SUMMARY
The integration of biophysical data from multiple sources is critical for developing accurate structural models of large multiprotein systems and their regulators. Mass spectrometry (MS) can be used to measure the insertion location for a wide range of topographically sensitive chemical probes, and such insertion data provide a rich, but disparate set of modeling restraints. We have developed a software platform that integrates the analysis of label-based MS data with protein modeling activities (Mass Spec Studio). Analysis packages can mine any labeling data from any mass spectrometer in a proteomics-grade manner, and link labeling methods with data-directed protein interaction modeling using HADDOCK. Support is provided for hydrogen/ deuterium exchange (HX) and covalent labeling chemistries, including novel acquisition strategies such as targeted HX-tandem MS (MS2) and data-independent HX-MS2. The latter permits the modeling of highly complex systems, which we demonstrate by the analysis of microtubule interactions.
INTRODUCTION
Integrative methods in structural biology are delivering impressive visualizations of higher-order multiprotein associations. The functional data gained from such representations are essential for understanding the properties emergent from self-assembling protein “building blocks”. The integrative concept involves a high-resolution structural analysis of these building blocks through conventional means, and then leapfrogging their inherent limitations by completing the structure-building exercise using biophysical methods, which may be of lower resolution, but can be applied to the assembled state (Karaca and Bonvin, 2013; Thalassinos et al., 2013; Ward et al., 2013). The potential of this approach has been portrayed through the modeling of a growing number of complex states, built from fitting the refined structures of individual components into cryoelectron microscopy reconstructions (Schraidt and Marlovits, 2011; Topf et al., 2008), as well as small-angle X-ray scattering envelopes (Devarakonda et al., 2011; Putnam et al., 2007). Models can generate testable mechanisms even when the structures of all the building blocks are not fully available, as shown in a recent structure for a membrane-bound proton-driven ATP synthase (Lau and Rubinstein, 2012). Any technology that contributes spatial or conformational information on the free and bound states adds considerable value to accurate model building, and when chosen carefully, technologies with complementary attributes can overcome deficiencies in any one approach (Alber et al., 2007; Lasker et al., 2012).
As we continue to image molecular events at wider spatial and temporal scales, we require methods that can provide restraint data under a wide range of conditions. Biological mass spec-trometry (MS) is moving to support such activities and is quite likely the most promising technology for generating residue-level topographical data in the least restrictive manner (Politis et al., 2014). Numerous recent examples have begun to incorporate MS for structure-building activities. Crosslink detection by proteomic methods and the computational tools developed for them are useful for coarse positioning (Ciferri et al., 2008; Greber et al., 2014; Kahraman et al., 2013; Merkley et al., 2014; Walzthoeni et al., 2013), but a wealth of “single-point” chemistries are available to monitor conformational dynamics and map proteins more completely and at higher resolution (Konermann et al., 2011; Mendoza and Vachet, 2009). MS methods developed to monitor site-specific labeling kinetics can define interfaces at a resolution approaching individual residues (Bennett et al., 2010; Landgraf et al., 2012; Melero et al., 2012; Pan et al., 2012; Roberts et al., 2012). Labeling chemistries are available for both the protein backbone (hydrogen/deuterium exchange) and amino acid side chains (covalent methods like hydroxyl radical labeling).
Label detection by MS shares certain features with MS-driven proteomics. Both invoke enzymatically driven workflows to generate large sets of peptides. These peptides need to be identified and then quantified using either label-based or label-free methods. However, the experiments are structured quite differently and the data are used in much different ways. MS-based integrative methods begin with a known set of proteins, often use different proteases (Ahn et al., 2013), and need to quantify chemical modifications at every residue in a sequence. The data are then interpreted for structural or conformational meaning. A number of software tools support the basics of hydrogen/deuterium exchange (HX) analysis, for example HDX Workbench (Pascal et al., 2012), Hydra (Slysz et al., 2009), and Hexicon (Lindner et al., 2014), but no platform is sufficiently generic to accommodate any labeling chemistry, or support the ultimate goal of the integrative approach, namely, the restraint-based modeling of molecular structures. Here we present the Mass Spec Studio (the Studio), an adaptable framework designed to support the varied demands of MS-based integrative structural biology. The Studio incorporates efficient processing of liquid chromatography-tandem mass spectrometry (LC-MS/ MS) data with workflows designed to support the unique challenges of integrative methods, which includes the extraction of modeling restraints and structure-building activities.
