Summary
Recently, the Surface (S)-layer glycoprotein of the thermoacidophilic crenarchaeote Sulfolobus acido-caldarius was found to be N-glycosylated with a heterogeneous family of glycans, with the largest having a composition Glc1Man2GlcNAc2 plus 6-sulfoquinovose. However, genetic analyses of genes involved in the N-glycosylation process in Crenarchaeota were missing so far. In this study we identify a gene cluster involved in the biosynthesis of sulfoquinovose and important for the assembly of the S-layer N-glycans. A successful markerless in-frame deletion of agl3 resulted in a decreased molecular mass of the S-layer glycoprotein SlaA and the flagellin FlaB, indicating a change in the N-glycan composition. Analyses with nanoLC ES-MS/MS confirmed the presence of only a reduced trisaccharide structure composed of Man1GlcNAc2, missing the sulfoquinovose, a mannose and glucose. Biochemical studies of the recombinant Agl3 confirmed the proposed function as a UDP-sulfoquinovose synthase. Furthermore, S. acidocaldarius cells lacking agl3 had a significantly lower growth rate at elevated salt concentrations compared with the background strain, underlining the importance of the N-glycosylation to maintain an intact and stable cell envelope, to enable the survival of S. acidocaldarius in its extreme environment.
Introduction
Glycosylation is one of the most predominant post-translational protein modifications, which plays a critical role in a variety of biological functions. Glycosylation affects not only the folding, stability and half-life time of proteins, but is also important in surface- and cell adhesion, and enables biofilm formation (Peng et al., 2008). Further, the attached glycan facilitates the interaction of cell surfaces with their environment, therefore, influencing pathogenicity (Szymanski et al., 1999; 2002) and the host invasion ability of an organism (Upreti et al., 2003).
Historically, it was long believed that protein N-glycosylation is a unique phenomenon restricted to the Eukarya and that a single, defined glycosylation pathway can lead to the attachment of a glycan to a nascent protein. However, in the last decade this has been disproven, evoking questions about how glycosylation occurs in Bacteria and Archaea. The N-linked glycosylation pathways, in which an oligosaccharide is bound to an asparagine (N) residue in the glycosylation consensus sequence (Asn-X-Thr/Ser; X ≠ Pro) onto a nascent protein via a β-glycosylamide linkage, share some similarities among the three domains of life. This includes the step-wise assembly of sugars, donated by either nucleotide activated or lipid activated sugars, onto a lipid carrier by specific glycosyltransferases. Although Eukarya, Bacteria and Archaea all seem to have certain characteristics of the N-glycosylation pathway in common, the glycosylation in Bacteria and Archaea are more diverse. For instance, a variety of monosaccharides is used, which leads to versatile structures of the oligosaccharides (Calo et al., 2010).
In recent years substantial progress in analysing the archaeal N-glycosylation pathway of three distinct Euryarchaea Haloferax volcanii (Abu-Qarn et al., 2007; Magidovich et al., 2010; Yurist-Doutsch et al., 2010), Methanococcus maripaludis (Kelly et al., 2009; VanDyke et al., 2009) and Methanococcus voltae (Voisin et al., 2005; Chaban et al., 2009) has been made. Most of the glycosyltransferases playing a role in the N-glycosylation pathways have been identified and described in these organisms. Moreover, it was demonstrated that the oligosaccharyltransferase, AglB in euryarchaeota, is a non-essential protein (Chaban et al., 2006; Abu-Qarn et al., 2007). However, no information about the N-glycosylation pathway of Crenarcheaota exists. Many extracellular proteins such as the surface (S-) layer protein, sugar binding proteins, cytochromes, pilins, and flagellins of crenarchaea have been shown to be sugar-modified (Albers and Meyer, 2011). However, very little is known about the structures and biosynthesis of these modifications. The only well-characterized glycoproteins in crenarchaea are the cytochrome b558/566 (Zahringer et al., 2000) and the S-layer SlaA (Peyfoon et al., 2010) from the thermoacidophilic archaeon Sulfolobus acidocaldarius. They share a hexasaccharide structure that exhibits very interesting features: (i) in contrast to the linear euryarchaeal glycans this hexasaccharide is tri-branched; (ii) it contains the typical eukaryal chitobiose core; and (iii) it contains 6-sulfoquinovose (6-deoxy-6-sulfoglucose), a sugar that is commonly only found in photosynthetic membranes of plants and phototrophic bacteria.
Sulfoquinovose (QiuS) is found as the head group of the nonphosphorous lipid sulfoquinovosyldiacylglycerol (SQDG), which is localized exclusively in the photosynthetic membrane of all higher plants, mosses, ferns, algae, as well as in most photosynthetic bacteria (Heinz, 1993; Benning, 1998). SQDG is a relatively minor lipid compared with the other characteristic lipids monogalactosyldiacylglycerol, digalactosyldiacylglycerol and phosphatidylglycerol, comprising only 4–7% of total leaf lipids. Although there seems to be a tight correlation between SQDG and phototropic organisms, which implies that SQDG might play an important role for photosynthesis, the presence of SQDG does not always correlate with photosynthetic ability. Thus, some phototrophic bacteria lack or possess only small amounts of SQDG (Sallal et al., 1990; Merritt et al., 1991) while non-photosynthetic organisms like Rhizobium meliloti and the extreme thermoacidophilic Alicyclobacillus acidocaldarius (Wisotzkey et al., 1992) possesses SQDG (Langworthy et al., 1976; Cedergreen and Hollingsworth, 1994). Deletion studies revealed that SQDG is important only under certain conditions such as phosphate limitation (Benning et al., 1993); furthermore, it was recently demonstrated that the UDP-sulfoquinovose synthase was upregulated under salt stress conditions, implying a important role for maintaining the integrity and stability of the thylakoid membrane under these stress conditions (Xu et al., 2010).
Chemical mutagenesis screening for SQDG deficit mutants and genetic complementation revealed three essential genes for the synthesis of SQDG (Benning and Somerville, 1992a,b) designed sqdA, sqdB and sqdC. Later, SqdD was also identified as a participant in the biosyntheses pathway for SQDG. From all of these genes sqdB is the only gene that has apparent orthologues in every SQDG producing organism.
We have now identified a sulfoquinovose biosynthesis operon in S. acidocaldarius, Saci0421-Saci0424 and designated these genes involved in archaeal glycosylation agl1 to agl4. Taking advantage of the genetic tools that are now available for this crenarchaeon (Wagner et al., 2009), we used genetic, biochemical and mass spectrometric methods to elucidate the function of agl3, a predicted UDP-sulfoquinovose synthase.
Results
Biosynthesis of UDP-sulfoquinovose in S. acidocaldarius
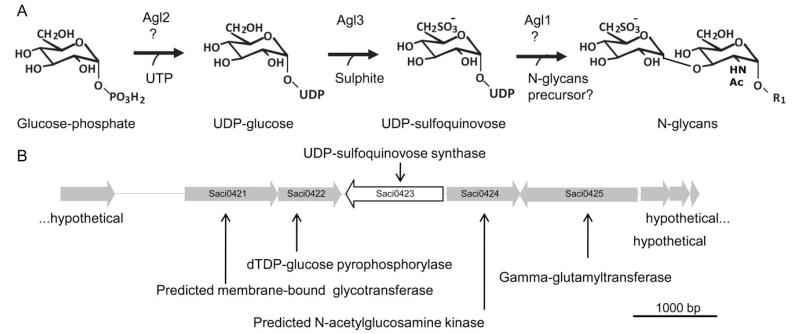
To elucidate the biosynthesis pathway of sulfoquinovose the genome of S. acidocaldarius was scanned for the presence of sequence homologues to the known bacterial sqdB or eukaryal sqd1 genes encoding the UDP-sulfoquinovose synthase, involved in the biosynthesis of the sulfolipid sulfoquinovosyldiacylglycerol (Fig. S1). This analysis revealed a homologue Saci0423, which has 40% sequence identity. The gene agl3 (Saci0423), annotated as a sugar epimerase, shows the conserved Rossman-fold motifs at the N-terminus of the protein sequence as well as conserved amino acids proposed to bind NAD+ (Mulichak et al., 1999) (Fig. S2).
Upstream of the gene agl3, two genes are localized, which might also be involved in the biosynthesis of UDP-sulfoquinovose, namely agl1 (Saci0421) predicted to code for a membrane bound glycosyltransferase and agl2 (Saci0422) annotated as a dTDP-glucose pyrophosphorylase. Additionally, directly downstream of agl3, the gene agl4 (Saci0424) annotated as a glucokinase might be also involved, phosphorylating glucose as the first reaction step. The proposed biosynthesis pathway for UDP-sulfoquinovose in S. acidocaldarius is given in Fig. 1. Here, phosphorylated glucose may be generated by agl4, which presumably acts as a precursor for UDP-glucose synthesis by agl2. In the second step, UDP-glucose will be further modified with sulphite by the action of agl3 to create UDP-sulfoquinovose. Eventually, the activated sulfoquinovose is transferred onto the nascent glycan structure (Fig. 1).
