Abstract
In the biosphere, sucrose is mainly synthesized in oxygenic photosynthetic organisms, such as cyanobacteria, green algae and land plants, as part of the carbon dioxide assimilation pathway. Even though its central position in the functional biology of plants is well documented, much less is known about the role of sucrose in cyanobacteria. In those prokaryotes, sucrose accumulation has been associated with salt acclimation, and considered as a compatible solute in low-salt tolerant strains. In the last years, functional characterizations of sucrose metabolizing enzymes, metabolic control analysis, cellular localization of gene expressions, and reverse genetic experiments have revealed that sucrose metabolism is crucial in the diazotrophic growth of heterocystic strains, and besides, that it can be connected to glycogen synthesis. This article briefly summarizes the current state of knowledge of sucrose physiological functions in modern cyanobacteria and how they might have evolved taking into account the phylogenetic analyses of sucrose enzymes.
Keywords: sucrose metabolism, compatible solutes, salt tolerance, glycogen, nitrogen fixation, signal molecule
1. Introduction
Cyanobacteria are among the most diverse groups of prokaryotic organisms that perform oxygenic photosynthesis. In a long evolutionary history, cyanobacterial diversification was one of the most important increases in physiological and morphological complexity of the prokaryotes [1]. As the result of their remarkable capacity to adapt to environmental changes by the acquisition of elaborate growth strategies [2], modern cyanobacteria exhibit a wide range of morphologies (i.e., unicellular to various multicellular organizations) and can be found occupying marine or freshwater aquatic environments or terrestrial ecosystems [2]. Particularly, the majority of free-living forms are abundant in waters with rapid and significant salinity fluctuations and water status [3]. To cope with these variations, cyanobacteria have developed salt acclimation mechanisms that involve the active extrusion of toxic ions and the accumulation of compatible solutes, such as sucrose, trehalose, glucosylglycerol, glucosylglycerate and glycine betaine [4]. The first recognized physiological function for sucrose was its salt stress-induced accumulation, which was well documented in the early 80s [5,6,7]. An extensive screening of cyanobacteria revealed that freshwater strains with rather low salt tolerance mainly accumulate sucrose. An additional role was proposed for sucrose as a carbon carrier molecule from the vegetative cell to the heterocyst in filamentous nitrogen-fixing strains [8]. However, a better understanding of its functions was gained after the functional identification of sucrose metabolism-related genes in unicellular and heterocyst-forming strains [9,10,11,12].
This review summarizes the current state of knowledge of sucrose roles in cyanobacteria, as a stress-response molecule, its relationship with glycogen metabolism, and most outstandingly, as a crucial molecule in filamentous heterocyst-forming strains.
2. Sucrose Metabolism in Cyanobacteria
2.1. Sucrose Enzymes
In contrast to plant enzymes [13,14], the proteins involved in sucrose metabolism in cyanobacteria, more recently described, have been studied to a lesser extent.
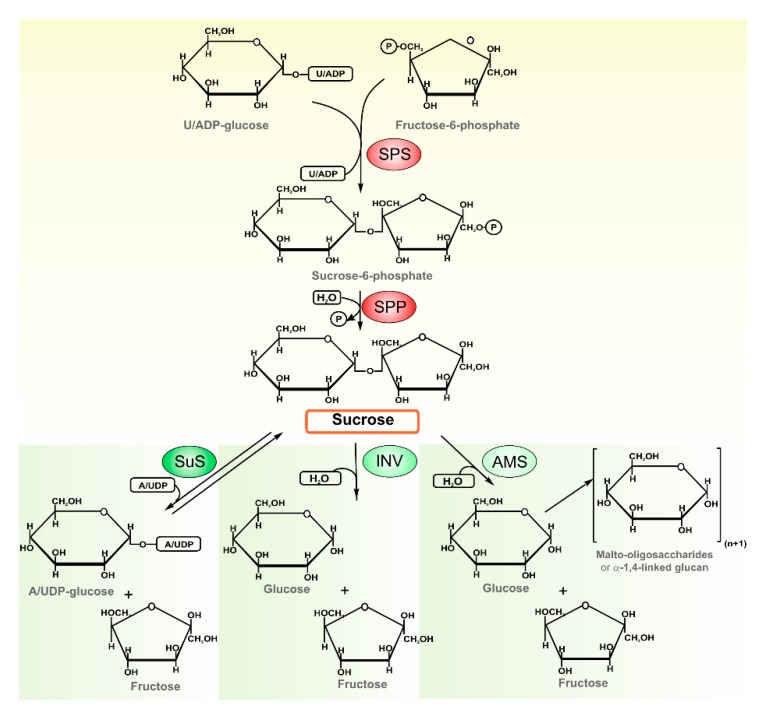
The identification and functional characterization of sucrose-synthesis related enzymes was first described in Anabaena sp. PCC 7119, a filamentous heterocyst-forming strain [15]. Further studies were carried out in other model cyanobacteria, including Synechocystis sp. PCC 6803 (a freshwater unicellular strain) [16,17], Anabaena sp. PCC 7120 [18,19], Synechococcus sp. PCC 7002 (unicellular marine strain) [10], Microcystis aeruginosa PCC 7806 (a bloom-forming strain) [11], and Synechococcus elongatus PCC 7942 [12]. Basically, for sucrose-biosynthesis, it was shown a similar route to that of plants involving the sequential action of sucrose-phosphate synthase (SPS, U/ADP-glucose: d-fructose-6-phosphate 2-α-d-glucosyltransferase, EC 2.4.1.14) and sucrose-phosphate phosphatase (SPP, sucrose-6F-phosphate-phosphohydrolase, EC 3.1.3.24), yielding free sucrose and Pi (Figure 1). Cyanobacterial SPSs display important biochemical differences in comparison with the orthologous plant proteins. Thus, SPSs are not specific for UDP–glucose and most SPSs can accept ADP–glucose and, to a minor extent, other sugar nucleotides as substrates [15,16,19]. The hydrolysis of the intermediate by SPP leads to an essentially irreversible pathway providing an efficient production of sucrose even at low substrate concentrations [9,13].
Figure 1.
Schematic representation of sucrose metabolism in cyanobacteria. Sucrose biosynthesis involves the sequential action of SPS and SPP, yielding free sucrose and inorganic phosphate. Cyanobacterial SPSs preferentially use ADP-glucose or UDP-glucose, as substrates. The disaccharide degradation can be carried out by the activities of the three different enzymes: (i) SuS that catalyzes a readily reversible reaction but that in vivo acts in the cleavage of sucrose, supplying ADP-glucose, a precursor for glycogen synthesis; however in vitro, SuS can also accept other sugar nucleotides (i.e., UDP) as substrate; (ii) A/N-Inv that irreversible hydrolyzes sucrose into glucose and fructose; and (iii) AMS that is able to catalyze not only sucrose hydrolysis to hexoses, but also to transfer the glucose moiety to a soluble maltooligosaccharide or to an insoluble α 1,4-glucan.
On the other hand, sucrose utilization depends on the activity of three different enzymes: (i) sucrose synthase (SuS, A/UDP-glucose: d-fructose 2-α-d-glucosyltransferase, EC 2.4.1.13) that catalyzes a ready reversible reaction, but that, in vivo, is primarily involved in sucrose cleavage to supply sugar-nucleotide molecules and fructose [14,20,21]; (ii) alkaline/neutral invertase (A/N-Inv, an α-glycosidase, no EC number assigned yet), grouped in the glycoside family 100, which irreversibly hydrolyze the disaccharide into glucose and fructose [22]; and (iii) amylosucrase (AMS, EC 2.4.1.4), a member of family 13 of the glycoside hydrolases, described in bacteria, and recently reported in Synechococcus sp. PCC 7002 [23]. AMS is able to hydrolyze sucrose to glucose and fructose, and/or to transfer the glucose moiety to a maltooligosaccharide or α 1,4-glucan (amylose-like polymers).
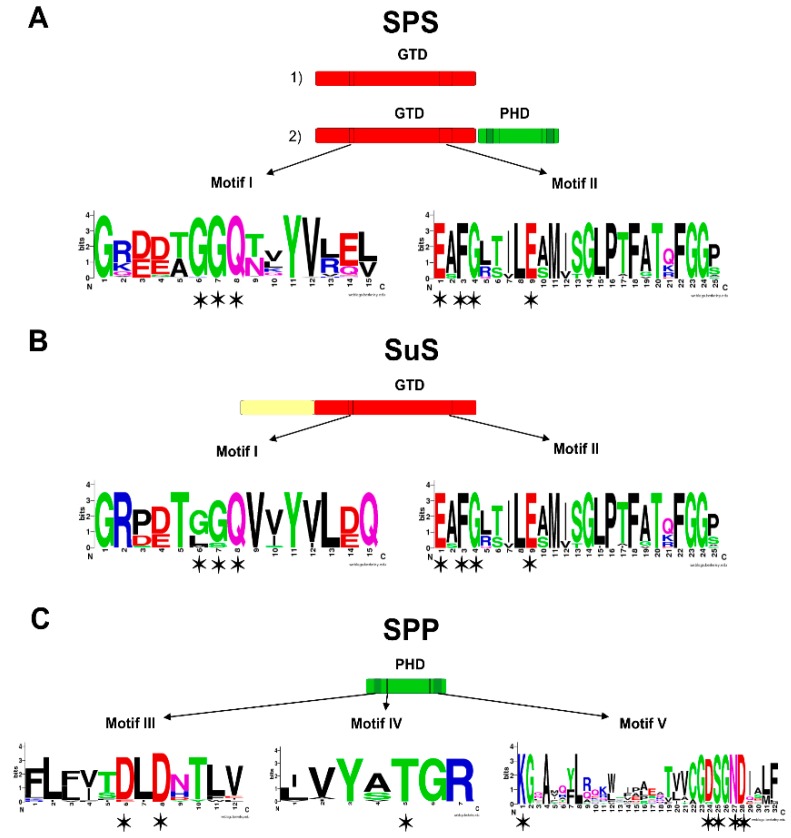
The functional characterization of the genes related to sucrose biosynthesis in unicellular and filamentous heterocyst-forming cyanobacteria have contributed to new insights into their structure, disclosing that SPS, SPP and SuS have a modular architecture [9]. The analysis of the two SPSs of Anabaena sp. PCC 7120 (SPS-A and SPS-B) uncovered an approximately 400 amino-acid region shared by all SPSs, allowing to define a functional glucosyltransferase domain (GTD) [19] (Figure 2). Similarly, the Anabaena SPP characterization [19] defined a phosphohydrolase domain (PHD) sharing conserved residues with other phosphohydrolases (Figure 2). SPSs support modularity since two different SPS domainal arrangements could be identified: the minimal SPS unit (GTD), coincidental with Anabaena SPSs, and the bidomainal SPS prototype (GTD-PHD), present in Synechocystis SPS, where the PHD is non-functional [19]. Additionally, the existence of bidomainal/bifunctional SPSs (exhibiting SPS and SPP activity) was demonstrated [12]. On the other hand, the analysis of SuS sequences also revealed that these proteins featured a GTD with a distinctive C-terminal extension [9].