RESULTS
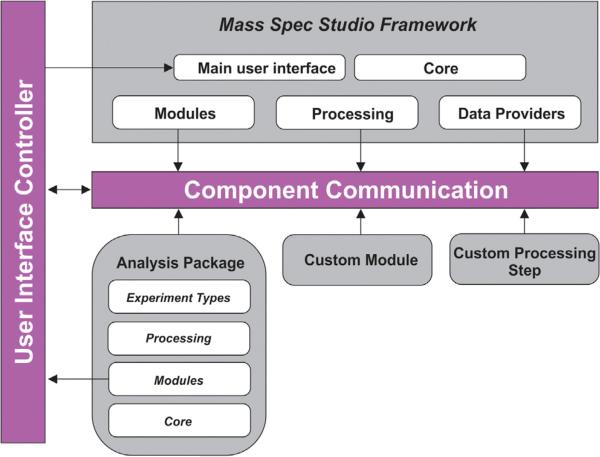
A Framework for Rapid Application Development
Most software for the processing of MS data is vendor-supplied and inflexibly tied to a set of industry-driven applications. Support for structural biology is lacking, both in terms of quantitating chemical labeling events in proteins, and mining the data for structural restraints. The Studio represents a new extensible architecture for the analysis of MS data (Figure 1) and is specifically designed to foster the development of innovative structure-based methods involving MS data. It supports a plugin model, designed to capture and reuse components for a variety of applications. Analysis packages are assembled from a base of components, and novel components are added to a repository for reuse. The Studio presents a flexible framework that automatically links components in the correct fashion, and communication is through high-level interfaces rather than at a low level through source code. An efficient communications protocol allows us to build and concatenate application packages to support entire workflows, spanning data processing to structure building. Essential elements of the design and workflow are provided in Supplemental Experimental Procedures (available online). The Studio offers a series of prebuilt application packages for structural biologists as described below and functions with data from all major instrument platforms. Additional details on application functionality are supplied in Supplemental Experimental Procedures.
Figure 1. A Conceptual Overview of the Component-Based Architecture of the Studio Framework.
The encapsulation model for the core library allows for easy adding, swapping, and removing of components. The component communication model combined with the user interface controller allows seamless linkage of components with each other, as well as with the core library.
Hydrogen/Deuterium Exchange Analysis Package
There are three types of experiments that are supported by the HX analysis package, all based on peptide-level deuteration analysis, or the bottom-up approach (Marcsisin and Engen, 2010). The method involves continuous labeling of proteins and protein complexes using D2O. Labeled samples are quenched to arrest the exchange, and then digested with a nonselective protease. The rate of deuterium labeling is measured at various time-points, and a common goal for each experiment type is to determine where and how a complexation event alters the rate of deuterium incorporation at locations in the protein backbone. The experiments differ in how they support complex sample types and in how they use MS/MS data for label measurement.
