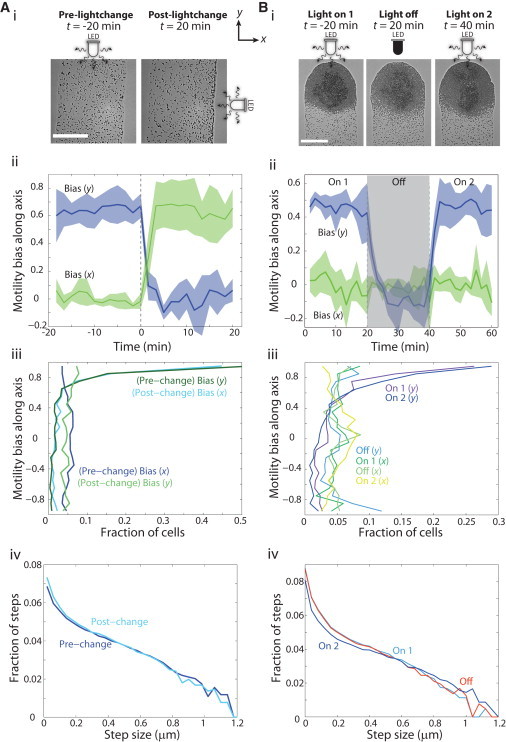
Cells rapidly reorient and respond to new light conditions, with no detectable memory of light or movement directionality. t = 0 min denotes the time of the first change in light condition. (A) The direction of light incidence on an inoculation of cells was rotated by 90°, from the y axis to the x axis. (i) Phase-contrast images of cells within the midfinger, before and after the rotation. (ii) After the direction change (dotted line), the cells reequilibrated their movement biases along the directions parallel and perpendicular to the light to their prechange values within minutes. (iii) The distribution of bias values also reequilibrated rapidly. (iv) Step-size distributions were similar pre- and post-light change. (B) Cells were imaged for 20 min with incident light (“On 1”), after which the light source was switched off for 20 min (“Off”), and then switched back on for 20 min (“On 2”). (i) Phase-contrast images of a fingertip at the start of imaging and at the end of the “Off” and “On 2” intervals. (ii) The motility bias parallel to the light quickly dropped to a slightly negative value once the light was turned off, and then reestablished its original value on restoration of incident light with minutes. (iii) The distributions of bias values along each axis were the same before and after the “Off” condition, whereas the distribution in the perpendicular direction was similar to the parallel direction in the “Off” condition. Distributions were computed from every 100 s interval over 20 min of imaging. (iv) Step-size distributions were similar with the light on and off. Distributions were compiled using n = 4 independent inoculations.
Figure 4.
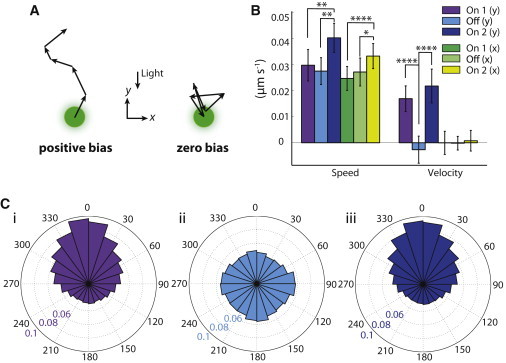
Cells maintain their speed and polarize their motion toward the light during phototaxis. (A) Schematic of two cells with similar speeds, moving with positive bias (left) and zero bias (right). (B) The average speed of single cells was similar along the two axes, before and after the light was turned off. Data are mean ± SE of n = 4 independent inoculations. Asterisks represent statistically significant differences, with ∗p < 0.05 and ∗∗p < 10−4. There was a small but significant increase in the average speed of single cells upon restoration of light (t(8) = 4.0, p < 0.01; t(8) = 5.0, p < 0.01; t(8) = 4.7, p < 10−4; t(8) = 2.7, p < 0.05, respectively). Similar to the parallel bias (Fig. 2Bii), once the light was turned off, the parallel velocity decreased approximately to zero (t(8) = 11.6, p < 10−4). Upon renewal of light, the velocity reverted to a value similar to its initial value (t(8) = 11.6, p < 10−4). (C) The distributions of step angles under the respective light conditions (color scheme as in B) illustrate an increase in the fraction of steps taken toward the light when light is present.
Cell tracking and analysis
Cell tracking was performed using custom MATLAB (The Mathworks, Natick, MA) software to quantify the positions and velocities of individual cells over time. In each frame, individual cells were segmented using thresholding and a watershed transform, and the locations of their centers of mass were computed. To avoid artifacts because of inconsistent segmentation of cell doublets, centroids that were closer than 0.93 μm (four pixels) were removed from subsequent analyses, and the tracks of the associated cells were terminated. The track of each cell was identified using nearest-neighbor distance across frames. The step size taken over each 1 s interval between frames was defined as the Euclidean distance moved by the cell over that time. To avoid incorrect assignment of tracks at high cell density, step sizes > 1.1625 μm (five pixels) were ignored and the tracks were terminated. For calculations of motility characteristics, only cells that traversed a net Euclidean distance of > 2 μm over the 10 min interval were used. The step angle was calculated relative to the light axis by taking the inverse tangent of the ratio between the displacements along the perpendicular and parallel axes. Steps taken directly toward or away from the light were defined as 0° and 180°, respectively. The speed, velocity, and bias values of each cell were calculated over 100 s intervals, unless otherwise indicated. These parameters were measured separately along each axis (parallel and perpendicular to the light), and included the times during those intervals when cells appeared stationary (for < 10 min). Speed and velocity were calculated by dividing the total path length of displacement by the length of the time interval of measurement (Fig. 1 D). Bias was calculated by dividing the resultant displacement by the total path length. The statistical significance of differences in the means was determined using Student’s t-test.
