Abstract
Zoonotic viruses, such as HIV, Ebola virus, coronaviruses, influenza A viruses, hantaviruses, or henipaviruses, can result in profound pathology in humans. In contrast, populations of the reservoir hosts of zoonotic pathogens often appear to tolerate these infections with little evidence of disease. Why are viruses more dangerous in one species than another? Immunological studies investigating quantitative and qualitative differences in the host-virus equilibrium in animal reservoirs will be key to answering this question, informing new approaches for treating and preventing zoonotic diseases. Integrating an understanding of host immune responses with epidemiological, ecological, and evolutionary insights into viral emergence will shed light on mechanisms that minimize fitness costs associated with viral infection, facilitate transmission to other hosts, and underlie the association of specific reservoir hosts with multiple emerging viruses. Reservoir host studies provide a rich opportunity for elucidating fundamental immunological processes and their underlying genetic basis, in the context of distinct physiological and metabolic constraints that contribute to host resistance and disease tolerance.
Zoonotic viruses are emerging from the wild and causing serious disease in humans. Understanding how host reservoirs are able to tolerate the infection and spread the virus is a key element to informing new approaches for treating and preventing zoonotic diseases.
Main Text
Introduction
Emerging infectious diseases have an enormous impact on human health (Marston et al., 2014). Viruses account for a significant proportion of emerging infections, and the majority have zoonotic origins, including ebolaviruses, human immunodeficiency virus (HIV), hantaviruses, Hendra and Nipah viruses, severe acute respiratory syndrome (SARS) coronavirus, and influenza A viruses (Jones et al., 2008, Taylor et al., 2001). Transmission can occur directly to people from live reservoir hosts (e.g., bat shedding of Nipah virus into date palm collection vessels [Luby et al., 2006]). In other instances, exposures to novel viruses have been associated with the butchering of reservoir hosts, such as bush meat in SIV or simian foamy virus transmission (Hahn et al., 2000, Wolfe et al., 2004) and, recently, in the index case of the ebolavirus outbreak in the Democratic Republic of Congo (WHO, 2014). Alternatively, transmission can be facilitated by intermediate hosts (e.g., Nipah virus infection of pigs in Malaysia resulting in pig-to-pig and pig-to-human transmission by aerosol [Parashar et al., 2000]) or can be transferred via insect vectors, as is the case for Dengue fever and West Nile virus (Mackenzie and Jeggo, 2013). Substantial growth in size and mobility of human populations, along with environmental and climate changes, and the spread of agricultural practices promoting human-animal contact has led to an increased frequency of pathogen emergence and potential for rapid dissemination (Karesh et al., 2012). Novel viruses are being described that cause disease in humans, such as the recently identified Middle East respiratory syndrome coronavirus (MERS-CoV) associated with acute respiratory illness and renal failure (Zaki et al., 2012). Other zoonotic viruses continue to spread into new populations, as is the case for a current outbreak of Ebola virus in western Africa, where this virus was not previously documented and where it is having an unprecedented societal, economic, and public health impact (Pandey et al., 2014, WHO Ebola Response Team, 2014).
Tools to rapidly detect and sequence novel viruses have greatly improved in recent years, facilitating their detection and diagnosis in humans (Marston et al., 2014) and simplifying the identification of putative reservoir hosts. For instance, the origins of Ebola virus, although first identified in 1976, were only recently tied to bats (Biek et al., 2006, Pigott et al., 2014). These tools are enabling initiatives to monitor viruses in wildlife populations in their natural habitat before they emerge in humans and other animals (Mokili et al., 2012, Morse et al., 2012). Ecological, epidemiological, and evolutionary processes involved in the introduction and spread of pathogens in novel host populations are the subject of intensive research (Antia et al., 2003, Holmes and Drummond, 2007, Woolhouse et al., 2012). However, there is little understanding of the within-host immunological processes underlying reservoir host-virus interactions, and this issue is rarely addressed in studies of emerging viral diseases. Yet within-host processes are ultimately critical in determining the outcome of infection, the balance between limiting infection-associated pathology and clearing the virus, and therefore the likelihood of transmission.
Upon cross-species jumps, viruses can result in severe or fatal disease in the novel, non-natural hosts, while these same viruses often appear to cause only mild infections in their reservoir hosts. Examples exist in which circulating viruses can be lethal in reservoir host populations, as is the case for rabies virus in bat populations. But even rabies virus mortality rates were recently estimated using epidemiological models as being much lower in their natural bat reservoir (∼10%) than in other mammals (Blackwood et al., 2013). As we will discuss, there have been few detailed direct studies of the pathogenesis of emerging viruses in their natural wildlife hosts, and given the notorious difficulty of measuring mortality rates in wildlife populations, some reservoir host populations could be affected to a greater degree than we currently understand. Longitudinal studies may reveal fitness costs, even when the symptoms of infection are far less pronounced than what is observed in non-natural human hosts. One documented example is simian immunodeficiency virus in chimpanzees (SIVcpz), the viral precursor of HIV-1 in humans. SIVcpz leads to detectable depletion of CD4+ T cells and is associated with shorter life spans and reduced reproductive success of wild chimpanzees (Keele et al., 2009). Although SIVcpz infection is less pathogenic in chimpanzees than HIV-1 in humans, it is not apathogenic. Whether this is related to the more recent acquisition of SIV by chimpanzees as compared to the older association of SIV with other African nonhuman primates is still unclear (Bailes et al., 2003). In contrast, SIV infection of sooty mangabeys and African green monkeys, two natural reservoir hosts of SIV, despite a high prevalence in the wild and persistent high-level viral replication, has no detectable impact on host survival or health (Keele et al., 2009, Pandrea and Apetrei, 2010, Silvestri et al., 2003). In fact, in many cases, available observations suggest that zoonotic viruses are able to persist in natural reservoir populations for significant periods of time with no overt signs of pathology.
Different viral zoonoses affecting humans are characterized by distinct biological and clinical manifestations, with many of them displaying a significant immunopathological component. Here, we argue that elucidating the nature of immune responses in individual natural hosts may inform our understanding of how virus-host equilibria are established without substantially impacting host health. Furthermore, this may provide insight into the mechanisms of disease pathogenesis and immunity in humans. We ask whether there are fundamental immunologic properties that govern different infection outcomes of natural reservoir species compared to non-natural human hosts while acknowledging that other aspects of host physiology, behavior, or population biology may also play fundamentally important roles. Further, we examine whether there are any immunological principles underlying the association of specific reservoir host species with multiple emerging viruses. Eventually, characterizing the host-pathogen interface in key reservoir hosts may enable us to predict which novel viruses are more likely to pose threats to human health.
Pathogenesis of Emerging Viral Infections in Humans
The pathogenic mechanisms underlying disease manifestations arising in humans infected with specific emerging viruses—whether acute or chronic in nature—are diverse, complex, and incompletely understood. Infection outcomes may vary from resolving with only mild symptoms (e.g., simian foamy virus [Wolfe et al., 2004]) to rapid developing severe disease that is either fatal or cleared (e.g., ebolaviruses), whereas others are persistent and lead to disease only after a prolonged period (e.g., HIV). The balance between protective and pathogenic immune responses is critical (Rouse and Sehrawat, 2010). A pivotal role in this balance is played by innate immune effectors that detect the presence of viral products by pattern recognition receptors, thus initiating the host response (Iwasaki, 2012). The specific cytokines, chemokines, and lipid mediators produced in response to immune activation, as well as downstream adaptive responses that themselves can modulate the innate response, can skew the relative balance between aggressive responses that rapidly clear infection and responses that minimize the extent of immunopathology. Exuberant innate immune activation can also play a direct role in precipitating host organ tissue damage (Cameron et al., 2008, Kuiken et al., 2012).
Despite a range of virus-specific mechanisms for pathogenesis, some common themes exist. In emerging viral infections causing disease in humans in whom host responses have been studied, innate immune responses are often thought to underlie severe disease manifestations. In the acute respiratory infections with SARS CoV and influenza A, excessive innate immune activation causes local tissue damage and compromises the generation of protective adaptive immune responses (Kash et al., 2006, Peiris et al., 2010). During infection with highly pathogenic influenza A viruses, production of inflammatory cytokines and chemokines responsible for the recruitment of neutrophils and mononuclear cells to the site of infection contributes to the severity of the pulmonary disease and systemic complications (Kuiken et al., 2012, Peiris et al., 2010). In addition, other mediators of inflammation, including sphingosine phosphates, also play an important role in the immunopathogenesis of influenza tissue damage and disease (Oldstone and Rosen, 2014). Specific virus strains such as H5N1 and the 1918 H1N1 viruses are particularly potent in activating high-level sustained proinflammatory cytokine production following infection (Kash et al., 2006, Peiris et al., 2010). Similarly, the pathogenesis of SARS involves infection of important cell populations in the lung, along with the elicitation of high levels of a range of inflammatory mediators (Totura and Baric, 2012). Importantly, in individuals who resolve SARS CoV infections, evidence of active innate responses wane, and effective adaptive responses develop that clear the infection. By contrast, individuals with poor outcomes exhibit persistently elevated type I interferon (IFN) production, IFN-stimulated gene (ISG) expression, and chemokine production in association with impaired antiviral antibody responses (Cameron et al., 2008, Totura and Baric, 2012).
For emerging viruses that cause hemorrhagic fever, dysregulation of the host response, the induction of inflammatory mediators, and the impairment of adaptive immunity, in addition to direct viral damage of host tissues, are central to pathogenesis (Geisbert and Jahrling, 2004). In fatal human filovirus infections (of which Ebola virus is the best studied), antigen-presenting cells, including dendritic cells and macrophages, represent important targets for virus infection in vivo. High levels of proinflammatory cytokines, chemokines, and tissue factor lead to lymphocyte apoptosis, pathologic activation of coagulation cascades, and widespread compromise of vascular integrity that results in multiorgan failure and a septic shock-like syndrome (Martinez et al., 2012, Misasi and Sullivan, 2014, Zampieri et al., 2007). In infections (for instance, with West Nile virus) in which encephalitis due to viral penetration of the blood-brain barrier leads to neuropathology, inflammatory responses may be responsible for blood-brain barrier compromise (Wang et al., 2004).
In HIV infection, a persistent yet ineffective host response in the face of chronic viral replication contributes substantially to pathogenesis. Erosion of immunocompetence was initially thought to arise from active virus replication causing progressive depletion of CD4+ T cell populations. However, more recent evidence indicates that chronic, pleiotropic immune activation (evidenced by increased activation, proliferation, apoptosis, and dysfunction across diverse immune effector cell populations) during HIV infection is the major driver of progressive immune deficiency leading to AIDS (Moir et al., 2011). A key contributor to chronic immune activation in HIV is the ability of unrelenting virus replication to stimulate host innate immune responses and the production of type I IFNs and other proinflammatory cytokines (Miedema et al., 2013). The resulting compromised infection resistance may further amplify systemic immune activation and exacerbate its damaging effects by resulting in the expansion of the enteric virome, damaging the intestinal epithelium and leading to the translocation of intestinal bacteria, viruses, and the antigens derived from them (Brenchley et al., 2006, Handley et al., 2012).