One-Dimensional Hydrogen/Deuterium Exchange-Mass Spectrometry
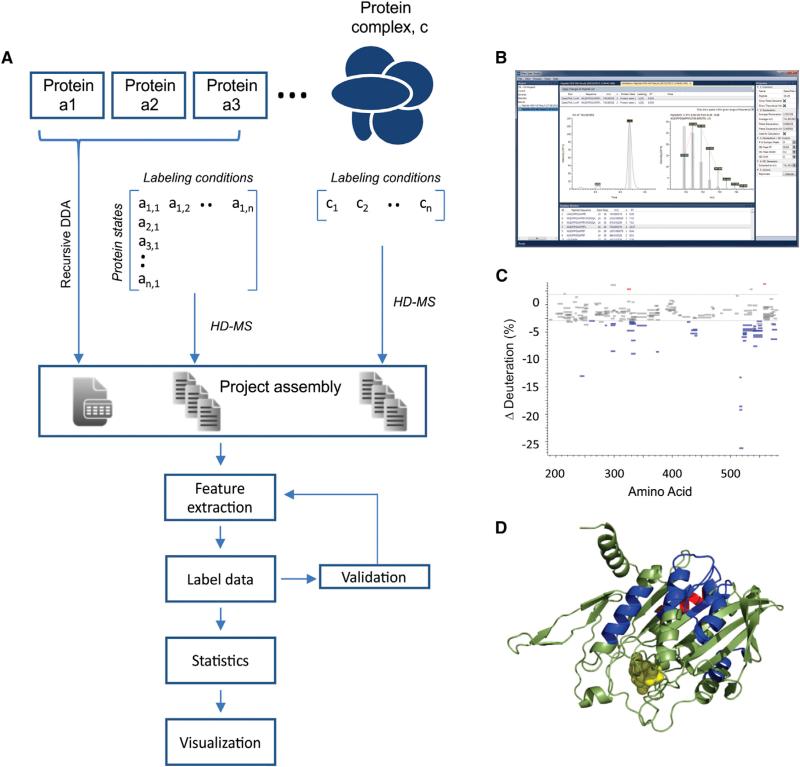
The one-dimensional (1D) HX-MS experiment supports the extraction of peptide-level deuteration data from large sets of LC-MS runs. It represents a 1D analysis, in that only MS spectra are used to quantify deuterium incorporation. The workflow is used to visualize binding-induced changes in deuteration kinetics for individual proteins or those involved in larger multiprotein assemblies (Figure 2A). Projects are assembled from all LC-MS data files, together with lists of peptides and their retention times identified in previous experiments using proteomics methods. The deuteration data are then extracted from all the peptides detected in each LC-MS run. The HX analysis package implements an interactive graph control module, to rapidly validate and correct peptide selections and isotope profile definitions (Figure 2B). Correcting peptide lists generated through proteomics experiments is necessary; as such experiments do not mesh cleanly with the demands of HX-MS analysis (Wales et al., 2013). Noisy spectra, strongly overlapped isotopic distributions, and nonapex chromatographic retention times can generate successful peptide identifications, but may not be useful for deuteration analysis. The set of LC-MS runs can be quickly reprocessed using the validated peptide list, to generate a validated data set of deuteration values.
Figure 2. Process Control in the Studio for Protein Conformational Analysis Using 1D HX-MS.
(A) Data from individual proteins or collections of proteins (in any state of assembly) are grouped according to the labeling conditions applied (e.g., different time-points) and associated with the feature lists assembled from data-dependent acquisitions of LC-MS/MS runs. The Studio automates the extraction of features from the full set, the essential deuteration data, and provides strong methodology for data validation and knowledge extraction.
(B) Validation is enabled using interactive spectral viewers.
(C) A Woods plot, providing a useful statistical output for comparing deuteration data for a protein in any two states, here the ADP and ATP loaded states for EGFP-MCAK at a given labeling time-point (300 s.)
(D) A dynamic molecular viewer linked to the statistical output, allowing for interactive labeling of a 3D structural representation with HX data, here showing the structural representation of MCAK (Protein Data Bank [PDB] 1V8K). Disordered N-terminal, C-terminal, and neck domains are expressed using colored boxes that correspond to the regions in (C).