Figure 1.
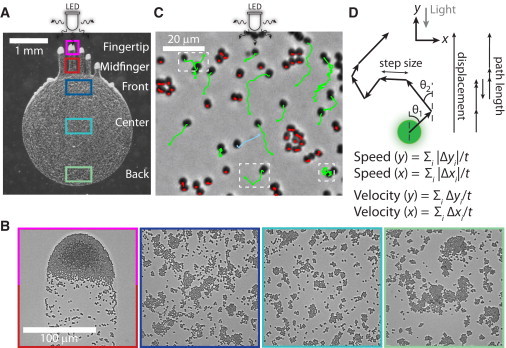
Quantification of the biased random walk behavior of Synechocystis cells during phototaxis. (A) Wild-type cells collectively moved toward a directed light source (white-light LED) and formed finger-like projections after ∼24 h. The community can be separated into at least five regions (colored boxes). (B) Example images of each of the regions defined in (A), with the fingertip and midfinger regions shown in the left-most box. (C) Single-cell tracks over a 200 s interval within the front region in (A); the phase-contrast image is the final frame. A fraction of the cells moved on average toward the light source (green tracks), although these cells also wandered from side to side and/or occasionally moved away from the light. Some cells moved away from the light on average (blue tracks) or remained approximately stationary (red tracks) for the duration of the tracking. The white, dashed boxes highlight cells that appeared to change direction during the imaging interval. (D) We quantified single-cell movement using a custom cell-tracking algorithm that extracts the length and angle relative to the light source (θ) of steps taken between imaging frames. Motility bias was defined as the ratio of the displacement over the total path length. Equations are for the speed and velocity in each direction, which are proportional to the path length and displacement, respectively.
Results
Single-cell time-lapse video microscopy reveals heterogeneity in phototactic behavior across a community
We used a standard phototaxis assay (7,10,17) in which a small volume (1 μL) of exponentially growing Synechocystis cells was spotted onto a low-concentration (0.4%) agarose plate, and then placed in the path of a directional, warm-white light-emitting diode (LED). Typically, within ∼24 h, the cells formed long (mm-scale) finger-like projections that moved in an approximately straight path toward the light source (Fig. 1 A). These projections had a high cell density at the tip. To determine the statistics of the motion of the cells within such a community, we used time-lapse microscopy and automated image analysis to track the movements of a large collection of single cells over a 10 min period at 1 s intervals. As previously reported (7,8,10), cells engaged in random walks with a bias toward the light source (Fig. 1 B). Single-cell tracking after 24 h revealed three patterns of behavior: 1) persistent motion over short timescales (∼10 s), 2) cells often changed their movement direction during the 10 min imaging interval, and 3) a fraction of the cells remained approximately stationary (defined as traversing < 2 μm net Euclidean distance over the 10 min interval). We consistently observed that the motile fraction varied spatially across the spot, with very few motile cells in the back of the drop as compared with the front (Fig. 2, A and B).
Figure 2.
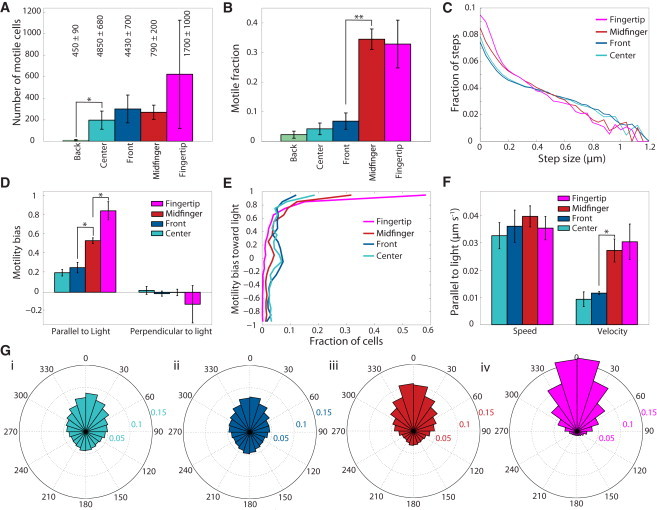
Cells exhibit a broad distribution of motility bias within and across community locations. (A) Number of cells that traversed more than 2 μm from their original position over a 10 min imaging interval (see Fig. 1A for location definitions). Relatively few cells were motile in the back region. The total number of cells tracked in each region, including nonmotile cells, is indicated. Data are mean ± SE of n = 4 independent inoculations. Asterisks represent statistically significant differences, with ∗p < 10−2 and ∗∗p < 10−4. The back region had a significantly lower number of motile cells than the center region Student's t-test with six degrees of freedom (t(6) = 4.4, p < 10−2). (B) Fraction of segmented cells in each region that was motile. The midfinger region had a significantly higher fraction of motile cells than the front region (t(6) = 12.5, p < 10−4). (C) The distribution of step sizes over 1 s intervals was similar in all regions, with a change in slope at ∼0.2 μm that is likely attributable to progressive pilus retraction (Fig. S2 A). (D) Mean bias values were significantly higher in the fingertip and midfinger regions. The bias of cells within the midfinger region was significantly higher than cells within the front and center regions (t(6) = 9.0, p < 10−3), whereas the bias within the fingertip region was significantly higher than in the midfinger (t(6) = 6.3, p < 10−3). Mean bias perpendicular to the light was approximately zero in all regions, where positive directionality is defined as 90° clockwise from the incident light direction. (E) Fingertip and midfinger regions had a higher fraction of cells that moved processively toward the light (parallel bias close to 1), whereas all regions had approximately zero bias in the perpendicular direction. (F) The cell speed parallel to the axis of light incidence was approximately constant across the regions, whereas the velocity reflected the increase in bias toward the front of the drop shown in (D). (G) Rose plots of the distribution of step angles in directions relative to the light source. Regions are colored as in (A)–(F). 0° and 180° denote steps taken directly toward and away from the light, respectively.