Figure 2.
Domainal arrangements of sucrose-synthesis related proteins. SPS, SPP and SuS (sucrose-synthesis related proteins) are modular proteins based on a glycosyltransferase domain (GTD, red box) and a phosphohydrolase domain (PHD, green box) [9,19]. (A) Two domain arrangements have been described for cyanobacterial SPSs: (1) the minimal SPS unit (GTD), or unidomainal SPS; and (2) the two-domain SPS prototype (GTD-PHD) or bidomainal SPS; (B) SuS presents a GTD, with a characteristic N-terminal extension (yellow box). The resolution of the crystallographic structure of Halothermothrix orenii SPS (2r66A and 2r68A) [24], and of Arabidopsis thaliana SuS1 (3s28A) [25] allowed the identification of the residues involved in the sugar and in the NDP-glucose binding sites, within motif I and II, respectively (denoted with asterisks); (C) SPPs exhibit only a PHD. Motives III to V are characteristic of proteins grouped in the phosphohrydrolase superfamily and related to SPP activity. The crystallization Synechocystis sp. PCC 6803 SPP (1s2oA) led to the identification of the residues involved in the catalytic activity [26]. The critical residues were found within PHD motives (denoted with asterisks). Logos were constructed using the above mentioned conserved motives (WebLogo server [27]).
2.2. The Ancestral Origin of Sucrose Biosynthesis
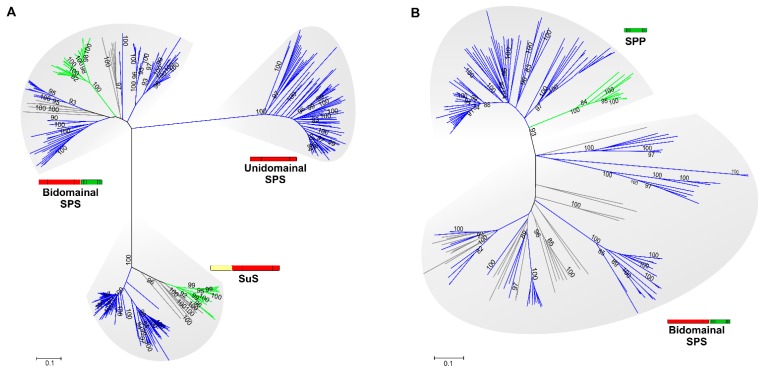
Earlier phylogenetic analyses based on GTD and PHD sequences revealed that sucrose biosynthesis proteins might have arisen from primordial functional domains shuffled during evolution [9], which was corroborated using sequences from 191 genomes available in May 2014 (Figure 3). The ancestral origin of sucrose metabolism postulated by Salerno and Curatti [9] was strongly supported by a recent study using ancestral sequence reconstruction coupled with phylogenetic analysis of sucrose synthesis genes [28]. In this report, it was hypothesized that sucrose synthesis in algae (chlorophytes and streptophytes) and land plants was likely inherited from cyanobacteria, and the chloroplast ancestor likely had the ability to synthesize sucrose [28,29]. However, sucrose metabolism genes were transferred to the nucleus, giving rise to a novel pathway in the plant lineage [9].
Figure 3.
Phylogenetic analysis of SPS, SPP and SuS proteins based on GTD and PHD sequences. Homologs were retrieved from public databases (JGI-DOE, http://www.jgi.doe.gov) by BLASTp searches using as query SPS and SuS of Anabaena sp PCC 7120, and SPS and SPP of Synechocystis sp. PCC 6803. Unrooted dendrograms were obtained using the maximum parsimony (1000 replicates). After sequence alignments, GTD (A) or PHD (B) regions described by Cumino et al. [19] were identified with ClustalW [30]. Trees were generated with the MEGA5 software [31]. Major groups are identified to give clues about their function, species, or taxonomic information: (A) GTDs corresponding to bidomainal and unidomainal SPSs and SuS; (B) PHDs, corresponding to SPP and to bidomainal SPSs. Cyanobacteria, blue lines; plants, green lines; bacteria, grey lines. Bootstrap results are not shown when values were lower than 90%.
Searches for homologs to sucrose-metabolism related genes revealed that they are not universal in modern strains (Table 1). While in most filamentous heterocyst-forming strains (Nostocales and Stigonematales), and in four out of six Pleurocapsales genomes, sucrose synthesis genes could be retrieved, they have lost in about 58% and 67% of the genomes of Chroococcales (unicellular strains) and Oscillatoriales (filamentous strains), respectively (Table 1). Blank [28] concluded that it is likely that multiple sucrose synthesis genes may have been present in the cyanobacterial ancestor, and that subsequent losses in many clades, gene duplication events in other groups (notably in Nostocales), and their regain by lateral gene transfer, might have occurred.
Table 1.
Occurrence of homologous sequences to SPS, SPP, SuS and A/N-Inv encoding genes present in cyanobacterial genomes. BLAST searches were carried out using as query Anabaena sp. PCC 7120 SPS (1), Synechocystis sp. PCC 6803 SPS (2), Anabaena sp. PCC 7120 SPP (3), SuS (4), A/N-Inv (5), and Synechococcus sp. PCC 7002 AMS (6).
| Order | Strain | Habitat | Place of origin | SPS (GTD) 1 | SPS (GTD-PHD) 2 | SPP 3 | SuS 4 | A/N-Inv 5 | AMS 6 |
|---|---|---|---|---|---|---|---|---|---|
| Chroococcales | Acaryochloris marina MBIC11017 | Marine | Pacific Ocean | + | - | + | + | + | - |
| Acaryochloris sp. CCMEE 5410 | Aquatic | USA | + | - | + | + | + | - | |
| Chamaesiphon minutus PCC 6605 | Aquatic | USA | 2+ | - | + | - | - | - | |
| Crocosphaera watsonii WH 0003 | Marine | Pacific Ocean | - | - | - | - | - | - | |
| Crocosphaera watsonii WH 0003 | Marine | Pacific Ocean | - | - | - | - | - | - | |
| Crocosphaera watsonii WH 0401 | Marine | Atlantic Ocean | - | - | - | - | - | - | |
| Crocosphaera watsonii WH 8501 | Marine | Atlantic Ocean | - | - | - | - | - | - | |
| Crocosphaera watsonii WH 8501 | Marine | Atlantic Ocean | - | - | - | - | - | - | |
| Cyanobacterium aponinum PCC 10605 | Freshwater | Italy | - | - | - | - | - | - | |
| Cyanobacterium sp. UCYN-A | Marine | Pacific Ocean | - | - | - | - | - | - | |
| Cyanobacterium stanieri PCC 7202 | Freshwater | Chad | - | - | - | - | + | - | |
| Cyanobium gracile PCC 6307 | Aquatic | USA | + | + | 2+ | - | - | + | |
| Cyanobium sp. PCC 7001 | Marine | USA | + | + | + | - | + | - | |
| Cyanothece sp. BH63E, ATCC 51472 | Aquatic | USA | - | - | + | - | - | - | |
| Cyanothece sp. BH68, ATCC 51142 | Marine | USA | - | - | + | - | - | - | |
| Cyanothece sp. CCY 0110 | Marine | Tanzania | - | - | + | - | - | - | |
| Cyanothece sp. PCC 7424 | Freshwater | Senegal | 2+ | - | + | + | + | - | |
| Cyanothece sp. PCC 7425 | Freshwater | Senegal | 2+ | - | + | 2+ | - | - | |
| Cyanothece sp. PCC 7822 | Freshwater | India | + | - | + | - | + | - | |
| Cyanothece sp. PCC 8801 | Freshwater | Taiwan | - | - | - | - | - | - | |
| Cyanothece sp. PCC 8802 | Freshwater | Taiwan | - | - | - | - | - | - | |
| Dactylococcopsis salina PCC 8305 | Aquatic | Israel | + | - | + | + | + | - | |
| Geminocystis herdmanii PCC 6308 | Aquatic | USA | - | - | - | - | - | - | |
| Gloeobacter kilaueensis JS1 | Freshwater | Hawaii | + | - | + | + | + | - | |
| Gloeobacter violaceus PCC 7421 | Freshwater | Switzerland | + | - | + | + | - | - | |
| Gloeocapsa sp. PCC 73106 | Freshwater | Switzerland | + | - | - | - | - | - | |
| Gloeocapsa sp. PCC 7428 | Freshwater | Sri Lanka | - | - | + | 2+ | + | - | |
| Halothece sp. PCC 7418 | Freshwater | Israel | 2+ | - | + | + | - | - | |
| Microcystis aeruginosa DIANCHI905/PCC7806 | Freshwater | China/ Netherlands | + | - | + | + | - | - | |
| Microcystis aeruginosa NIES-843 | Freshwater | Japan | - | - | - | - | - | - | |
| Microcystis aeruginosa PCC 7941 | Freshwater | Canada | - | - | - | - | - | - | |
| Microcystis aeruginosa PCC 9432 | Freshwater | Canada | - | - | - | - | - | - | |
| Microcystis aeruginosa PCC 9443 | Freshwater | Africa | - | - | - | - | - | - | |