Of note, in many instances, viral proteins produced by emerging viruses antagonize specific immune recognition mechanisms or circumvent cell-intrinsic restriction factors, thereby influencing host antiviral responses and viral replication kinetics. Given the central role of type I IFN in the direct inhibition of viral replication within cells and in the activation and execution of host innate and adaptive immune responses, it is not surprising that diverse viral gene products have been shown to specifically block the induction or the effects of the host IFN response. Ebolavirus VP35 and VP24, lassa virus NP, influenza virus NS1, SARS CoV NSP1, and ORF3b are just a few examples (Ayllon and Garcia-Sastre, 2015, Hastie et al., 2012, Totura and Baric, 2012). Viral products have also been demonstrated to intercept ISGs, other immune recognition pathways, restriction factors, or production of effector molecules (Menachery et al., 2014). Features of the virion, such as the complex structure and heavy glycosylation of the HIV-1 envelope protein gp120, can make them less effective targets for elicitation of neutralizing antibody responses or structurally impervious to them when they arise (Burton et al., 2005). Moreover, the viral genome itself plays a role in the rapid evasion of adaptive immune responses, with the generation of diverse viral quasispecies that is an inherent consequence of error-prone RNA replication mechanisms (Holmes and Drummond, 2007). Thus, the efficacy of the host response to infection depends on the complex interplay of beneficial activation of innate and adaptive immune responses, the potency and impact of viral strategies for immune evasion, and the deleterious consequences of protracted or misdirected immune responses.
Reservoir Host Infection and Disease Tolerance
A number of nonmutually exclusive hypotheses might explain why many reservoir hosts of viruses capable of causing disease in humans or other animal hosts do not show severe clinical or behavioral signs of infection (Table 1 ): (1) the virus is cytopathic in the nonnatural host, but not in its reservoir, (2) viral tropism differs between natural and non-natural hosts, (3) differences in the interaction between the viral genome or viral gene products and host resistance mechanisms alter the infection outcome between natural and non-natural hosts, (4) the virus confers a benefit to the host that is not conferred to humans or other animal host species, (5) interactions within the zoonotic host between the virus and other elements of the microbiome alter the pathogenesis of infection in a manner that does not apply to humans, (6) reservoir host responses control viral replication more effectively, or (7) viral infection is better tolerated by reservoir hosts even when viral loads are high. In most cases, knowledge about the features of viral infections in their reservoir hosts and immune responses elicited is incomplete, and it remains unclear which of these explanations holds for particular zoonotic viruses (Table 1). Moreover, as-yet-undiscovered mechanisms may exist which the study of reservoir hosts could reveal.
Table 1.
Features of Emerging Viruses in the Natural Reservoir Hosts from which They Originated
| Virusa (Family) | Genome | Pathogenesis in Humans | Natural Host | Features of Infection and Immune Response in Natural Host |
||||
|---|---|---|---|---|---|---|---|---|
| Virus Replication | Innate Response | Ab/B Cell | T Cell | Treg | ||||
| Bas-Congo (Rhabdoviridae) | −ssRNA | acute hemorrhagic fever | ? | ? | ? | ? | ? | ? |
| MERS Coronavirus (Coronaviridae) | +ssRNA | acute pneumonia, renal failure | bats, camels | ? | ? | ? | ? | ? |
| Chikungunya (Togaviridae) | +ssRNA | high fever, skin rash, arthralgia | African primates | ? | ? | ? | ? | ? |
| Crimean-Congo hemorrhagic fever (Bunyaviridae) | −ssRNA | hemorrhagic fever | hares, large herbivores? | ? | ? | + | ? | ? |
| Ebola (Filoviridae) | −ssRNA | hemorrhagic fever | fruit bats | ? | ? | + | ? | ? |
| Hanta (Bunyaviridae) | −ssRNA | hemorrhagic fever with renal syndrome, cardiopulmonary syndrome | rodents, shrews, and bats | P | anti-inflammatory | + | (+) | + |
| Hendra and Nipah (Paramyxoviridae) | −ssRNA | severe acute encephalitis, respiratory disease, systemic vasculitis | fruit bats | ? | ? | + T/P | ? | ? |
| Hepatitis E (Hepeviridae) | +ssRNA | hepatitis | pigs | ? | ? | ? | ? | ? |
| HIV (Retroviridae) | −ssRNA | AIDS | African primates | P (high) | diminished chronic type I IFN | (+) (low titres) | (+) | + |
| Influenza A (Orthomyxoviridae) | −ssRNA | respiratory disease | aquatic birds | T? | variations in innate signaling pathways | + | ? | ? |
| bats? | ||||||||
| Lassa (Arenaviridae) | −ssRNA | mild febrile illness but can result in hemorrhagic fever | rodents | P | ? | + | ? | ? |
| Lymphocytic choriomeningitis (Arenaviridae) | −ssRNA | mild febrile illness to meningeal symptoms; fatal in immunocompromised | rodentsb | P, T? | ? | + | ? | ? |
| Menangle (Paramyxoviridae) | −ssRNA | influenza-like illness and rash | fruit bats | ? | ? | + | ? | ? |
| SARS coronavirus (Coronaviridae) | +ssRNA | progressive atypical pneumonia | horseshoe bats | ? | ? | + | ? | ? |
| Rabies (Rhabdoviridae) | −ssRNA | neurological disease | bats | T? | ? | + T? | ? | ? |
| Rift Valley fever (Bunyaviridae) | −ssRNA | hemorrhagic fever | ? | ? | ? | ? | ? | ? |
| West Nile (Flaviviridae) | +ssRNA | fever, meningoencephalitis | birds? | ? | ? | + | ? | ? |
| Yellow fever (Flaviviridae) | +ssRNA | hemorrhagic fever, jaundice | African primates | T | ? | + | ? | ? |
T, transient; P, persistent; ?, unknown;+, present; –, undetectable; (+), present but weak/low compared to non-natural hosts.
The list is not exhaustive but highlights representative examples of emerging viral infections. See also Table S1 for a complete list of references.
Note that, although LCMV infection of inbred laboratory mice is an extremely well-studied pathogen-host system and a lot is known about the immunological responses during chronic and acute LCMV infections, very little is known about LCMV infection in wild rodents, circulating strains, whether virus is cleared or persistent, and how much this depends on the rodent species and virus strain in question.
In some cases, differences in viral tropism may influence the host-virus interface, resulting in reduced pathogenesis. For instance, in the sooty mangabey, a natural host for the SIV that is the origin of HIV-2, long-lived memory T cells may be more resistant to infection due to reduced CCR5 expression (a coreceptor for SIV) such that T cell homeostasis is preserved (Paiardini et al., 2011). Similarly, memory CD4+ T cells in African green monkeys downmodulate CD4 expression, rendering them resistant to SIV infection and to infection-induced dysregulation of their homeostasis (Beaumier et al., 2009).
Alternatively, the role of viral genetic functions and structural attributes in shaping the nature and consequences of host-virus interactions may be distinct in natural versus non-natural hosts. Interestingly, most studies thus far have used non-natural hosts or their cells to explore the interaction of viral products with host resistance mechanisms and impact on viral virulence. In fact, much of our current understanding of viral immune evasion and restriction factor inhibition strategies has come from studies of emerging viruses. However, although these strategies can be fit in a logical framework in the context of a pathogenic infection, it is not clear how such regulatory and structural mechanisms function or evolved within a nonpathogenic reservoir host environment, whether they promote virus persistence or limit pathologic consequences of infection, and how they impact cross-species jumps. For example, the type I IFN-inducible host cell restriction factor tetherin, identified as a cellular factor that acts to restrict the release of newly produced HIV-1 particles from the surface of human cells, is antagonized by the HIV-1 accessory protein Vpu. Of the four groups of HIV-1 (M, N, O, and P) that arose via independent transmission events from chimpanzees, only group M viruses have become pandemic—the group shown to possess the most potent anti-tetherin activities (Sauter, 2014). Tetherin also inhibits the egress of arenavirus, filovirus, and rhabdovirus particles from infected cells, and proteins such as Ebola virus GP1,2 counteract the effects of tetherin (Misasi and Sullivan, 2014, Sauter, 2014). But whether these have a similar role in their reservoir hosts remains to be examined.
Innate and adaptive immune cell development and their activation thresholds are calibrated by interactions with commensals or previous pathogen encounters. Thus, it is possible that the overall microbial environment of a reservoir host species contributes to altering the outcome of a viral infection. For example, helminth infection can either reactivate herpesviruses from latency through generation of cytokines that activate or repress viral promoters (Reese et al., 2014), or it can directly inhibit T cell responses (Osborne et al., 2014). Multiple studies now indicate that the microbiome alters host responses to viral and parasite infection (Virgin, 2014). In some cases, these prolonged relationships may confer a mutualistic symbiotic advantage to the host species (Barton et al., 2007). Thus, a reservoir host may tolerate a given chronic infection and offset any fitness cost of this infection by the benefit of altered host resistance to other pathogens. However, there are as yet few studies that explore this set of issues that might distinguish the outcome of infection between a natural host and humans (Virgin, 2014, Virgin et al., 2009).
Lastly, an important set of mechanisms that determine the capacity of a reservoir host to carry a dangerous virus may be the host’s ability to tolerate the damage caused by a specific pathogen or by the immune responses raised to it. This parameter, termed disease or infection tolerance, is measured as the rate of change in disease severity as a function of pathogen loads and is in contrast to host resistance mechanisms, which reduce pathogen burden (Schneider and Ayres, 2008). Interestingly, evolutionary theory predicts that, because pathogens will engage in an arms race with the host to evade resistance mechanisms but there is no selection on pathogens to evade disease tolerance mechanisms, resistance traits will be more polymorphic in a population than tolerance traits (Roy and Kirchner, 2000). We note that immune tolerance as defined by diminished immune responses to a given antigen is only one component of disease tolerance. The concept of disease tolerance was first examined in the context of selective pressures exerted by parasites and herbivores on plant evolution, and it is only recently that studies have begun to examine disease tolerance in mammalian host-parasite interactions (Råberg et al., 2007, Regoes et al., 2014) and investigate the pathways involved (Figueiredo et al., 2013, Jamieson et al., 2013, Jeney et al., 2014, Rodrigue-Gervais et al., 2014, Weber et al., 2014). Characterizing disease tolerance mechanisms is of particular importance to understanding the absence of severe disease in reservoir hosts and in the development of methods able to modulate the human response to the same virus without the risk of selecting for viral evasion mechanisms. An immunologic understanding of reservoir host species biology will include the study of all mechanisms by which the host species minimizes the overall fitness costs to the individual or the population, attendant on carrying a given pathogen in a form that is transmissible to humans.
