SUMMARY
There is a lack of effective predictive biomarkers to precisely assign optimal therapy to cancer patients. While most efforts are directed at inferring drug response phenotype based on genotype, there is very focused and useful phenotypic information to be gained from directly perturbing the patient’s living cancer cell with the drug(s) in question. To satisfy this unmet need we developed the Dynamic BH3 Profiling technique to measure early changes in net pro-apoptotic signaling at the mitochondrion (‘priming’) induced by chemotherapeutic agents in cancer cells, not requiring prolonged ex vivo culture. We find in cell line and clinical experiments that early drug-induced death signaling measured by Dynamic BH3 Profiling predicts chemotherapy response across many cancer types and many agents, including combinations of chemotherapies. We propose that Dynamic BH3 Profiling can be used as a broadly applicable predictive biomarker to predict cytotoxic response of cancers to chemotherapeutics in vivo.
INTRODUCTION
A fundamental challenge across medicine is to assign to a patient the drug or combination of drugs that will be of greatest benefit. In oncology, this choice has historically been driven by the anatomic location and histology of the tumor. Later, therapeutic decision-making was assisted by immunohistochemistry, cytogenetics, flow cytometric analysis of cell surface antigens. In more recent years, there are examples where gene expression signatures and specific genetic alterations have been essential to therapeutic decisions (Chapman et al., 2011; Paez et al., 2004). However, true personalization of therapy remains an elusive goal in most cases. In all too many cases, cancer patients show little benefit from therapy. Moreover, it is likely that many tumors have unrecognized sensitivity to agents for which there is simply no useful predictive biomarker to inform therapy decisions (Garraway and Janne, 2012; Haibe-Kains et al., 2013). In this era of growing therapeutic options, there is a comparable growing need for predictive biomarkers (Sawyers, 2008; Yaffe, 2013).
A feature common to nearly all of the biomarkers in use or in development in oncology is that they are studies performed on dead cancer cells. They are attempts to predict cancer cell behavior based on detailed analysis of components of the cell, such as DNA, RNA, or proteins (Barretina et al., 2012). In some cases, abnormalities in single genes are studied. There are spectacular examples of success with this approach, such as the use of EGFR mutations to guide treatment with EGFR inhibitors in lung cancer (Paez et al., 2004), or BRAF mutations to guide treatment with vemurafenib in melanoma (Chapman et al., 2011), or c-Kit mutations to guide treatment with imatinib in GIST (Joensuu et al., 2001). However, most drugs in development or approved for cancer lack a simple genetic predictor, which impedes their clinical development (Sikorski and Yao, 2010). One popular approach to this problem is to identify signatures based on huge amounts of information based on genomes, transcriptomes, or proteomes (Barretina et al., 2012; Garraway and Janne, 2012). These strategies are relatively early in development and their power remains to be seen. Despite the abundance of information these strategies provide, they still share a weakness, that they are all studies of dead cancer cells. They lack a measure of cancer cell function or response to perturbation. Studies of complex systems in and out of biology are often greatly augmented by observations of responses to strategic perturbations. Here we present results of strategic perturbations of cancer cells with drugs and their mitochondria with peptides in a strategy we call Dynamic BH3 Profiling (DBP).
DBP interrogates the BCL-2 family of proteins that regulates commitment to the mitochondrial pathway of apoptosis, the program of cell death that is commonly used by cancer cells in response to most chemotherapeutic agents. The BCL-2 family of proteins controls mitochondrial outer membrane permeabilization (MOMP) (Certo et al., 2006; Chipuk et al., 2010). The effector proteins BAX and BAK, when activated, oligomerize to form pores in the mitochondrial outer membrane that induce release of cytochrome c and the loss of mitochondrial transmembrane potential, as well as release of SMAC/DIABLO and other proteins that trigger apoptosome formation, caspase activation and finally apoptosis (Kluck et al., 1997; Wei et al., 2001). These effector proteins can be activated by the BH3-only proteins BIM, BID (and perhaps PUMA), also known as activators (Sarosiek et al., 2013). Both effectors and activators can be inhibited by the anti-apoptotic members of the family, including BCL-2, BCL-XL, MCL-1 and others (Certo et al., 2006). There is a fourth group of proteins, called sensitizers (comprising proteins like BAD, BMF, NOXA, HRK and others) that by themselves are not able to induce BAX and BAK oligomerization, but instead selectively inhibit the anti-apoptotic members of the family, thus indirectly promoting MOMP (Letai et al., 2002). The BH3 domain is a roughly 20-amino acid amphipathic alpha helix that is necessary for most of the hetero-dimeric interactions of BCL-2 family proteins that regulate apoptosis. Synthetic BH3 domain oligopeptides can execute most of the pro-apoptotic functions of pro-apoptotic BCL-2 family proteins (Certo et al., 2006).
BH3 peptides are thus a convenient, titratable reagent that can be exploited to systematically study mitochondrial readiness to undergo apoptosis. This understanding of the BCL-2 family of proteins and their interactions allowed the development of the BH3 profiling technique (Ryan et al., 2010) that identifies cancer cells’ selective dependence on anti-apoptotic proteins, and also measures overall apoptotic sensitivity or ‘priming for death’ (Deng et al., 2007a). “Priming” is a measure of how close a cell is to the threshold of apoptosis. Procedurally, priming corresponds to the sensitivity of mitochondria to BH3 peptides. The more sensitive mitochondria are to BH3 peptides, the more primed they are. We have previously found that the state of “priming’ prior to therapy was an excellent predictor of chemotherapeutic response in vivo (Ni Chonghaile et al., 2011; Vo et al., 2012). Differences in priming between cancer cells and normal tissues also provide an explanation for the therapeutic index of conventional chemotherapeutic drugs that target ubiquitous elements such as DNA and microtubules.
The main principle of Dynamic BH3 Profiling is to expose cancer cells to short incubations with drugs of interest and measure whether the drug exposure induces an increase in priming compared to an untreated control. In this paper we use DBP to test the hypothesis that early death signaling predicts cytotoxicity, even when the cell death does not occur until days after the death signaling is measured. Our results support the model that initiation of death signaling is the main regulator of eventual commitment to cell death. Moreover, we show that we can perform these measurements on primary patient cancer cells in a way that predicts clinical response to therapy.
RESULTS
Dynamic BH3 profiling predicts chemotherapy sensitivity in Non-Small Cell Lung Cancer cell lines
Our strategy rests upon the hypothesis that it is the initiation of death signaling that distinguishes cells destined to be killed by an agent from those destined to survive.
We therefore rigorously tested the hypothesis that measurement of early death signaling by Dynamic BH3 Profiling (DBP) (Figure 1A) could predict a cytotoxic response that did not occur until several days later. We first used Non-Small Cell Lung Cancer (NSCLC) cell lines derived from PC9. This cell line has an exon 19 deletion in the EGFR gene rendering it sensitive to EGFR-specific tyrosine kinase inhibitors (TKI) like erlotinib or gefitinib. PC9GR was obtained by continuously exposing PC9 to increasing concentrations of gefitinib (Ercan et al., 2010), selecting for a T790M mutation in EGFR that renders it non-sensitive to gefitinib, but still sensitive to the mutant selective EGFR TKI WZ4002 (Zhou et al., 2009). A third cell line, PC9WZR, was similarly selected for resistance to WZ4002. It possesses an EGFR T790M mutation and a MAPK1 amplification conferring resistance to both gefitinib and WZ4002. However, PC9WZR is sensitive to the combination of WZ4002 with the MEK inhibitor CI-1040, by completely blocking the MAPK pathway (Ercan et al., 2012). This set of cell lines provided a useful initial model of differential sensitivity to targeted therapies upon which to test our strategy.