| Microcystis aeruginosa PCC 9701 | Freshwater | France | - | - | - | - | - | - | |
| Microcystis aeruginosa PCC 9717 | Freshwater | France | - | - | - | - | - | - | |
| Microcystis aeruginosa PCC 9806 | Freshwater | USA | - | - | - | - | - | - | |
| Microcystis aeruginosa PCC 9807 | Freshwater | South Africa | - | - | - | - | - | - | |
| Microcystis aeruginosa PCC 9808 | Freshwater | Australia | - | - | - | - | - | - | |
| Microcystis aeruginosa PCC 9809 | Freshwater | USA | - | - | - | - | - | - | |
| Microcystis aeruginosa SPC777 | Freshwater | Brazil | - | - | - | - | - | - | |
| Microcystis aeruginosa TAIHU98 | Freshwater | China | - | - | - | - | - | - | |
| Microcystis sp. T1-4 | Freshwater | Thailand | - | - | - | - | - | - | |
| Prochlorococcus marinus AS9601 | Marine | Arabian Sea | + | - | - | - | - | - | |
| Prochlorococcus marinus CCMP 1375 | Marine | Atlantic Ocean | + | - | - | - | + | - | |
| Prochlorococcus marinus CCMP 1986 | Marine | Mediterranean Sea | + | - | - | - | + | - | |
| Prochlorococcus marinus MIT 9211 | Marine | Pacific Ocean | + | - | - | - | + | - | |
| Prochlorococcus marinus MIT 9215 | Marine | Pacific Ocean | + | - | - | - | + | - | |
| Prochlorococcus marinus MIT 9301 | Aquatic | Sargasso Sea | + | - | - | - | + | - | |
| Prochlorococcus marinus MIT 9303 | Aquatic | Sargasso Sea | - | + | - | - | + | - | |
| Prochlorococcus marinus MIT 9312 | Marine | Gulf Stream | + | - | - | - | + | - | |
| Prochlorococcus marinus MIT 9313 | Marine | Gulf Stream | - | + | - | - | + | - | |
| Prochlorococcus marinus MIT 9515 | Marine | Pacific Ocean | + | - | - | - | + | - | |
| Prochlorococcus marinus MIT9202 | Marine | Pacific Ocean | + | - | - | - | + | - | |
| Prochlorococcus marinus NATL1A | Marine | Atlantic Ocean | - | + | - | - | + | - | |
| Prochlorococcus marinus NATL2A | Marine | Atlantic Ocean | - | + | - | - | + | - | |
| Prochlorococcus sp. CC9311 | Marine | USA | - | + | - | - | + | - | |
| Prochlorococcus sp. CC9605 | Marine | USA | - | + | - | - | + | - | |
| Prochlorococcus sp. CC9902 | Marine | USA | - | + | - | - | + | - | |
| Prochlorococcus sp. W10 | Marine | Pacific Ocean | - | - | - | - | + | - | |
| Prochlorococcus sp. W11 | Marine | Pacific Ocean | - | - | - | - | + | - | |
| Prochlorococcus sp. W12 | Marine | Pacific Ocean | - | - | - | - | - | - | |
| Prochlorococcus sp. W2 | Marine | Pacific Ocean | + | - | - | - | - | - | |
| Prochlorococcus sp. W3 | Marine | Pacific Ocean | - | - | - | - | - | - | |
| Prochlorococcus sp. W4 | Marine | Pacific Ocean | - | - | - | - | + | - | |
| Prochlorococcus sp. W5 | Marine | Pacific Ocean | - | - | - | - | - | - | |
| Prochlorococcus sp. W6 | Marine | Pacific Ocean | - | - | - | - | - | - | |
| Prochlorococcus sp. W7 | Marine | Pacific Ocean | + | - | - | - | + | - | |
| Prochlorococcus sp. W8 | Marine | Pacific Ocean | + | - | - | - | + | - | |
| Prochlorococcus sp. W9 | Marine | Pacific Ocean | - | - | - | - | + | - | |
| Prochlorococcus sp. WH 7803 | Marine | Sargasso Sea | - | + | - | - | + | - | |
| Prochlorococcus sp. WH8102 | Marine | Atlantic Ocean | - | + | - | - | + | - | |
| Prochlorothrix hollandica PCC 9006 | Freshwater | Netherlands | - | - | - | - | - | - | |
| Rubidibacter lacunae KORDI 51-2 | Marine | Micronesia | + | - | + | + | - | - | |
| Synechococcus elongatus PCC 6301 | Freshwater | USA | - | + | - | - | + | - | |
| Synechococcus elongatus PCC 7942 | Freshwater | USA | - | + | - | - | + | - | |
| Synechococcus sp. BL107 | Marine | Mediterranean Sea | - | + | - | - | + | - | |
| Synechococcus sp. CB0101 | Marine | USA | - | + | + | - | + | - | |
| Synechococcus sp. CB0205 | Marine | USA | - | + | + | - | + | - | |
| Synechococcus sp. CC9616 | Marine | Pacific Ocean | - | - | - | - | + | - | |
| Synechococcus sp. JA-2-3B'a(2-13) | Freshwater | USA | - | - | + | - | + | - | |
| Synechococcus sp. JA-3-3Ab | Freshwater | USA | - | - | + | - | + | - | |
| Synechococcus sp. KORDI-100 | Marine | South Korea | + | - | - | - | + | -- | |
| Synechococcus sp. KORDI-49 | Marine | South Korea | + | - | - | - | + | - | |
| Synechococcus sp. KORDI-52 | Marine | South Korea | - | + | - | - | + | - | |
| Synechococcus sp. PCC 6312 | Marine | USA | - | + | - | - | + | - | |
| Synechococcus sp. PCC 7002 | Marine | Atlantic Ocean | - | + | - | - | - | + | |
| Synechococcus sp. PCC 7003 | Marine | USA | - | + | + | - | - | + | |
| Synechococcus sp. PCC 7117 | Marine | Asustralia | - | + | + | - | - | + | |
| Synechococcus sp. PCC 73109 | Marine | USA | - | + | + | - | - | + | |
| Synechococcus sp. PCC 7335 | Marine | Mexico | - | - | - | - | - | - | |
| Synechococcus sp. PCC 7336 | Marine | USA | - | - | - | - | - | - | |
| Synechococcus sp. PCC 7502 | Freshwater | Switzerland | + | - | + | - | + | - | |
| Synechococcus sp. PCC 8807 | Freshwater | Gabon | - | + | - | - | - | + | |
| Synechococcus sp. RCC 307 | Marine | Mediterranean Sea | + | - | + | - | + | - | |
| Synechococcus sp. RS9916 | Marine | Israel | + | + | - | - | + | - | |
| Synechococcus sp. RS9917 | Marine | Israel | - | + | - | - | + | - | |
| Synechococcus sp. WH 8016 | Marine | USA | - | + | - | - | + | - | |
| Synechococcus sp. WH 8109 | Marine | Sargasso Sea | - | + | - | - | + | - | |
| Synechococcus sp. WH5701 | Marine | USA | + | - | + | - | + | + | |
| Synechococcus sp. WH7805 | Marine | Sargasso Sea | - | + | - | - | - | - | |
| Synechocystis sp. PCC 6308 | Freshwater | USA | - | + | + | - | - | - | |
| Synechocystis sp. PCC 6803 | Freshwater | USA | - | + | + | - | + | - | |
| Synechocystis sp. PCC 7509 | Freshwater | Switzerland | - | - | + | + | + | - | |
| Thermosynechococcus elongatus BP-1 | Freshwater | Japan | - | + | + | + | - | - | |
| Oscillatoriales | Arthrospira maxima CS-328 | Freshwater | Chad | - | - | - | + | - | - |
| Arthrospira platensis C1 | Freshwater | Chad | - | - | - | + | - | - | |
| Arthrospira platensis NIES-39 | Freshwater | Chad | - | - | - | + | - | - | |
| Arthrospira platensis Paraca | Freshwater | Peru | - | - | - | + | - | - | |
| Arthrospira sp. PCC 8005 | Freshwater | India, Kenya, Mexico or Peru | - | - | - | 2+ | - | - | |
| Crinalium epipsammum PCC 9333 | Unknown | Unknown | 2+ | - | + | + | + | - | |
| Cyanobacterium sp. ESFC-1 | Freshwater | USA | 2+ | - | + | + | - | - | |
| Geitlerinema sp. PCC 7105 | Freshwater | USA | 2+ | - | + | + | - | - | |
| Geitlerinema sp. PCC 7407 | Freshwater | Unknown | + | - | 2+ | + | + | - | |
| Leptolyngbya boryana PCC 6306 | Freshwater | USA | 2+ | - | + | 2+ | 2+ | - | |
| Leptolyngbya sp. 2LT21S03 | Desert soil | Israel | - | - | + | - | - | - | |
| Leptolyngbya sp. PCC 6406 | Freshwater | USA | - | - | - | - | - | - | |
| Leptolyngbya sp. PCC 7375 | Freshwater | USA | - | - | - | - | - | - | |
| Leptolyngbya sp. PCC 7376 | Freshwater | USA | - | - | - | - | - | - | |
| Lyngbya majuscula 3L | Marine | Netherlands Antilles | + | - | + | 2+ | + | - | |
| Lyngbya sp. CCY 8106 | Freshwater | Germany | - | - | - | - | - | - | |
| Microcoleus chthonoplastes PCC 7420 | Marine | USA | + | - | + | + | + | - | |
| Microcoleus sp. PCC 7113 | Soil | USA | 4+? | - | + | 2+ | 2+ | - | |