Disease Tolerance and Transmission of Zoonotic Viruses
Determining the kinetics of viral replication, the extent of immune control, and parameters that might impact these, such as genetic variation, hibernation, pregnancy, or stress, may provide key insights into why some viruses are more likely to make cross-species jumps and why some of these cause severe disease in humans. Indeed, for many reservoir hosts, immune responses have not been characterized beyond demonstrating seroconversion (Table 1). Further, population-level persistence of emerging pathogens in their reservoirs has primarily been investigated from an ecological perspective, and the contribution of the host response to determining population-level infection dynamics within reservoirs is unclear. Experimental studies will provide critical data to improve field ecological studies by identifying which age classes, gender, or status of individual to focus on in studying transmission events. Experimental infections will also form the basis for improving dynamic modeling of infectious diseases, which is used to assess infection risk and develop better controls to prevent spillovers (Lloyd-Smith et al., 2009). A substantial list of important biological questions remains to be answered through focused experimental studies of the interfaces between various reservoir hosts and the specific viruses they harbor (Box 1 ).
Box 1. A Checklist for Studying Emerging Viruses in Reservoir Hosts.
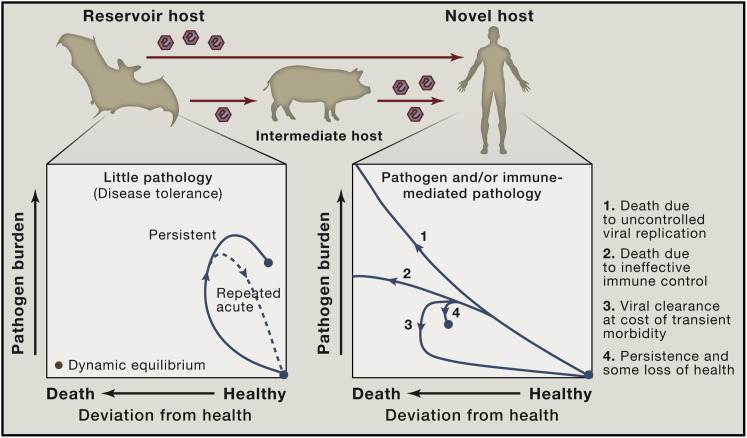
The extent to which immune responses limit viral replication in reservoir hosts is likely an important factor in the probability of cross-species transmission. Chronic or repeated virus shedding in infected reservoirs increases the likelihood of transmission due to the greater number of virus particles in the inoculum for each exposed individual. In addition, immunological effector mechanisms that substantially restrict virus replication will affect viral genetic diversity and evolution in ways that may impact the spillover probability and the strain structure present in the reservoir host population (Box 1). A greater viral diversity may increase the chances that at least a few viral variants are able to successfully establish an infection in the novel host. Thus, we speculate that viruses that have either chronically high virus levels or repeated transient high pathogen burdens in their natural hosts may be overrepresented among emerging viral pathogens in humans (Figure 1 ).
Figure 1.
Reservoir Host Infection and Disease Tolerance
A feature of zoonotic infections in an individual natural host may be the tolerance of high pathogen burdens in the absence of substantial deviation from health. Phase plots (adapted from Schneider and Ayres [2008]) illustrating possible infection trajectories following infection to highlight differences between novel and reservoir hosts in the extent of viral replication, viral kinetics, and associated disease burden. Transmission of zoonotic viruses may be more likely in cases in which there are persistently high viral loads or repeated acute infections of reservoir hosts. Within a population of the reservoir host, repeated infections with a pathogen may occur as a result of waning immunological memory within individuals, circulation of diverse strains to which there is minimal cross-reactive immunity, or the introduction of new susceptible individuals (juveniles). To what extent and in which instances features of the immune response in reservoir hosts contribute to pathogen maintenance in a population is an important open question.
Interestingly, experimental data has suggested that, in the case of malaria infection, increased tolerance may come at the cost of reduced resistance (Råberg et al., 2007). Such a tradeoff arises if resistance and tolerance are mechanistically connected. For instance, more aggressive effector responses that result in superior pathogen replication control may also be the responses that lead to increased collateral damage. Alternatively, such a tradeoff might be observed if resistance and tolerance are energetically costly, and it is only feasible to invest resources in one of the two strategies. Moreover, attenuated immune responses may result in waning immunologic memory. For instance, anti-yellow fever virus (YFV) antibody titers following YFV vaccination declined far more rapidly post infection in sooty mangabeys, a primate reservoir for YFV, than in YFV-disease susceptible rhesus macaques and humans (Mandl et al., 2011). Whether a tradeoff between resistance and tolerance is a generalized phenomenon remains to be investigated. A study of the rate of decline in CD4+ T cells across different HIV viral load set points in humans found no relationship between resistance and tolerance, although it may be the case that such a link would only become apparent over a longer period of host-pathogen coevolution (Regoes et al., 2014).
Several examples have been described in which zoonotic viruses replicate at persistently high levels in their reservoirs. SIV establishes chronic infections in African primate species with high levels of viremia and little evidence of effective immune control (Pandrea and Apetrei, 2010, Silvestri et al., 2003). Hantavirus infections of rodent reservoirs result in persistence even in the presence of high antibody titers (Easterbrook and Klein, 2008, Schountz and Prescott, 2014). For both of these viruses, extensive within-host and between-host viral diversity has also been described in their natural hosts, and distinct strains are associated with distinct host species, suggestive of a coevolutionary history (Demma et al., 2005, Feuer et al., 1999).
In contrast, infections of wild waterfowl (Anas platyrhynchos) with influenza A virus were shown to be transient (Jourdain et al., 2010). Given the high prevalence of seropositivity and the presence of an enormous diversity of influenza A strains present in birds (Verhagen et al., 2012), this might be an example in which reservoir hosts previously infected with one strain are only partially protected from infection with co-circulating viral strains, but this requires further investigation. In pteropid fruit bat species, infections with Hendra and Nipah viruses are likely also transient, but few experimental infection studies have been done so far and the infection incidence is currently unknown (Halpin et al., 2011, Middleton et al., 2007). These viruses have been detected at a prevalence of around 1% or less, although seroprevalence can be as high as 60% (Breed et al., 2013, Rahman et al., 2013). Given the seasonal and synchronous breeding of these bats, it is difficult to distinguish whether there are seasonal increases in incidence due to infections of susceptible juveniles whose maternal antibodies have waned (Epstein et al., 2013), or changes in bat immunity that enable reinfections or the reactivation of low-level infections (Sohayati et al., 2011).
Clinical manifestations of infection, such as sneezing, coughing, or diarrhea, can facilitate transmission between humans for certain zoonotic viruses where between-human secondary transmissions occur (Wolfe et al., 2007). SARS-CoV, for example, is transmitted by aerosolization and droplet infection between humans (Peiris et al., 2004), and Ebola virus spreads via contact with blood or body fluids (Dowell et al., 1999). In the absence of symptoms in disease-tolerant reservoir hosts, the routes of viral infection may be distinct from those observed in non-natural hosts. Whereas Nipah and Hendra virus is primarily respiratory in horses and pigs, transmitting oro-nasally (Geisbert et al., 2012), in bats, these viruses are shed in saliva and urine and may also be transmitted vertically (Halpin et al., 2000). For most zoonotic viruses, the transmission mode between reservoir hosts remains unknown.
Mechanisms of Disease Tolerance and Innate Immunity
Studying immune responses of reservoir hosts to their viruses provides a unique opportunity to define molecular mechanisms underlying disease tolerance and to identify therapeutic targets to prevent the dysregulated immunopathology that often characterizes infections of novel hosts. Mechanisms of disease tolerance (Schneider and Ayres, 2008) must have some degree of pathogen specificity to maintain immunocompetence. Innate immunity and mechanisms of recognizing viral invasion of cells are an important first line of host defense, and signals downstream of innate immune receptors play a key role in the magnitude of the immune response initiated (Iwasaki, 2012). Viruses can trigger endosomal, cell surface, or cytosolic innate immune receptors in a nucleic-acid-dependent manner (by their whole-genome, replication products or intermediates) or in a nucleic-acid-independent manner by virion glycoproteins or by more general changes associated with cellular stress (Iwasaki, 2012). It follows that investigating the innate immune recognition of zoonotic viruses in their reservoir hosts will provide important insight into how pathogen-specific disease tolerance is achieved.
Though innate recognition mechanisms are evolutionarily conserved, their complexity is becoming increasingly apparent. Regulatory factors are being identified that contribute to amplifying or dampening signaling pathways downstream of innate receptors and impact the type and amount of cytokines produced (Qian and Cao, 2013). In addition, the existence of genetic heterogeneity within and between populations is emerging as an important factor contributing to host variation in innate immune responses to infection and susceptibility to disease (Lee et al., 2014, Pothlichet and Quintana-Murci, 2013). In contrast, an understanding of variation in quantitative parameters, such as measures of maximum response, half-maximal effective concentration, the dose of stimuli at which a response is initiated, the kinetics of the response, and the total magnitude of the response (by measures such as area under the curve), is still lagging behind, even in humans and mice, and may well turn out to be a critical aspect in explaining differences between hosts. Ultimately, heterogeneity in qualitative and quantitative aspects of an immune response and the selection pressure exerted by specific pathogens shapes the innate immune system of a species and thus impacts responses to future encounters with novel infectious agents (Barreiro and Quintana-Murci, 2010), as well as perhaps baseline cellular states (for which the relevant measures are not always clear). Yet, species-specific differences in responses to immune stimuli are only beginning to be described (Barreiro et al., 2010, Brinkworth et al., 2012, Seok et al., 2013).
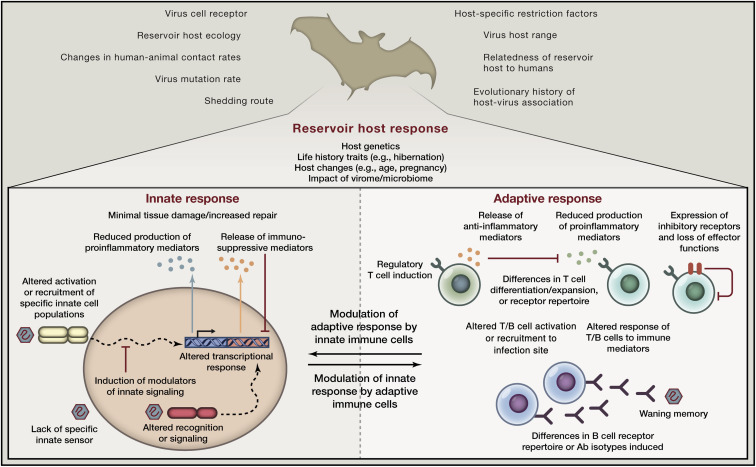
In reservoir hosts, specific changes in innate immune recognition, innate signaling pathways, cross-talk with the adaptive immune system, or the generation and differentiation of B and T cells may play a key role in the disease tolerance to viruses that they harbor (Figure 2 ). Understanding which receptors, modulating factors, or deleterious molecular mediators are involved will allow us to target these in humans to alter infection outcome. For instance, blocking specific immune signaling pathways during pathogenic ebolavirus or henipavirus infection through which signaling is reduced in bat reservoir hosts upon infection may reduce tissue damage in humans. Alternatively, inhibiting specific ISGs found to be activated in non-natural hosts but that are quiescent in natural hosts may reduce downstream detrimental proinflammatory effects in infections such as HIV without preventing the antiviral effects of other ISGs induced, rather than blocking type I IFN completely (Sandler et al., 2014, Schoggins et al., 2011).