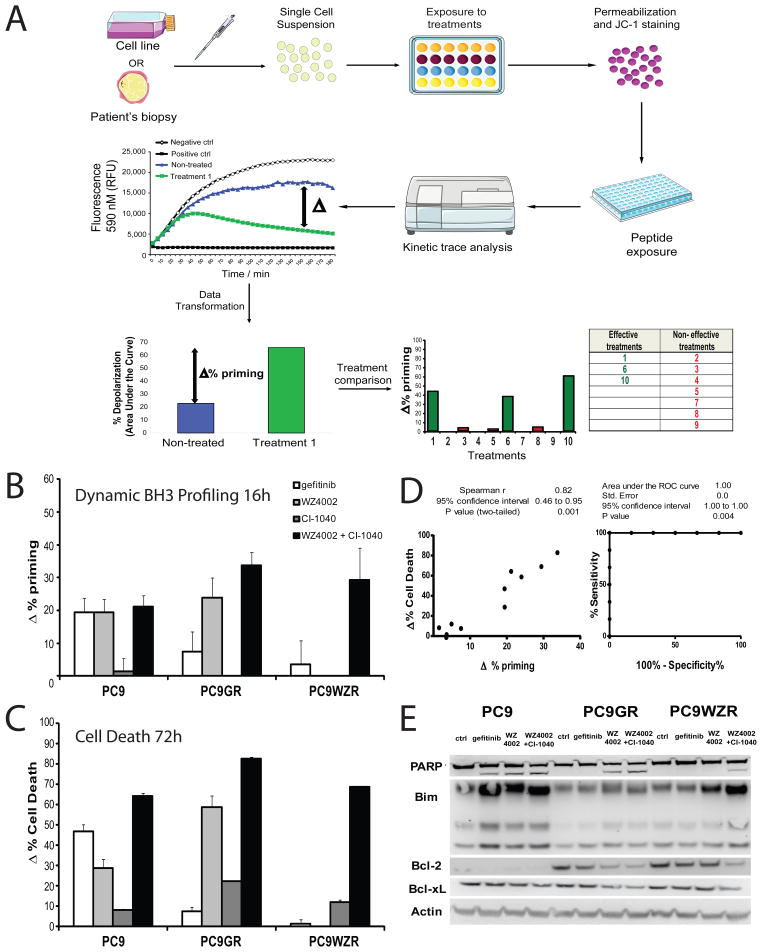
Figure 1. Dynamic BH3 profiling predicts chemotherapy sensitivity in PC9 cell lines.
(A) To perform Dynamic BH3 profiling (DBP) we obtain a single cell suspension from a cell line or a primary sample, and we expose the cells to the different drug treatments to be tested. After this incubation, we permeabilize, stain with the fluorescent dye JC-1 and expose the cells to different BH3 peptides that will promote mitochondrial depolarization and MOMP, the ultimate event that triggers apoptosis. By comparing the non-treated cells with the treated ones, DBP will determine the Δ% priming for each agent and identify which are most effective to induce apoptosis in that particular sample. All this analysis is performed in less than 24 hours, minimizing ex vivo culture. (B) DBP was performed on three different PC9 cell lines: parental PC9, PC9GR (gefitinib resistant, T790M mutation present) and PC9WZR (gefitinib and WZ4002 resistant, T790M mutation present), using a 16 hour incubation of: gefitinib 1μM, WZ4002 100 nM, CI-1040 3 μM (MEK inhibitor) and WZ4002+CI-1040. Results expressed as Δ% priming (increase in priming compared to non-treated cells). Values indicate mean values ± SEM, at least three independent experiments were performed (N≥3). (C) Cell death measurements at 72 hours for the same cell lines under the same treatments by FACS using Annexin V/PI staining. Results are expressed as increase on cell death or Δ%Cell Death, compared to non-treated cells. Values indicate mean values ± SEM, at least three independent experiments were performed (N≥3). (D) Plot showing correlation between Δ% priming at 16h and Δ%Cell Death at 72h. Receiver Operating Characteristic curve analysis at right. (E) Western Blot analysis, showing changes in the BCL-2 family of proteins. See also Figures S1, S2 and S3.
We performed DBP on each of the cell lines using a 16 hour treatment with gefitinib, WZ4002, CI-1040 or the combination WZ4002 plus CI-1040. 16 hours was chosen after empirically testing 4, 8, 16, 24, and 48 hours as it was the earliest time point that reliably provided a significant change in priming in PC9 cells treated with gefitinib. After testing several BH3 peptides, including BIM, HRK, and PUMA BH3, we found that BIM BH3 concentrations of 0.3 and 1 μM provided the most useful dynamic range (Figure 1 and Figure S1). Drug concentrations were chosen based on our and others’ prior experience, and the dose required for a complete blockade of the MAPK pathway (Ercan et al., 2012; Ercan et al., 2010). We observed an increase of priming induced in PC9 by gefitinib, WZ4002 and WZ4002 + CI-1040, as shown by the increase in BIM BH3-induced mitochondrial depolarization (Δ% priming). In PC9GR cells, WZ4002, but not gefinitib, increased priming. In PC9WZR cells, only the WZ4002 + CI-1040 increased mitochondrial priming (Figure 1B). We next measured cell death at 72 hours for the same cell lines following the same treatments using FACS analysis of Annexin V/PI staining (Figure 1C). When we compared Δ% priming and Δ% cell death, we observed an excellent correlation between both measurements (Figure 1D. left). The Receiver Operating Characteristic was also excellent, performing perfectly in this small number of tests.(Figure 1D, right). Note that DBP was performed at 16 hours when no significant cell death was evident, whereas cell death was analyzed more than two days later (Figure S2). Thus, the early priming increase measured by DBP provided accurate, drug-specific predictions about cytotoxicity even though the death took place days later.
DBP should only be predictive if the mitochondrial apoptosis pathway is being engaged. To confirm this engagement, we analyzed PARP cleavage, as well as levels of BIM, BCL-2 and BCL-XL proteins following 24h of drug treatment. When cytotoxicity was observed, PARP cleavage was detected. In addition, cytotoxicity correlated with either increases in BIM, decreases in anti-apoptotic proteins, or a combination of both effects, supporting the simultaneous participation of multiple BCL-2 family proteins in the determination of cell fate.(Deng et al., 2007b; Faber et al., 2011) (Figure 1E).
In order to determine if this predictive capacity of DBP could be generalized to other NSCLC models, we treated 8 different NSCLC cell lines with gefitinib, WZ4002, AZD6244 (MEK inhibitor), BEZ235 (PI3K/mTOR inhibitor) and the combination AZD6244 + BEZ235, that was previously described to treat murine lung cancers harboring the KRas G12D mutation (Engelman et al., 2008; Faber et al., 2009). We chose drug concentrations that had previously demonstrated in vitro cytotoxicity. Again, we compared the priming increase measured by DBP after 16h of treatment with cell death observed at 72 h (Figure S3A). Some of the cell lines analyzed had a tendency to show less cytotoxicity than would be expected by DBP for a few drugs. It is possible that measurement of cell death at longer time points would reduce such disagreements. Nonetheless, we observed a significant correlation between Δ% priming and Δ% cell death when all cell lines and treatments were considered (Figure S3B). To assess if DBP provided a useful binary predictor of cytotoxicity, we performed Receiver Operating Characteristic (ROC) curve analysis (Pencina et al., 2008). Typically, a random classifier would present an AUC of 0.5, while a perfect classifier would have a AUC of 1. In this case, the area under the ROC curve is 0.895 (Figure S3C), comparing favorably with the ROC performance of many clinically used predictors (Burstein et al., 2011). Note that this analysis relies not simply on measurements of the baseline priming, but rather on the degree to which drugs increase priming from that baseline.