| Microcoleus vaginatus FGP-2 | Desert soil | USA | + | - | + | + | - | - | |
| Microcoleus vaginatus PCC 9802 | Soil crusts | USA | + | - | + | + | - | - | |
| Nodosilinea nodulosa PCC 7104 | Soil | USA | - | - | - | - | - | - | |
| Oscillatoria acuminata PCC 6304 | Soil | USA | 2+ | + | + | + | 2+ | - | |
| Oscillatoria formosa PCC 6407 | Freshwater | USA | - | - | - | - | - | - | |
| Oscillatoria nigro-viridis PCC 7112 | Soil | USA | + | - | + | + | - | - | |
| Oscillatoria sp. PCC 10802 | Freshwater | Unknown | - | - | - | - | - | - | |
| Oscillatoria sp. PCC 6506 | Freshwater | Unknown | - | - | - | - | - | - | |
| Oscillatoriales sp. JSC-1 | Freshwater | USA | - | - | - | - | - | - | |
| Oscillatoriales sp. JSC-12 | Freshwater | USA | 2+ | - | + | + | 2+ | - | |
| Planktothrix agardhii NIVA-CYA 126/8 | Freshwater | Finland | - | - | - | - | - | - | |
| Planktothrix agardhii NIVA-CYA 34 | Freshwater | Norway | - | - | - | - | - | - | |
| Planktothrix agardhii NIVA-CYA 56/3 | Freshwater | Norway | - | - | - | - | - | - | |
| Planktothrix NIVA-CYA405 | Freshwater | Norway | - | - | - | - | - | - | |
| Planktothrix NIVA-CYA406 | Freshwater | Norway | - | - | - | - | - | - | |
| Planktothrix prolifica NIVA-CYA 540 (Draft1) | Freshwater | Norway | - | - | - | - | - | - | |
| Planktothrix rubescens NIVA-CYA 98 | Freshwater | Norway | - | - | - | - | - | - | |
| Planktothrix sp. 585 | Freshwater | Germany | - | - | - | - | - | - | |
| Planktothrix sp. NIVA CYA 15 | Freshwater | Norway | - | - | - | - | - | - | |
| Planktothrix sp. NIVA-CYA 407 | Freshwater | Norway | - | - | - | - | - | - | |
| Planktothrix sp. st147 | Freshwater | Germany | - | - | - | - | - | - | |
| Pseudanabaena sp. PCC 6802 | Freshwater | USA | + | - | + | + | + | - | |
| Pseudanabaena sp. PCC 7367 | Marine | USA | - | - | + | + | - | - | |
| Pseudanabaena sp. PCC 7429 | Freshwater | Switzerland | - | - | + | - | - | - | |
| Spirulina major PCC 6313 | Brackish water | USA | - | - | - | - | - | - | |
| Spirulina subsalsa PCC 9445 | Freshwater | Italy | - | - | - | - | - | - | |
| Trichodesmium erythraeum IMS101 | Marine | USA | + | - | - | - | - | - | |
| Pleurocapsales | Chroococcidiopsis sp. PCC 6712 | Freshwater | USA | - | - | + | - | - | - |
| Chroococcidiopsis thermalis PCC 7203 | Soil | Germany | 2+ | - | + | + | 2+ | - | |
| Pleurocapsa sp. PCC 7319 | Freshwater | Mexico | + | - | + | - | - | - | |
| Pleurocapsa sp. PCC 7327 | Freshwater | USA | + | - | + | 2+ | 2+ | - | |
| Stanieria cyanosphaera PCC 7437 | Freshwater | Cuba | 2+ | - | - | + | 2+ | - | |
| Xenococcus sp. PCC 7305 | Marine | USA | - | - | + | - | - | - | |
| Nostocales | Anabaena circinalis AWQC131C | Freshwater | Australia | + | - | + | - | - | - |
| Anabaena cylindrica PCC 7122 | Freshwater | United Kingdom | + | + | + | 2+ | + | - | |
| Anabaena sp. 90 | Freshwater | Finland | + | + | 2+ | + | + | - | |
| Anabaena sp. PCC 7108 | Freshwater | USA | + | + | + | + | + | - | |
| Anabaena variabilis ATCC 29413 | Freshwater | USA | 2+ | - | + | 2+ | + | - | |
| Calothrix desertica PCC 7102 | Sand | Chile | + | + | + | 2+ | 4+ | + | |
| Calothrix sp. PCC 6303 | Freshwater | USA | + | + | + | + | + | - | |
| Calothrix sp. PCC 7103 | Freshwater | USA | + | + | + | 2+ | 4+ | + | |
| Calothrix sp. PCC 7507 | Freshwater | Switzerland | 2+ | - | + | 2+ | + | - | |
| Cylindrospermopsis raciborskii CS-505 | Freshwater | Australia | + | - | + | - | + | - | |
| Cylindrospermopsis raciborskii CS-506 | Freshwater | Australia | + | - | + | - | + | - | |
| Cylindrospermopsis raciborskii CS-509 | Freshwater | Australia | + | - | + | - | + | - | |
| Cylindrospermum stagnale PCC 7417 | Soil | Sweden | + | + | + | 2+ | + | - | |
| Fremyella diplosiphon UTEX 481 | Freshwater | USA | - | + | 2+ | 3+ | 2+ | - | |
| Microchaete sp. PCC 7126 | Freshwater | USA | 2+ | + | 2+ | + | - | ||
| Nodularia spumigena CCY9414 | Brackish water | Denmark | + | + | + | 2+ | 2+ | - | |
| Nostoc azollae 0708 | Symbiont | Unkown | 2+ | - | + | + | + | - | |
| Nostoc punctiforme PCC 73102 | Freshwater | Australia | 2+ | - | 2+ | 2+ | 2+ | - | |
| Nostoc sp. PCC 7107 | Freshwater | USA | + | 2+ | 2+ | 2+ | - | ||
| Nostoc sp. PCC 7120 | Freshwater | USA | 2+ | - | + | 2+ | 2+ | - | |
| Nostoc sp. PCC 7524 | Freshwater | Sri Lanka | + | + | + | 2+ | 2+ | - | |
| Raphidiopsis brookii D9 | Freshwater | Brazil | - | - | - | - | - | - | |
| Rivularia sp. PCC 7116 | Freshwater | USA | + | - | + | 2+ | + | - | |
| Scytonema hofmanni PCC 7110 | Freshwater | Bermuda | 2+ | - | + | 2+ | 2+ | - | |
| Scytonema hofmanni UTEX 2349 | Freshwater | USA | + | - | + | 2+ | + | - | |
| Stigonematales | Chlorogloeopsis fritschii PCC 6912 | Freshwater | India | + | - | + | 2+ | 2+ | - |
| Chlorogloeopsis fritschii PCC 7702 | Soil | India | + | - | + | 2+ | - | ||
| Chlorogloeopsis sp. PCC 9212 | Freshwater | Spain | + | - | + | 2+ | 2+ | - | |
| Fischerella muscicola PCC 7414 | Freshwater | New Zealand | + | - | 2+ | 3+ | 2+ | - | |
| Fischerella muscicola SAG 1427-1 | Freshwater | India | - | - | - | 2+ | + | - | |
| Fischerella sp. JSC-11 | Freshwater | USA | + | - | + | 2+ | + | - | |
| Fischerella sp. PCC 9339 | Freshwater | Unknown | + | - | + | - | 2+ | - | |
| Fischerella sp. PCC 9431 | Freshwater | Unknown | 2+ | - | 2+ | + | + | - | |
| Fischerella sp. PCC 9605 | Freshwater | Israel | + | - | 2+ | 3+ | 2+ | - | |
| Fischerella thermalis PCC 7521 | Freshwater | USA | + | - | + | 3+ | + | - | |
| Mastigocladopsis repens PCC 10914 | Soil | Spain | 2+ | - | + | 2+ | + | - | |
| Mastigocoleus testarum BC008 | Marine | Puerto Rico | + | - | + | + | 2+ | - |
Regarding sucrose breakdown, homologs to SuS and A/N-Inv encoding genes are present in about 31% and 52%, respectively, of the genomes analyzed. In Chroococcales, there is a clear predominance of A/N-Inv for sucrose degradation with some particular exceptions (e.g., Microcystis strains). In contrast, homologs to SuS encoding genes could be retrieved mostly from genomes of heterocyst-forming cyanobacteria, and from a very few genomes of unicellular strains. Such is the case of the Chroococcales strains: Gloebacter violaceus PCC 7421, Microcystis aeruginosa PCC 7806, and Thermosynechococcus elongatus BP-1, where SuS encoding genes were functionally characterized [11]. The ancient origin of SuS in the cyanobacteria [9,28] is likely followed by gene duplications in Nostocales and several secondary gains in other strains. Thus, the occurrence of SuS in G. violaceus, which is the most deeply rooted cyanobacterium, seems to be a more recent lateral gene transfer event [11,28]. The fixation and long term persistence suggest that SuS might confer a selective advantage [32], such as the contribution to the ADP-glucose pool, which will be discussed in Section 3.2. In addition, the increase in SuS transcripts in M. aeruginosa and G. violaceus cells under hypoxic conditions led to speculate that this enzyme could be involved in the response to low oxygen conditions in some strains, which was also found in Arabidopsis [11,33].