To further characterize the cellular trajectories, we quantified the distance traversed by each nonstationary cell and its angle relative to the light source over each 1 s interval (Fig. 1 D). The distribution of step sizes ranged from 0 to ∼1.2 μm and initially decayed approximately linearly, with a shift in the slope at ∼0.2 μm (Fig. 2 C). Previous studies of Type IV pilus retraction in P. aeruginosa indicated a retraction speed of ∼0.5 μm/s (20), suggesting that this shift might be produced by partial pilus retractions over the 1 s intervals. When we plotted the sizes and angles of steps that followed those taken between 0.2 and 0.6 μm (Fig. S2), we detected a strong relationship between successive steps such that the subsequent step was taken at approximately the same angle as the preceding step (Fig. S2 A). This result is consistent with a progression of pilus retraction events, although there was little correlation in the sizes of subsequent steps (Fig. S2 B). This suggests that the degree of pilus retraction is more likely to be a random process, which is also consistent with the lack of a peak in the step size distribution (Fig. 2 C).
To determine the motility bias of the cells, we computed the ratio of the displacement and the total path length in the direction of the light during subintervals ranging from 10 to 300 s (Figs. 1 C and S3). We defined our axes such that overall movement toward the light source translates to positive displacement. Because of the stochastic nature of the motion and the broad distribution of step sizes, we found that over shorter subintervals, the fraction of cells with small bias was lower and cells exhibited more processive motion in both directions (see Methods and Fig. S3, A and B). We determined that consistent definitions of bias statistics required long measurement intervals (> 100 s subintervals, Fig. S3 A) and high data acquisition frequency (Fig. S3 C).
Given the heterogeneity in bias within a single field of view, we examined cells in different spatial locations to see if they exhibited different motility characteristics. We measured the number and fraction of motile cells within various regions of the fingered community at least 24 h after initial inoculation (Figs. 1 A and 2 A and B). For multiple independent inoculations (n = 4), very few ( 10) cells were motile in the back region, and hence we omitted this region from further analysis. We noted that the lack of motility in cells at the back of the drop persisted for days, as evidenced by the inability of these cells to move toward a rotated light source (Fig. S4 H). The heterogeneity in the motility characteristics was not merely because of differences in light flux; the flux of light incident on the different regions of the droplet was 18 μmol photons m-2s−1 in the back region and 26 μmol photons m-2s−1 at the fingertip region, a range over which drops placed at different intensities did not exhibit substantial variation in average bias (Fig. S4, A and B). The fractions of motile cells within the midfinger and fingertip (∼35% each) were significantly higher (p < 2 × 10−5) than in the center and front (∼5%) (Fig. 2 B). We measured the distribution of bias values in the four regions and found that cells in the center and front had significantly lower bias than cells within the fingers (Fig. 2, D and E). The distribution of positive bias values shifted upward in the regions with increased mean bias (Fig. 2, D and E), indicating that an increased fraction of cells moved more processively during the 100 s subintervals. These bias increases were also reflected in a higher frequency of steps taken toward the light (Fig. 2 Gi–iv). As expected of a phototactic response, the average motility bias perpendicular to the light source was approximately zero in all regions of the drop, where we define positive perpendicular bias as net motion toward the direction 90° clockwise from the incident light direction (Fig. 2 D). The average speed parallel to the light axis (Fig. 1 D), however, was approximately similar across the regions (Fig. 2 F). Thus, cells exhibit distinct motility behaviors across the community, with greater processivity displayed by cells that have moved out of the original inoculation.