Fig. 1.
Probable sulfoquinovose biosynthesis pathway in S. acidocaldarius.
A. Proposed biosynthetic pathway of the UDP-sulfoquinovose in S. acidocaldarius. R1, GlcNAc-P-Dol; UTP, uridine triphosphate; agl2 encoding for an UDP-glucose pyrophosphorylase; agl3 coding for an UDP-sulfoquinovose synthase; agl1 coding for a membrane bound glucosyltransferase. Biosynthetic pathway in agreement with (Shimojima, 2011).
B. Physical map of the gene region of S. acidocaldarius, in which the gene coding for UDP-sulfoquinovose synthase is located. Illustrated are the genes Saci0420 until Saci0428. The red displayed gene encodes the UDP-sulfoquinovose synthase. The genes Saci0421 und Saci0422 are most likely also involved in the biosynthesis of sulfoquinovose (see A).
Markerless in-frame deletion of Agl3
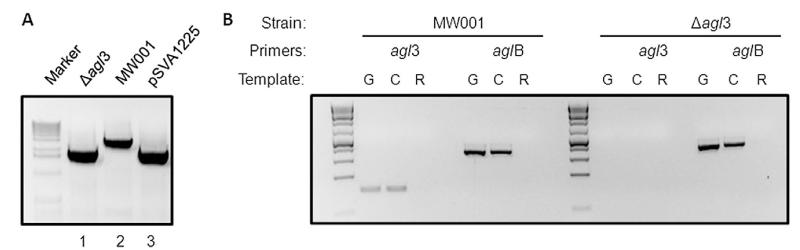
To confirm the proposed function of agl3 as an UDP-sulfoquinovose synthase, a markerless in-frame deletion mutant was created. This mutant was first confirmed by PCR amplification of the gene area with the up-for and down-rev primers, resulting either in a 3028 bp or a 1876 bp fragment for the background strain MW001 or the Δagl3 mutant respectively (Fig. 2).
Fig. 2.
Confirmation of the in-frame deletion mutant Δagl3 (Saci0423).
A. Gene deletion was confirmed by PCR using the outside primers against the flanking regions of agl3, DNA isolated from the background strain MW001 (lane 2), plasmid pSVA1225 used for the homologous recombination incorporating the up- and downstream region of deleted agl3 (lane 3), or the mutant lacking agl3 (lane 1).
B. RT-PCR confirms the deletion of the gene agl3 presumably encoding the UDP-sulfoquinovose. The cDNA (C) served as a template in PCR amplification using primers against the internal region of either the agl3 or aglB. In each case, PCR amplifications were also performed using genomic DNA (G) as a positive control and total RNA (R) as a negative control.
To confirm that in the mutant agl3 had been deleted, reverse transcription polymerase chain reaction (RT-PCR) was performed. For this experiment, RNA was isolated at mid-exponential growth phase of the background strain MW001 and Δagl3 mutant cells. Isolated RNA was used as a template for the synthesis of single-stranded cDNA, using reverse transcriptase. The cDNA was used as a template for the PCR using primers against the internal region of either the agl3 or aglB, encoding the oligosaccharyl transferase. The control experiments using genomic DNA, cDNA and RNA as templates for the PCR using primers against the internal region aglB (Fig. 2), confirmed for the mutant as well as for the background strain MW001, the absence of genomic DNA in the RNA sample as well as the functional synthesis of the cDNA. The PCR using primers against the internal region of agl3 confirmed the absence of agl3 in the deletion mutant strain.
Deletion of agl3 showed a significant change in the size of the S-layer SlaA and the flagellin FlaB glycoproteins
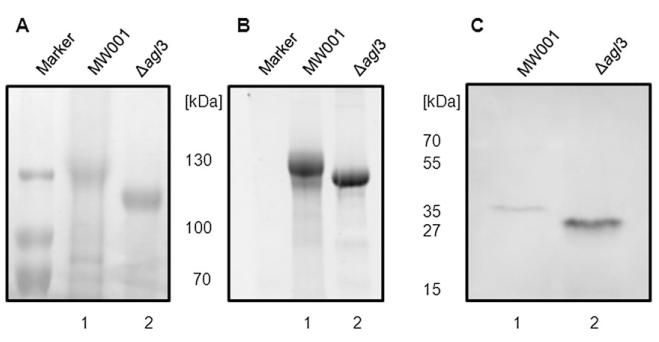
To determine the influence of Agl3 on the biogenesis of the S-layer glycoprotein, the S-layer proteins SlaA of strain MW001 and of the Δagl3 mutant were isolated and separated on a 8% SDS-PAGE (Fig. 3A and B). Coomassie staining revealed that both samples contained similar amounts of the S-layer glycoprotein, although the protein sample from Δagl3 revealed a slightly better staining, implying a better accessibility of the staining reagent to the protein. Moreover, SlaA from the Δagl3 deletion mutant migrated faster than the wild-type protein (Fig. 3A and B), implying a significant change in the composition of the SlaA glycan. Both S-layer protein samples were stained for carbohydrate using Pro-Q Emerald 300 demonstrating that both S-layer proteins were still modified by the attachment of glycans.
Fig. 3.
Effect of the agl3 deletion on S. acidocaldarius S-layer protein glycosylation. Equivalent amounts of the S-layer protein SlaA isolated from S. acidocaldarius MW001 (lane 1) and Δagl3 (lane 2) were separated by 8% SDS-PAGE and either (A) Coomassie blue-stained or (B) stained for carbohydrates using Emerald 300 (B).
C. Effect on S. acidocaldarius flagella protein FlaB glycosylation of the agl3 deletion. Equivalent amounts of cells from S. acidocaldarius MW001 (lane 1) and Δagl3 (lane 2) were separated by 11% SDS-PAGE and immunoblotted with antibodies raised against FlaB.
To determine whether also the glycan composition of the flagellin FlaB had been altered by the deletion of agl3, resuspended cell pellets from S. acidocaldarius MW001 and the Δagl3 strain were separated by 11% SDS-PAGE and immunoblotted with antibodies raised against FlaB. FlaB from the Δagl3 deletion mutant migrated significantly faster than the wild-type protein (Fig. 3C). Although equivalent amounts of cells were used as confirmed by Coomassie staining (data not shown), the antibody was binding with a higher affinity to the flagellin of the deletion strain, implying a better accessibility to the protein.
The SlaA glycan composition is changed in the Δagl3 deletion mutant
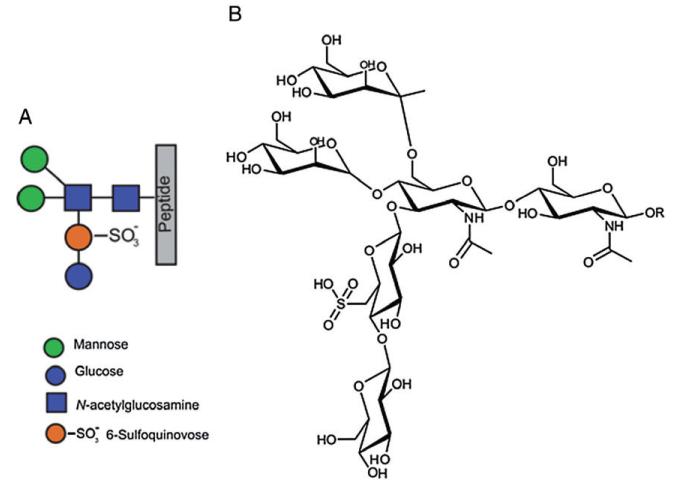
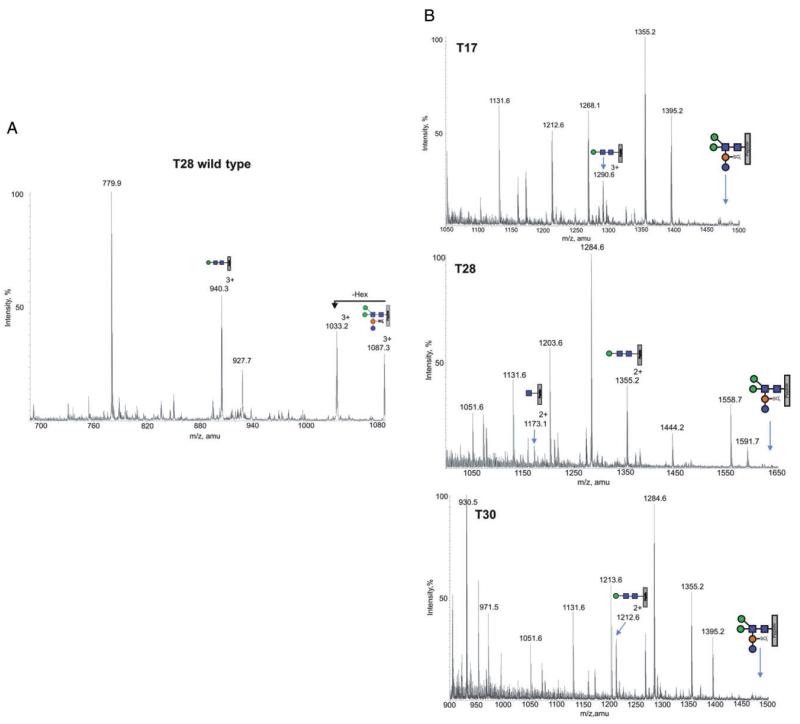
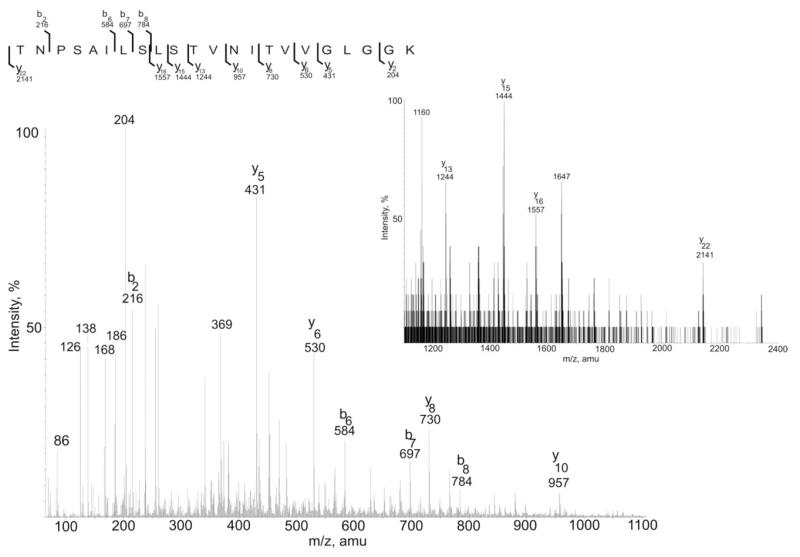