DBP predicts cytotoxicity in breast cancer cells
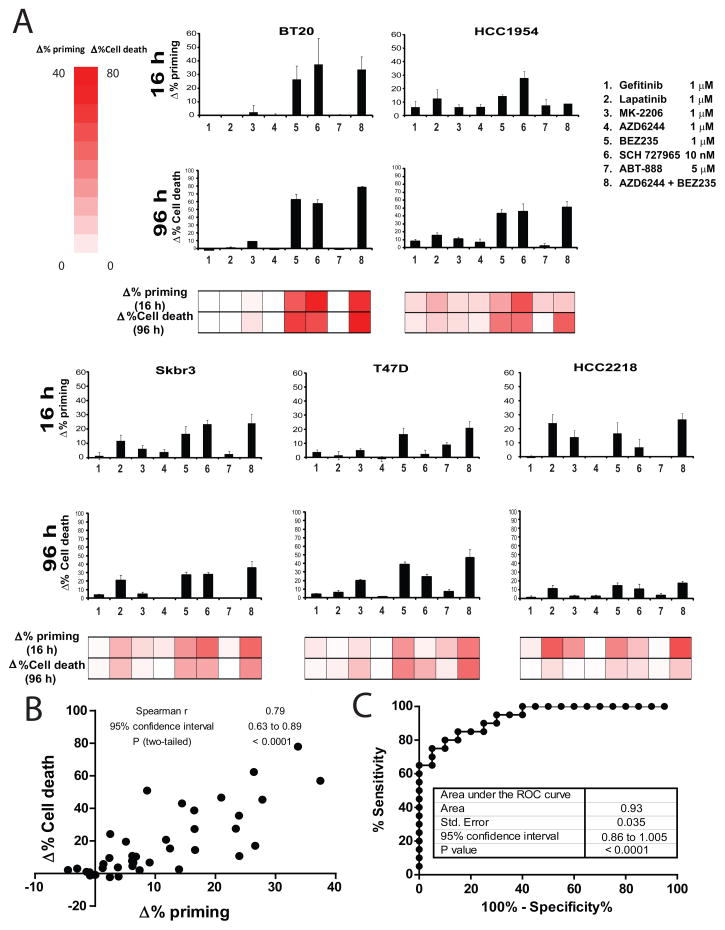
To test the generalizability of our hypothesis in a different type of cancer, we performed a similar set of experiments with 5 different human breast cancer cell lines treated with gefitinib, lapatinib (HER2 inhibitor), MK-2206 (AKT inhibitor), AZD6244 (MEK inhibitor), BEZ235 (PI3K/mTOR inhibitor), dinaciclib (SCH 727965, CDK inhibitor), ABT-888 (PARP inhibitor) and the combination AZD6244 + BEZ235, as previously described (Faber et al., 2009). Again we observed a significant correlation between Δ% priming after 16h of treatment and Δ% cell death at 96 h (Figures 2A, 2B). The area under the ROC curve for this set of cell lines is 0.93 (Figure 2C), thus objectively DBP is an excellent binary predictor for breast cancer cell lines’ response to chemotherapy.
Figure 2. DBP predicts chemotherapy sensitivity in breast cancer cell lines.
(A) DBP was performed in five breast cancer cell lines: BT20, HCC1954, SKBR3, T47D and HCC2218 showing different pattern of response to the treatments tested (16 hour incubation): 1) gefitinib 1μM, 2) lapatinib 1μM, 3) MK-2206 1μM, 4) AZD6244 1μM, 5) BEZ235 1μM, 6) dinaciclib 10 nM (SCH 727965), 7) ABT-888 5μM and the combination 8) AZD6244 + BEZ235. Results expressed as Δ% priming (increase in priming compared to non-treated cells). Values indicate mean values ± SEM, at least three independent experiments were performed (N≥3). Cell death measurements at 72 hours for the same cell lines under the same treatments by FACS using Annexin V/PI staining. Results are expressed as increase on cell death or Δ%Cell Death, compared to non-treated cells. Values indicate mean values ± SEM, at least three independent experiments were performed (N≥3). (B) Plot showing the significant correlation between Δ% priming at 16h and Δ%Cell Death at 72h. (C) Receiver Operating Characteristic (ROC) curve analysis.
Selecting the optimal kinase inhibitor using Dynamic BH3 profiling
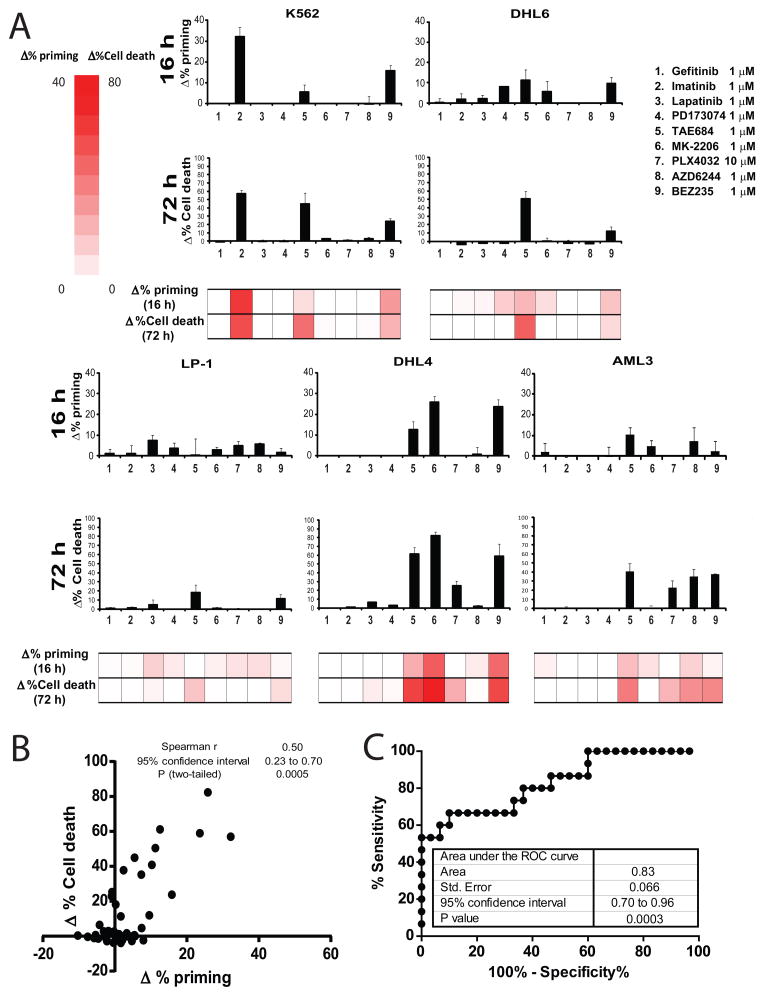
In clinical practice, an important application of a potentially powerful, widely applicable predictive biomarker would be to choose from among a panel of possible therapies (Sawyers, 2008). This is the central goal of what is currently commonly termed “precision medicine”. We hypothesized that if we could compare the death signaling induced by several different agents in a cancer cell, we could pick the ones that would work best. To test this principle, we selected 10 different cancer cell lines, chosen simply by variety and availability. For drugs, we chose 9 kinase inhibitors, for their diversity of targets and known in vivo activity. We chose kinase inhibitors because of their known use of the mitochondrial apoptotic pathway to kill cancer cells (Bhatt et al., 2010; Faber et al., 2011). Our question was, among these diverse cell lines and drugs, could DBP at an early time point be used to make individualized choices of the drugs most likely to kill each cancer cell line.
For this purpose we selected drugs targeting either key membrane receptor tyrosine kinases like geftinib (EGFR inh), imatinib (ABL inh.), lapatinib (HER2 inh.), PD173074 (FGFR inh.) and TAE684 (ALK inh.); or important intracellular serine/threonine kinases including MK-2206 (AKT inh.), PLX4032 (BRAFV600E inh.), AZD6244 (MEK inh.) and BEZ235 (PI3K/mTOR inh.). All of the compounds tested previously demonstrated cytotoxicty in cancer cell lines and/or murine cancer models, including hematological malignancies (Bhatt et al., 2010) and solid tumors (Maertens et al., 2013). We tested the panel of kinase inhibitors on several human hematological cancer cell lines: K562 (acute myelogenous leukemia), DHL6 (diffuse large B-cell lymphoma), LP1 (multiple myeloma), DHL4 (diffuse large B-cell lymphoma) and AML3 (acute myelogenous leukemia). First we performed DBP after 16 hour exposure to the different treatments (Figure 3A). We compared the DBP results to cell death achieved at 72 hours, expressed as Δ% Cell death (Figure 3A). Each cell line demonstrated a distinct pattern of drug induced priming increase, a distinct fingerprint of pathway addiction just as there was a distinct pattern of cytotoxic response to the drug panel. Most importantly for our question however, there was an excellent correlation of DBP with cytotoxicity days later (Figure 3B). For this set of hematological cell lines, predictive power of DBP was demonstrated by an AUC of the ROC curve of 0.83 (Figure 3C). Note that DBP identified the agent causing greatest cytotoxicity in 4 out of 5 cell lines. In the one exception, LP-1, there was little cytotoxicity induced by any of the drugs.