It should be underlined that several Synechococcus and Prochloroccocus genomes only have bidomainal SPS (GTD-PHD) gene sequences, and lack an independent SPP sequence. As mentioned above, bidomainal SPSs with conserved key catalytic residues in the PHD module (e.g., S. elongatus PCC 7942) [12], could exhibit both SPS and SPP activity.
Regarding gene locations in cyanobacterial genomes, sucrose synthesis-related genes are separately located on different regions of the chromosome. However, sucrose transcriptional units were found in a few strains, such as in Synechococcus sp. PCC 7002 [10,23] and Microcystis aeruginosa [34]. These “sucrose clusters” are transcriptional units that contain sequences coding for SPS and SPP, as well as for sucrose breakdown proteins (SuS or AMS).
3. Sucrose Roles in Cyanobacteria
3.1. Sucrose as a Compatible Solute
Salinity is a key abiotic factor in aquatic ecosystems. In response to changes in external salt concentration, cyanobacteria have developed different protective mechanisms to maintain internal osmotic potential and to cope with the rise of cell ion concentration [35]. The two basic physiological responses for salt acclimation include the extrusion of toxic inorganic anions and the accumulation of organic compounds of low molecular mass, so-called compatible solutes for not interfering with cell metabolism. These osmolytes reduce the internal osmotic cell potential and prevent the denaturation of macromolecules induced by low water or high ionic concentrations [4]. Also, these molecules were shown to protect the cell from other types of desiccation, as well as from cold and heat stress [35,36,37].
Among cyanobacteria, a correlation between the nature of the principal organic solute and the strain-specific salt resistant level has been established [38]. In general, freshwater strains with low halotolerance (up to 0.7 M·NaCl), accumulate disaccharides such as sucrose and/or trehalose, as their major compatible solute, while cyanobacteria of moderate salt tolerance (up to 1.7 M·NaCl) synthesize mainly glucosylglycerol and glucosylglycerate. In some of these strains, sucrose can also be produced as a minor osmolyte [35,39]. Finally, halophilic strains tolerate up to 3 M·NaCl, and accumulate glycine betaine and glutamate betaine as compatible solutes [4].
The origin of the different osmolytes has been investigated by ancestral sequence reconstruction and phylogenetic analysis of the genes underlying salinity preferences in cyanobacteria [28]. This study concludes that the emergence of sucrose synthesis is likely to be ancestral in cyanobacteria, and associated with growth in a low salinity environment. Other osmolytes (glucosylglycerol, glucosylglycerate and glycine betaine) have emerged afterwards. The conclusions drawn also support the hypothesis of the freshwater origin of cyanobacteria followed by subsequent independent divergences into the marine environment.
As an osmoprotectant compound, sucrose (a polyhydroxyl molecule) can directly interact with macromolecules to achieve their stabilization through a mechanism accounted by the water replacement hypothesis, i.e., by replacing at least part of the shell water around macromolecules [40]. The relevance of sucrose accumulation in salt acclimation was first shown in Nostoc muscorum, Synechococcus sp. PCC 6301 and Anabaena variabilis [5,41,42]. To date, the presence of sucrose as the main compatible osmolyte has been reported in many other freshwater strains [39] as well as in marine picocyanobacteria of the genera Prochlorococcus and Synechococcus [39].
Increase in sucrose accumulation in response to salt is due to higher SPS expression, as shown in filamentous nitrogen-fixing and unicellular strains [11,43,44,45]. For example, in Anabaena sp. PCC 7119 and 7120 cells, a short-term NaCl treatment resulted in a three-fold increase of SPS activity, which paralleled the rise of its polypeptide and transcript level [46,45]. Similar results were obtained in Synechocystis sp. PCC 6803 and M. aeruginosa PCC 7806 [34,47]. Interestingly, in the marine strain Synechococcus sp. PCC 7002, the addition of 684 mM·NaCl increased the expression of SPS and SPP encoding genes organized in a transcriptional unit (sucrose cluster), whose promoter region contains a consensus motif characteristic of osmotic and salt-activated genes [10]. Recently, in Anabaena sp. PCC 7120, it has been shown that OrrA, a NarL-type response regulator [48], is necessary to induce the genes involved in sucrose synthesis in response to salt stress [49].
Remarkably, after salt addition, not only the expression of sucrose synthesis proteins but also that of sucrose degradation enzymes increased in Anabaena sp. filaments, and in M. aeruginosa sp. PCC 7806 and G. violaceus PCC 7421 cells. Such effect on both, sucrose synthesis and breakdown, could be ascribed to a “sucrose cycling”, as reported by Cumino et al. [11,34,45].
It is worth noticing that, in addition to sucrose, novel compatible solutes (soluble polymers named sucroglucans) have been recently identified in Anabaena sp. PCC 7119, PCC 7120, and A. variabilis. These oligosaccharides reversibly accumulate in the first hours after exposure to NaCl [46], and constitute a series of non-reducing sucrose derivatives, where glucose is linked, through its hemiacetalic hydroxyl, to the 2 position of the glucose moiety of sucrose. The members of the sucroglucan series should be included in the repertoire of osmolytes synthesized in response to salt in low halotolerance filamentous heterocyst-forming strains [50].
However, what is more surprising is that sucrose appears to have a more intricate function than to being an osmolyte according to experiments reported for Synechocystis sp. PCC 6803. In this unicellular moderately halotolerant strain, the dominating osmoprotective compound is glucosylglycerol and sucrose was considered as a secondary osmolyte, either in salt-adapted or salt-shocked cells [35,44,51]. However, by examining the time-course of sucrose production in cells subject to a salt stress, a transient accumulation of the disaccharide was shown at the onset of treatment [47]. Intracellular sucrose concentration increased immediately after the NaCl shock, reaching its peak between 4 h and 6 h after the onset, and decreasing sharply in the following 20 h up to approximately the initial level. In line with that result, the steady-state amount of SPS gene transcripts increased very quickly after the salt shock, reaching a maximum after 30 min and returning to the initial levels during the following hour. It should be stressed that sucrose production kinetics contrasts with the typical accumulation pattern of an osmolyte like glucosylglycerol. Moreover, the analysis of a Synechocystis mutant strain impaired in sucrose synthesis showed that sucrose presence was essential in stationary phase cells to overcome a subsequent salt stress, but that it was dispensable for growth under standard conditions. In few words, Desplats et al. [47] accounted for the first demonstration of sucrose playing a role other than being a compatible osmolyte for salt tolerance and suggested that the disaccharide was likely acting as a signal molecule.
3.2. Sucrose and Nitrogen Fixation
Many cyanobacteria are able to simultaneous and independently produce photosynthetic molecular oxygen and fix atmospheric nitrogen (N2). Certainly, nitrogen fixation into ammonia is an energetically expensive process always linked to carbohydrate metabolism [52]. Because nitrogenase, the enzymatic system for N2 fixation, is inactivated upon exposure to oxygen, cyanobacteria have evolved different adaptations that include either temporal or spatial separation of the two processes [53]. Particularly, under aerobiosis and combined-nitrogen withdrawn, cyanobacteria clustered in phylogenetically-coherent groups of filamentous strains (Nostocales and Stigonematales) are able to differentiate a photosynthetic (vegetative) cell into a specialized cell called heterocyst, through a variety of structural, biochemical, and genetic changes, allowing the nitrogenase to be active [54,55,56]. Heterocysts, distributed in a semiregular pattern along the filaments [57], contain the oxygen-sensitive enzyme complex nitrogenase and lack photosystem II activity and ribulose-1,5-diphosphatecarboxylase (RuBisCo), a key enzyme in CO2 fixation during photosynthesis. Consequently, heterocysts are limited to a heterotrophic metabolism and depend on vegetative cells for the generation of carbon skeletons and reducing power [58,59,60].
Despite the fact that several attempts have been made to elucidate the carrier molecule/s responsible for transporting the reduced carbon from vegetative cells to heterocysts, it remains to be precisely identified. Several carbohydrates, including fructose, erythrose and sucrose, have been suggested as possible carriers [8,59,61]. Sucrose, as a transport molecule, was proposed based on SuS and invertase activities measured in Anabaena variabilis cell extracts [8]. However, this transport was not demonstrated and the function of SuS was shown to be sucrose cleavage [20,62].
The critical role of sucrose in carbon flux modulation in the nitrogen-fixing filaments of Anabaena sp. was undoubtedly proven in subsequent studies. Curatti et al. [63] showed that diazotrophic growth was impaired in an Anabaena mutant strain overexpressing the SuS encoding gene (susA), in which the disaccharide was not detectable. These results support the SuS involvement in the control of carbon flux in vegetative cells through the cleavage of sucrose. Indeed, it was a clear demonstration that SuS preferentially catalyzes the cleavage of the disaccharide in vivo. Afterwards, it was shown in heterocysts that the expression of SuS and RuBisCo is similarly down-regulated by a nitrogen source-dependent developmental program [62].