In order to verify changes in the glycan modification of the mutant, in terms of the glycan composition and structure, a tryptic digests of wild type and mutant were analysed by nanoLC ES-MS/MS. Wild-type S. acidocaldarius glycopeptides were previously shown by mass spectrometry to carry a hexasaccharide with the composition Glc1Man2GlcNAc2QuiS (Fig. 4) with some glycoforms being truncated at presumed intermediate steps of biosynthesis, with the trisaccharide Man1GlcNAc2 being the dominant truncated structure (Peyfoon et al., 2010). As shown in Fig. 5 for the tryptic peptides T17, T28 and T30 [see (Peyfoon et al., 2010) for designation of the tryptic peptides], the intact hexasaccharide is no longer present in the mutant. Instead, the largest molecular ions for each glycopeptide (1290.63+, 1355.22+ and 1212.62+ for T17, T28 and T30, respectively) correspond to glycosylation with the trisaccharide Hex1HexNAc2. MS/MS analyses confirmed the identity of the peptide in all three glycopeptides (T28 MS/MS data are presented as an example in Fig. 6). Significantly, the fingerprint fragment ions for HexNAc (m/z 204, 186, 168) consistently appeared in all MSMS data from the Δagl3 mutant, while the m/z 430 ion, which is diagnostic for the HexNAcQuiS moiety [see (Peyfoon et al., 2010)], was absent in all MS/MS spectra. Thus, the mass spectrometric data confirmed the absence of the sulfoquinovose residue in the mutant. Moreover, the MS data show that glycan biosynthesis in the mutant terminates at the trisaccharide sequence, suggesting that attachment of the second peripheral mannose (Fig. 4) requires prior addition of sulfoquinovose.
Fig. 4.
Graphic and chemical representation of the S-layer N-glycan.
A. The full glycan structure shown by the colour-coded symbols.
B. Chemical structure of the Glc1Man2GlcNAc2QuiS hexasaccharide observed in the wild type S. acidocaldarius.
Fig. 5.
Detailed analysis of the Δagl3 deletion mutant S-layer glycop peptide.
A. Total ion chromatogram of glycan profile for the T28 peptide in S. acidocaldarius wild type with the annotated peaks showing the glycoforms observed (M+3H)3+.
B. Total ion chromatogram for three selected glycopeptides in S. acidocaldarius lacking agl3. The annotated peaks show the Hex1HexNAc2 glycoform (M+2H)2+ is preserved on all glycopeptides, whereas the full glycan is completely missing. For reference, each spectra is annotated with the absent full glycan for each glycopeptide.
Fig. 6.
MSMS data of the T28 glycopeptide with the Hex1HeNAc2 residue. Both b and y theoretical ions firmly matched with the experimental MSMS data. Inset figure shows MS spectra with larger m/z-values.
The alteration of the N-glycan revealed a change in the phenotype under environmental stress conditions
Because the S-layer is the sole surface envelope in S. acidocaldarius, composed of two S-layer proteins, a large outer protein SlaA and the small membrane-bound protein SlaB, any changes in the biogenesis of these glycoproteins presumably have an effect on the interaction and the stability of the protective S-layer. Therefore, S. acidocaldarius most likely has to ensure the maintenance and integrity of its cell envelope. Due to the deletion of agl3, important for the biosynthesis of the sulfoquinovose incorporated in the N-glycan, the stability of the S-layer might be altered. To analyse any effect of the S-layer structural stability and arrangement, cells lacking agl3 and MW001 were grown under salt stress conditions.
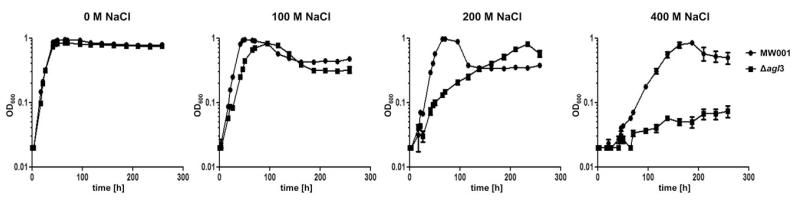
Under normal growth conditions (79°C and pH 3) no significant changes in the doubling times during growth of the mutants (td = 8.1 h ± 0.5 h) compared with the background strain MW001 (td = 7.9 h ± 0.02 h) was observed (Fig. 7). The effect of the reduced glycan structure to the S-layer stability became more obvious at elevated salt concentration. At 200 mM NaCl, MW001 showed no significant changes of the growth parameters (td = 8.8 h ± 0.02 h) compared with the standard condition. However, the mutant showed an increased lag-phase at 200 mM NaCl and a slightly prolonged doubling time (td = 13.1 h ± 0.4 h). At 300 mM NaCl, the growth of the deletion mutant is profoundly affected. Under these conditions the doubling time of the mutant (td = 63.9 h ± 2 h) is 4.5 times longer than that of the background strain (td = 14 h ± 0.4 h). At 400 mM NaCl, only the background strain MW001 is capable of sustaining growth after a long lag-phase (td = 32.5 h ± 2 h), whereas the mutant showed no growth. In the mutant sample the slight increased OD of 0.07 after 260 h was caused by the evaporation of the medium.
Fig. 7.
Response to salt stress in the Δagl3 strain. S. acidocaldarius MW001 or Δagl3 were grown in Brock medium at different salt concentrations (0–400 mM NaCl). Growth was measured at an optical density of 600 nm (OD600). Shown are values obtained from two independent repeats.
Expression, purification, activity of the Agl3 and complementation of the deletion mutant
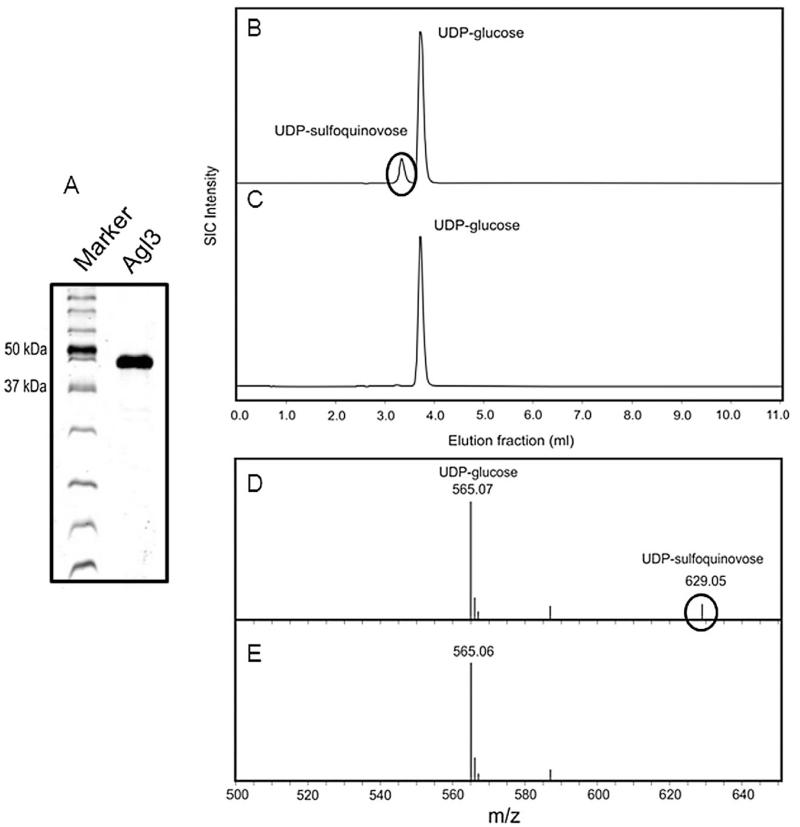
To unequivocally establish that Saci0423 is an UDP-sulfoquinovose synthase (Agl3), the 48 kDa enzyme was cloned into an expression vector, produced in E. coli and purified by His-tag affinity chromatography (Fig. 8A). The enzyme was purified at a final yield of 1.25 mg l−1 of E. coli expression culture. By Western blot analysis and MS the identity of the recombinant protein was confirmed (results not shown).