Cells respond quickly to changes in light directionality and flux
Given that cells within the finger regions displayed higher motility bias than cells in the center or front regions, we reasoned that surface characteristics, such as EPS laid down by the cells, might confer information about the directionality of the light. To test this hypothesis, we carried out two types of experiments in which the incident light was perturbed. First, we rotated the plate relative to the light source so that the incidence direction shifted by 90° (light was incident along the x axis instead of the y axis) (Fig. 3 Ai). We imaged the same group of cells in the midfinger, where cells are well separated, easy to track, and not subject to spatial constraints as in the fingertip, for 20 min before and after the plate rotation. Because the light shift can be carried out rapidly, we were able to track cells soon (< 30 s) after the change. We observed that the cells reoriented their movement toward the new light direction within 100 s. This reorientation manifested as a sharp decrease in motility bias along the y axis to zero and as a corresponding sharp increase in the bias along the x axis (Fig. 3 Aii). Within < 5 min, the collection of cells reached mean values (Fig. 3 Aii) and distributions of the bias (Fig. 3 Aiii) in the directions parallel and perpendicular to the light source that were similar to those measured before the rotation. The bias in the x direction then decreased slightly over the next 15 min, coincident with cells clustering at the edge of the finger. The ability of the cells to rapidly reorient their motility indicates that the light direction is the major determinant of motility behavior and other factors do not appear to confer any long-term memory of light directionality.
In a complementary approach, we examined the behavior of cells placed in incident light, then subjected to 20 min of darkness, and restoration of incident light afterward (Fig. 3 B). Similar to the directionality-change experiment (Fig. 3 A), cells responded rapidly to the removal and restoration of light, reaching a stable mean value and distribution of the bias in < 5 min (Fig. 3 Bii). The cells did not display any noticeable persistence in movement toward the original light direction once the stimulus was removed, and they lost their positive mean bias along the x axis within 120 s (Fig. 3 Bii). The bias in this direction decreased reproducibly to a slightly negative value (Fig. 3 Bii). We attribute this to the slight loss of cohesion experienced by the cells in the cell-dense fingertip as the bias decreased in the dark, resulting in cells at the interface between the midfinger and fingertip moving back into the midfinger (Fig. 3 Bi). Consequently, these cells occupied space and therefore negatively displaced the cells that were being tracked in the midfinger region. Perpendicular bias along the y axis remained essentially zero throughout the experiment (Fig. 3 Bii). Upon restoration of the light source, the mean bias (Fig. 3 Bii) and bias distribution (Fig. 3 Biii) along the y axis rapidly returned to the value before the light was turned off.
In each of the three light conditions, we measured cell speed (path length/time) and velocity (displacement/time) (Fig. 1 D) to determine whether light increased motility in addition to introducing a bias. As expected, the velocity was approximately zero in the direction perpendicular to the light in all cases, and the velocity in the direction of the light was zero in the dark and attained a similar positive value in both light-on conditions (Fig. 4, A and B). Interestingly, the average speed was unaffected by the removal of the light (Fig. 4 B), indicating that removing the light mainly affected the bias. This observation was consistent with our observation that the step-size distributions differed negligibly across light conditions (Fig. 3, A and Biv), indicating that cells exhibited similar movements in the light and in the dark. Upon restoration of the light, there was a small but significant increase (p < 5×10−3) in speed along both the x and y axes relative to the previous light on period. The speed was generally the same in both the x and y directions (Fig. 4 B), indicating that positive bias was achieved by biasing the step angle (Fig. 4 C) rather than step size (Fig. 3, A and Biv), which did not change according to light condition. The step-angle distributions (Fig. 4 C) indicated that phototaxis involves an increase/decrease in the fraction of movement toward/away from the light, while maintaining the same fraction of movement in the perpendicular direction as in the dark. Taken together, our single-cell trajectory analysis suggests that the biased random walk in phototaxis is the result of a redistribution of steps toward the light source rather than an overall increase in the frequency or size of steps.
The photoreceptor TaxD1 promotes positive phototaxis by inhibiting sideways motion
Cells in which the gene encoding the photoreceptor TaxD1 has been inactivated have been shown to lose positive phototaxis and exhibit negative phototaxis (8,15). We observed that taxD1 communities moving away from the light source formed a single, wide front with a thick, cell-dense fingertip (Fig. 5, A and B). This morphology contrasted with the well-separated fingers observed in wild-type populations (Fig. 1 A) and was independent of initial inoculation cell densities (Fig. S5), suggesting a general difference in the motility behavior of taxD1 cells relative to that of wild-type. To determine whether changes in single-cell behavior could account for these different community morphologies, we tracked the motion of single taxD1 cells in the front region, directly behind the finger, in the presence of directional light. Although taxD1 cells exhibited biased random walks qualitatively similar to wild-type cells, there appeared to be a pronounced increase in motion perpendicular to the light source (Fig. 5 C); taxD1 cells took steps in the direction perpendicular to the light approximately as often as they stepped away from the light (Fig. 5 D).
Figure 5.