The analysis of complete sequenced genomes indicates that most heterocyst-forming strains have at least two homologs to SPS genes and two homologs to A/N-Inv genes (Table 1). The existence of SPS-A and SPS-B, and Inv-A and Inv-B in Anabaena sp. PCC 7120 [15,19,64] has raised the question of whether each isoenzyme could play distinct roles in the nitrogen-fixing filaments. Analyses of cellular localizations of the isoforms, phenotypes of insertional mutants, and studies on the transcriptional regulation of the sucrose protein encoding genes, have provided conclusive evidence supporting the role of the disaccharide as an intermediate in the reduced carbon flux along the N2-fixing filaments. Furthermore, it was shown that while both SPSs and both A/N-Inv contribute to sucrose metabolism in vegetative cells, only SPS-B and Inv-B are active in heterocysts [20,65,66]. These enzymes are likely to be part of a sucrose cycling inside the heterocyst, playing essential functions in carbon–nitrogen balance. Cumino et al. [20] proposed that a sucrose cycling mechanism may be operating in the heterocysts, allowing cell metabolism to shift easily from sucrose production to degradation through A/N-Inv, hexokinase, hexose-P mutase, hexose-P isomerase, ADP-glucose pyrophosphorylase (AGPase, a key enzyme involved in glycogen synthesis), SPS-B, and sucrose-phosphate phosphatase activities. Even though the role of glycogen in nitrogen fixation has been studied for several decades [67,68,69,70,71,72], the interconnection between glycogen metabolism and sucrose in nitrogen-fixing filaments was demonstrated by the integration of results of expression and metabolic flux analyses of sucrose metabolism enzymes. This allowed proposing that a sucrose cycling is linked to glycogen metabolism and respiration [20]. According to sucrose metabolic network modeling in nitrogen-fixing Anabaena filaments, AGPase flux alone (calculated by metabolic simulation) was insufficient to supply the substrate (ADP-glucose) for glycogen and sucrose synthesis. Then, it was proposed that sucrose cleavage by SuS in the vegetative cells would contribute to the ADP-glucose pool [20]. Moreover, SuS was shown to be involved in the sucrose to insoluble polysaccharides conversion according to nutritional and environmental signals in Anabaena filamentous under diazotrophic growth [21].
The coordination at the transcriptional level of sucrose metabolism with nitrogen assimilation was supported by experimental evidence indicating that NtcA (a global nitrogen regulator in cyanobacteria, required for the expression of proteins subject to ammonium repression [73]) also regulates sucrose metabolism genes in Anabaena sp. PCC 7120. NtcA acts as a transcriptional activator of the encoding genes of SPS-B and Inv-B (both proteins located into heterocyst), and as an inhibitor of SuS encoding gene [65,66,74]. NtcA is required to maintain a high sucrose biosynthesis and a low rate of the disaccharide cleavage in the vegetative cells. Also, in the heterocyst, it regulates both sucrose synthesis by SPS-B and its hydrolysis by Inv-B. In mutants where the Inv-B encoding gene (invB) was knocked out, the filaments were unable to grow on diazotrophic conditions and the accumulation of sucrose and glycogen was altered [65]. These results demonstrate an essential role for Inv-B for diazotrophic growth and that Inv-B plays a key part in the coordination of sucrose and glycogen metabolism. It appears that NtcA integrates signals from carbon and nitrogen metabolism, and regulates gene expression accordingly to redirect metabolism as a function of the carbon/nitrogen status of the cell. Therefore, an expanded role as a global metabolism regulator was proposed for NtcA [74].
4. Final Remarks
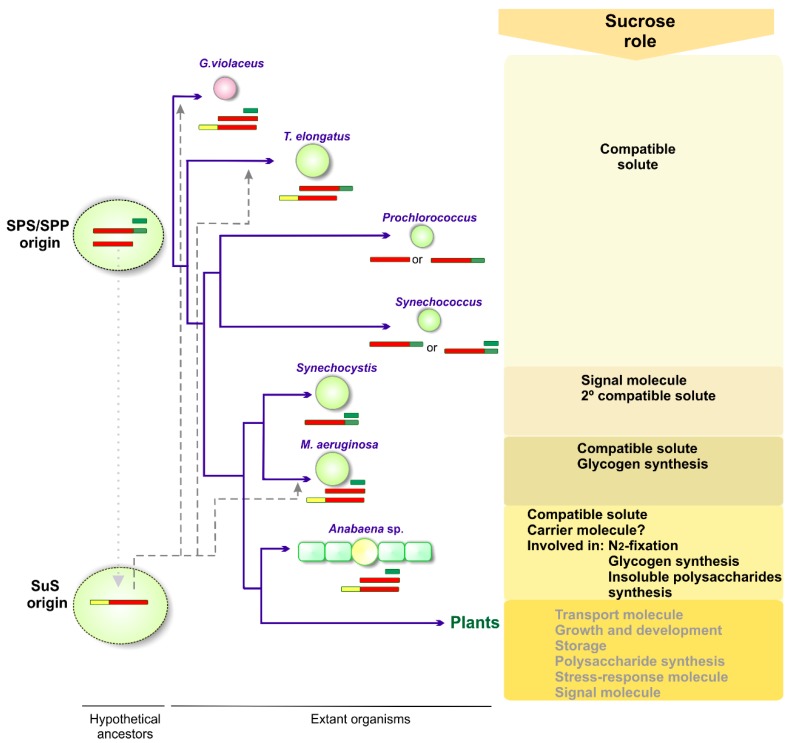
In the last two decades, a comprehensive set of data has contributed to reveal that sucrose, aside from being a compatible solute in response to salinity, can play other crucial roles in the life of many cyanobacteria. Figure 4 summarizes the current state of knowledge of sucrose physiological functions in modern cyanobacteria and how they are related to cyanobacterial phylogeny and to the occurrence of sucrose synthesis proteins.
Figure 4.
Schematic representation of sucrose roles along the hypothetical evolutionary pathway of cyanobacteria. The phylogenetic relationships among species are depicted according to rDNA 16S sequence analysis. Sucrose metabolism is likely to be originated in freshwater habitat and multiple sucrose synthesis genes might have been present in a cyanobacterial ancestor [27]. Sucrose synthesis is found in G. violaceus that has ancestral characteristics and diverged early within the radiation of cyanobacteria. A fusion of primordial GTD and PHD might have given rise to a hypothetical common-ancestral SPS (GTD-PHD) gene, which is found mostly in the marine Prochlorococcus/Synechococcus clade. Sucrose has been identified as a primary compatible solute in Prochlorococcus, and as secondary osmolyte in Synechococcus strains and in Synechocystis sp. PCC 6803 [36]. The involvement of sucrose in glycogen and polysaccharides production seems to be due to the emergence of SuS (dotted line), crucial in filamentous heterocyst-forming strains [20,21], as well as in strains (such as G. violaceus, Thermosynechococcus elongatus and Microcystis aeruginosa PCC 7806), where SuS are likely to be acquired by lateral gene transfer (dashed lines). In heterocystic strains, sucrose is a key molecule during nitrogen fixation and it was proposed as a carrier molecule to transport carbon along the filament. It is also involved in glycogen synthesis and in other polysaccharide accumulation. Plant sucrose metabolism has been acquired during the endosymbiotic origin of the chloroplast at the time of the cyanobacterial phylogenetic radiation.
The emergence of sucrose as a compatible solute is likely to be ancestral and to have occurred in freshwater strains where cyanobacteria might have originated and the synthesis of the disaccharide was sufficient to withstand low salinity environments. The subsequent acquisition of the synthesis of new compatible solutes by unicellular strains conferred them moderate tolerance to salinity, and it is likely that sucrose was shifted into a secondary role as osmolyte. This would be the case of Synechocystis sp. PCC 6803, where glucosylglycerol is the main osmolyte. However, in this strain a new function for sucrose was made evident. Whether the low and transient accumulation of sucrose (typical of a signal molecule) in Synechocystis could be a more general feature in cyanobacteria should be further explored.
In addition to its role as a stress-response molecule, in filamentous nitrogen-fixing strains, sucrose metabolism is crucial for the heterocyst function, in glycogen accumulation, and in the flux of carbon between sucrose and polysaccharides.
Despite the fact that the capability of sucrose synthesis is likely to be ancestral and mostly universal in extant cyanobacteria, it seems not to be essential for the survival of many strains, as it was lost in many clades and mutants impaired in sucrose synthesis could be isolated. However, the acquisition of sucrose synthesis genes by lateral transfer could point to some adaptive advantage to occupy new ecological niches.
We wonder which might have been the selective advantage of sucrose that led to such an evolutionary choice in the cyanobacterial lineage.
Acknowledgments
This work was supported by the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, PIP 134), Universidad Nacional de Mar del Plata, and Fundación para Investigaciones Biológicas Aplicadas (FIBA).