Fig. 8.
Agl3 is a UDP-sulfoquinovose synthase.
A. SDS-PAGE analysis (12% gel) of the expression and purification of sulfoquinovose synthase (Agl3) from S. acidocaldarius in E. coli.
B and C. RP-HPLC analysis of the conversion of UDP-glucose and sulphite into UDP-sulfoquinovose catalysed by (B) Agl3 and the (C) negative control.
D and E. PGC-ESI-MS analysis showing the conversion of UDP-glucose and sulphite into UDP-sulfoquinovose catalysed by Agl3 (D) and negative control (E).
For activity assays, the purified Agl3 was incubated overnight with UDP-glucose in the presence of sodium sulphite. As calculated from the RP-HPLC analysis, under the chosen assay conditions the conversion rate of UDP-Glc and sulphite into UDP-sulfoquinovose was 11% (Fig. 8B), whereas in the control sample without enzyme under the same conditions no UDP-sulfoquinovose was formed (Fig. 8C). PcG-ESI-MS analysis of the product confirmed its identity as UDP-sulfoquinovose (Fig. 8D/E).
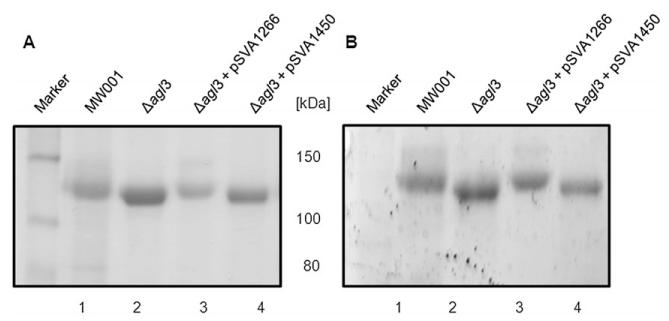
To prove that the deletion of agl3 did not have secondary effects on other enzymes involved in the sulfoquinovose production, we cloned the agl3 gene in a S. acidocaldarius vector pSVA1450, a variant of the pCmalLacS (Berkner et al., 2010), and expressed it in the agl3 deletion mutant under the control of a maltose inducible promoter. S-layer proteins were isolated from the background strain MW001, the Δagl3 deletion mutant, and from the Δagl3 deletion mutant transformed with either a control plasmid (pSVA1450) not containing any gene in the expression cassette or the plasmid expressing the agl3 gene (pSVA1266). Figure 9 shows the different S-layer proteins separated on an SDS-PAGE and stained by Coomassie. It was clear, that the expression of agl3 led to the restoration of the production of full-length glycan as the S-layer protein of the complemented Δagl3 strain was running at the same molecular weight as the S-layer protein isolated from the MW001 strain. The presence of the control plasmid had no effect on the running behaviour of the isolated S-layer protein.
Fig. 9.
Complementation of the Δagl3 deletion mutant. Equivalent amounts of the S-layer protein SlaA isolated from S. acidocaldarius MW001 (lane 1), Δagl3 (lane 2), Δagl3 complemented with pSVA1266 (lane 3), and Δagl3 with control plasmid pSVA1450 (lane 4) were separated by 8% SDS-PAGE and either (A) Coomassie blue-stained or (B) stained for carbohydrates using Emerald 300 (B).
Discussion
So far knowledge of archaeal N-glycosylation pathway is still restricted to three distinct euryarchaeota; these are, M. voltae, M. maripaludis and H. volcanii (Voisin et al., 2005; Abu-Qarn et al., 2007; Chaban et al., 2009; Kelly et al., 2009; VanDyke et al., 2009; Magidovich et al., 2010; Yurist-Doutsch et al., 2010). Furthermore structural as well as in vitro activity assays of the key enzyme AglB from Pyrococcus furiosus, revealed new insides in the catalysing amino acids as well as the preferred N-glycosylation motifs (Igura et al., 2007; Igura and Kohda, 2011). It is astonishing that the effect of glycosylation in archaea is not well characterized, despite the fact that most of the surface proteins, e.g. archaeal flagellins (Ng et al., 2006), pilins (Ng et al., 2011), S-layer proteins (Mescher and Strominger, 1976; Kessel et al., 1988; 1990; Sumper et al., 1990; Zeitler et al., 1998; Voisin et al., 2005; Peyfoon et al., 2010), and transport proteins (Albers et al., 1999; Elferink et al., 2001; Palmieri et al., 2006) from diverse archaea being known to be modified by O- and/or N-glycosylation.
The S. acidocaldarius S-layer protein SlaA is highly glycosylated, incorporating one N-glycosylation motif for each stretch of 30–40 amino acid residues. Although not all predicted glycosylation sites were shown to be modified, analysis of the C-terminus of the protein confirmed the high glycosylation density, which at least nine (and possibly all) of the 11 N-glycosylation motifs were found to be modified (Peyfoon et al., 2010). The identification of N-glycosyltransferases in S. acidocaldarius is not trivial due to the limited sequence similarity of glycosyltransferases (GTases) compared with GTases participating in the eukaryal or bacterial N-glycosylation process, and the fact that the GTases are not clustered near the aglB as is the case in euryarchaeota. The scattering of the GTases all over the genome seems to be an adaptation of the thermophilic lifestyle as it is found also in other thermophilic archaea (Magidovich and Eichler, 2009). The knowledge that sulfoquinovose is incorporated in the N-glycan of the S-layer enabled us to identify the sulfoquinovose biosynthesis gene cluster; however, no additional gene possibly involved in protein glycosylation could be detected near this cluster. A BLAST search of known bacterial sqdB or eukaryal sqd1 genes encoding the UDP-sulfoquinovose synthase against the genome of S. acidocaldarius revealed a protein with ~ 40% sequence identity. The gene Saci0423 coding for the predicted UDP-sulfoquinovose synthase is localized in a cluster comprising further genes presumably also participating in the biosynthesis of UDP-sulfoquinovose (Fig. 1). We designated these genes agl1 to agl4 (archaeal glycosylation) with agl3 being the potential UDP-sulfoquinovose synthase.
The analysis of S-layer proteins from a Δagl3 deletion mutant showed enhanced migration in SDS-PAGE compared to the S-layer protein of the background strain MW001. Although the same protein amount was loaded onto the gel, a significantly higher Coomassie Brilliant Blue staining of the S-layer protein from the Δagl3 strain was observed, most probably due to better accessibility of the protein by the reduced glycan composition. The same held true for the flagellin FlaB. This result indicated that the sulfoquinovose glycan decorating the S-layer protein is likely to also be attached to the FlaB protein. The presence of the same glycan structure linked to different surface proteins is common in Archaea. In M. voltae, the flagella and the S-layer protein, carry the same N-glycans (Voisin et al., 2005). Also in M. maripaludis the pilin subunit and the flagella subunit share the same base glycan structure, on which an additional hexose is attached on the pilin subunit in a mutant unable to assemble flagella (VanDyke et al., 2009; Ng et al., 2011). However, there are also reports of Archaea in which the glycan composition structure differs even within one protein. The S-layer protein from Hb. salinarum has been shown to be modified by two different N-glycan moieties and one O-glycan structure (Paul et al., 1986; Lechner and Sumper, 1987). However, it should be mentioned that in general the N- and O-linked glycans differ, as it was shown, e.g. in S-layer of H. volcanii (Sumper et al., 1990; Abu-Qarn et al., 2008) or the cytochrome b558 of S. acidocaldarius (Hettmann et al., 1998; Zahringer et al., 2000).
To verify whether the different migration behaviour of the S-Layer protein isolated from the wild type and the agl3 deletion mutant did indeed correspond to the missing sulfoquinovose residue in the glycan structure, isolated S-layer proteins were analysed by a nanoLC ES-MS/MS. These analyses confirmed that a trisaccharide corresponding to Man1GlcNAc2 was the most abundant as well as the largest N-linked glycan in the deletion mutant, whereas the full-length hexasaccharide (Glc1QuiS1Man2GlcNAc2) was no longer present (Fig. 5). These results confirmed that the sulfoquinovose is absent in the N-glycans of the Δagl3 deletion mutant and that the alteration in the migration behaviour of the S-layer protein on SDS-PAGE corresponds to the S-layer proteins carrying the truncated version of the N-glycan. Furthermore, the analysis implies a highly ordered assembly of the sugar precursor during the N-glycosylation process. The lack of the second Man residue connected directly to the second GlcNAc residue shows that the unknown GTase transferring the second Man requires a sulfoquinovose attached to the glycan to either recognize the reaction site or to catalyse/fulfil the transferring step onto the second GlcNAc residue.