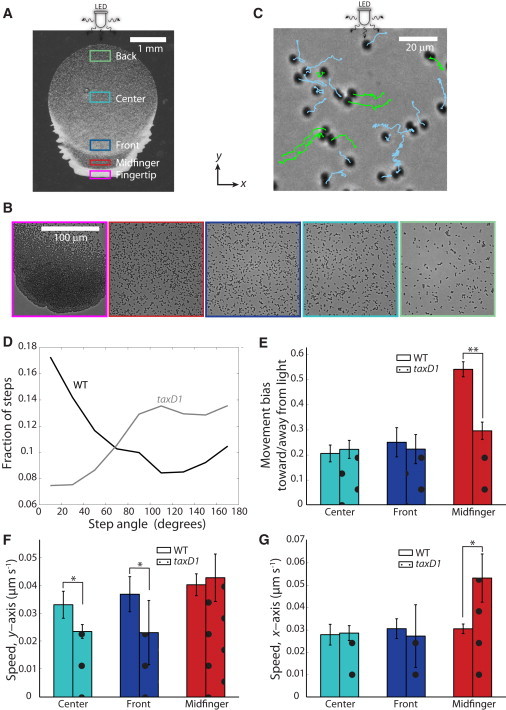
TaxD1 enforces directionality of movement. (A) taxD1 cells collectively moved away from the light with a single, wide front. Thin, cell-dense projections also extended outward from the sides of the progressing front. (B) Single cells exhibited pronounced sideways and backward movement (color scheme as inFig. 1B). (C) Example images of each of the regions defined in (A). (D) taxD1 cells took a higher fraction of steps in the perpendicular direction and a lower fraction in the parallel direction than wild-type cells. (E) Motility bias values of taxD1 cells moving away from the light were similar to wild-type cells moving toward the light in the center and front regions. However, taxD1 cells in the midfinger region exhibited lower motility bias than wild-type cells (t(6) = 36.4, p < 10−4). Data are mean ± SE of n = 4 independent inoculations. Asterisks represent statistically significant differences, with ∗p < 0.02 and ∗∗p < 10−4. (F) The speed along the axes parallel to the light was higher in wild-type compared with taxD1 cells in the center (t(6) = 3.5, p < 0.02) and front regions (t(6) = 3.4, p < 0.02), whereas in (G), the speed perpendicular to the light was higher in taxD1 cells within the midfinger (t(6) = 4.1, p < 0.01).
taxD1 cells took a lower fraction of steps away from the light (step angle ∼180°) than the wild-type cells did toward the light (step angle ∼0°) (Fig. 5 D), resulting in a lower motility bias in taxD1 cells in the midfinger region (p < 0.0001) compared with wild-type cells (Fig. 5 E). The speed of taxD1 cells along the light direction remained similar to that of wild-type cells (Fig. 5 E), although there was a significant increase in the speed along the direction perpendicular to incident light in the midfinger region (p < 0.01) (Fig. 5 G). On the other hand, the speed of taxD1 cells along the light direction was lower in the center (p < 0.02) and front (p < 0.02) regions but similar along the perpendicular direction. Therefore, we conclude that TaxD1 is responsible for both promoting movement toward the light and suppressing sideways movement.
To determine whether the increased sideways movement of taxD1 cells is sufficient to explain the change from a fingered community to a single front, we modified our previous reaction-diffusion model (11) to include an anisotropic diffusion tensor:
| (1) |
where the light is incident along the y axis, C is the cellular concentration field, S is the EPS concentration field, M = 1-e-S is the mobility, β is the light-induced bias, and λ is the anisotropy in diffusion; λ = 1 corresponds to our previous model (11), whereas λ < 1 and λ > 1 correspond to more rapid diffusion in the x and y directions, respectively. Simulations based on this modified model showed a relatively sharp transition from a community with many finger-like projections for λ = 1 (Fig. 6 A) to one with a single front for λ = 0.5 (Fig. 6 B), similar to our experimental observations (Fig. 5 A). Overall progression of the front slowed with smaller values of λ (Fig. 6 B), suggesting that sideways movement has a negative impact on collective motility. Thus, the observed changes in single-cell movements are sufficient to switch the community architecture from finger-like patterns (Fig. 1 A) to a single front (Fig. 5 A).
Figure 6.
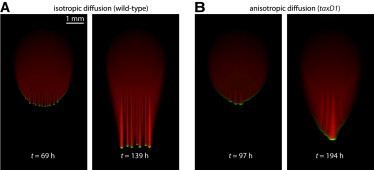
Biophysical model predicts a transition in front morphology due to changes in single-cell motility in taxD1 cells. Simulations are based on the model from (11), along with the modifications described in Eq. 1, with and . Cellular and EPS concentrations are shown in shades of green and red, respectively. (A) Isotropic diffusion (λ = 1) generates fingering patterns similar to that of wild-type communities (Fig. 1A). (B) Anisotropic diffusion (λ = 0.5) with more motion perpendicular to the direction of incident light was sufficient to eliminate fingering, instead resulting in a single, broad front, similar to Fig. 5A.