Author Contributions
G. L. Salerno and M. A. Kolman performed literature research, planned the organization and layout, and wrote the manuscript. C. N. Nishi and M. Perez-Cenci contributed to the writing of part of the manuscript. All authors have read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Rippka R., Deruelles J., Waterbury J.B., Herdman M., Stanier R.Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1979;111:1–61. doi: 10.1099/00221287-111-1-1. [DOI] [Google Scholar]
- 2.Whitton B.A., Potts M. Introduction to the cyanobacteria. In: Whitton B.A., editor. Ecology of Cyanobacteria II. Springer; Dordrecht, The Netherlands: 2012. pp. 1–13. [Google Scholar]
- 3.Reed R.H., Stewart W.D.P. The responses of cyanobacteria to salt stress. In: Rogers L.J., Gallon J.R., editors. Biochemistry of the Algae and Cyanobacteria. Clarendon Press; Oxford, UK: 1988. pp. 217–231. Chapter 12. [Google Scholar]
- 4.Hagemann M. Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol. Rev. 2011;35:87–123. doi: 10.1111/j.1574-6976.2010.00234.x. [DOI] [PubMed] [Google Scholar]
- 5.Blumwald E., Tel-Or E. Osmoregulation and cell composition in salt-adaptation of Nostoc. Muscorum. Arch. Microbiol. 1982;132:168–172. doi: 10.1007/BF00508725. [DOI] [Google Scholar]
- 6.Reed R.H., Richardson D.L., Warr S.R.C., Stewart W.D.P. Carbohydrate accumulation and osmotic stress in cyanobacteria. J. Gen. Microbiol. 1984;130:1–4. [Google Scholar]
- 7.Reed R.H., Chudek J.A., Foster R., Stewart W.D.P. Osmotic adjustment in cyanobacteria from hypersaline environments. Arch. Microbiol. 1984;138:333–337. doi: 10.1007/BF00410900. [DOI] [Google Scholar]
- 8.Schilling N., Ehrnsperger K. Cellular differentiation of sucrose metabolism in Anabaena variabilis. Z. Naturforsch. 1985;40:776–779. [Google Scholar]
- 9.Salerno G.L., Curatti L. Origin of sucrose metabolism in higher plants: When, how and why? Trends Plant Sci. 2003;8:63–69. doi: 10.1016/S1360-1385(02)00029-8. [DOI] [PubMed] [Google Scholar]
- 10.Cumino A.C., Perez-Cenci M., Giarrocco L.E., Salerno G.L. The proteins involved in sucrose synthesis in the marine cyanobacterium Synechococcus sp. PCC 7002 are encoded by two genes transcribed from a gene cluster. FEBS Lett. 2010;584:4655–4660. doi: 10.1016/j.febslet.2010.10.040. [DOI] [PubMed] [Google Scholar]
- 11.Kolman M.A., Torres L.L., Martin M.L., Salerno G.L. Sucrose synthase in unicellular cyanobacteria and its relationship with salt and hypoxic stress. Planta. 2012;235:955–964. doi: 10.1007/s00425-011-1542-5. [DOI] [PubMed] [Google Scholar]
- 12.Martinez-Noel G., Cumino A.C., Kolman M.A., Salerno G.L. First evidence of sucrose biosynthesis by single cyanobacterial bimodular proteins. FEBS Lett. 2013;587:1669–1674. doi: 10.1016/j.febslet.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 13.Winter H., Huber S.C. Regulation of sucrose metabolism in higher plants: Localization and regulation of activity of key enzymes. Crit. Rev. Biochem. Mol. Biol. 2000;35:253–289. doi: 10.1080/10409230008984165. [DOI] [PubMed] [Google Scholar]
- 14.Koch K. Sucrose metabolism: Regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr. Opin. Plant. Biol. 2004;7:235–246. doi: 10.1016/j.pbi.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 15.Porchia A.C., Salerno G.L. Sucrose biosynthesis in a prokaryotic organism: Presence of two sucrose-phosphate synthases in Anabaena with remarkable differences compared with the plant enzymes. Proc. Natl. Acad. Sci. USA. 1996;93:13600–13604. doi: 10.1073/pnas.93.24.13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curatti L., Folco E., Desplats P., Abratti G., Limones V., Herrera-Estrella L., Salerno G. Sucrose-phosphate synthase from Synechocystis sp. strain PCC 6803: Identification of the spsA gene and characterization of the enzyme expressed in Escherichia coli. J. Bacteriol. 1998;180:6776–6779. doi: 10.1128/jb.180.24.6776-6779.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lunn J.E., Price G.D., Furbank R.T. Cloning and expression of a prokaryotic sucrose-phosphate synthase gene from the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 1999;40:297–305. doi: 10.1023/A:1006130802706. [DOI] [PubMed] [Google Scholar]
- 18.Cumino A., Ekeroth C., Salerno G.L. Sucrose-phosphate phosphatase from Anabaena sp. strain PCC 7120: Isolation of the protein and gene revealed significant structural differences from the higher-plant enzyme. Planta. 2001;214:250–256. doi: 10.1007/s004250100608. [DOI] [PubMed] [Google Scholar]
- 19.Cumino A., Curatti L., Giarrocco L., Salerno G.L. Sucrose metabolism: Anabaena sucrose-phosphate synthase and sucrose-phosphate phosphatase define minimal functional domains shuffled during evolution. FEBS Lett. 2002;517:19–23. doi: 10.1016/S0014-5793(02)02516-4. [DOI] [PubMed] [Google Scholar]
- 20.Cumino A.C., Marcozzi C., Barreiro R., Salerno G.L. Carbon cycling in Anabaena sp. PCC 7120. Sucrose synthesis in the heterocysts and possible role in nitrogen fixation. Plant Phys. 2007;143:1385–1397. doi: 10.1104/pp.106.091736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curatti L., Giarrocco L.E., Cumino A.C., Salerno G.L. Sucrose synthase is involved in the conversion of sucrose to polysaccharides in filamentous nitrogen-fixing cyanobacteria. Planta. 2008;228:617–625. doi: 10.1007/s00425-008-0764-7. [DOI] [PubMed] [Google Scholar]
- 22.Vargas W.A., Salerno G.L. The Cinderella story of sucrose hydrolysis: Alkaline/neutral invertases, from cyanobacteria to unforeseen roles in plant cytosol and organelles. Plant Sci. 2010;178:1–8. doi: 10.1016/j.plantsci.2009.09.015. [DOI] [Google Scholar]
- 23.Perez-Cenci M., Salerno G.L. Functional characterization of Synechococcus amylosucrase and fructokinase encoding genes discovers two novel actors on the stage of cyanobacterial sucrose metabolism. Plant Sci. 2014;224:95–102. doi: 10.1016/j.plantsci.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 24.Chua T.K., Bujnicki J.M., Tan T.C., Huynh F., Patel B.K., Sivaraman J. The structure of sucrose phosphate synthase from Halothermothrix orenii reveals its mechanism of action and binding mode. Plant Cell. 2008;20:1059–1072. doi: 10.1105/tpc.107.051193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng Y., Anderson S., Zhang Y., Garavito R.M. The structure of sucrose synthase-1 from Arabidopsis thaliana and its functional implications. J. Biol. Chem. 2010;286:36108–36118. doi: 10.1074/jbc.M111.275974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fieulaine S., Lunn J.E., Borel F., Ferrer J. The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell. Plant Cell. 2005;17:2049–2058. doi: 10.1105/tpc.105.031229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. Weblogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blank C.E. Phylogenetic distribution of compatible solute synthesis genes support a freshwater origin for cyanobacteria. J. Phycol. 2013;49:880–895. doi: 10.1111/jpy.12111. [DOI] [PubMed] [Google Scholar]
- 29.Nagao M., Uemura M. Sucrose phosphate phosphatase in the green alga Klebsormidium flaccidum (Streptophyta) lacks an extensive C-terminal domain and differs from that of land plants. Planta. 2012;235:851–861. doi: 10.1007/s00425-011-1550-5. [DOI] [PubMed] [Google Scholar]
- 30.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acid Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. Mega5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koonin E.V., Makarova K.S., Aravind L. Horizontal gene transfer in prokaryotes: Quantification and classification. Ann. Rev. Microbiol. 2001;55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barratt D.H.P., Derbyshire P., Findlay K., Pike M., Wellner N., Lunn J., Feil R., Simpson C., Maule A.J., Smith A.M. Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proc. Natl. Acad. Sci. USA. 2009;106:13124–13129. doi: 10.1073/pnas.0900689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolman M.A., Salerno G.L. Sucrose in bloom-forming cyanobacteria: Gain and loss of genes involved in its biosynthesis. Environ. Microbiol. 2015 doi: 10.1111/1462-2920.13071. submitted for publication. [DOI] [PubMed] [Google Scholar]
- 35.Hagemann M., Erdmann N. Environmental stresses. In: Rai A.K., editor. Cyanobacterial Nitrogen Metabolism and Environmental Biotechnology. Springer Verlag Narosa Publishing House; New Delhi, India: 1997. pp. 155–221. [Google Scholar]
- 36.Oren A. Salts and Brines. In: Whitton B.A., editor. Ecology of Cyanobacteria II. Springer; Dordrecht, The Netherlands: 2012. pp. 401–426. [Google Scholar]
- 37.Higo A., Katoh H., Ohmori K., Ikeuchi M., Ohmori M. The role of a gene cluster for trehalose metabolism in dehydration tolerance of the filamentous cyanobacterium Anabaena sp. PCC 7120. Microbiology. 2006;152:979–987. doi: 10.1099/mic.0.28583-0. [DOI] [PubMed] [Google Scholar]
- 38.Reed R.H., Stewart W.D.P. Osmotic adjustment and organic solute accumulation in unicellular cyanobacteria from freshwater and marine habitats. Mar. Biol. 1985;88:1–9. doi: 10.1007/BF00393037. [DOI] [Google Scholar]
- 39.Klahn S., Hagemann M. Compatible solute biosynthesis in cyanobacteria. Environ. Microbiol. 2011;13:551–562. doi: 10.1111/j.1462-2920.2010.02366.x. [DOI] [PubMed] [Google Scholar]
- 40.Potts M. Desiccation tolerance: A simple process? Trends Microbiol. 2001;9:553–559. doi: 10.1016/S0966-842X(01)02231-4. [DOI] [PubMed] [Google Scholar]
- 41.Blumwald E., Mehlhorn R.J., Packer L. Studies of osmoregulation in salt adaptation of cyanobacteria with ESR spin-probe techniques. Proc. Natl. Acad. Sci. USA. 1983;80:2599–2602. doi: 10.1073/pnas.80.9.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Erdmann N. Organic osmoregulatory solutes in blue-green algae. Z. Pflanzenphysiol. 1983;110:147–155. doi: 10.1016/S0044-328X(83)80161-5. [DOI] [Google Scholar]
- 43.Page-Sharp M., Behm C.A., Smith G.D. Involvement of the compatible solutes trehalose and sucrose in the response to salt stress of a cyanobacterial Scytonema species isolated from desert soils. Biochim. Biophys. Acta. 1999;1472:519–528. doi: 10.1016/S0304-4165(99)00155-5. [DOI] [PubMed] [Google Scholar]
- 44.Hagemann M., Marin K. Salt-induced sucrose accumulation is mediated by sucrose-phosphate-synthase in cyanobacteria. J. Plant. Phys. 1999;155:424–430. doi: 10.1016/S0176-1617(99)80126-6. [DOI] [Google Scholar]
- 45.Salerno G.L. (Instituto de Investigaciones en Biodiversidad y Biotecnología (INBIOTEC-CONICET) and Fundación para Investigaciones Biológicas Aplicadas (FIBA), Mar del Plata, Argentina). Personal Communication. 2014.