Figure 2.
Differences in Immune Responses of Reservoir Hosts Impact Disease Tolerance, Infection Outcome, and the Probability of Emergence in Humans
Qualitative or quantitative aspects of innate or adaptive immune responses and their cross-talk may differ in reservoir hosts compared to novel hosts, impacting viral replication kinetics, decreasing pathology, and/or increasing the likelihood of transmission. Thus, reservoir host antiviral immunity may be one factor impacting the probability of emergence of a zoonotic virus in humans, in addition to other ecological, evolutionary, or virological risk factors.
Innate immune responses can play a directly beneficial role in effecting early viral control following infection and are of fundamental importance in generating adaptive cellular and humoral immune responses required to limit virus replication. Thus, the absence of RIG-I, a sensor of viral RNA ligands, has been hypothesized to make chickens less resistant to influenza viruses as compared to natural bird reservoirs such as ducks (Barber et al., 2010). However, specific innate immune responses may also precipitate immunopathologic consequences. For example, the absence of TLR3 leads to a reduced production of inflammatory mediators and improved survival upon infection with phlebovirus or influenza A viruses, despite similar or increased levels of viral replication (Gowen et al., 2006, Le Goffic et al., 2006). IL-17RA knockout mice infected with influenza A virus also display lower expression of inflammatory cytokines and chemokines, reduced lung inflammation, and improved survival despite having higher levels of virus replication as compared to wild-type laboratory inbred mice (Peiris et al., 2010). In addition, studies of different inbred mouse strains that are either susceptible or resistant to influenza-associated mortality have demonstrated greater activation of genes associated with inflammatory responses in susceptible mice and have begun to define the underlying genetic determinants of divergent infection outcomes (Peiris et al., 2010). Importantly, differences in responses between natural and non-natural hosts may not simply be due to the presence or absence of specific receptors or the switching on or off of specific signaling cascades. Rather, in the context of complex immune processes and cellular interactions, even small differences in the magnitude or timing of responses can substantially alter infection outcome. The use of mathematical models and careful measurements of rates of change of key response parameters may be required to ensure that such explanations are not missed.
Currently, the best-studied reservoir host responses are those of African primate species to SIV and of rodent species to hantavirus (Easterbrook and Klein, 2008, Miedema et al., 2013, Pandrea and Apetrei, 2010, Schountz and Prescott, 2014). Unlike humans infected with HIV, African primates do not develop the severe immune dysfunction and susceptibility to opportunistic infections that characterize AIDS. Pathogenic lentivirus infections in non-natural human or rhesus macaque hosts are associated with an elevated expression of ISGs and increased levels of immune activation, which includes increases in the activation and proliferation of T cells (Moir et al., 2011). During HIV infection, the majority of type I IFN production is likely due to the recognition of HIV by TLR7 expressed by pDCs (Beignon et al., 2005). However, in marked contrast to pathogenic HIV and SIV infections, an ISG expression signature is absent in sooty mangabeys during chronic SIV infection (Mandl et al., 2008). Further, chronically SIV-infected sooty mangabeys do not display elevations in aberrant T cell activation and its consequences (Silvestri et al., 2003). Whether the absence of chronic type I IFN production in sooty mangabeys is a result of reduced pDC activation by SIV or a function of actively suppressive mechanisms remains debated (Bosinger et al., 2013, Mandl et al., 2008). In African green monkeys, another reservoir host for SIV, a similar absence of an ISG expression signature during chronic SIV infection has been described (Jacquelin et al., 2009). In addition, SIV infection of African green monkeys is associated with an anti-inflammatory gene expression signature (Kornfeld et al., 2005). Interestingly, baseline cellular activation states, as measured by Ki67 on NK and T cells, are significantly lower in sooty mangabeys and African green monkey reservoir hosts than in the non-natural, AIDS-susceptible rhesus macaque host, though the relevance of this for immune response parameters upon infection has not yet been explored (Mandl et al., 2008).
The situation in hantavirus-infected rodents has some notable parallels to host-virus interactions in SIV-infected primates. Rodents infected with hantavirus do not show signs of disease or of discernable fitness costs (Easterbrook and Klein, 2008, Schountz and Prescott, 2014). As in SIV-infected AGMs, induction of regulatory T cells at later stages of hantavirus infection, while resulting in viral persistence, reduces effector T cell responses and is thought to limit immunopathology (Table 1) (Easterbrook and Klein, 2008, Schountz and Prescott, 2014. Exactly how the switch from initial immune activation to a dampened response is mediated in rodents infected with hantavirus is still unclear. Specific viral proteins may interact with rodent immune pathways to limit proinflammatory responses while at the same time preventing clearance. Alternatively, or in addition, as may be the case for African primates infected with SIV, changes in innate recognition could directly lead to reduced pathology due to the reduction in the production of inflammatory mediators or due to the induction of responses that actively suppress damaging responses. Adaptive immune responses may also play a major role in modulating innate immunity in reservoir hosts. For example, cytokines secreted by activated T cells can activate or suppress innate cells, and antigen-antibody complexes impact innate cell function via Fc receptor signaling. The induction of inhibitory receptors on T cells, such as PD-1, can directly contribute to limiting T-cell-mediated pathology while preventing viral clearance (Barber et al., 2006, Frebel et al., 2012) (Figure 2). Recent data from chronically lymphocytic choriomeningitis virus (LCMV)-infected mice has highlighted that an expanded population of regulatory T cells dampens CD8+ T cell responses and that the depletion of regulatory T cells in conjunction with the blockade of PD-1 signaling substantially reduces viral load (Penaloza-MacMaster et al., 2014).
The innate sensors of nucleic acid engaged by a specific virus are a function of its genome. Therefore, the innate pathways important to sensing viruses with the same genomic composition or replication scheme often overlap substantially (Iwasaki, 2012). It has been hypothesized that viruses sharing cell tropism and features that result in their recognition by the same innate receptor, termed “virotypes,” will trigger the same host transcriptional response (Virgin, 2014). As a consequence of this, it is possible that changes in a discrete innate pathway predispose certain reservoir hosts to asymptomatically harbor multiple viruses recognized by the same pathway if such changes increase disease tolerance to these virotypes in similar ways. Data from human studies have shown that deficiencies—in particular, innate receptor signaling molecules—can result in very defined and narrow susceptibilities to only a few specific pathogens (Al-Herz et al., 2014). Redundancies in immune recognition explain why reservoir host-pathogen associations might be very specific and thus not impair resistance to other pathogens. Sooty mangabeys, in addition to being reservoirs for SIV, are also natural hosts for yellow fever virus (YFV) and remain disease free upon infection. Studies of immune responses of sooty mangabeys to YFV have shown that, like for SIV, they mount reduced effector T cell responses following YFV infection compared to humans and macaques (Mandl et al., 2011). Both YFV and SIV stimulate type I IFN upon recognition by TLR7, so this raises the intriguing possibility that specific changes in TLR7 signaling in sooty mangabeys impact their adaptive immune responses to these viruses.
Indeed, patterns of reservoir host and viral associations have been observed, but their biological significance is unclear, given the only very recent increase in comprehensive studies of species’ viromes. For instance, bats are the reservoir of several emerging infectious agents, the majority of which are single-stranded RNA viruses, such as SARS coronavirus, MERS coronavirus, Nipah and Hendra viruses, rabies, and related lyssaviruses Menangle virus, Marburg, and Ebola viruses (Table 1). Bats may also be the ancient natural host reservoirs of the flavivirus hepatitis C (Quan et al., 2013). Is this merely a result of biases in sampling and analysis, or does the coevolution of a host with one virus of a given virotype open a window to infection with others of the same virotype? Is this a result of life history traits of bats and/or a function of species-specific variations in innate immune recognition of RNA viruses between humans and bats? Some studies have suggested that bats are disproportionately responsible for viral emergence relative to species diversity, but this conclusion and the reasons for the association of bats with zoonotic viruses remain a matter of debate (Luis et al., 2013, Olival et al., 2013). Interestingly, many viruses that cause persistent infections in humans use DNA as genomic material (Virgin et al., 2009). Is this entirely a property of these viruses or a result of a coevolution of our immune response with DNA virotypes that is different in other mammalian species with a distinct history of virus encounters? Zoonotic DNA viruses that cause disease in humans certainly exist, including monkeypox and herpes B viruses. A comparative and immune-centric understanding of virus-host relationships between virotypes and species will be required to investigate such questions. As our data set on host-virus associations increases, so will our ability to discern patterns and investigate their causes.
Reservoir Host Physiology, Metabolism, and Infection Tolerance
Studying reservoir host responses will enable us to more accurately measure the true physiological impact of emerging viruses on their coevolved host. The immune system is embedded in the complex physiology of an organism, and as such, infections and the immune responses that they elicit trigger behavioral, autonomic, endocrine, and metabolic effects. Soluble immune mediators like IL-1 and TNFα result in “sickness behavior,” such as social withdrawal and decreases in food intake and motor activity in infected individuals (Dantzer et al., 2008). Specific changes in metabolism during inflammation can lead to cachexia, which is particularly well documented in Mycobacterium tuberculosis (once termed “consumption”), or HIV (once termed “slim disease”) infections or to local changes in tissue microenvironment. In turn, metabolic processes and changes in body temperature can impact host immunity. Metabolic byproducts can recruit immune cells to sites of infection, activate innate immune cells via recognition through the pattern recognition receptors, which also sense microbial products, and initiate the resolution of inflammation (Jin et al., 2013, Kominsky et al., 2010). Nutrients, metabolites, and oxygen availability within tissues can impact immune cell effector functions and immune homeostasis (Pearce and Pearce, 2013, Sitkovsky and Lukashev, 2005).
It is possible that fundamental differences in reservoir host physiology and the circuitry that interfaces with the immune system are the consequence of distinct evolutionary tradeoffs between species and impact infection outcome or the resulting disease manifestations. For instance, bats have evolved unique life history traits that may have influenced their exposure to and interaction with pathogens in ways that have led to distinct virus-host détentes. Bats are capable of sustained flight and have exceptionally long lifespans despite their high metabolic rates and small body size (Wilkinson and South, 2002). Consistent with the idea that changes in energy metabolism were required to meet the extraordinary cost of flight, mitochondrial and nuclear genes involved in oxidative phosphorylation have undergone positive selection in bat lineages (Shen et al., 2010). Concomitant selection pressure on DNA damage repair pathway genes to prevent negative physiological effects of high metabolic rates caused by the release of reactive oxygen species may have had as yet undefined consequences for innate immune responses to viruses (Zhang et al., 2013). Hibernation has also been associated with altered immune cell function and reduced lymphocyte trafficking, an understanding of which could be key to explaining the devastating impact of White-nose syndrome in North American bat populations (Bouma et al., 2010, Meteyer et al., 2012).