Discussion
In this study, we used quantitative analysis of single-cell trajectories to characterize phototactic motility, which helped reveal links between single-cell behavior and community organization. To our knowledge, this is the first study to consider the effects of movement noise on quantitative motility characteristics, providing an important benchmark for future studies involving single-cell tracking. Our measurements of speed and bias also illustrate the importance of using a rapid acquisition frequency and a long imaging interval (Fig. S3). Although all moving cells in the monitored communities engaged in biased random walks (Fig. 1), our data revealed spatial heterogeneities in motility bias (Fig. 2). Moreover, each region of the community contained both motile and nonmotile cells, with the fraction of nonmotile cells being greatest at the back of the drop (Figs. 2, A and B, and S4). Taken together, our data suggest that motile cells from the initial inoculation move toward the light and aggregate at the front until enough EPS accumulates to allow for the formation of fingering projections. Conversely, a fraction of cells remain at the back of the drop, and appear to be nonmotile over several days, as demonstrated in experiments in which we rotated the light source by 90° (Fig. S4 H). However, it remains unclear whether the lack of motility in this subpopulation is attributable to heterogeneities in pilus abundance (4,12–14,22), cyclic AMP levels (19,22,36,37), the extracellular environment (11,33,34,38,39), or other factors.
Synechocystis cells responded rapidly (within minutes) to changes in light conditions by adjusting the distribution of step angles, with little to no memory about previous light conditions (Fig. 3). Moreover, our observation that cells in the fingers did not move with a bias when the light was turned off (Figs. 3 B and 4 C) indicates that the extracellular environment does not provide any memory of past movement or light directionality. These results support our previous modeling of phototaxis as a simple random walk with an instantaneous light bias (11). Furthermore, the speed of movement was unaltered by the presence of light (Fig. 4 B). These data suggest that the extent of pilus retraction across the cell surface is conserved in all conditions; motility bias likely results from the polarization of pilus activity with an increase in the force and/or frequency of pilus retraction on the side of the cell facing the light, and vice versa on the side facing away from the light (Fig. 7). In Neisseria gonorrhoeae, a tug-of-war mechanism with Type IV pili has been proposed to drive persistent motility (40); in Synechocystis, our data indicate that motility persistence lasts no more than a few minutes during changes in light incidence. This may allow cells in an unpredictable, fluctuating natural light environment to rapidly respond to changes in light flux and direction, so as to move into an optimal location for photosynthesis (6).
Figure 7.
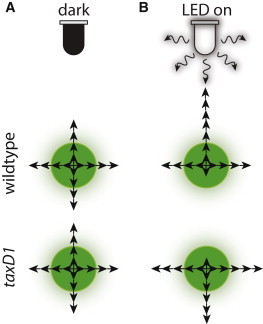
Model of single-cell motility. (A) Wild-type and taxD1 cells move equally in all directions in the dark. (B) Incident light from the top of the figure induces wild-type cells to take more steps toward the light than away from the light, whereas taxD1 cells also take more steps perpendicular to the light. Across all cases, the distribution of step sizes is essentially the same; instead, the distribution of step angles is affected.
Dissection of the complex integration of signals from the multiple photoreceptors in Synechocystis requires the elucidation of wild-type and mutant phenotypes that encompass multiple behaviors. Our single-cell tracking data complements an earlier study in which single cells on a glass slide move away from the light on average, albeit with lower displacements than what wild-type cells exhibit toward light of the same flux (8). Our results also indicate that taxD1 cells exhibited increased sideways movement, suggesting that TaxD1 reinforces directional movement by reducing the fraction of steps taken perpendicular to the light source. Importantly, this sideways movement manifested in differences at the community scale, with a single, wide front and small sideways protrusions (Figs. 5 A and S5). The consistency between our computational predictions for taxD1 cells and our experimental observations (Fig. 6) provide further support for our previously studied minimal phototaxis model (11). Further single-cell studies of other mutants and in various light conditions should help to clarify the roles of each photoreceptor and the classes of single-cell and community behaviors.
Conclusions
Our results illustrate the utility of quantitative, single-cell measurements for characterizing complex behavior. In the case of phototaxis, simple manipulations of the light conditions or genetic perturbations can be used to determine the factors regulating motility bias and speed. Our observations of both spatial and population heterogeneity should provide useful constraints for refining models of phototaxis (10,41,42) with the aim of capturing and predicting single-cell dynamics. In addition, interpretations of phototactic behavior at the community scale are greatly enhanced by single-cell phenotypes; for example, the negative phototaxis phenotype of taxD1 populations belies more subtle alterations at the single-cell level. Although we speculate that it is the activity rather than the localization of the pilus motors that is altered in response to changing light conditions, further studies that explicitly measure the intracellular localization patterns of the photoreceptors and pili (14,21,43) will yield information about the mechanisms by which cells adapt to light stimuli. The application of a combination of genetic, imaging, and computational analysis tools to phototaxis provides a general paradigm for dissecting regulatory networks that effect robust responses from input signals.
Contributor Information
Kerwyn Casey Huang, Email: kchuang@stanford.edu.
Devaki Bhaya, Email: dbhaya@carnegiescience.edu.