Figure 3. Identifying the optimal treatment in hematological malignancies using DBP.
We selected several drugs targeting either key membrane receptors: 1) gefitinib 1μM, 2) imatinib 1μM, 3) lapatinib 1μM, 4) PD173074 1μM and 5) TAE684 1μM; or important intracellular kinases: 6) MK-2206 1μM, 7) PLX4032 10μM, 8) AZD6244 1μM and 9) BEZ235 1μM, and we tested them with several human hematological cancer cell lines: K562, DHL6, LP1, DHL4 and AML3. (A) DBP (16 hour incubation) results expressed as Δ%priming and cell death measurements at 72 hours using Annexin V/PI staining expressed as Δ%Cell Death. Values indicate mean values ± SEM, at least three independent experiments were performed (N≥3) (B) Plot showing the significant correlation between Δ% priming at 16h and Δ%Cell Death at 72h. (C) ROC curve analysis shows AUC=0.83.
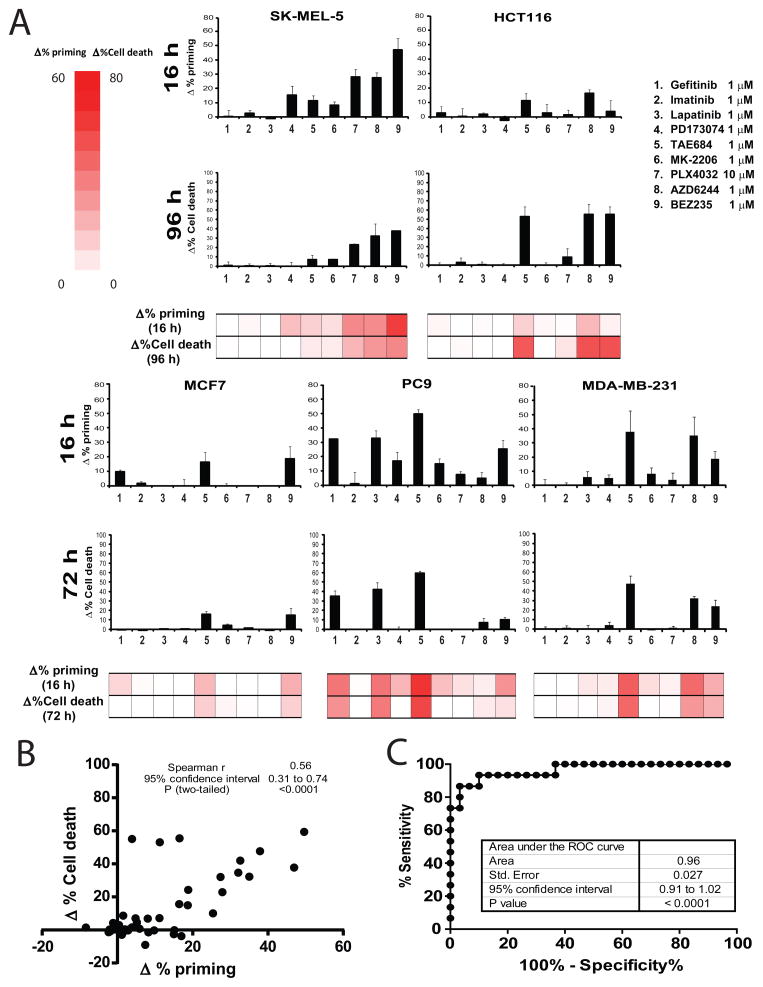
We next examined the predictive capacity of DBP with several diverse human solid tumor cell lines: MCF7 (Breast Cancer), PC9 (Non-Small Cell Lung Cancer), Sk-mel-5 (Melanoma), HCT116 (Colon carcinoma) and MDA-MB-231 (Breast Cancer). We exposed the cells to the different treatments for 16 hours and performed DBP (Figure 4A), comparing it with the cell death observed at 72–96 hours (Figure 4A). In some cases, a 96 hour time point was required due to slow kinetics of cytotoxicity. Similarly as observed for hematological malignancies, the different cell lines responded differently to the drugs tested, but a significant correlation between DBP and cytotoxicity was detected (Figure 4B). Sk-mel-5 was the only one sensitive to PLX4032, as expected for a BRAFV600E expressing melanoma cell line, but was also sensitive to MEK (AZD6244) and PI3K/mTOR (BEZ235) inhibition, correlating with the cell death detected three days later, at 96 hours. On the other hand, PC9, as shown previously (Figure 1 B–C), responded to gefitinib (Ercan et al., 2010; Faber et al., 2011), but also to lapatinib and TAE684; correlating with cell death at 72 hours. For this set of solid tumor cell lines the AUC of the ROC curve was 0.96 (Figure 4C). In three out of five cell lines, DBP clearly predicted the most cytotoxic drug. In the other two MCF7 and HCT116, there was nearly equal maximum response of the same two drugs in both DBP and cytotoxicity.
Figure 4. Identifying the optimal treatment in solid tumors using DBP.
We tested the same panel of kinase inhibitors on several human solid tumor cell lines: MCF7, PC9, Sk5mel, HCT116 and MDA-MB-231. (A) DBP (16 hour incubation) results expressed as Δ%priming and cell death measurements at 72 or 96 hours (as indicated) using Annexin V/PI staining expressed as Δ%Cell Death. Values indicate mean values ± SEM, at least three independent experiments were performed (N≥3) (B) Plot showing the significant correlation between Δ% priming at 16h and Δ%Cell Death at 72/96h. (C) The ROC curve analysis has an AUC=0.96, indicating that DBP is an excellent binary predictor for chemotherapy response in solid tumor cell lines. See also Figure S4.
Throughout this paper, we use loss of fluorescence from an indicator compound, JC-1, that is sensitive to the electropotential gradient across the inner mitochondrial membrane. We have previously shown that this JC-1 signal provides a good surrogate for permeabilization of the outer mitochondrial membrane (Ryan et al., 2010). To verify that this surrogacy is maintained in DBP, we compared measuring MOMP by JC-1 or by efflux of cytochrome c as read on a flow cytometer (Ryan and Letai, 2013) (Figure S4). Our results show good agreement between the two techniques, supporting the use of JC-1 fluorescence as a surrogate for MOMP in the context of DBP.