An Experimental Toolkit for Reservoir Host Immunology
Detailed assessments of immune responses in reservoir host species have been challenging for a number of reasons. One practical constraint has been establishing colonies of reservoir host species to enable longitudinal experiments under controlled conditions. Immunological field studies, though valuable in specific instances, can be difficult to interpret given that pathogen exposure status between sampling time points is unknown and it may not always be possible to recapture the same individuals. However, keeping reservoir host colonies can require specific animal husbandry techniques, may involve training animals to accept available food sources not eaten in the wild, and can require access to special facilities to maintain the animals (such as aviaries to harbor bats or birds) and to perform infections with viruses that are often lethal to humans. Moreover, importing reservoir hosts from their country of origin can be costly, and quarantines are required to ascertain that they are free of the zoonotic pathogens dangerous to humans. The lack of experimental studies of wildlife reservoirs is also a product of our poor understanding of the true wildlife reservoirs for a number of recently emerged viruses. However, such studies, if conducted correctly, are critical to understanding pathogenesis and the risk of exposure for humans.
A second technical challenge to studies of reservoir host immunity has been the establishment of viral infection models that mimic infection in the wild and that are useful for investigating pathogen kinetics and immune responses over time in a setting where the time of infection is known and other environmental parameters can be controlled (Box 1). Extensive viral passaging in vitro, especially in cells lines that, while permissive for infection, may be quite distinct from cells infected in vivo, can lead to the selection of viral variants that can no longer replicate effectively in the host of origin. Similarly, the use of clonal viruses can be problematic in recapitulating typical outcomes of natural infections should initiation of infection with diverse viral quasispecies be important. As such, the choice of viral inoculum can greatly influence the course of infection. For example, in studies of sooty mangabeys in which highly pathogenic SIV variants passaged in macaques were used, the infection was rapidly controlled, quite unlike the persistent high-level replication seen in natural infections (Kaur et al., 1998). Subsequent infection studies have used plasma samples harvested from SIV-infected sooty mangabeys (Mandl et al., 2008). In other instances, whether experimental infections of reservoir hosts recapitulate features of natural infection in the wild remains to be validated. Infections of bats with Nipah and Hendra viruses resulted in very few bats with detectable levels of viral replication, although in some instances seroconversion was seen nonetheless, and it is unclear whether this very low infection level mimics infections in the wild (Halpin et al., 2011, Middleton et al., 2007). Experimental rabies infections of bat reservoir hosts led to muscle weakness, paralysis, and reductions in body weight and were usually lethal (Turmelle et al., 2010), but it is difficult to extrapolate such studies to an understanding of the mortality rate of rabies in wild bat populations. In fact, epidemiological models of data gathered in field studies suggest that lethality upon rabies infection may be a far rarer outcome in wild bat populations than in the lab (Blackwood et al., 2013). Similarly, whereas LCMV infection of inbred mouse strains is one of the best-studied systems for viral immunity, surprisingly little is known about the immunologic and virologic characteristics of the host-virus interface in wild mouse species that are reservoir hosts for LCMV and other arenaviruses (Table 1) (Wade et al., 2002). Host genetic differences or small changes in the LCMV viral sequence can affect whether infections are chronic or acute (Ahmed et al., 1988). In addition, vertical transmission can lead to the establishment of a life-long viral carrier state with little adaptive immune responses raised. The dependence on the host-pathogen balance established on both strain and route of infection means that, without much information on this, it is currently difficult to extrapolate insights from laboratory experiments to infections of mice in the wild.
Notably, developing an infection model that recapitulates human disease and can be used to study host responses during pathogenic infections to perform comparative studies with natural hosts can also be challenging if laboratory mice are not susceptible to infection or are resistant to disease. In the former case, identification of the viral host cell receptor in humans can enable the development of mouse strains expressing this human protein on relevant cell types, thus enabling viral replication in mice. For instance, the expression of human dipeptidyl peptidase 4, the host cell receptor used by MERS coronovirus, in mice using adenovirus vector-mediated transduction successfully rendered mice susceptible to infection with MERS coronavirus (Zhao et al., 2014). In the case in which mice are permissive for infection but distinct host responses lead to only mild disease, the genetic traits contributing to disease susceptibility differences can be explored using a set of well-defined recombinant inbred mouse strains, the Collaborative Cross (Churchill et al., 2004), as was recently done for Ebola virus infection (Rasmussen et al., 2014).
Investigating reservoir host immunity has also been hampered by the paucity of available immunological tools. This includes sequences of immunologically important genes, markers for cell populations of interest, and available monoclonal antibodies specific for such markers, as well as antibody tools to identify cytokines or chemokines produced by immune cell populations in vitro or in vivo. The development of such tools will be essential to increasing the kinds of questions that can be answered. Despite the difficulties that remain, recent methodological advances have enormous potential in their application to studies of immune responses of natural hosts and have opened new avenues of research.
First, there has been a rapid expansion in genomes sequenced for different reservoir host species. These sequences are themselves already informative. For instance, they allow insight into the presence or absence of specific innate receptors or cytokine mediators and provide clues as to the mechanism of B or T cell repertoire diversity generation and B cell isotypes present. Furthermore, comparative analyses can highlight genomic regions with changes of possible relevance to disease outcome upon infection and can generate hypotheses that can be tested further. A comparison between the Pteropus alecto and Myotis davidii bat genomes showed that several innate immune genes, including TLR7, were under positive selection in bat ancestors and that both species lack NK cell receptors for MHC-I, KLRs, and KIRs (Zhang et al., 2013). Genomic approaches complement the targeted sequencing of specific genes hypothesized to play a role in viral control and determination of their tissue distribution—for instance, the sequencing of specific cytokine genes in deer mice (P. maniculatus) reservoirs of hantavirus (Herbst et al., 2002) or interferon system genes and innate receptors in bat species (Baker et al., 2013). In addition, next-generation sequencing is rapidly emerging as a way to define the viruses that we may have to worry about as future emerging infections (Mokili et al., 2012) and when comparing the baseline microbiome or virome of different species, as well as documenting infection-related changes in their composition (Handley et al., 2012, Virgin, 2014).
Second, unbiased approaches can be used to probe both quantitative and qualitative changes in the transcriptome or proteome. The availability of reservoir host genomes or related species makes the use of microarray technology for gene expression profiling possible. Furthermore, even in the absence of a reference genome, next generation sequencing and the de novo assembly of transcripts, while computationally more challenging, now allows the assessment of gene expression. Such methods provide a promising means by which to illuminate gene expression changes—for instance, in infected versus uninfected reservoir hosts—and to allow comparisons with changes seen in infected humans. This approach has already been successfully applied to characterize responses of non-natural hosts to a number of pathogens, including infection with influenza A, SARS, and HIV (Cameron et al., 2008, Kash et al., 2006, Moir et al., 2011). Although such methods applied to bulk tissue samples may not be able to distinguish between the induction of genes from changes in cell distribution, they may identify specific targets for further study. Eventually, the development of single-cell transcriptome analyses and computational tools to infer the presence of specific cell populations and their phenotype within bulk samples may provide additional insight when flow cytometric identification of immune cell populations using monoclonal antibodies is not possible.
Lastly, our increased understanding of the various innate immune receptors and the distinct components of pathogens that they recognize has resulted in the availability of tools with which to probe responses to specific receptor agonists in vivo and in vitro. Our knowledge of innate immunity enables us to make reasonable guesses as to which types of innate receptors might be relevant to the host-pathogen interface of particular viruses. This is particularly useful when the pathogen in question requires biosafety level 4 laboratories, thus making direct infection studies challenging. Together with the methods outlined above, it is now possible to determine which genes change their expression early on in response to the administration of, for instance, defined TLR agonists targeting the specific TLRs involved in innate recognition of the virus of interest. We can characterize specific gene circuits and their temporal regulation, comparing responses to what is seen in more well-studied species in that regard. It is also possible to measure differences in basal gene expression levels, as well as quantitative changes in response to different doses of relevant stimuli to obtain measures of the dynamic range of the response, the dose at which a response is initiated, and its duration. In addition, innate immunity can be probed in cross-sectional studies of animals, which can provide information on how infection itself can modulate immune responsiveness.
Conclusions
Given the threats to human health and economic development posed by emerging viral diseases, better understanding the nature and diversity of mechanisms responsible for the ability of reservoir hosts to tolerate viral infections and facilitate transmission to other host species is critical. Studies to date have focused primarily on viral detection and diversity, rather than on illuminating the nature of the host-virus equilibrium extant in their wild animal reservoirs. Achieving the goal of a holistic approach to understanding viral emergence will require the integration of immunological studies with investigations of animal physiology and metabolism, as well as the synthesis of such studies with epidemiological, ecological, and evolutionary insights of viral emergence. Cross-disciplinary collaborations between mechanism-targeted immunology and microbiology laboratories, veterinarians with experience in establishing colonies of wild species, and ecologists able to perform in-country field studies will be especially important in advancing our knowledge in this field. The enormous value of cross-geographical and interdisciplinary initiatives in understanding viral emergence has already been demonstrated by programs such as PREDICT, established by the U.S. Agency for International Development to detect emerging pandemic threats in partnership with local governments and scientists (Morse et al., 2012). It is also apparent in other cross-continental research teams’ efforts, such as the multinational collaboration that promptly tracked the emergence and evolution of the Ebola virus variants responsible for the ongoing outbreak in West Africa (Gire et al., 2014). The development of immunological reagents, in addition to the application of recently developed methods to define host genome sequences and to characterize patterns of host gene expression following infection or other experimental intervention, will yield valuable insights into immune responses elicited by emerging viruses in their natural hosts. Such studies may identify important pathways of pathogenesis in humans by elucidating why these infections result in such distinct infection outcomes in natural hosts and may indicate novel approaches for the treatment and prevention of zoonotic infections. They may also establish whether there are fundamental immunologic reasons—in addition to proposed behavioral, ecological, and evolutionary ones—to explain why certain animals, including bats, primates, and rodents, are most often implicated as the reservoir hosts for emerging viral zoonoses. Finally, studies of reservoir hosts stand to significantly advance our understanding of fundamental immunological processes contributing to host resistance and disease tolerance.
Acknowledgments
We would like to thank Nienke Vrisekoop, Roland Regoes, and Ronald Germain for insightful comments, support, and advice during the preparation of this manuscript. We are also extremely grateful to Tony Schountz and our anonymous reviewers for their reading of the manuscript and their very thoughtful feedback. This work was supported, in part, by the Intramural Research Program of NIAID, NIH.
Footnotes
Supplemental Information includes one table and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2014.12.003.