Supporting Material
References
- 1.Mullineaux C.W. How do cyanobacteria sense and respond to light? Mol. Microbiol. 2001;41:965–971. doi: 10.1046/j.1365-2958.2001.02569.x. [DOI] [PubMed] [Google Scholar]
- 2.Van Haastert P.J., Devreotes P.N. Chemotaxis: signalling the way forward. Nat. Rev. Mol. Cell Biol. 2004;5:626–634. doi: 10.1038/nrm1435. [DOI] [PubMed] [Google Scholar]
- 3.Schweinitzer T., Josenhans C. Bacterial energy taxis: a global strategy? Arch. Microbiol. 2010;192:507–520. doi: 10.1007/s00203-010-0575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhaya D. Light matters: phototaxis and signal transduction in unicellular cyanobacteria. Mol. Microbiol. 2004;53:745–754. doi: 10.1111/j.1365-2958.2004.04160.x. [DOI] [PubMed] [Google Scholar]
- 5.Yoshihara S., Ikeuchi M. Phototactic motility in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Photochem. Photobiol. Sci. 2004;3:512–518. doi: 10.1039/b402320j. [DOI] [PubMed] [Google Scholar]
- 6.Bhaya D., Brahamsha B. The Cell Biology of Cyanobacteria. Caister Academic Press; Poole, UK: 2014. Motility and the regulation of phototaxis in cyanobacteria; pp. 233–262. [Google Scholar]
- 7.Choi J.S., Chung Y.H., Park Y.M. Photomovement of the gliding cyanobacterium Synechocystis sp. PCC 6803. Photochem. Photobiol. 1999;70:95–102. doi: 10.1562/0031-8655(1999)070<0095:potgcs>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- 8.Ng W.O., Grossman A.R., Bhaya D. Multiple light inputs control phototaxis in Synechocystis sp. strain PCC6803. J. Bacteriol. 2003;185:1599–1607. doi: 10.1128/JB.185.5.1599-1607.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moon Y.J., Kim S.I., Chung Y.H. Sensing and responding to UV-A in cyanobacteria. Int. J. Mol. Sci. 2012;13:16303–16332. doi: 10.3390/ijms131216303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burriesci M., Bhaya D. Tracking phototactic responses and modeling motility of Synechocystis sp. strain PCC6803. J. Photochem. Photobiol. B. 2008;91:77–86. doi: 10.1016/j.jphotobiol.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 11.Ursell T., Chau R.M.C., Huang K.C. Motility enhancement through surface modification is sufficient for cyanobacterial community organization during phototaxis. PLOS Comput. Biol. 2013;9:e1003205. doi: 10.1371/journal.pcbi.1003205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhaya D., Watanabe N., Grossman A.R. The role of an alternative sigma factor in motility and pilus formation in the cyanobacterium Synechocystis sp. strain PCC6803. Proc. Natl. Acad. Sci. USA. 1999;96:3188–3193. doi: 10.1073/pnas.96.6.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhaya D., Bianco N.R., Grossman A. Type IV pilus biogenesis and motility in the cyanobacterium Synechocystis sp. PCC6803. Mol. Microbiol. 2000;37:941–951. doi: 10.1046/j.1365-2958.2000.02068.x. [DOI] [PubMed] [Google Scholar]
- 14.Mattick J.S. Type IV pili and twitching motility. Annu. Rev. Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 15.Bhaya D., Takahashi A., Grossman A.R. Novel motility mutants of Synechocystis strain PCC 6803 generated by in vitro transposon mutagenesis. J. Bacteriol. 2001;183:6140–6143. doi: 10.1128/JB.183.20.6140-6143.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshihara S., Suzuki F., Ikeuchi M. Novel putative photoreceptor and regulatory genes required for the positive phototactic movement of the unicellular motile cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2000;41:1299–1304. doi: 10.1093/pcp/pce010. [DOI] [PubMed] [Google Scholar]
- 17.Fiedler B., Börner T., Wilde A. Phototaxis in the cyanobacterium Synechocystis sp. PCC 6803: role of different photoreceptors. Photochem. Photobiol. 2005;81:1481–1488. doi: 10.1562/2005-06-28-RA-592. [DOI] [PubMed] [Google Scholar]
- 18.Song J.Y., Cho H.S., Park Y.I. Near-UV cyanobacteriochrome signaling system elicits negative phototaxis in the cyanobacterium Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. USA. 2011;108:10780–10785. doi: 10.1073/pnas.1104242108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhaya D., Nakasugi K., Burriesci M.S. Phototaxis and impaired motility in adenylyl cyclase and cyclase receptor protein mutants of Synechocystis sp. strain PCC 6803. J. Bacteriol. 2006;188:7306–7310. doi: 10.1128/JB.00573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skerker J.M., Berg H.C. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. USA. 2001;98:6901–6904. doi: 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiang P., Habash M., Burrows L.L. Disparate subcellular localization patterns of Pseudomonas aeruginosa Type IV pilus ATPases involved in twitching motility. J. Bacteriol. 2005;187:829–839. doi: 10.1128/JB.187.3.829-839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burrows L.L. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu. Rev. Microbiol. 2012;66:493–520. doi: 10.1146/annurev-micro-092611-150055. [DOI] [PubMed] [Google Scholar]