- 46.Salerno G.L., Porchia A.C., Vargas W., Abdian P.L. Fructose-containing oligosaccharides: Novel compatible solutes in Anabaena cells exposed to salt stress. Plant Sci. 2004;167:1003–1008. doi: 10.1016/j.plantsci.2004.05.029. [DOI] [Google Scholar]
- 47.Desplats P., Folco E., Salerno G.L. Sucrose may play an additional role to that of an osmolyte in Synechocystis sp. PCC 6803 salt-shocked cells. Plant Phys. Biochem. 2005;43:133–138. doi: 10.1016/j.plaphy.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 48.Schwartz S.H., Black T.A., Jager K., Panoff J.-M., Wolk C.P. Regulation of an osmoticum-responsive gene in Anabaena sp. strain PCC 7120. J. Bacteriol. 1998;180:6332–6337. doi: 10.1128/jb.180.23.6332-6337.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ehira S., Kimura S., Miyazaki S., Ohmori M. Sucrose synthesis is controlled by the two-component response regulator OrrA in the nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. App. Environ. Microbiol. 2014:5672–5679. doi: 10.1128/AEM.01501-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pontis H.G., Vargas W.A., Salerno G.L. Structural characterization of the members of a polymer series, compatible solutes in Anabaena cells exposed to salt stress. Plant Sci. 2007;172:29–35. doi: 10.1016/j.plantsci.2006.07.007. [DOI] [Google Scholar]
- 51.Miao X., Wu Q., Wu G., Zhao N. Sucrose accumulation in salt-stressed cells of agp gene deletion mutant in cyanobacterium Synechocystis sp. PCC 6803. FEMS Microbiol. Lett. 2003;218:71–77. doi: 10.1111/j.1574-6968.2003.tb11500.x. [DOI] [PubMed] [Google Scholar]
- 52.Ludden P., Barris R.H. Biochemical basis of plant breeding. In: Neyra C.A., editor. Nitrogen Metabolism. Volume 2. CRC Press; Boca Raton, FL, USA: 1986. pp. 41–59. [Google Scholar]
- 53.Berman-Frank I., Lundgren P., Falkowski P. Nitrogen fixation and photosynthetic oxygen evolution in cyanobacteria. Res. Microbiol. 2003;154:157–164. doi: 10.1016/S0923-2508(03)00029-9. [DOI] [PubMed] [Google Scholar]
- 54.Buikema W.J., Haselkorn R. Isolation and complementation of nitrogen fixation mutants of the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 1991;173:1879–1885. doi: 10.1128/jb.173.6.1879-1885.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolk P.C. Heterocyst formation in Anabaena. In: Brun Y.V., Shimkets L.J., editors. Prokaryotic Development. American Society for Microbiology; Washington, DC, USA: 2000. pp. 83–104. [Google Scholar]
- 56.Yoon H.-S., Golden J.W. PatS and products of nitrogen fixation control heterocyst pattern. J. Bacteriol. 2001;183:2605–2613. doi: 10.1128/JB.183.8.2605-2613.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Golden J.W., Yoon H.-S. Heterocyst development in Anabaena. Curr. Opin. Microbiol. 2003;6:557–563. doi: 10.1016/j.mib.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 58.Wolk C.P. Movement of carbon from vegetative cells to heterocysts in Anabaena cylindrica. J. Bacteriol. 1968;96:2138–2143. doi: 10.1128/jb.96.6.2138-2143.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolk C.P., Ernst A., Elhai J. Heterocyst metabolism and development. In: Whitton B.A., Potts M., editors. The Molecular Biology of Cyanobacteria. Springer; Dordrecht, The Netherlands: 2004. pp. 769–823. [Google Scholar]
- 60.Zhang C.C., Laurent S., Sakr S., Peng L., Bedu S. Heterocyst differentiation and pattern formation in cyanobacteria: A chorus of signals. Mol. Biol. 2006;59:367–375. doi: 10.1111/j.1365-2958.2005.04979.x. [DOI] [PubMed] [Google Scholar]
- 61.Privalle L.S., Burris R.H. Adenine nucleotide levels in and nitrogen fixation by the cyanobacterium Anabaena sp. strain 7120. J. Bacteriol. 1983;154:351–355. doi: 10.1128/jb.154.1.351-355.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Curatti L., Giarrocco L., Salerno G.L. Sucrose synthase and RuBisCo expression is similarly regulated by the nitrogen source in the nitrogen-fixing cyanobacterium Anabaena sp. Planta. 2006;223:891–900. doi: 10.1007/s00425-005-0142-7. [DOI] [PubMed] [Google Scholar]
- 63.Curatti L., Flores E., Salerno G. Sucrose is involved in the diazotrophic metabolism of the heterocyst-forming cyanobacterium Anabaena sp. FEBS Lett. 2002;513:175–178. doi: 10.1016/S0014-5793(02)02283-4. [DOI] [PubMed] [Google Scholar]
- 64.Vargas W., Cumino A., Salerno G.L. Cyanobacterial alkaline/neutral invertases. Origin of sucrose hydrolysis in the plant cytosol? Planta. 2003;216:951–960. doi: 10.1007/s00425-002-0943-x. [DOI] [PubMed] [Google Scholar]
- 65.Vargas W.A., Nishi C.N., Giarrocco L.E., Salerno G.L. Differential roles of alkaline/neutral invertases in Nostoc sp. PCC 7120: Inv-B isoform is essential for diazotrophic growth. Planta. 2011;233:153–162. doi: 10.1007/s00425-010-1288-5. [DOI] [PubMed] [Google Scholar]
- 66.Lopez-Igual R., Flores E., Herrero A. Inactivation of a heterocyst-specific invertase indicates a principal role of sucrose catabolism in heterocysts of Anabaena sp. J. Bacteriol. 2011;192:5526–5533. doi: 10.1128/JB.00776-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fay P. Factors influencing dark nitrogen fixation in a blue-green alga. App. Environ. Microbiol. 1976;31:376–379. doi: 10.1128/aem.31.3.376-379.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lockau W., Peterson R.B., Wolk C.P., Burris R.H. Modes of reduction of nitrogenase in heterocysts isolated from Anabaena species. Biochimica et Biophysica Acta (BBA)—Bioenergetics. 1978;502:298–308. doi: 10.1016/0005-2728(78)90051-8. [DOI] [PubMed] [Google Scholar]
- 69.Ernst A., Boger P. Glycogen accumulation and the induction of nitrogenase activity in the heterocyst-forming cyanobacterium Anabaena variabilis. J. Gen. Microbiol. 1985;131:3147–3153. [Google Scholar]
- 70.Ernst A., Reich S., Boger P. Modification of dinitrogenase reductase in the cyanobacterium Anabaena variabilis due to C starvation and ammonia. J. Bacteriol. 1990;172:748–755. doi: 10.1128/jb.172.2.748-755.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vargas W., Martin M.L., Salerno G. Myths and Facts on Cytosolic Sucrose Hydrolysis. In: Magazu S., editor. Sucrose: Properties, Biosynthesis and Health Implications. Nova Science Publishers Inc.; Hauppauge, NY, USA: 2013. pp. 155–176. [Google Scholar]
- 72.Jensen B.B., Cox R.P., Burris R.H. Isolation of cyanobacterial heterocysts with high and sustained dinitrogen-fixation capacity supported by endogenous reductants. Arch. Microbiol. 1986;145:241–247. doi: 10.1007/BF00443652. [DOI] [PubMed] [Google Scholar]
- 73.Luque I., Forchhammer K. Nitrogen Assimilation and C/N Balance Sensing. In: Herrero A., Flores E., editors. The Cyanobacteria: Molecular Biology, Genomics and Evolution. Caister Academic Press; Norfolk, UK: 2008. pp. 335–382. [Google Scholar]
- 74.Marcozzi C., Cumino A.C., Salerno G.L. Role of NtcA, a cyanobacterial global nitrogen regulator, in the regulation of sucrose metabolism gene expression in Anabaena sp. PCC 7120. Arch. Microbiol. 2009;191:255–263. doi: 10.1007/s00203-008-0450-y. [DOI] [PubMed] [Google Scholar]