Supplemental Information
References
- Ahmed R., Simon R.S., Matloubian M., Kolhekar S.R., Southern P.J., Freedman D.M. Genetic analysis of in vivo-selected viral variants causing chronic infection: importance of mutation in the L RNA segment of lymphocytic choriomeningitis virus. J. Virol. 1988;62:3301–3308. doi: 10.1128/jvi.62.9.3301-3308.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Herz W., Bousfiha A., Casanova J.L., Chatila T., Conley M.E., Cunningham-Rundles C., Etzioni A., Franco J.L., Gaspar H.B., Holland S.M. Primary immunodeficiency diseases: an update on the classification from the international union of immunological societies expert committee for primary immunodeficiency. Front. Immunol. 2014;5:162. doi: 10.3389/fimmu.2014.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antia R., Regoes R.R., Koella J.C., Bergstrom C.T. The role of evolution in the emergence of infectious diseases. Nature. 2003;426:658–661. doi: 10.1038/nature02104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayllon J., Garcia-Sastre A. The NS1 protein: A multitasking virulence factor. Curr. Top. Microbiol. Immunol. 2015;386:73–107. doi: 10.1007/82_2014_400. [DOI] [PubMed] [Google Scholar]
- Bailes E., Gao F., Bibollet-Ruche F., Courgnaud V., Peeters M., Marx P.A., Hahn B.H., Sharp P.M. Hybrid origin of SIV in chimpanzees. Science. 2003;300:1713. doi: 10.1126/science.1080657. [DOI] [PubMed] [Google Scholar]
- Baker M.L., Schountz T., Wang L.F. Antiviral immune responses of bats: a review. Zoonoses Public Health. 2013;60:104–116. doi: 10.1111/j.1863-2378.2012.01528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber D.L., Wherry E.J., Masopust D., Zhu B., Allison J.P., Sharpe A.H., Freeman G.J., Ahmed R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- Barber M.R., Aldridge J.R., Jr., Webster R.G., Magor K.E. Association of RIG-I with innate immunity of ducks to influenza. Proc. Natl. Acad. Sci. USA. 2010;107:5913–5918. doi: 10.1073/pnas.1001755107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro L.B., Quintana-Murci L. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat. Rev. Genet. 2010;11:17–30. doi: 10.1038/nrg2698. [DOI] [PubMed] [Google Scholar]
- Barreiro L.B., Marioni J.C., Blekhman R., Stephens M., Gilad Y. Functional comparison of innate immune signaling pathways in primates. PLoS Genet. 2010;6:e1001249. doi: 10.1371/journal.pgen.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton E.S., White D.W., Cathelyn J.S., Brett-McClellan K.A., Engle M., Diamond M.S., Miller V.L., Virgin H.W., 4th Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447:326–329. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- Beaumier C.M., Harris L.D., Goldstein S., Klatt N.R., Whitted S., McGinty J., Apetrei C., Pandrea I., Hirsch V.M., Brenchley J.M. CD4 downregulation by memory CD4+ T cells in vivo renders African green monkeys resistant to progressive SIVagm infection. Nat. Med. 2009;15:879–885. doi: 10.1038/nm.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beignon A.S., McKenna K., Skoberne M., Manches O., DaSilva I., Kavanagh D.G., Larsson M., Gorelick R.J., Lifson J.D., Bhardwaj N. Endocytosis of HIV-1 activates plasmacytoid dendritic cells via Toll-like receptor-viral RNA interactions. J. Clin. Invest. 2005;115:3265–3275. doi: 10.1172/JCI26032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biek R., Walsh P.D., Leroy E.M., Real L.A. Recent common ancestry of Ebola Zaire virus found in a bat reservoir. PLoS Pathog. 2006;2:e90. doi: 10.1371/journal.ppat.0020090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood J.C., Streicker D.G., Altizer S., Rohani P. Resolving the roles of immunity, pathogenesis, and immigration for rabies persistence in vampire bats. Proc. Natl. Acad. Sci. USA. 2013;110:20837–20842. doi: 10.1073/pnas.1308817110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosinger S.E., Johnson Z.P., Folkner K.A., Patel N., Hashempour T., Jochems S.P., Del Rio Estrada P.M., Paiardini M., Lin R., Vanderford T.H. Intact type I Interferon production and IRF7 function in sooty mangabeys. PLoS Pathog. 2013;9:e1003597. doi: 10.1371/journal.ppat.1003597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouma H.R., Carey H.V., Kroese F.G. Hibernation: the immune system at rest? J. Leukoc. Biol. 2010;88:619–624. doi: 10.1189/jlb.0310174. [DOI] [PubMed] [Google Scholar]
- Breed A.C., Meers J., Sendow I., Bossart K.N., Barr J.A., Smith I., Wacharapluesadee S., Wang L., Field H.E. The distribution of henipaviruses in Southeast Asia and Australasia: is Wallace’s line a barrier to Nipah virus? PLoS ONE. 2013;8:e61316. doi: 10.1371/journal.pone.0061316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley J.M., Price D.A., Schacker T.W., Asher T.E., Silvestri G., Rao S., Kazzaz Z., Bornstein E., Lambotte O., Altmann D. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat. Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- Brinkworth J.F., Pechenkina E.A., Silver J., Goyert S.M. Innate immune responses to TLR2 and TLR4 agonists differ between baboons, chimpanzees and humans. J. Med. Primatol. 2012;41:388–393. doi: 10.1111/jmp.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton D.R., Stanfield R.L., Wilson I.A. Antibody vs. HIV in a clash of evolutionary titans. Proc. Natl. Acad. Sci. USA. 2005;102:14943–14948. doi: 10.1073/pnas.0505126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron M.J., Bermejo-Martin J.F., Danesh A., Muller M.P., Kelvin D.J. Human immunopathogenesis of severe acute respiratory syndrome (SARS) Virus Res. 2008;133:13–19. doi: 10.1016/j.virusres.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G.A., Airey D.C., Allayee H., Angel J.M., Attie A.D., Beatty J., Beavis W.D., Belknap J.K., Bennett B., Berrettini W., Complex Trait Consortium The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- Dantzer R., O’Connor J.C., Freund G.G., Johnson R.W., Kelley K.W. From inflammation to sickness and depression: when the immune system subjugates the brain. Nat. Rev. Neurosci. 2008;9:46–56. doi: 10.1038/nrn2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demma L.J., Logsdon J.M., Jr., Vanderford T.H., Feinberg M.B., Staprans S.I. SIVsm quasispecies adaptation to a new simian host. PLoS Pathog. 2005;1:e3. doi: 10.1371/journal.ppat.0010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell S.F., Mukunu R., Ksiazek T.G., Khan A.S., Rollin P.E., Peters C.J. Transmission of Ebola hemorrhagic fever: a study of risk factors in family members, Kikwit, Democratic Republic of the Congo, 1995. Commission de Lutte contre les Epidémies à Kikwit. J. Infect. Dis. 1999;179(Suppl 1):S87–S91. doi: 10.1086/514284. [DOI] [PubMed] [Google Scholar]
- Easterbrook J.D., Klein S.L. Immunological mechanisms mediating hantavirus persistence in rodent reservoirs. PLoS Pathog. 2008;4:e1000172. doi: 10.1371/journal.ppat.1000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein J.H., Baker M.L., Zambrana-Torrelio C., Middleton D., Barr J.A., Dubovi E., Boyd V., Pope B., Todd S., Crameri G. Duration of Maternal Antibodies against Canine Distemper Virus and Hendra Virus in Pteropid Bats. PLoS ONE. 2013;8:e67584. doi: 10.1371/journal.pone.0067584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuer R., Boone J.D., Netski D., Morzunov S.P., St Jeor S.C. Temporal and spatial analysis of Sin Nombre virus quasispecies in naturally infected rodents. J. Virol. 1999;73:9544–9554. doi: 10.1128/jvi.73.11.9544-9554.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo N., Chora A., Raquel H., Pejanovic N., Pereira P., Hartleben B., Neves-Costa A., Moita C., Pedroso D., Pinto A. Anthracyclines induce DNA damage response-mediated protection against severe sepsis. Immunity. 2013;39:874–884. doi: 10.1016/j.immuni.2013.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frebel H., Nindl V., Schuepbach R.A., Braunschweiler T., Richter K., Vogel J., Wagner C.A., Loffing-Cueni D., Kurrer M., Ludewig B., Oxenius A. Programmed death 1 protects from fatal circulatory failure during systemic virus infection of mice. J. Exp. Med. 2012;209:2485–2499. doi: 10.1084/jem.20121015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert T.W., Jahrling P.B. Exotic emerging viral diseases: progress and challenges. Nat. Med. 2004;10(Suppl):S110–S121. doi: 10.1038/nm1142. [DOI] [PubMed] [Google Scholar]
- Geisbert T.W., Feldmann H., Broder C.C. Animal challenge models of henipavirus infection and pathogenesis. Curr. Top. Microbiol. Immunol. 2012;359:153–177. doi: 10.1007/82_2012_208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gire S.K., Goba A., Andersen K.G., Sealfon R.S., Park D.J., Kanneh L., Jalloh S., Momoh M., Fullah M., Dudas G. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science. 2014;345:1369–1372. doi: 10.1126/science.1259657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowen B.B., Hoopes J.D., Wong M.H., Jung K.H., Isakson K.C., Alexopoulou L., Flavell R.A., Sidwell R.W. TLR3 deletion limits mortality and disease severity due to Phlebovirus infection. J. Immunol. 2006;177:6301–6307. doi: 10.4049/jimmunol.177.9.6301. [DOI] [PubMed] [Google Scholar]
- Hahn B.H., Shaw G.M., De Cock K.M., Sharp P.M. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- Halpin K., Young P.L., Field H.E., Mackenzie J.S. Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. J. Gen. Virol. 2000;81:1927–1932. doi: 10.1099/0022-1317-81-8-1927. [DOI] [PubMed] [Google Scholar]
- Halpin K., Hyatt A.D., Fogarty R., Middleton D., Bingham J., Epstein J.H., Rahman S.A., Hughes T., Smith C., Field H.E., Daszak P., Henipavirus Ecology Research Group Pteropid bats are confirmed as the reservoir hosts of henipaviruses: a comprehensive experimental study of virus transmission. Am. J. Trop. Med. Hyg. 2011;85:946–951. doi: 10.4269/ajtmh.2011.10-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley S.A., Thackray L.B., Zhao G., Presti R., Miller A.D., Droit L., Abbink P., Maxfield L.F., Kambal A., Duan E. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 2012;151:253–266. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie K.M., Bale S., Kimberlin C.R., Saphire E.O. Hiding the evidence: two strategies for innate immune evasion by hemorrhagic fever viruses. Curr. Opin. Virol. 2012;2:151–156. doi: 10.1016/j.coviro.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst M.M., Prescott J., Palmer A.D., Schountz T. Sequence and expression analysis of deer mouse interferon-gamma, interleukin-10, tumor necrosis factor, and lymphotoxin-alpha. Cytokine. 2002;17:203–213. doi: 10.1006/cyto.2001.0998. [DOI] [PubMed] [Google Scholar]