To understand the activity of the Agl3 enzyme we performed an in vitro assay, which showed that the recombinant Agl3 indeed catalyses the biosynthesis of UDP-sulfoquinovose by the transfer of sulphite to UDP-glucose (Fig. 9). However, the reaction rate of this enzyme was quite low, transforming only 11% of the UDP-glucose to UDP-sulfoquinovose at the preferred temperature of 70°C in an over night reaction. Also the eukaryal homologue SQD1 from Arabidopsis thaliana recombinant expressed in E. coli showed low enzyme activity (Sanda et al., 2001). It was proposed that the limited enzyme activity is caused either by a non-perfect or missing cofactor or that the enzyme is part of a multiprotein complex. Although SQD1 normally contains NAD+ in its binding site, the addition of NAD+ in the in vitro assay did not affect product formation (Sanda et al., 2001). Further, different sulphonic acid substrates (sulphate, APS, PAPS, sulphite, thiosulphate, sulphide, or sulfoglutathione) were tested and it was shown that sulphite was the best substrate for the biosynthesis of QuiS (Sanda et al., 2001).
Purification of the spinach SQD1 from chloroplast indeed showed that SQD1 is part of a 250 kDa protein complex (Shimojima and Benning, 2003), in which ferredoxin-dependent glutamate synthase (FdGOGAT) was co-purified. In a tentative model the flavin containing protein FdGOGAT is thought to deliver the sulphide directly to SQD1, mitigating the potential problem of the low concentration of sulphite in plant cells and enhancing the enzyme activity (Shimojima et al., 2005). Whether also in S. acidocaldarius the UDP-sulfoquinovose synthase is forming a complex with the archaeal version of FdGOGAT and whether this is increasing activity of the enzyme in vivo is not known yet.
Although the glycosylation defect in Δagl3 showed no alteration of the phenotype under normal growth condition, under elevated salt concentration (200, 300 and 400 mM NaCl), the deletion mutant showed a significant growth retardation. Furthermore, the deletion strain showed a reduced maximum OD600 (Fig. 7). Due to the missing sulfoquinovose, which enhances the negative charge of the surface creating a hydrated shell, the stability of the S-layer is clearly affected. A comparison of the glycosylation of related S-layer glycoproteins from moderate and extreme halophilic archaea revealed drastic differences in the glycosylation patterns. Neutral oligosaccharides are synthesized in the moderate halophile H. volcanii, whereas in the extreme halophile Hbt. salinarum, these sugars are replaced by glucuronic acid, which are additionally sulphated, introducing another negative moiety. The increased negative charge character had been proposed to represent an adaptation to the enhanced saline environment (Mengele and Sumper, 1992).
In conclusion, we have identified the first player involved in the N-glycosylation pathway of a crenarchaeon. Using deletion mutant analysis and enzyme assays we demonstrated that Agl3 is the UDP-sulfoquinovose synthase of S. acidocaldarius and plays an important role in building up the glycans, which are attached to the S-layer and the flagellin proteins. Further studies will be undertaken to uncover the complete N-glycosylation pathway in S. acidocaldarius.
Experimental procedures
Strains and growth conditions
Sulfolobus acidocaldarius MW001(ΔpyrE) (M. Wagner and S.V. Albers, unpublished) and the mutant Δsaci0423 (Δagl3) were grown in Brock medium at 79°C, pH adjusted to 3 using sulphuric acid, and supplemented with 0.1% w/v NZamine and 0.1% w/v dextrin as carbon and energy source (Brock et al., 1972). Gelrite (0.6%) plates were supplemented with the same nutrients (as shown above), with the addition of 10 mM MgCl2 and 3 mM CaCl2. For second selection plates 10 μg ml−1 uracil and 100 μg ml−1 5-fluoroorotic acid (5-FOA) were added. For the growth of the uracil auxotrophic mutants MW001 and Δagl3, 10 μg ml−1 uracil was added to the medium. Growth of cells was monitored by measuring the optical density at 600 nm.
Construction of deletion plasmid
To test the functional properties of the predicted UDP-sulfoquinovose-synthase, a markerless deletion mutant of the gene was created as previously described (Wagner et al., 2009). Briefly, the MW001 strain, in which the pyrE genes were disrupted, resulting in an auxotrophic phenotype for uracil, was transformed with plasmid pSVA1225. For the construction of this plasmid 800–1000 bp fragments up- and downstream of Saci0423 were PCR amplified. For the upstream fragment the forward primer introduced an ApaI restriction site at the 5′ end, the reverse primer was designed to incorporate 15 bp at his 5′ end, originated from the reverse complement 5′ downstream fragment (primers: 5′ CCCA CTGGGCCCTCTCAGATGTTTACGTGCCTATCTC 3′ and 5′ GTAATGAGGATTCTAGTTAAAAGAGTAAGATAAACATAAA TCTCATG 3′, respectively).
For the downstream fragment the forward primer designed to incorporate 15 bp at his 5′ end, with the reverse complement of the 5′ upstream fragment, the reversed primer introduced a BamHI restriction site at the 5′ end (primers: 5′ TCTTACTCTTTTAACTAGAATCCTCATTACGCTTCGAG 3′ and 5′ CGCCGAGGATCCCTGCACCCTAATTCCTTGAAT GC 3′, respectively).
The up- and downstream fragment are overlapping with 30 bp and were fused by an overlapping PCR, using the 3′ ends of the up- and downstream fragments as primers. The amplified PCR fragment was purified by electrophoresis in 0.8% agarose gel using a Nucleospin Extract kit (Macherey-Nagel, Düren, Germany), and digested with ApaI and BamHI. The repurified fragment was ligated into plasmid pSVA407, containing pyrEF (M. Wagner and S.V. Albers, unpublished), predigested with the same restriction enzymes. The ligation was transformed into E. coli DH5α and plated on LB-plates containing 50 μg ml−1 ampicillin; the identity of the plasmid was confirmed by sequencing. In order to methylate the plasmid to avoid restriction in S. acidocaldarius, pSVA1225 was transformed in E. coli ER1821 cells containing pM.EsaBC4I (available from NEB), which expresses a methylase.
Transformation and selection of the deletion mutant in S. acidocaldarius
Competent cells were generated according to the protocol of Kurosawa and Grogan (Kurosawa and Grogan, 2005). S. acidocaldarius strain MW001 was grown in Brock medium containing 0.1% w/v NZamine and 0.1% dextrin until an OD600 between 0.1 and 0.3. The cells were cooled on ice for 20 min and harvested by centrifugation (2000 g at 4°C for 20 min). Cell pellets were washed three times in 50 ml, 10 ml and 1 ml of cold 20 mM sucrose (in demineralized water) applying mild centrifugation (2000 g at 4°C for 20 min). Finally the cell pellet was suspended in 20 mM sucrose to an OD600 of 10. Cells were stored in 50 μl aliquots at −80°C.
For electroporation, a 50 μl aliquot of competent MW001 cells was mixed on ice with 400–600 ng of methylated pSVA1225 plasmid and incubated for 5 min on ice. Samples were transferred into a 1 mm gap electroporation cuvette. The electroporation was performed at 1250 V, 1000 Ω, 25 μF using a Biorad gene pulser II (Biorad, X, USA). Immediately after the pulse 50 μl of a 2× concentrated recovery solution (1% sucrose, 20 mM β-alanine, 20 mM malate buffer pH 4.5, 10 mM MgSO4) was added. After 30 min of incubation at 75°C under mild shaking conditions (150 r.p.m.), additionally 100 μl of heated 2× concentrated recovery solution were added, and 2 × 100 μl were spread to Gelrite plates containing Brock medium supplemented with 0.1% NZamine and 0.1% dextrin. Plates were incubated for 5–7 days at 78°C. Large brownish colonies were used to inoculate 50 ml of Brock medium containing 0.1% NZ-amine. After 3 days of incubation at 78°C, each culture was screened for the presence of the genomic integrated plasmid pSVA1225 by PCR. Positive tested cultures were grown in Brock medium supplemented with 0.1% NZamine and 0.1% dextrin until an OD of 0.4, from which 40 μl aliquots were spread on second selection plates. Plates were incubated for 5–7 days at 78°C, colonies were picked and streaked out on new second selection plates to ensure that they were formed from single colonies. These colonies were screened for the absence or presence of the agl3 gene by PCR. PCR results were confirmed by sequencing and RT-PCR.