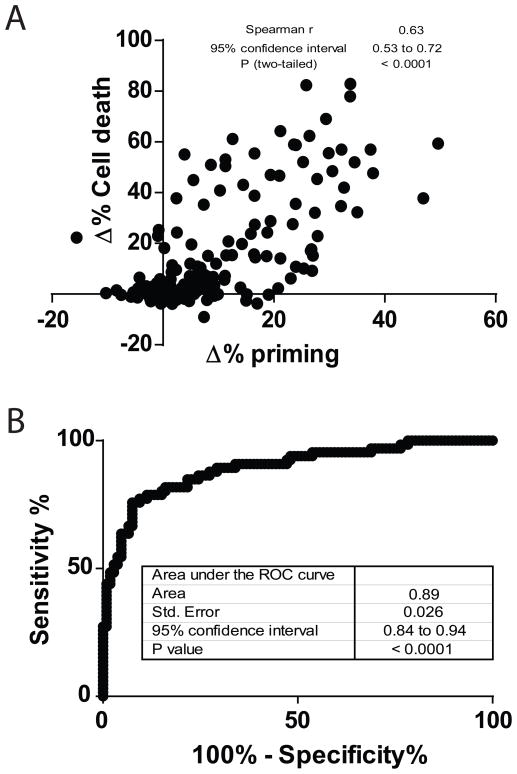
To test the generalizability of the principle that early drug-induced priming changes predict eventual cytotoxicity across a wide variety of both solid and liquid cancers and a wide variety of agents, we combined the data of Figure 1, 2, 3, 4 and Figure S3. We observed that there is a significant correlation between Δ% priming and Δ% Cell Death (figure 5A). Note that liquid tumors in general have a greater cytotoxic response per change in priming, perhaps explained by the higher baseline mitochondrial apoptotic priming we observe in hematologic cancer cell lines compared to solid tumor cell lines. In addition, the ROC analysis suggests that DBP could be a good binary predictor for cytotoxicity across a wide range of pathway inhibitors and cancer, with an AUC for the ROC curve of 0.89 (Figure 5B). These results suggest the most significant hurdle that must be cleared for a drug to cause cytotoxicity is simply the initiation of death signaling. Regardless of the pathway inhibited and regardless of the cell of origin of the cancer, early drug-induced death signaling predicts later cytotoxicity.
Figure 5. Dynamic BH3 profiling is a good binary predictor for cell lines.
(A) Compilation of Figures 1, 2, 3, 4 and Suppl. Figure 3 results, showing a significant correlation between Δ% priming and Δ% Cell Death for all cell lines analyzed. (B) The total area under the ROC curve is 0.89, indicating that is a good binary predictor for chemotherapy response in all the cell lines and treatments tested.
Choosing the best treatment among several options
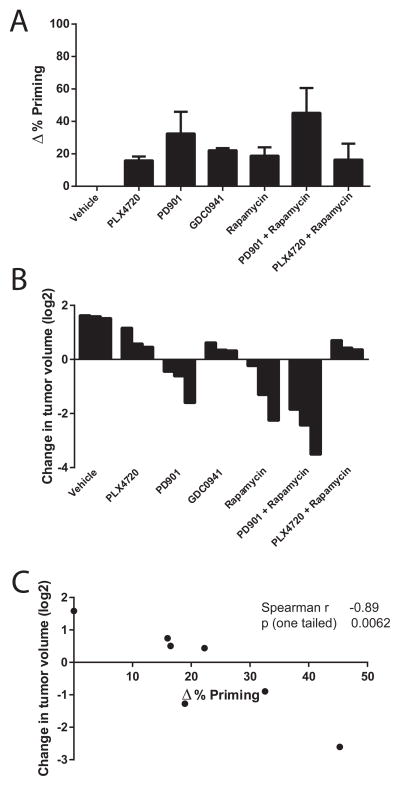
A predictive biomarker can be used to identify the best therapy among many treatment options for a single patient. To test the ability of DBP to identify the most effective therapy among a myriad of treatment options we turned to an allograft melanoma model. Mouse melanomas harboring compound mutations in Braf and Nf1 readily grow as allografts and are resistant to selective BRAF inhibitors, but sensitive to (combined) MEK/mTORC1 inhibition (Maertens et al., 2013). To ask whether DBP could discriminate among the in vivo efficacy of several therapies on the same tumor model, we exposed Braf/Nf1-mutant melanoma cells to different targeted agents for 16 hours: PLX4720 (a PLX4032 analogue that inhibits mutant BRAFV600E), PD0325901 (referred to as PD-901, a MEK inhibitor), GDC-0941 (a PI3K inhibitor) and rapamycin (an mTORC1 inhibitor), as single agents or in combination. Of all the treatments tested, PD-901 in combination with rapamycin induced the greatest increase in priming (Figure 6A). These findings correlate well with the preclinical data previously generated using this tumor model (Figure 4C Maertens et al., 2013). More specifically, of all (combination) therapies tested in vivo the PD-901/rapamycin combination caused the greatest tumor shrinkage, as summarized in Figure 6B. Across all of the treatments, we observed a significant correlation between DBP results and the in vivo data obtained in the Braf/Nf1-mutant allografts (Figure 6C). These results suggest that DBP can be used as a predictive biomarker to select among treatment options to identify treatments that will provide best in vivo benefit.
Figure 6. DBP can identify the best in vivo treatment among several options.
Braf/Nf1-mutant melanoma cells were treated ex vivo with PLX4720 1 μM, PD0325901 (referred to as PD-901) 0.25 μM, GDC-0941 1 μM, rapamycin 0.1 μM, PD-901 + rapamycin and PLX4720 + rapamycin. (A) DBP (16 hour incubation) results expressed as Δ%priming. Values indicate mean values ± SEM, at least three independent experiments were performed (N≥3) (B) In vivo response for this Braf/Nf1-mutant allograft melanoma model (adapted from Figure 4C Maertens et al., 2013), expressed as change in tumor volume (log2) after 7 days of treatment (C) Correlation between Δ%priming and change in tumor volume.
Identifying the best-responding patients to a single therapy in a patient cohort
Predictive biomarkers can also be used to stratify likelihood of response to a single therapy among many patients. This can be described as a companion diagnostic use. Having thoroughly supported the hypothesis that early death signaling detected by DBP predicts cytotoxicity in vitro, it was important to test whether our tool can likewise discriminate between clinical sensitivity and resistance to anti-cancer therapies using primary patient samples. We chose treatment of CML with imatinib as a first test of this principle. CML cells possess a t(9;22) translocation creating a BCR-ABL fusion protein that results in constitutive active ABL kinase activity. CML is typically sensitive to inhibitors of ABL kinase including imatinib (Sawyers, 1999).
To demonstrate the correlation between imatinib’s inhibition of ABL and an increase in apoptotic priming, we treated two human CML cell lines with different concentrations of imatinib. After 16 hours of treatment, we observed that the dephosphorylation of ABL, and its downstream target CRKL correlated with an increase in priming. Note that frank cell death began days later, at 72 hours (Figure S5).
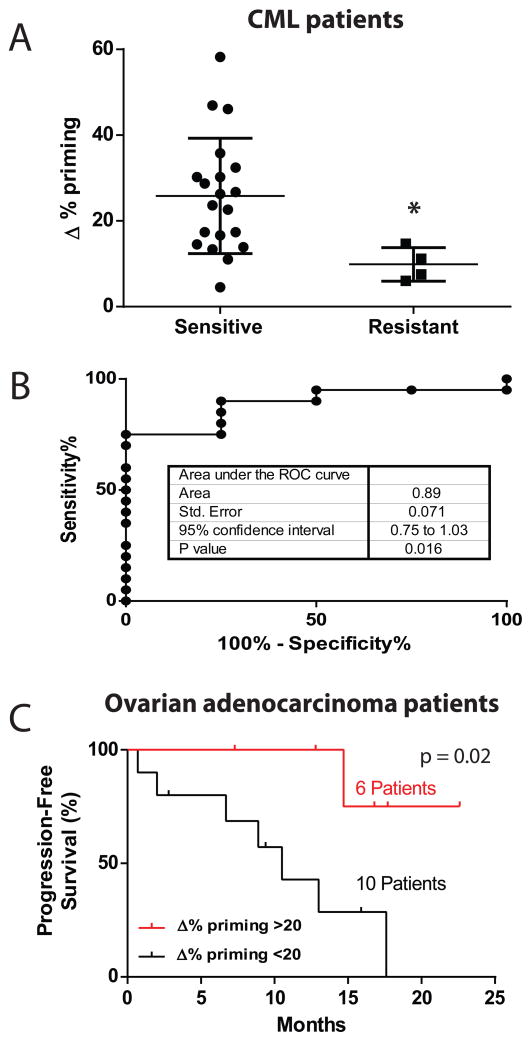
We treated bone marrow cells obtained from 24 CML patients for 16 hours with imatinib, performed DBP, and recorded the change in priming induced. Initial resistance to imatinib is very rare in CML, so we compared samples of patients who were newly started on imatinib, all of whom entered at least a complete hematologic remission (“sensitive”, Figure 7A) with samples from patients obtained when they were known to be refractory to imatinib (“resistant”, Figure 7A). Samples from patients that were sensitive to imatinib showed a significantly higher Δ% priming compared to those that did not respond (Figure 7A). We next tested the ability of DBP to segregate clinical sensitivity and resistance in a binary fashion. with ROC analysis (Figure 7B). The area under the ROC curve was 0.89, p=0.016, supporting the ability of DBP to discriminate clinical sensitivity and resistance. There was variability in the quality of the tracings obtained, likely due to variability in the viability of the thawed patient samples. When we applied criteria only to accept tracings for which there was at least a difference of 100 relative fluorescent units between our positive control (FCCP) and negative control, we observed similar results, with an AUC of 0.88. This came at the cost of excluding 7 samples from analysis based on the criteria (Figure S6). Basically, every newly diagnosed patient with CML will be started on imatinib or another tyrosine kinase inhibitor, and nearly all will have at least a complete hematologic remission. Thus, there is little need for a new predictive biomarker to guide administration of tyrosine kinase inhibitors in CML. Nonetheless, this study demonstrates the principal that DBP can distinguish clinical sensitivity and resistance to a targeted agent.