- 23.Lamparter T., Esteban B., Hughes J. Phytochrome Cph1 from the cyanobacterium Synechocystis PCC6803. Purification, assembly, and quaternary structure. Eur. J. Biochem. 2001;268:4720–4730. doi: 10.1046/j.1432-1327.2001.02395.x. [DOI] [PubMed] [Google Scholar]
- 24.Yoshihara S., Katayama M., Ikeuchi M. Cyanobacterial phytochrome-like PixJ1 holoprotein shows novel reversible photoconversion between blue- and green-absorbing forms. Plant Cell Physiol. 2004;45:1729–1737. doi: 10.1093/pcp/pch214. [DOI] [PubMed] [Google Scholar]
- 25.Okajima K., Yoshihara S., Ikeuchi M. Biochemical and functional characterization of BLUF-type flavin-binding proteins of two species of cyanobacteria. J. Biochem. 2005;137:741–750. doi: 10.1093/jb/mvi089. [DOI] [PubMed] [Google Scholar]
- 26.Moon Y.J., Kim S.J., Chung Y.H. Sensing UV/blue: pterin as a UV-A absorbing chromophore of cryptochrome. Plant Signal. Behav. 2010;5:1127–1130. doi: 10.4161/psb.5.9.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zirak P., Penzkofer A., Hegemann P. Absorption and emission spectroscopic characterization of blue-light receptor Slr1694 from Synechocystis sp. PCC6803. J. Photochem. Photobiol. B. 2007;86:22–34. doi: 10.1016/j.jphotobiol.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 28.Chen M., Blankenship R.E. Expanding the solar spectrum used by photosynthesis. Trends Plant Sci. 2011;16:427–431. doi: 10.1016/j.tplants.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 29.Moon Y.J., Lee E.M., Chung Y.H. The role of cyanopterin in UV/blue light signal transduction of cyanobacterium Synechocystis sp. PCC 6803 phototaxis. Plant Cell Physiol. 2010;51:969–980. doi: 10.1093/pcp/pcq059. [DOI] [PubMed] [Google Scholar]
- 30.Wilde A., Fiedler B., Börner T. The cyanobacterial phytochrome Cph2 inhibits phototaxis towards blue light. Mol. Microbiol. 2002;44:981–988. doi: 10.1046/j.1365-2958.2002.02923.x. [DOI] [PubMed] [Google Scholar]
- 31.Ikeuchi M., Ishizuka T. Cyanobacteriochromes: a new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochem. Photobiol. Sci. 2008;7:1159–1167. doi: 10.1039/b802660m. [DOI] [PubMed] [Google Scholar]
- 32.Waters C.M., Bassler B.L. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 33.Lu A., Cho K., Shi W. Exopolysaccharide biosynthesis genes required for social motility in Myxococcus xanthus. Mol. Microbiol. 2005;55:206–220. doi: 10.1111/j.1365-2958.2004.04369.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhao K., Tseng B.S., Wong G.C. Psl trails guide exploration and microcolony formation in Pseudomonas aeruginosa biofilms. Nature. 2013;497:388–391. doi: 10.1038/nature12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stanier R.Y., Kunisawa R., Cohen-Bazire G. Purification and properties of unicellular blue-green algae (order Chroococcales) Bacteriol. Rev. 1971;35:171–205. doi: 10.1128/br.35.2.171-205.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshimura H., Yoshihara S., Ohmori M. A cAMP receptor protein, SYCRP1, is responsible for the cell motility of Synechocystis sp. PCC 6803. Plant Cell Physiol. 2002;43:460–463. doi: 10.1093/pcp/pcf050. [DOI] [PubMed] [Google Scholar]
- 37.Terauchi K., Ohmori M. Blue light stimulates cyanobacterial motility via a cAMP signal transduction system. Mol. Microbiol. 2004;52:303–309. doi: 10.1111/j.1365-2958.2003.03980.x. [DOI] [PubMed] [Google Scholar]
- 38.Ali A., Johnson J.A., Sozhamannan S. Mutations in the extracellular protein secretion pathway genes (eps) interfere with rugose polysaccharide production in and motility of Vibrio cholerae. Infect. Immun. 2000;68:1967–1974. doi: 10.1128/iai.68.4.1967-1974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berleman J.E., Vicente J.J., Zusman D.R. FrzS regulates social motility in Myxococcus xanthus by controlling exopolysaccharide production. PLoS ONE. 2011;6:e23920. doi: 10.1371/journal.pone.0023920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marathe R., Meel C., Klumpp S. Bacterial twitching motility is coordinated by a two-dimensional tug-of-war with directional memory. Nat. Commun. 2014;5:3759. doi: 10.1038/ncomms4759. [DOI] [PubMed] [Google Scholar]
- 41.Galante A., Wisen S., Levy D. Modeling local interactions during the motion of cyanobacteria. J. Theor. Biol. 2012;309:147–158. doi: 10.1016/j.jtbi.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levy D., Requeijo T. Stochastic models for phototaxis. Bull. Math. Biol. 2008;70:1684–1706. doi: 10.1007/s11538-008-9314-5. [DOI] [PubMed] [Google Scholar]
- 43.Kondou Y., Mogami N., Manabe K. Bipolar localization of putative photoreceptor protein for phototaxis in thermophilic cyanobacterium Synechococcus elongatus. Plant Cell Physiol. 2002;43:1585–1588. doi: 10.1093/pcp/pcf176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