RT-PCR
RNA isolation was carried out using an RNAeasy Plus Mini extraction kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. RNA concentration was analysed by spectrophotometrically. Contaminating genomic DNA was eliminated by a repetitive 2 h incubation with DNase I (Fermentas, St. Leon-Rot, Germany), followed by a repurification step with the RNAeasy Plus Mini extraction kit. Before cDNA preparation control experiments were performed using PCR amplification on total RNA to exclude any contribution from contaminating genomic DNA. First Strand cDNA synthesis kit (Fermentas) was used for cDNA synthesis, according to the manufacturer’s instructions. To confirm the deletion of the gene agl3 by RT-PCR, specific forward and reverse primers amplifying the internal region of agl3 (primers: 5′GACGAAGATGCCCTCATGCTC 3′ and 5′GTTGGGATGTTACTAATGGGCAC 3′) or as a control the internal region of aglB (primers: 5′CTGCTGCAATTACAGCGTTC 3′ and 5′ AACCGTGAGCTACTTCAGAC 3′) were designed and the single-stranded cDNA served as a template in the PCR reaction.
S-layer Isolation
Fresh cells or frozen cell pellets from a 50 ml culture were resuspended in 40 ml of buffer A (10 mM NaCl, 1 mM PMSF, 0.5% Na-lauroylsarcosine) with the addition of a spatula tip of DNase. The samples were shaken for 45 min at 37°C and centrifuged for 20 min in an Optima Max-XU Ultracentrifuge (Beckman Coulter) at 21 000 r.p.m., yielding a brownish tan pellet. The pellet was resuspended in 1.5 ml of buffer A and incubated for 30 min at 37°C. After centrifugation in a tabletop centrifuge at 14 000 r.p.m. the pellet was purified by repeated washes in buffer B (10 mM NaCl, 0.5 mM MgSO4, 0.5% SDS), incubation for 20 min at 37°C and subsequent centrifugation, until a translucent tan pellet was obtained. Once the pellet was translucent the S-layer proteins were once washed with water and then stored in water at 4°C.
Immunostaining of the flagellin FlaB
Fresh cells from 50 ml MW001 and Δagl3 cultures, respectively, were resuspended in 40 ml of Brock medium without NZamine as a C- and energy source and grown for 4 h at 79°C to induce the production of flagella (K. Lassak, unpublished). After centrifugation, cells were resuspended in 1 ml of buffer (20 mM TrisHCl, 500 mM NaCl). From these samples, 30 μl was loaded on a 11% SDS-PAGE and run at 100 V. Transfer to a PVDF membrane and blotting were performed as commonly done. The primary antibody was an antibody raised in rabbits against a FlaB peptide (Eurogentech) and for detection an anti rabbit IgG–alkaline phosphatase coupled antibody (Sigma Aldrich, St Louis, USA) was used. Chemifluorescence was measured in a Fujifilm LAS-4000 Luminescent image analyzer (Fujifilm, Duesseldorf, Germany).
Tryptic digest for the mutant S-layer samples
Purified S-layer mutants were run on a 2–8% precast gel (Nitrogen, Paisley, UK) and stained with Novotex Colloidal blue stain (Invitrogen). The stained band was excised from the gel and cut into relatively small pieces and destained, using 400 μl of 50% (v/v) acetonitrile in 0.1 M ammonium bicarbonate (pH 8.4) and dried in a SpeedVac. Dried gel pieces were reswollen in AMBIC solution and incubated in 25 ng μl−1 trypsin (20 μl) (Promega cat V5111) at 37°C, overnight. The supernatant was stored in a clean Eppendorf tube. In order to stop the tryptic reaction, the gel pieces were incubated (37°C for 10 min) in 0.1% TFA (50 μl) followed by incubation (37°C for 10 min) in acetonitrile (100 μl) to shrink the gel pieces. The supernatant was then added to the previously stored glycopeptides and reduced to a volume of 35 μl for nanoLC-ES-MS analysis.
ES-MS analysis
The tryptic peptides and glycopeptides were analysed by nano-LC-ES-MS/MS coupled to quadruple TOF analyser (Q-STAR Pulsar I, MDS Sciex). Peptides/glycopeptides were separated using a nano-LC gradient method, generated by an Ultimate pump fitted with a Famos autosampler and a Switchos microcolumn switching module (LC Packings, Amesterdam, The Netherlands). An analytical C18 nanocapillary column (75 m inside diameter ×15 cm, PepMap) with a microprecolumn C18 cartridge for online peptide/glycopeptides separation was coupled to the system. Samples were injected onto the precolumn and eluted with 0.1% formic acid (Sigma) in water (HPLC grade, Purite) for 4 min. The eluate was then transferred onto the nanocapillary column with a flow rate of 150 nl min−1 using the following gradient program; Solvent A [0.05% (v/v) formic acid in a 95:5 (v/v) water/acetonitrile/water mixture] and solvent B [0.04% formic acid in a 95:5 (v/v) acetonitrile/water mixture]: 99% A from 0 to 5 min, 99 to 90% A from 5 to 10 min, 90 to 60% A from 10 to 70 min, 60 to 50% A from 70 to 71 min, 50 to 5% A from 71 to 75 min, 5% A from 75 to 85 min, 5 to 95% A from 85 to 86 min, and 95% A from 86 to 90 min. Data acquisition was performed using Analyst QS software with an automatic information-dependant-acquisition (IDA) function.
Cloning and purification of the UDP-sulfoquinovose synthase
The gene coding for a predicted UDP-sulfoquinovose synthase (Saci0423; agl3) was amplified from the genomic DNA of S. acidocaldarius introducing NdeI and XhoI restriction sites (primers: 5′ CCCCCCCATATGAGGATTCTAGTACTAG GAATT 3′ and 5′ CCCCCCCTCGAGACCACCCGCACC ACCTCTTACTCTTTTAACGTATTGTGGTTT3′) and the fragment was cloned into the pET 30a(+) vector (Qiagen) using standard procedures. Protein expression in E. coli Walker cells C41(DE3) was induced by addition of 1 mM isopropyl thio-beta-d-galactoside in 400 ml of LB medium for 3 h at 37°C. The cells were harvested and washed once with 50 mM sodium-phosphate buffer, pH 6.5, and resuspended in same buffer. The cell suspension was supplemented with PMSF and disrupted by incubation at 70°C for 20 min, followed by centrifugation at 13.000 g, allowing the precipitation of the majority of E. coli proteins and partial purification of the hyperthermophilic enzyme. The Agl3 was purified by Ni-affinity chromatography according to a standard protocol. Briefly, the protein was applied on Hi-trap Ni–NTA column (Qiagen) in 50 mM Tris/HCl (pH 8), containing 150 mM NaCl, washed with 10 volumes of the Hitrap column and eluted with Tris/HCl (pH 6.5), supplemented with 200 mM imidazole. The eluted fraction was dialysed against Tris/HCl, pH 6.5. The protein expression and purification was analysed by SDS-PAGE upon Coomassie Brilliant Blue staining.
Activity analysis of the sulfoquinovose synthase Agl3
The conversion of UDP-glucose and sulphite to UDP-sulfoquinovose catalysed by Agl3 was carried out in Tris/HCl (pH 6.5) containing 5 mM UDP-glucose, 20 mM sodium sulphite and 18 μg of purified Agl3 in total volume of 140 μl. The reaction was carried out overnight at 70°C. The samples were filtrated with 0.5 ml centrifugal filter units (Amicon Ultra) and the reaction product was analysed by RP-HPLC on a Nucleosil 120–3 C18 column (Macherey-Nagel) with a flow rate of 0.6 ml min−1 and 0.4 M Tris/HCl (pH 6.1) as eluent (Ultimate U3000 System, Dionex) (Martin et al., 1989; Zayni et al., 2007). The formation of UDP-sulfoquinovose was verified by PGC-ESI-MS(MS) analysis after prepurification of the reaction mixture on a porous graphitized carbon cartridge (Pabst et al., 2010).
Cloning of the agl3 into the S. acidocaldarius expression vector
The gene of agl3 was amplified from genomic DNA of S. acidocaldarius introducing BamHI restriction sites at the 5′ ends (primers: 5′ GCAGGGATCC TCTTACTCTTTTAACG TATTGTGG 3′ and 5′ GCAG GGATCC ATGAGGATTCTAG TACTAGGAATTGATGGTC 3′, respectively) and the fragment was cloned into pMZ1 (Zolghadr et al., 2007) yielding vector pSVA1257. To eliminate an internal NcoI restriction site in the agl3 sequence a silent point mutation PCR using the following primers was performed (primers: 5′ GATGGGTACTATGGGTGAGTATG 3′ and 5′ CACCCATAGTACCCATCTTC 3′, respectively). The resulting plasmid was digested with NcoI and EagI and the insert containing the agl3 gene was ligated into pSVA1450, an improved expression vector (M. Wagner and S.V. Albers, unpublished) based on pCmalLacS (Berkner et al., 2010) yielding pSVA1266. pCmalLacS contains a maltose inducible promoter that can be used for the expression of any gene of interest in S. acidocaldarius (Berkner et al., 2010). The plasmid was methylated and then transformed into the Δagl3 deletion strain as described in Berkner et al. Cultures grown from single colonies were induced with 0.1% maltose, grown until an OD of 0.8, pelleted and then used for the S-layer isolation.