- Holmes E.C., Drummond A.J. The evolutionary genetics of viral emergence. Curr. Top. Microbiol. Immunol. 2007;315:51–66. doi: 10.1007/978-3-540-70962-6_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A. A virological view of innate immune recognition. Annu. Rev. Microbiol. 2012;66:177–196. doi: 10.1146/annurev-micro-092611-150203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquelin B., Mayau V., Targat B., Liovat A.S., Kunkel D., Petitjean G., Dillies M.A., Roques P., Butor C., Silvestri G. Nonpathogenic SIV infection of African green monkeys induces a strong but rapidly controlled type I IFN response. J. Clin. Invest. 2009;119:3544–3555. doi: 10.1172/JCI40093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson A.M., Pasman L., Yu S., Gamradt P., Homer R.J., Decker T., Medzhitov R. Role of tissue protection in lethal respiratory viral-bacterial coinfection. Science. 2013;340:1230–1234. doi: 10.1126/science.1233632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeney V., Ramos S., Bergman M.L., Bechmann I., Tischer J., Ferreira A., Oliveira-Marques V., Janse C.J., Rebelo S., Cardoso S. Control of disease tolerance to malaria by nitric oxide and carbon monoxide. Cell Rep. 2014;8:126–136. doi: 10.1016/j.celrep.2014.05.054. [DOI] [PubMed] [Google Scholar]
- Jin C., Henao-Mejia J., Flavell R.A. Innate immune receptors: key regulators of metabolic disease progression. Cell Metab. 2013;17:873–882. doi: 10.1016/j.cmet.2013.05.011. [DOI] [PubMed] [Google Scholar]
- Jones K.E., Patel N.G., Levy M.A., Storeygard A., Balk D., Gittleman J.L., Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdain E., Gunnarsson G., Wahlgren J., Latorre-Margalef N., Bröjer C., Sahlin S., Svensson L., Waldenström J., Lundkvist A., Olsen B. Influenza virus in a natural host, the mallard: experimental infection data. PLoS ONE. 2010;5:e8935. doi: 10.1371/journal.pone.0008935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karesh W.B., Dobson A., Lloyd-Smith J.O., Lubroth J., Dixon M.A., Bennett M., Aldrich S., Harrington T., Formenty P., Loh E.H. Ecology of zoonoses: natural and unnatural histories. Lancet. 2012;380:1936–1945. doi: 10.1016/S0140-6736(12)61678-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kash J.C., Tumpey T.M., Proll S.C., Carter V., Perwitasari O., Thomas M.J., Basler C.F., Palese P., Taubenberger J.K., García-Sastre A. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature. 2006;443:578–581. doi: 10.1038/nature05181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur A., Grant R.M., Means R.E., McClure H., Feinberg M., Johnson R.P. Diverse host responses and outcomes following simian immunodeficiency virus SIVmac239 infection in sooty mangabeys and rhesus macaques. J. Virol. 1998;72:9597–9611. doi: 10.1128/jvi.72.12.9597-9611.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele B.F., Jones J.H., Terio K.A., Estes J.D., Rudicell R.S., Wilson M.L., Li Y., Learn G.H., Beasley T.M., Schumacher-Stankey J. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature. 2009;460:515–519. doi: 10.1038/nature08200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominsky D.J., Campbell E.L., Colgan S.P. Metabolic shifts in immunity and inflammation. J. Immunol. 2010;184:4062–4068. doi: 10.4049/jimmunol.0903002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld C., Ploquin M.J.Y., Pandrea I., Faye A., Onanga R., Apetrei C., Poaty-Mavoungou V., Rouquet P., Estaquier J., Mortara L. Antiinflammatory profiles during primary SIV infection in African green monkeys are associated with protection against AIDS. J. Clin. Invest. 2005;115:1082–1091. doi: 10.1172/JCI23006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken T., Riteau B., Fouchier R.A., Rimmelzwaan G.F. Pathogenesis of influenza virus infections: the good, the bad and the ugly. Curr. Opin. Virol. 2012;2:276–286. doi: 10.1016/j.coviro.2012.02.013. [DOI] [PubMed] [Google Scholar]
- Le Goffic R., Balloy V., Lagranderie M., Alexopoulou L., Escriou N., Flavell R., Chignard M., Si-Tahar M. Detrimental contribution of the Toll-like receptor (TLR)3 to influenza A virus-induced acute pneumonia. PLoS Pathog. 2006;2:e53. doi: 10.1371/journal.ppat.0020053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.N., Ye C., Villani A.C., Raj T., Li W., Eisenhaure T.M., Imboywa S.H., Chipendo P.I., Ran F.A., Slowikowski K. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science. 2014;343:1246980. doi: 10.1126/science.1246980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Smith J.O., George D., Pepin K.M., Pitzer V.E., Pulliam J.R., Dobson A.P., Hudson P.J., Grenfell B.T. Epidemic dynamics at the human-animal interface. Science. 2009;326:1362–1367. doi: 10.1126/science.1177345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby S.P., Rahman M., Hossain M.J., Blum L.S., Husain M.M., Gurley E., Khan R., Ahmed B.N., Rahman S., Nahar N. Foodborne transmission of Nipah virus, Bangladesh. Emerg. Infect. Dis. 2006;12:1888–1894. doi: 10.3201/eid1212.060732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis A.D., Hayman D.T., O’Shea T.J., Cryan P.M., Gilbert A.T., Pulliam J.R., Mills J.N., Timonin M.E., Willis C.K., Cunningham A.A. A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proc. Biol. Sci. 2013;280:20122753. doi: 10.1098/rspb.2012.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie J.S., Jeggo M. Reservoirs and vectors of emerging viruses. Curr. Opin. Virol. 2013;3:170–179. doi: 10.1016/j.coviro.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandl J.N., Barry A.P., Vanderford T.H., Kozyr N., Chavan R., Klucking S., Barrat F.J., Coffman R.L., Staprans S.I., Feinberg M.B. Divergent TLR7 and TLR9 signaling and type I interferon production distinguish pathogenic and nonpathogenic AIDS virus infections. Nat. Med. 2008;14:1077–1087. doi: 10.1038/nm.1871. [DOI] [PubMed] [Google Scholar]
- Mandl J.N., Akondy R., Lawson B., Kozyr N., Staprans S.I., Ahmed R., Feinberg M.B. Distinctive TLR7 signaling, type I IFN production, and attenuated innate and adaptive immune responses to yellow fever virus in a primate reservoir host. J. Immunol. 2011;186:6406–6416. doi: 10.4049/jimmunol.1001191. [DOI] [PubMed] [Google Scholar]
- Marston H.D., Folkers G.K., Morens D.M., Fauci A.S. Emerging viral diseases: confronting threats with new technologies. Sci. Transl. Med. 2014;6 doi: 10.1126/scitranslmed.3009872. 53ps10. [DOI] [PubMed] [Google Scholar]
- Martinez O., Leung L.W., Basler C.F. The role of antigen-presenting cells in filoviral hemorrhagic fever: gaps in current knowledge. Antiviral Res. 2012;93:416–428. doi: 10.1016/j.antiviral.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Eisfeld A.J., Schafer A., Josset L., Sims A.C., Proll S., Fan S., Li C., Neumann G., Tilton S.C. Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. Mbio. 2014;5:e01174–e01214. doi: 10.1128/mBio.01174-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meteyer C.U., Barber D., Mandl J.N. Pathology in euthermic bats with white nose syndrome suggests a natural manifestation of immune reconstitution inflammatory syndrome. Virulence. 2012;3:583–588. doi: 10.4161/viru.22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton D.J., Morrissy C.J., van der Heide B.M., Russell G.M., Braun M.A., Westbury H.A., Halpin K., Daniels P.W. Experimental Nipah virus infection in pteropid bats (Pteropus poliocephalus) J. Comp. Pathol. 2007;136:266–272. doi: 10.1016/j.jcpa.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Miedema F., Hazenberg M.D., Tesselaar K., van Baarle D., de Boer R.J., Borghans J.A. Immune activation and collateral damage in AIDS pathogenesis. Front. Immunol. 2013;4:298. doi: 10.3389/fimmu.2013.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misasi J., Sullivan N.J. Camouflage and misdirection: the full-on assault of ebola virus disease. Cell. 2014;159:477–486. doi: 10.1016/j.cell.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir S., Chun T.W., Fauci A.S. Pathogenic mechanisms of HIV disease. Annu. Rev. Pathol. 2011;6:223–248. doi: 10.1146/annurev-pathol-011110-130254. [DOI] [PubMed] [Google Scholar]
- Mokili J.L., Rohwer F., Dutilh B.E. Metagenomics and future perspectives in virus discovery. Curr. Opin. Virol. 2012;2:63–77. doi: 10.1016/j.coviro.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse S.S., Mazet J.A., Woolhouse M., Parrish C.R., Carroll D., Karesh W.B., Zambrana-Torrelio C., Lipkin W.I., Daszak P. Prediction and prevention of the next pandemic zoonosis. Lancet. 2012;380:1956–1965. doi: 10.1016/S0140-6736(12)61684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldstone M.B., Rosen H. Cytokine storm plays a direct role in the morbidity and mortality from influenza virus infection and is chemically treatable with a single sphingosine-1-phosphate agonist molecule. Curr. Top. Microbiol. Immunol. 2014;378:129–147. doi: 10.1007/978-3-319-05879-5_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olival K.J., Epstein J.H., Wang L.-F., Field H.E., Daszak P. Are bats unique virus reservoirs? In: Aguirre A.A., Ostfeld R.S., Daszak P., editors. New Directions in Conservation Medicine Applied Cases of Ecological Health. Oxford University Press; New York: 2013. pp. 195–212. [Google Scholar]
- Osborne L.C., Monticelli L.A., Nice T.J., Sutherland T.E., Siracusa M.C., Hepworth M.R., Tomov V.T., Kobuley D., Tran S.V., Bittinger K. Coinfection. Virus-helminth coinfection reveals a microbiota-independent mechanism of immunomodulation. Science. 2014;345:578–582. doi: 10.1126/science.1256942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiardini M., Cervasi B., Reyes-Aviles E., Micci L., Ortiz A.M., Chahroudi A., Vinton C., Gordon S.N., Bosinger S.E., Francella N. Low levels of SIV infection in sooty mangabey central memory CD4+ T cells are associated with limited CCR5 expression. Nat. Med. 2011;17:830–836. doi: 10.1038/nm.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A., Atkins K.E., Medlock J., Wenzel N., Townsend J.P., Childs J.E., Nyenswah T.G., Ndeffo-Mbah M.L., Galvani A.P. Strategies for containing Ebola in West Africa. Science. 2014;346:991–995. doi: 10.1126/science.1260612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandrea I., Apetrei C. Where the wild things are: pathogenesis of SIV infection in African nonhuman primate hosts. Curr. HIV/AIDS Rep. 2010;7:28–36. doi: 10.1007/s11904-009-0034-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parashar U.D., Sunn L.M., Ong F., Mounts A.W., Arif M.T., Ksiazek T.G., Kamaluddin M.A., Mustafa A.N., Kaur H., Ding L.M. Case-control study of risk factors for human infection with a new zoonotic paramyxovirus, Nipah virus, during a 1998-1999 outbreak of severe encephalitis in Malaysia. J. Infect. Dis. 2000;181:1755–1759. doi: 10.1086/315457. [DOI] [PubMed] [Google Scholar]