Figure 7. DBP can stratify in vivo drug response to imatinib in a cohort of CML patients and to carboplatin in ovarian adenocarcinoma patients.
(A) 24 Frozen Ficoll purified Bone Marrow primary CML samples were treated for 16 hour with imatinib 1 and 5 μM, and DBP was then performed. Results are expressed as Δ%priming. * p<0.05. (B) A ROC curve analysis for this set of samples. The AUC is 0.89. (C) 16 ovarian adenocarcinoma patient samples were analyzed by DBP with carboplatin. We treated the samples for 16 hours with carboplatin 100 μg/mL, and DBP was then performed. Shown is a Kaplan -Meier plot of the patients’ Progression-Free survival in response to carboplatin and taxol. A significant difference was observed between those patients whose samples showed a Δ%priming >20% from those that were <20%, as assessed by Mantel-Cox statistical analysis. See also Figures S5 and S6, and Table 1.
Dynamic BH3 profiling predicts carboplatin response in ovarian cancer patients
Although our testing was focused on using DBP with targeted agents, pro-death signaling resulting from treatment with classical cytotoxic chemotherapies should be predictive of cellular response since these drugs also largely kill via the mitochondrial apoptotic pathway. We obtained 16 primary ovarian adenocarcinomas from surgical resection. We treated a single cell suspension of these tumors with carboplatin, the standard front-line therapy, ex vivo for 16 hours and performed DBP. We detected a robust Δ% priming (≥ 20%) in 6 of the patient specimens.
All analyzed patients were then treated with carboplatin in combination with taxol in the clinic. We then collected and analyzed the clinical data on the patients to assess progression free survival using an abnormal and rising CA-125 as an index for progression. Patients with ovarian adenocarcinomas that exhibited a robust Δ% priming (≥ 20%) experienced a significantly longer progression free survival to those patients who did not (Figure 7C).
DISCUSSION
Here we tested and supported the hypothesis that the initiation of death signaling is sufficient to determine eventual commitment to cell death. By detecting early death signaling, DBP can predict in vitro and in vivo cytotoxic response in varied cancers to varied classes of chemotherapeutic agents, agents which have in common only their ability to kill cancer cells via the mitochondrial pathway of apoptosis. While this provides basic mechanistic information about the events between drug treatment and commitment to cell death, we anticipate that its greatest utility might be in prediction of cancer patients’ response to therapy in the clinic. Over the past decade, an ever-growing number of therapies have been approved for use in medical oncology. But every tumor is distinct, with its own particular signaling network and pathway addiction yielding a distinct pattern of sensitivity to cancer therapeutics. The task of precision cancer medicine is to match a tumor to those agents that will most effectively eliminate it (Garraway and Janne, 2012; Sawyers, 2008; Yaffe, 2013).
An analogous problem was faced in the previous century in the world of clinical microbiology. As the number of antibiotics proliferated, it became more challenging to identify the best drug for a particular isolate of bacteria. The very practical solution that emerged was to simply grow a lawn of bacteria and expose the isolate to all available antibiotics in the form of drug-soaked disks. Antibiotics were then chosen from those that caused the greatest elimination of bacteria. This practice is still the standard and has not been displaced by any modern technology, including genomics, proteomics, or systems biology. While this method reveals little about signaling pathways and genetics of bacteria, it is supremely useful because it functionally summarizes the contribution of many genes and pathways to the phenotype that is most pertinent, the response of the viable bacterium to antibiotics. A version of this assay has been the mainstay of clinical microbiology for many decades.
Analogous ex vivo approaches have been attempted in oncology, but with little success. A typical strategy was to expose a patient’s tumor to drugs and place it into ex vivo culture for 3–14 days followed by evaluation of cell death, proliferation, or colony formation (Burstein et al., 2011). The biggest difficulty was the requirement for ex vivo culture of cancer cells. Many cancer cells simply rapidly die in ex vivo culture. Those that survive can undergo arrest or other phenotypic changes that accompany the transfer from a comfortable in vivo niche to an ex vivo plastic dish in 21% oxygen. In addition, if the culture is prolonged, there can be selection for non-tumor cells or clones that are poorly representative of the patient’s tumor. The result, in any case, was a series of studies that did not provide sufficient predictive power to be clinically useful. Exciting new ex vivo cell culture strategies using more modern techniques require weeks to months (Crystal et al., 2014). Their utility in guiding patient care will doubtless be tested in the coming years. Patient-derived xenograft (PDX) mouse models are being tested as a newer venue for functional assessment of tumor cell response to –– therapy (Hidalgo et al., 2014). However, the time (months) and expense that are required to establish PDX models may limit their utility in clinical medicine.
Here we have taken a different approach. Appreciating the tremendous advantages of perturbing the actual patient tumor cell with the actual therapy of interest, we instead have prioritized making observations early enough that long term ex vivo culture is not needed. While we have found that death signaling can be detected as early as 4 hours after treatment, depending on the drug, we have found that a sixteen hour incubation is sufficient for most agents to produce measurable death signaling in responding cells. We demonstrated that DBP can be exploited to select among many therapies the one that is best for a single tumor (Figure 6). We also demonstrated that DBP can select among many patients those that are most likely to respond to a single therapy (Figure 7). These are the two major functions of a clinically useful predictive biomarker, and it is notable that DBP can perform them both. Of equal importance, the clinical and in vivo experiments of Figure 7 demonstrate that useful predictive observations of both liquid (CML) and solid (ovarian) primary human tumors is consistent with a simple 16 hour monolayer culture.
We anticipate that DBP may be used to make personalized choices of therapy for patients. One could use DBP to choose agents among a panel of candidate drugs for one individual patient. Alternatively, one could use DBP to stratify a panel of patients to identify those most likely to respond to a individual drug. In the case of drugs that have activity only in a subset of a particular disease, we believe DBP can more efficiently stratify patient selection for clinical trials or clinical use by prospectively identifying those whose tumors are most likely to respond. In addition, while our focus here was on cancer cells, it is important to realize that this approach is also applicable to the study of non-malignant cells. As such, it can be used as a probe of sensitivities of cells in normal biology to a variety of insults, or as a toxicology tool to predict the toxicity of novel agents to normal tissues.
While we have focused mainly on single agent therapies in our proofs of principle studies, a strength of this approach is that it should work for both single agent and combination therapies. In fact, we explicitly demonstrated this in Figures 1, 2 and S3. Given the nearly universal emergence of resistance to single agent targeted therapies, even when there is an excellent initial response, strategies for the rational choice of personalized combination therapies is of great importance. We can envision two ways DBP could be used to fashion such strategies. One is to simply expose tumor cells to the combinations as we did in Figures 1, 2 and S3. Another is to test a panel of single agents via DBP, and combine two or more with good single agent activity.