Supplementary Material
Acknowledgement
B.H.M. and S.V.A. were supported by intramural funds of the Max Planck Society. B.Z. was supported by the Austrian Science Fund FWF P22791-B11 (to P.M.). The Imperial College authors were supported by the Biotechnology and Biological Sciences Research Council (BBF0083091, BBC5196701) and the UK Research Councils’ Basic Technology Initiative Translational Grant (EP/G037604/1). The authors wish to thank Sonja Zayni for assisting with the RP-HPLC analysis of UDP-sulfoquinovose.
Footnotes
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Abu-Qarn M, Yurist-Doutsch S, Giordano A, Trauner A, Morris HR, Hitchen P, et al. Haloferax volcanii AglB and AglD are involved in N-glycosylation of the S-layer glycoprotein and proper assembly of the surface layer. J Mol Biol. 2007;374:1224–1236. doi: 10.1016/j.jmb.2007.10.042. [DOI] [PubMed] [Google Scholar]
- Abu-Qarn M, Giordano A, Battaglia F, Trauner A, Hitchen PG, Morris HR, et al. Identification of AglE, a second glycosyltransferase involved in N- glycosylation of the Haloferax volcanii S-layer glycoprotein. J Bacteriol. 2008;190:3140–3146. doi: 10.1128/JB.00056-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers SV, Meyer BH. The archaeal cell envelope. Nat Rev Microbiol. 2011;9:414–426. doi: 10.1038/nrmicro2576. [DOI] [PubMed] [Google Scholar]
- Albers SV, Elferink MG, Charlebois RL, Sensen CW, Driessen AJ, Konings WN. Glucose transport in the extremely thermoacidophilic Sulfolobus solfataricus involves a high-affinity membrane-integrated binding protein. J Bacteriol. 1999;181:4285–4291. doi: 10.1128/jb.181.14.4285-4291.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning C. Biosynthesis and function of the sulfolipid sulfoquinovosyl diacylglycerol. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:53–75. doi: 10.1146/annurev.arplant.49.1.53. [DOI] [PubMed] [Google Scholar]
- Benning C, Somerville CR. Identification of an operon involved in sulfolipid biosynthesis in Rhodobacter sphaeroides. J Bacteriol. 1992a;174:6479–6487. doi: 10.1128/jb.174.20.6479-6487.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning C, Somerville CR. Isolation and genetic complementation of a sulfolipid deficient mutant of Rhodobacter sphaeroides. J Bacteriol. 1992b;174:2352–2360. doi: 10.1128/jb.174.7.2352-2360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning C, Beatty JT, Prince RC, Somerville CR. The sulfolipid sulfoquinovosyldiacylglycerol is not required for photosynthetic electron-transport in Rhodobacter sphaeroides but enhances growth under phosphate limitation. Proc Natl Acad Sci USA. 1993;90:1561–1565. doi: 10.1073/pnas.90.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkner S, Wlodkowski A, Albers SV, Lipps G. Inducible and constitutive promoters for genetic systems in Sulfolobus acidocaldarius. Extremophiles. 2010;14:249–259. doi: 10.1007/s00792-010-0304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock TD, Brock KM, Belly RT, Weiss RL. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch Mikrobiol. 1972;84:54–68. doi: 10.1007/BF00408082. [DOI] [PubMed] [Google Scholar]
- Calo D, Kaminski L, Eichler J. Protein glycosylation in Archaea: sweet and extreme. Glycobiology. 2010;20:1065–1076. doi: 10.1093/glycob/cwq055. [DOI] [PubMed] [Google Scholar]
- Cedergreen R, Hollingsworth R. Occurrence of sulfoquinovosyl diacyl-glycerol in some members of the family Rhizobiaceae. J Lipid Res. 1994;35:1452–1461. [PubMed] [Google Scholar]
- Chaban B, Voisin S, Kelly J, Logan SM, Jarrell KF. Identification of genes involved in the biosynthesis and attachment of Methanococcus voltae N-linked glycans: insight into N-linked glycosylation pathways in Archaea. Mol Microbiol. 2006;61:259–268. doi: 10.1111/j.1365-2958.2006.05226.x. [DOI] [PubMed] [Google Scholar]
- Chaban B, Logan SM, Kelly JF, Jarrell KF. AglC and AglK are involved in biosynthesis and attachment of diacetylated glucuronic acid to the N-glycan in Methanococcus voltae. J Bacteriol. 2009;191:187–195. doi: 10.1128/JB.00885-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elferink MG, Albers SV, Konings WN, Driessen AJ. Sugar transport in Sulfolobus solfataricus is mediated by two families of binding protein-dependent ABC transporters. Mol Microbiol. 2001;39:1494–1503. doi: 10.1046/j.1365-2958.2001.02336.x. [DOI] [PubMed] [Google Scholar]
- Heinz E. Recent investigations on the biosynthesis of the plant sulfolipid. In: Kok LJD, editor. Sulfur Nutrition and Assimilation in Higher Plants. SPB Academic Publishing; Den Haag, the Netherlands: 1993. pp. 163–178. [Google Scholar]
- Hettmann T, Schmidt CL, Anemuller S, Zahringer U, Moll H, Petersen A, Schafer G. Cytochrome b558/566 from the archaeon Sulfolobus acidocaldarius. A novel highly glycosylated, membrane-bound b-type hemoprotein. J Biol Chem. 1998;273:12032–12040. doi: 10.1074/jbc.273.20.12032. [DOI] [PubMed] [Google Scholar]
- Igura M, Kohda D. Quantitative assessment of the preferences for the amino acid residues flanking archaeal N-linked glycosylation sites. Glycobiology. 2011;21:575–583. doi: 10.1093/glycob/cwq196. [DOI] [PubMed] [Google Scholar]
- Igura M, Maita N, Obita T, Kamishikiryo J, Maenaka K, Kohda D. Purification, crystallization and preliminary X-ray diffraction studies of the soluble domain of the oligosaccharyltransferase STT3 subunit from the thermophilic archaeon Pyrococcus furiosus. Acta Crystallograph Sect F Struct Biol Cryst Commun. 2007;63:798–801. doi: 10.1107/S1744309107040134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J, Logan SM, Jarrell KF, VanDyke DJ, Vinogradov E. A novel N-linked flagellar glycan from Methanococcus maripaludis. Carbohydr Res. 2009;344:648–653. doi: 10.1016/j.carres.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Kessel M, Wildhaber I, Cohen S, Baumeister W. 3-Dimensional structure of the regular surface glycoprotein layer of Halobacterium volcanii from the Dead-Sea. EMBO J. 1988;7:1549–1554. doi: 10.1002/j.1460-2075.1988.tb02974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessel M, Volker S, Santarius U, Huber R, Baumeister W. 3-Dimensional reconstruction of the surface protein of the extremely thermophilic archaebacterium Archaeoglobus fulgidus. Syst Appl Microbiol. 1990;13:207–213. [Google Scholar]
- Kurosawa N, Grogan DW. Homologous recombination of exogenous DNA with the Sulfolobus acidocaldarius genome: properties and uses. FEMS Microbiol Lett. 2005;253:141–149. doi: 10.1016/j.femsle.2005.09.031. [DOI] [PubMed] [Google Scholar]
- Langworthy TA, Mayberry WR, Smith PF. A sulfonolipid and novel glucosamidyl glycolipids from the extreme thermoacidophile Bacillus acidocaldarius. Biochim Biophys Acta. 1976;431:550–569. doi: 10.1016/0005-2760(76)90220-4. [DOI] [PubMed] [Google Scholar]
- Lechner J, Sumper M. The primary structure of a procaryotic glycoprotein. Cloning and sequencing of the cell surface glycoprotein gene of halobacteria. J Biol Chem. 1987;262:9724–9729. [PubMed] [Google Scholar]
- Magidovich H, Eichler J. Glycosyltransferases and oligosaccharyltransferases in Archaea: putative components of the N-glycosylation pathway in the third domain of life. FEMS Microbiol Lett. 2009;300:122–130. doi: 10.1111/j.1574-6968.2009.01775.x. [DOI] [PubMed] [Google Scholar]
- Magidovich H, Yurist-Doutsch S, Konrad Z, Ventura VV, Dell A, Hitchen PG, Eichler J. AglP is a S-adenosyl-L-methionine-dependent methyltransferase that participates in the N-glycosylation pathway of Haloferax volcanii. Mol Microbiol. 2010;76:190–199. doi: 10.1111/j.1365-2958.2010.07090.x. [DOI] [PubMed] [Google Scholar]
- Martin A, Ruggiero-Lopez D, Broquet P, Richard M, Louisot P. High performance liquid chromatographic study of GDP-mannose and GDP-fucose metabolism. J Chromatogr. 1989;497:319–325. doi: 10.1016/0378-4347(89)80036-2. [DOI] [PubMed] [Google Scholar]