To illustrate the functionality of the Studio for this experiment type, we explored the effect of nucleotide exchange on the conformational status of mitotic centromere associated kinesin (MCAK; see Supplemental Experimental Procedures). MCAK depolymerizes microtubules in a process that is essential for the detection and capture of sister chromatids in the developing mitotic spindle (Wordeman and Mitchison, 1995; Wordeman et al., 2007). This process is driven by conformational changes in the kinesin upon the exchange of ADP for ATP. MCAK conformational stability is strongly regulated by the exchange, promoting a transition between an open (ADP) and a closed (ATP) state (Ems-McClung et al., 2013). Replicate deuteration data for the ADP and ATP loaded forms of a truncated EGFP-MCAK were measured. The statistical analysis package in the Studio was used to mine the HX output and identify regions of perturbed labeling by comparing the ADP-loaded state to the ATP-loaded state. The typical pairwise comparison for a given time-point can be displayed using a Woods plot (Figure 2C), which is fully interactive with PyMOL for molecular visualization of the data (Figure 2D). Using a corrected feature list, the project output was generated on a modest computer in a few minutes. Output options include kinetic labeling plots for all detected peptides and alternative statistical representations. We have included a detailed tutorial for this experiment type, in addition to a functional description (Supplemental Experimental Procedures). The HX package supports high-resolution MS. Deuteration data and deuterium distribution values can be extracted from any expanded isotopic envelope, and the framework supports the incorporation of alternative algorithms for deuteration analysis (Liu et al., 2014).
Targeted Hydrogen/Deuterium Exchange-Mass Spectrometry2
In many instances a peptide-level analysis is sufficiently informative for conformational studies, for example, when monitoring folding events or perturbations of secondary structural units. However, defining binding sites and allosteric effects often demands a higher structural resolution, and this requires tandem MS (MS2). A potent method involves a targeted MS2 reanalysis of peptides that show evidence of perturbed deuteration kinetics in previous 1D HX-MS runs (Landgraf et al., 2012). To support this method, we built an application in the HX package for measuring deuteration data from peptide fragments, generated either by collisionally induced dissociation (CID) or electron-mediated events like electron-transfer dissociation (ETD). ETD is most useful for such purposes as sequence reads are longer, and this fragmentation technology is effective at minimizing deuterium scrambling within the peptide sequence. Scrambling erodes structural resolution and must be avoided (Jørgensen et al., 2005).
The targeted HX-MS2 experiment has two central components. The first defines a subset of peptides to explore at higher structural resolution by graphically interacting with the HX-MS data through the Woods plots (Figure 3). Areas of protein sequence that require an MS2 analysis in a subsequent experiment are marked. A selection algorithm interacts with the HX-MS data and returns a set of peptides that can be used directly to build an MS2 experiment. The algorithm prioritizes the selections for ETD analysis by selecting the optimal peptide charge states, and a scheduling function ensures that as much time as possible is spent generating ETD data on each peptide. The approach is similar to strategies found in targeted proteomics (Bertsch et al., 2010). This utility allows the user to generate high-quality fragmentation data sufficient for measuring deuteration at the fragment level. An example of the targeted high-resolution workflow is provided as a tutorial in Supplemental Experimental Procedures.
Figure 3. The Interactive Targeted HX-MS2 Experiment.
(A) The workflow involves a collection of data using the conventional HX-MS method, and then identifying areas of interest in the statistical output that may require advanced, nonscrambling fragmentation methods such as ETD for deuteration analysis at higher structural resolution in a subsequent experiment.
(B) The data from this second-pass analysis can be interrogated in both MS and MS/MS space with the interactive validation viewers.
(C) The per-residue deuteration data can be integrated with the statistical output of the first HX-MS analysis.
The second component interprets the results of the targeted MS2 runs by visualizing the fragment deuteration levels. The data may be validated using interactive graphs and visualized with a scatterplot. The scatterplot is designed to summarize all fragment-level deuteration for the peptide (Figure 3). It is referenced against a deuterium-scrambled state, which helps the user confirm if fragmentation has preserved the location of the deuteriums or if scrambling has occurred. The high-resolution data can be meshed with the Woods plot from the HX-MS analysis through the statistics package. This allows the user to maintain a full view of the data and interactively access the mass shift data at higher structural resolution (Figure 3).