- Pearce E.L., Pearce E.J. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38:633–643. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Guan Y., Yuen K.Y. Severe acute respiratory syndrome. Nat. Med. 2004;10(Suppl):S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Hui K.P., Yen H.L. Host response to influenza virus: protection versus immunopathology. Curr. Opin. Immunol. 2010;22:475–481. doi: 10.1016/j.coi.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penaloza-MacMaster P., Kamphorst A.O., Wieland A., Araki K., Iyer S.S., West E.E., O’Mara L., Yang S., Konieczny B.T., Sharpe A.H. Interplay between regulatory T cells and PD-1 in modulating T cell exhaustion and viral control during chronic LCMV infection. J. Exp. Med. 2014;211:1905–1918. doi: 10.1084/jem.20132577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigott D.M., Golding N., Mylne A., Huang Z., Henry A.J., Weiss D.J., Brady O.J., Kraemer M.U., Smith D.L., Moyes C.L. Mapping the zoonotic niche of Ebola virus disease in Africa. eLife. 2014;3 doi: 10.7554/eLife.04395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pothlichet J., Quintana-Murci L. The genetics of innate immunity sensors and human disease. Int. Rev. Immunol. 2013;32:157–208. doi: 10.3109/08830185.2013.777064. [DOI] [PubMed] [Google Scholar]
- Qian C., Cao X. Regulation of Toll-like receptor signaling pathways in innate immune responses. Ann. N Y Acad. Sci. 2013;1283:67–74. doi: 10.1111/j.1749-6632.2012.06786.x. [DOI] [PubMed] [Google Scholar]
- Quan P.L., Firth C., Conte J.M., Williams S.H., Zambrana-Torrelio C.M., Anthony S.J., Ellison J.A., Gilbert A.T., Kuzmin I.V., Niezgoda M. Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc. Natl. Acad. Sci. USA. 2013;110:8194–8199. doi: 10.1073/pnas.1303037110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Råberg L., Sim D., Read A.F. Disentangling genetic variation for resistance and tolerance to infectious diseases in animals. Science. 2007;318:812–814. doi: 10.1126/science.1148526. [DOI] [PubMed] [Google Scholar]
- Rahman S.A., Hassan L., Epstein J.H., Mamat Z.C., Yatim A.M., Hassan S.S., Field H.E., Hughes T., Westrum J., Naim M.S., Henipavirus Ecology Research Group Risk Factors for Nipah virus infection among pteropid bats, Peninsular Malaysia. Emerg. Infect. Dis. 2013;19:51–60. doi: 10.3201/eid1901.120221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen A.L., Okumura A., Ferris M.T., Green R., Feldmann F., Kelly S.M., Scott D.P., Safronetz D., Haddock E., LaCasse R. Host genetic diversity enables Ebola hemorrhagic fever pathogenesis and resistance. Science. 2014;346:987–991. doi: 10.1126/science.1259595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese T.A., Wakeman B.S., Choi H.S., Hufford M.M., Huang S.C., Zhang X., Buck M.D., Jezewski A., Kambal A., Liu C.Y. Coinfection. Helminth infection reactivates latent γ-herpesvirus via cytokine competition at a viral promoter. Science. 2014;345:573–577. doi: 10.1126/science.1254517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regoes R.R., McLaren P.J., Battegay M., Bernasconi E., Calmy A., Günthard H.F., Hoffmann M., Rauch A., Telenti A., Fellay J., Swiss HIV Cohort Study Disentangling human tolerance and resistance against HIV. PLoS Biol. 2014;12:e1001951. doi: 10.1371/journal.pbio.1001951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue-Gervais I.G., Labbé K., Dagenais M., Dupaul-Chicoine J., Champagne C., Morizot A., Skeldon A., Brincks E.L., Vidal S.M., Griffith T.S., Saleh M. Cellular inhibitor of apoptosis protein cIAP2 protects against pulmonary tissue necrosis during influenza virus infection to promote host survival. Cell Host Microbe. 2014;15:23–35. doi: 10.1016/j.chom.2013.12.003. [DOI] [PubMed] [Google Scholar]
- Rouse B.T., Sehrawat S. Immunity and immunopathology to viruses: What decides the outcome? Nat. Rev. Immunol. 2010;10:514–526. doi: 10.1038/nri2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B.A., Kirchner J.W. Evolutionary dynamics of pathogen resistance and tolerance. Evolution. 2000;54:51–63. doi: 10.1111/j.0014-3820.2000.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Sandler N.G., Bosinger S.E., Estes J.D., Zhu R.T., Tharp G.K., Boritz E., Levin D., Wijeyesinghe S., Makamdop K.N., del Prete G.Q. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature. 2014;511:601–605. doi: 10.1038/nature13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter D. Counteraction of the multifunctional restriction factor tetherin. Front. Microbiol. 2014;5:163. doi: 10.3389/fmicb.2014.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D.S., Ayres J.S. Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat. Rev. Immunol. 2008;8:889–895. doi: 10.1038/nri2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P., Rice C.M. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schountz T., Prescott J. Hantavirus immunology of rodent reservoirs: current status and future directions. Viruses. 2014;6:1317–1335. doi: 10.3390/v6031317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok J., Warren H.S., Cuenca A.G., Mindrinos M.N., Baker H.V., Xu W., Richards D.R., McDonald-Smith G.P., Gao H., Hennessy L., Inflammation and Host Response to Injury, Large Scale Collaborative Research Program Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. USA. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y.Y., Liang L., Zhu Z.H., Zhou W.P., Irwin D.M., Zhang Y.P. Adaptive evolution of energy metabolism genes and the origin of flight in bats. Proc. Natl. Acad. Sci. USA. 2010;107:8666–8671. doi: 10.1073/pnas.0912613107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestri G., Sodora D.L., Koup R.A., Paiardini M., O’Neil S.P., McClure H.M., Staprans S.I., Feinberg M.B. Nonpathogenic SIV infection of sooty mangabeys is characterized by limited bystander immunopathology despite chronic high-level viremia. Immunity. 2003;18:441–452. doi: 10.1016/s1074-7613(03)00060-8. [DOI] [PubMed] [Google Scholar]
- Sitkovsky M., Lukashev D. Regulation of immune cells by local-tissue oxygen tension: HIF1 alpha and adenosine receptors. Nat. Rev. Immunol. 2005;5:712–721. doi: 10.1038/nri1685. [DOI] [PubMed] [Google Scholar]
- Sohayati A.R., Hassan L., Sharifah S.H., Lazarus K., Zaini C.M., Epstein J.H., Shamsyul Naim N., Field H.E., Arshad S.S., Abdul Aziz J., Daszak P., Henipavirus Ecology Research Group Evidence for Nipah virus recrudescence and serological patterns of captive Pteropus vampyrus. Epidemiol. Infect. 2011;139:1570–1579. doi: 10.1017/S0950268811000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor L.H., Latham S.M., Woolhouse M.E.J. Risk factors for human disease emergence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001;356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totura A.L., Baric R.S. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr Opin Virol. 2012;2:264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmelle A.S., Jackson F.R., Green D., McCracken G.F., Rupprecht C.E. Host immunity to repeated rabies virus infection in big brown bats. J. Gen. Virol. 2010;91:2360–2366. doi: 10.1099/vir.0.020073-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen J.H., Munster V.J., Majoor F., Lexmond P., Vuong O., Stumpel J.B., Rimmelzwaan G.F., Osterhaus A.D., Schutten M., Slaterus R., Fouchier R.A. Avian influenza a virus in wild birds in highly urbanized areas. PLoS ONE. 2012;7:e38256. doi: 10.1371/journal.pone.0038256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin H.W. The virome in mammalian physiology and disease. Cell. 2014;157:142–150. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin H.W., Wherry E.J., Ahmed R. Redefining chronic viral infection. Cell. 2009;138:30–50. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- Wade C.M., Kulbokas E.J., 3rd, Kirby A.W., Zody M.C., Mullikin J.C., Lander E.S., Lindblad-Toh K., Daly M.J. The mosaic structure of variation in the laboratory mouse genome. Nature. 2002;420:574–578. doi: 10.1038/nature01252. [DOI] [PubMed] [Google Scholar]
- Wang T., Town T., Alexopoulou L., Anderson J.F., Fikrig E., Flavell R.A. Toll-like receptor 3 mediates West Nile virus entry into the brain causing lethal encephalitis. Nat. Med. 2004;10:1366–1373. doi: 10.1038/nm1140. [DOI] [PubMed] [Google Scholar]
- Weber B., Schuster S., Zysset D., Rihs S., Dickgreber N., Schürch C., Riether C., Siegrist M., Schneider C., Pawelski H. TREM-1 deficiency can attenuate disease severity without affecting pathogen clearance. PLoS Pathog. 2014;10:e1003900. doi: 10.1371/journal.ppat.1003900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2014) (http://www.who.int/csr/don/2014_08_27_ebola/en/: World Health Organization).
- WHO Ebola Response Team Ebola virus disease in West Africa—the first 9 months of the epidemic and forward projections. N. Engl. J. Med. 2014;371:1481–1495. doi: 10.1056/NEJMoa1411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson G.S., South J.M. Life history, ecology and longevity in bats. Aging Cell. 2002;1:124–131. doi: 10.1046/j.1474-9728.2002.00020.x. [DOI] [PubMed] [Google Scholar]
- Wolfe N.D., Switzer W.M., Carr J.K., Bhullar V.B., Shanmugam V., Tamoufe U., Prosser A.T., Torimiro J.N., Wright A., Mpoudi-Ngole E. Naturally acquired simian retrovirus infections in central African hunters. Lancet. 2004;363:932–937. doi: 10.1016/S0140-6736(04)15787-5. [DOI] [PubMed] [Google Scholar]
- Wolfe N.D., Dunavan C.P., Diamond J. Origins of major human infectious diseases. Nature. 2007;447:279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse M., Scott F., Hudson Z., Howey R., Chase-Topping M. Human viruses: discovery and emergence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012;367:2864–2871. doi: 10.1098/rstb.2011.0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zampieri C.A., Sullivan N.J., Nabel G.J. Immunopathology of highly virulent pathogens: insights from Ebola virus. Nat. Immunol. 2007;8:1159–1164. doi: 10.1038/ni1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Cowled C., Shi Z., Huang Z., Bishop-Lilly K.A., Fang X., Wynne J.W., Xiong Z., Baker M.L., Zhao W. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science. 2013;339:456–460. doi: 10.1126/science.1230835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Li K., Wohlford-Lenane C., Agnihothram S.S., Fett C., Zhao J., Gale M.J., Jr., Baric R.S., Enjuanes L., Gallagher T. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc. Natl. Acad. Sci. USA. 2014;111:4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.