- Mengele R, Sumper M. Drastic differences in glycosylation of related S-layer glycoproteins from moderate and extreme halophiles. J Biol Chem. 1992;267:8182–8185. [PubMed] [Google Scholar]
- Merritt MV, Rosenstein SP, Loh C, Chou RHS, Allen MM. A comparison of the major lipid classes and fatty-acid composition of marine unicellular Cyanobacteria with fresh water species. Arch Microbiol. 1991;155:107–113. [Google Scholar]
- Mescher MF, Strominger JL. Purification and characterization of a prokaryotic glycoprotein from cell envelope of Halobacterium salinarium. J Biol Chem. 1976;251:2005–2014. [PubMed] [Google Scholar]
- Mulichak AM, Theisen MJ, Essigmann B, Benning C, Garavito RM. Crystal structure of SQD1, an enzyme involved in the biosynthesis of the plant sulfolipid headgroup donor UDP-sulfoquinovose. Proc Natl Acad Sci USA. 1999;96:13097–13102. doi: 10.1073/pnas.96.23.13097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SY, Chaban B, Jarrell KF. Archaeal flagella, bacterial flagella and type IV pili: a comparison of genes and posttranslational modifications. J Mol Microbiol Biotechnol. 2006;11:167–191. doi: 10.1159/000094053. [DOI] [PubMed] [Google Scholar]
- Ng SY, Wu J, Nair DB, Logan SM, Robotham A, Tessier L, et al. Genetic and mass spectrometry analyses of the unusual type IV-like pili of the archaeon Methanococcus maripaludis. J Bacteriol. 2011;193:804–814. doi: 10.1128/JB.00822-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst M, Grass J, Fischl R, Leonard R, Jin C, Hinterkorner G, et al. Nucleotide and nucleotide sugar analysis by liquid chromatography-electrospray ionizationmass spectrometry on surface-conditioned porous graphitic carbon. Anal Chem. 2010;82:9782–9788. doi: 10.1021/ac101975k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri G, Casbarra A, Fiume I, Catara G, Capasso A, Marino G, et al. Identification of the first archaeal oligopeptide-binding protein from the hyperthermophile Aeropyrum pernix. Extremophiles. 2006;10:393–402. doi: 10.1007/s00792-006-0508-1. [DOI] [PubMed] [Google Scholar]
- Paul G, Lottspeich F, Wieland F. Asparaginyl-N-acetylgalactosamine. Linkage unit of halobacterial glycosaminoglycan. J Biol Chem. 1986;261:1020–1024. [PubMed] [Google Scholar]
- Peng Z, Fives-Taylor P, Ruiz T, Zhou M, Sun B, Chen Q, Wu H. Identification of critical residues in Gap3 of Streptococcus parasanguinis involved in Fap1 glycosylation, fimbrial formation and in vitro adhesion. BMC Microbiol. 2008;8:52. doi: 10.1186/1471-2180-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyfoon E, Meyer B, Hitchen PG, Panico M, Morris HR, Haslam SM, et al. The S-layer glycoprotein of the crenarchaeote Sulfolobus acidocaldarius is glycosylated at multiple sites with chitobiose-linked N-glycans. Archaea. 2010;2010:754101. doi: 10.1155/2010/754101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallal AK, Nimer NA, Radwan SS. Lipid and fatty acid composition of fresh water Cyanobacteria. J Gen Microbiol. 1990;136:2043–2048. [Google Scholar]
- Sanda S, Leustek T, Theisen MJ, Garavito RM, Benning C. Recombinant Arabidopsis SQD1 converts udp-glucose and sulfite to the sulfolipid head group precursor UDP-sulfoquinovose in vitro. J Biol Chem. 2001;276:3941–3946. doi: 10.1074/jbc.M008200200. [DOI] [PubMed] [Google Scholar]
- Shimojima M. Biosynthesis and functions of the plant sulfolipid. Prog Lipid Res. 2011;50:234–239. doi: 10.1016/j.plipres.2011.02.003. [DOI] [PubMed] [Google Scholar]
- Shimojima M, Benning C. Native uridine 5′-diphosphate-sulfoquinovose synthase, SQD1, from spinach purifies as a 250-kDa complex. Arch Biochem Biophys. 2003;413:123–130. doi: 10.1016/s0003-9861(03)00112-7. [DOI] [PubMed] [Google Scholar]
- Shimojima M, Hoffmann-Benning S, Garavito RM, Benning C. Ferredoxin-dependent glutamate synthase moonlights in plant sulfolipid biosynthesis by forming a complex with SQD1. Arch Biochem Biophys. 2005;436:206–214. doi: 10.1016/j.abb.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Sumper M, Berg E, Mengele R, Strobel I. Primary structure and glycosylation of the S-layerprotein of Haloferax volcanii. J Bacteriol. 1990;172:7111–7118. doi: 10.1128/jb.172.12.7111-7118.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski CM, Yao R, Ewing CP, Trust TJ, Guerry P. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol Microbiol. 1999;32:1022–1030. doi: 10.1046/j.1365-2958.1999.01415.x. [DOI] [PubMed] [Google Scholar]
- Szymanski CM, Burr DH, Guerry P. Campylobacter protein glycosylation affects host cell interactions. Infect Immun. 2002;70:2242–2244. doi: 10.1128/IAI.70.4.2242-2244.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upreti RK, Kumar M, Shankar V. Bacterial glycoproteins: functions, biosynthesis and applications. Proteomics. 2003;3:363–379. doi: 10.1002/pmic.200390052. [DOI] [PubMed] [Google Scholar]
- VanDyke DJ, Wu J, Logan SM, Kelly JF, Mizuno S, Aizawa S, Jarrell KF. Identification of genes involved in the assembly and attachment of a novel flagellin N-linked tetrasaccharide important for motility in the archaeon Methanococcus maripaludis. Mol Microbiol. 2009;72:633–644. doi: 10.1111/j.1365-2958.2009.06671.x. [DOI] [PubMed] [Google Scholar]
- Voisin S, Houliston RS, Kelly J, Brisson JR, Watson D, Bardy SL, et al. Identification and characterization of the unique N-linked glycan common to the flagellins and S-layer glycoprotein of Methanococcus voltae. J Biol Chem. 2005;280:16586–16593. doi: 10.1074/jbc.M500329200. [DOI] [PubMed] [Google Scholar]
- Wagner M, Berkner S, Ajon M, Driessen AJ, Lipps G, Albers SV. Expanding and understanding the genetic toolbox of the hyperthermophilic genus Sulfolobus. Biochem Soc Trans. 2009;37:97–101. doi: 10.1042/BST0370097. [DOI] [PubMed] [Google Scholar]
- Wisotzkey JD, Jurtshuk P, Jr, Fox GE, Deinhard G, Poralla K. Comparative sequence analyses on the 16S rRNA (rDNA) of Bacillus acidocaldarius, Bacillus acidoterrestris, and Bacillus cycloheptanicus and proposal for creation of a new genus, Alicyclobacillus gen. nov. Int J Syst Bacteriol. 1992;42:263–269. doi: 10.1099/00207713-42-2-263. [DOI] [PubMed] [Google Scholar]
- Xu C, Sibicky T, Huang B. Protein profile analysis of salt-responsive proteins in leaves and roots in two cultivars of creeping bentgrass differing in salinity tolerance. Plant Cell Rep. 2010;29:595–615. doi: 10.1007/s00299-010-0847-3. [DOI] [PubMed] [Google Scholar]
- Yurist-Doutsch S, Magidovich H, Ventura VV, Hitchen PG, Dell A, Eichler J. N-glycosylation in Archaea: on the coordinated actions of Haloferax volcanii AglF and AglM. Mol Microbiol. 2010;75:1047–1058. doi: 10.1111/j.1365-2958.2009.07045.x. [DOI] [PubMed] [Google Scholar]
- Zahringer U, Moll H, Hettmann T, Knirel VA, Schafer G. Cytochrome b558/566 from the archaeon Sulfolobus acidocaldarius has a unique Asn-linked highly branched hexasaccharide chain containing 6-sulfoquinovose. Eur J Biochem. 2000;267:4144–4149. doi: 10.1046/j.1432-1327.2000.01446.x. [DOI] [PubMed] [Google Scholar]
- Zayni S, Steiner K, Pföstl A, Hofinger A, Kosma P, Schäffer C, Messner P. The dTDP-4-dehydro-6-deoxyglucose reductase encoding fcd gene is part of the surface layer glycoprotein glycosylation gene cluster of Geobacillus tepidamans GS5-97T. Glycobiology. 2007;17:433–443. doi: 10.1093/glycob/cwl084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitler R, Hochmuth E, Deutzmann R, Sumper M. Exchange of Ser-4 for Val, Leu or Asn in the sequon Asn-Ala-Ser does not prevent N-glycosylation of the cell surface glycoprotein from Halobacterium halobium. Glycobiology. 1998;8:1157–1164. doi: 10.1093/glycob/8.12.1157. [DOI] [PubMed] [Google Scholar]
- Zolghadr B, Weber S, Szabo Z, Driessen AJ, Albers SV. Identification of a system required for the functional surface localization of sugar binding proteins with class III signal peptides in Sulfolobus solfataricus. Mol Microbiol. 2007;64:795–806. doi: 10.1111/j.1365-2958.2007.05697.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.