A tremendous amount of information has been collected on cancer cells in the past few years, and the amount is likely to continue to grow exponentially. Much of this information is now genetic, with whole cancer genomes being sequenced (Barretina et al., 2012). In addition, there are technologies that garner an abundance of gene expression information, and those that capture protein expression (Kornblau et al., 2009). It remains to be seen how widely these technologies will be useful in better assigning therapy to patients. However, despite the huge amounts of information acquired, one common limitation of these studies is that they all represent static observations of dead cells. That means that a tremendous amount of the functional complexity of the cell has been lost to study. With DBP, we anticipate that a small number of strategic perturbations (drug and peptide exposures) on viable cells will yield vastly fewer bits of information, but that a great proportion of the bits will be clinically actionable.
EXPERIMENTAL PROCEDURES
Cell lines and treatments
RPMI 1640 media supplemented with 10% heat inactivated fetal bovine serum (Gibco) 10 mM L-Glutamine and 100 U/ml penicillin and 100 μg/ml streptomycin, was used for the culture of the cell lines used. The cells were cultured at 37°C in a humidified atmosphere of 5% CO2.
Isolation and Treatment of Primary CML Cells
30 primary CML samples from bone marrow biopsies viably frozen in 90% FBS/10%DMSO were obtained from the the Pasquarello Tissue Bank at Dana-Farber Cancer Institute and from Dr. Philip C. Amrein at the Massachusetts General Hospital. Cells were thawed and resuspended in complete RPMI media and washed with fresh media, counted by trypan blue exclusion and plated in a 12-well plate, 1 million cells/well and treated with imatinib 1 and 5 μM. DBP failed on 5 samples due to failure of mitochondria to maintain transmembrane polarization. After a 16 h incubation at 37°C in a humidified atmosphere of 5% CO2, Dynamic BH3 Profile analysis was performed. Clinical response data was compiled by clinicians; patients are considered responders when complete hematologic response was observed.
Ovarian primary tumors
Fresh primary tumors, prospectively obtained from routine resections after patients signed an informed consent approved by the Institutional Review Board (DFCI#02-051), were used for preparation of viable single cell suspensions. Tumors were first mechanically dissociated and digested for 1 hour at 37°C in 1 mg/mL collagenase/dispase (Roche Diagnostics). Cells were then filtered through a cell strainer and cell viability was assessed by trypan blue exclusion. Cells were then frozen in freezing buffer (fetal bovine serum with 10% DMSO). For DBP, cells were thawed and resuspended in complete RPMI media with 100 U/ml of DNAse I and incubated 15 min at room temperature. Then the cells were washed with fresh media, counted by trypan blue exclusion and plated in a 12-well plate, 0.2–0.5 M cells/well and treated with carboplatin 100 μg/ml. After a 16 h incubation at 37°C in a humidified atmosphere of 5% CO2. Dynamic BH3 Profile analysis was performed blinded to clinical outcome. Clinical response data was compiled by clinicians 6–24 months after sample acquisition.
Dynamic BH3 Profiling
2 × 104 cells/well were used for cell lines, 4 × 104 cells/well were used for primary CML and AML. 15 μL of BIM BH3 peptide (final concentration of 0.03, 0.1, 0.3, 1 and 3 μM) in T-EB (300 mM Trehalose, 10 mM hepes-KOH pH 7.7, 80 mM KCl, 1 mM EGTA, 1 mM EDTA, 0.1% BSA, 5 mM succinate) were deposited per well in a black 384-well plate (BD Falcon no. 353285). Single cell suspensions were washed in T-EB before being resuspended at 4× their final density. One volume of the 4× cell suspension was added to one volume of a 4× dye solution containing 4 μM JC-1, 40 μg/mL oligomycin, 0.02% digitonin, 20 mM 2-mercaptoethanol in T-EB. This 2× cell/dye solution stood at RT for 10 min to allow permeabilization and dye equilibration. A total of 15 μL of the 2× cell/dye mix was then added to each treatment well of the plate, shaken for 15 s inside the reader, and the fluorescence at 590 nm monitored every 5 min at RT. Percentage loss of Ψμ for the peptides is calculated by normalization to the solvent only control DMSO (0% depolarization) and the positive control FCCP (100% depolarization). Individual DBP analysis were performed using triplicates for DMSO, FCCP and the different BIM BH3 concentrations used, and the expressed values stand for the average of three different readings. In cases were Standard Deviation was >10%, the outlying reading was discarded. % priming stands for the maximum % depolarization obtained from the different BIM BH3 concentrations tested; typically 0.03, 0.1, 0.3, 1 and 3 μM. Δ% priming stands for the difference between treated cells minus non-treated cells (% primingtreated − % primingnon-treated). See also Figure S1.
Cell viability assays
Cells were stained with fluorescent conjugates of Annexin-V (BioVision) and/or propidium iodide (PI) and analyzed on a FACS Canto machine (BD). Viable cells are annexin-V negative and PI negative, and cell death is expressed are 100%-viable cells. Δ% Cell Death stands for the difference between treated cells minus non-treated cells (% Cell Deathtreated − % Cell Deathnon-treated).
Immunoblotting
Total cell lysates were prepared in 1% Chaps buffer [5 mM MgCl2, 137 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Chaps, 20 mM Tris-HCl (pH 7.5), and protease inhibitors (Complete, Roche)]. Cells were washed twice, resuspended with 50–100 μL of CHAPS lysis buffer, and kept on ice for 30 minutes. Then, the cellular suspension was centrifuged at 16.100g for 5 minutes, and the supernatant used to perform the immunoblotting analysis.
20 μg of protein was loaded on NuPAGE 10% Bis-Tris polyacrylamide gels (Invitrogen). The following antibodies were used to detect proteins on the membrane (dilution 1:1000): Actin (Chemicon, MAB1501); PARP-1 (cell signaling, #9542); BCL-2 (Epitomics, #1017-1); BIM (Cell Signaling, #2933); BCL-xL (Cell signaling, #2762).
Statistical analysis
Statistical significance of the results was analyzed using Student’s t-tail test using GraphPad Prism 5.0 software. *p < 0.05 and **p < 0.01 were considered significant. SEM stands for Standard Error of the Mean. For ROC curve analysis cell lines were considered responsive to treatment when Δ% cell death > 10 %; CML clinical samples when the patient achieved a complete hematologic response after treatment; for ovarian adenocarcinoma biopsies, clinical response data was compiled by clinicians 6–24 months after sample acquisition.
Supplementary Material
Acknowledgments
We gratefully acknowledge funding from the Beatriu de Pinós programme from la Generalitat de Catalunya in Spain (J.M), and NIH grants RO1CA129974, R01CA135257, P01CA068484, and P01CA139980. A.L. is a Leukemia and Lymphoma Society Scholar. R.D. would like to thank the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (AMRF) and NIH grant U01CA152990. We thank the Pasquarello Tissue Bank and the DFCI Flow Cytometry. We also thank Richard P. Oakley Jr. and http://www.servier.com/Powerpoint-image-bank (licensed under Creative Commons Attribution 3.0 Unported License) for allowing us to use some images to elaborate Figure 1 and the Graphical Abstract.
Footnotes
Supplemental Information includes Extended Experimental Procedures, one table and six supplementary figures that can be found with this article online.
AUTHOR CONTRIBUTION
The study was conceived and designed by A.L and J.M. Figure 2 analyses were performed by J.D.D. under supervision of J.M and A.L. P.C.A. provided CML patient samples and relevant clinical correlation. Ovarian adenocarcinoma patient samples and clinical correlation were obtained from N.S.H., H.P., R.S. B., U.M. and R.D; K.A.S. processed these samples. Drug-resistant NSCLC cell lines were obtained from D.E. and P.A.J.; melanoma murine allograft cells and relevant in vivo data were obtained from O.M. and K.C. All other experiments were performed by J.M. with technical assistance from J.R under supervision of A.L.. Figures 6 and 7C were prepared by K.A.S., all others by J.M. The manuscript was prepared by A.L and J.M. with input from most authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt AP, Bhende PM, Sin SH, Roy D, Dittmer DP, Damania B. Dual inhibition of PI3K and mTOR inhibits autocrine and paracrine proliferative loops in PI3K/Akt/mTOR-addicted lymphomas. Blood. 2010;115:4455–4463. doi: 10.1182/blood-2009-10-251082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein HJ, Mangu PB, Somerfield MR, Schrag D, Samson D, Holt L, Zelman D, Ajani JA. American Society of Clinical Oncology clinical practice guideline update on the use of chemotherapy sensitivity and resistance assays. J Clin Oncol. 2011;29:3328–3330. doi: 10.1200/JCO.2011.36.0354. [DOI] [PubMed] [Google Scholar]
- Certo M, Del Gaizo Moore V, Nishino M, Wei G, Korsmeyer S, Armstrong SA, Letai A. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell. 2006;9:351–365. doi: 10.1016/j.ccr.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crystal AS, Shaw AT, Sequist LV, Friboulet L, Niederst MJ, Lockerman EL, Frias RL, Gainor JF, Amzallag A, Greninger P, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014;346:1480–1486. doi: 10.1126/science.1254721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Carlson N, Takeyama K, Dal Cin P, Shipp M, Letai A. BH3 profiling identifies three distinct classes of apoptotic blocks to predict response to ABT-737 and conventional chemotherapeutic agents. Cancer Cell. 2007a;12:171–185. doi: 10.1016/j.ccr.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Deng J, Shimamura T, Perera S, Carlson NE, Cai D, Shapiro GI, Wong KK, Letai A. Proapoptotic BH3-only BCL-2 family protein BIM connects death signaling from epidermal growth factor receptor inhibition to the mitochondrion. Cancer Res. 2007b;67:11867–11875. doi: 10.1158/0008-5472.CAN-07-1961. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, Maira M, McNamara K, Perera SA, Song Y, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercan D, Xu C, Yanagita M, Monast CS, Pratilas CA, Montero J, Butaney M, Shimamura T, Sholl L, Ivanova EV, et al. Reactivation of ERK signaling causes resistance to EGFR kinase inhibitors. Cancer Discov. 2012;2:934–947. doi: 10.1158/2159-8290.CD-12-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercan D, Zejnullahu K, Yonesaka K, Xiao Y, Capelletti M, Rogers A, Lifshits E, Brown A, Lee C, Christensen JG, et al. Amplification of EGFR T790M causes resistance to an irreversible EGFR inhibitor. Oncogene. 2010;29:2346–2356. doi: 10.1038/onc.2009.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber AC, Corcoran RB, Ebi H, Sequist LV, Waltman BA, Chung E, Incio J, Digumarthy SR, Pollack SF, Song Y, et al. BIM expression in treatment-naive cancers predicts responsiveness to kinase inhibitors. Cancer Discov. 2011;1:352–365. doi: 10.1158/2159-8290.CD-11-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber AC, Li D, Song Y, Liang MC, Yeap BY, Bronson RT, Lifshits E, Chen Z, Maira SM, Garcia-Echeverria C, et al. Differential induction of apoptosis in HER2 and EGFR addicted cancers following PI3K inhibition. Proc Natl Acad Sci U S A. 2009;106:19503–19508. doi: 10.1073/pnas.0905056106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway LA, Janne PA. Circumventing cancer drug resistance in the era of personalized medicine. Cancer Discov. 2012;2:214–226. doi: 10.1158/2159-8290.CD-12-0012. [DOI] [PubMed] [Google Scholar]
- Haibe-Kains B, El-Hachem N, Birkbak NJ, Jin AC, Beck AH, Aerts HJ, Quackenbush J. Inconsistency in large pharmacogenomic studies. Nature. 2013 doi: 10.1038/nature12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo M, Amant F, Biankin AV, Budinska E, Byrne AT, Caldas C, Clarke RB, de Jong S, Jonkers J, Maelandsmo GM, et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 2014;4:998–1013. doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joensuu H, Roberts PJ, Sarlomo-Rikala M, Andersson LC, Tervahartiala P, Tuveson D, Silberman S, Capdeville R, Dimitrijevic S, Druker B, et al. Effect of the tyrosine kinase inhibitor STI571 in a patient with a metastatic gastrointestinal stromal tumor. N Engl J Med. 2001;344:1052–1056. doi: 10.1056/NEJM200104053441404. [DOI] [PubMed] [Google Scholar]
- Kluck RM, Bossy-Wetzel E, Green DR, Newmeyer DD. The release of cytochrome c from mitochondria: a primary site for Bcl-2 regulation of apoptosis. Science. 1997;275:1132–1136. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- Kornblau SM, Tibes R, Qiu YH, Chen W, Kantarjian HM, Andreeff M, Coombes KR, Mills GB. Functional proteomic profiling of AML predicts response and survival. Blood. 2009;113:154–164. doi: 10.1182/blood-2007-10-119438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2:183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- Maertens O, Johnson B, Hollstein P, Frederick DT, Cooper ZA, Messiaen L, Bronson RT, McMahon M, Granter S, Flaherty K, et al. Elucidating distinct roles for NF1 in melanomagenesis. Cancer Discov. 2013;3:338–349. doi: 10.1158/2159-8290.CD-12-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Chonghaile T, Sarosiek KA, Vo TT, Ryan JA, Tammareddi A, del Moore VG, Deng J, Anderson KC, Richardson P, Tai YT, et al. Pretreatment mitochondrial priming correlates with clinical response to cytotoxic chemotherapy. Science. 2011;334:1129–1133. doi: 10.1126/science.1206727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Pencina MJ, D’Agostino RB, Sr, D’Agostino RB, Jr, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med. 2008;27:157–172. doi: 10.1002/sim.2929. discussion 207–112. [DOI] [PubMed] [Google Scholar]
- Ryan J, Letai A. BH3 profiling in whole cells by fluorimeter or FACS. Methods. 2013;61:156–164. doi: 10.1016/j.ymeth.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan JA, Brunelle JK, Letai A. Heightened mitochondrial priming is the basis for apoptotic hypersensitivity of CD4+ CD8+ thymocytes. Proc Natl Acad Sci U S A. 2010;107:12895–12900. doi: 10.1073/pnas.0914878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarosiek KA, Chi X, Bachman JA, Sims JJ, Montero J, Patel L, Flanagan A, Andrews DW, Sorger P, Letai A. BID preferentially activates BAK while BIM preferentially activates BAX, affecting chemotherapy response. Mol Cell. 2013;51:751–765. doi: 10.1016/j.molcel.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyers CL. Chronic myeloid leukemia. N Engl J Med. 1999;340:1330–1340. doi: 10.1056/NEJM199904293401706. [DOI] [PubMed] [Google Scholar]
- Sawyers CL. The cancer biomarker problem. Nature. 2008;452:548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- Sikorski R, Yao B. Visualizing the landscape of selection biomarkers in current phase III oncology clinical trials. Sci Transl Med. 2010;2:34ps27. doi: 10.1126/scitranslmed.3001087. [DOI] [PubMed] [Google Scholar]
- Vo TT, Ryan J, Carrasco R, Neuberg D, Rossi DJ, Stone RM, Deangelo DJ, Frattini MG, Letai A. Relative mitochondrial priming of myeloblasts and normal HSCs determines chemotherapeutic success in AML. Cell. 2012;151:344–355. doi: 10.1016/j.cell.2012.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, Roth KA, MacGregor GR, Thompson CB, Korsmeyer SJ. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB. The scientific drunk and the lamppost: massive sequencing efforts in cancer discovery and treatment. Sci Signal. 2013;6:pe13. doi: 10.1126/scisignal.2003684. [DOI] [PubMed] [Google Scholar]
- Zhou W, Ercan D, Chen L, Yun CH, Li D, Capelletti M, Cortot AB, Chirieac L, Iacob RE, Padera R, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.