Data Independent Hydrogen/Deuterium Exchange-Mass Spectrometry2
High-resolution MS has allowed HX applications to tackle protein systems of moderately high complexity (Zhang et al., 2011), but many complexes that can be reconstituted or isolated have molecular weights exceeding the capacity of the 1D HX-MS method. When multiple proteins are processed using typical nonspecific proteases, the usable sequence coverage for any single protein begins to drop. MS signals start to overlap and the high peptide load reduces ionization efficiency in the mass spectrometer. Longer chromatographic runs cannot be used to solve these problems, because the deuterium label is transient and bleeds away within 10–30 min. Thus, the reduced intensity and quality of peptide isotopic distributions effectively limit the size of complexes addressable by HX-MS. Significant performance enhancement should be possible with quantitative MS2. A method of analysis has been proposed where unique fragment ions can be used as surrogates for deuteration analysis (Percy and Schriemer, 2011). The method allows for peptide deuteration to be measured in a less-noisy second MS dimension, and it increases the dynamic range of the deuteration measurement. However, the labor required to select surrogate fragments for each peptide in a large digest is prohibitively high. Generic methods for a complete and systematic collection of MS2 data for all peptides in an LC run would offer a significant improvement.
The advantages of the surrogate approach can be returned to complex analysis using data independent acquisition (DIA) modes of operation, such as the “sequential window acquisition of all theoretical spectra” (SWATH) mode (Chapman et al., 2013). In DIA, the entire m/z range is fragmented, and the associations between MS and MS2 peaks are rebuilt computationally by correlating the chromatographic properties of the data. The SWATH mode is appealing for HX applications, because the mass range is compartmentalized into moderately sized windows, and each compartment is scanned quickly enough to support liquid chromatography. In this way the entire peptide digest is represented in MS2 space, and all surrogates are present. We built a component in the HX package to support SWATH for deuteration analysis (Figure 4). Efficient data extraction is applied to both MS and MS2 domains, and an algorithm correlates peptide and fragment chromatographic retention times to define associations. Knowing the protein composition of the sample provides a natural advantage over related methods in proteomics, even though nonspecific proteases add complexity, each fragment ion can be assessed for uniqueness. This simplifies peptide identification, which we have based on a proteomics scoring algorithm (Craig et al., 2004), and uses only the unique fragments in the association groups. As shown in Figure 4, the DIA concept is very efficient and information-rich. It allows for peptide identification, peptide deuteration measurement in MS, and fragment deuteration measurement in MS2, in a single experiment. It simplifies the workflow, resolves peak conflicts, and extends the HX method to systems of greater complexity. The experiment supports SWATH-like implementations involving ETD fragmentation, although such modes have not yet been reported.
Figure 4. Data Independent Acquisition for the HX-MS2 Experiment.
The package quantifies deuterium uptake from peptide MS data and MS/MS data, after a correlation of spectral data.
(A) For high quality data, deuteration values can be measured for both the peptide and the corresponding fragment set.
(B) For lower quality MS data, either due to poor ion statistics or peak overlap, the fragments offer a surrogate for deuteration analysis. Although this mode is currently demonstrated for CID fragmentation, ETD fragmentation with no scrambling is possible as a means of returning higher structural resolution to the bottom-up method.
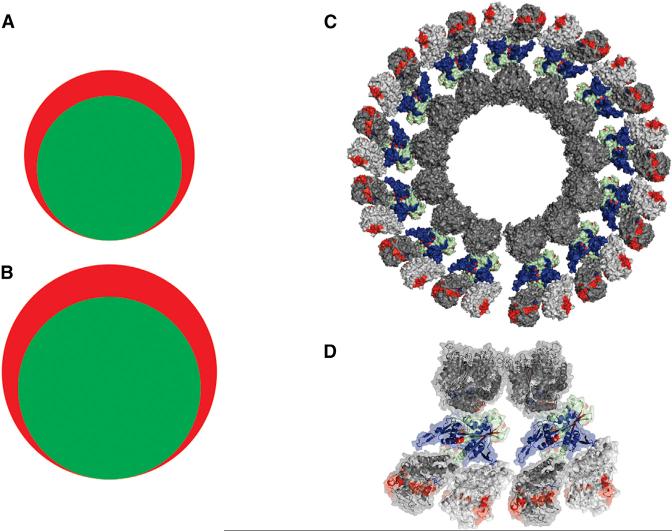
To test the data-independent HX-MS2 experiment type, we analyzed MCAK in complexation with microtubules. ATP-loaded MCAK preferentially interacts with microtubule plus ends, where it induces curvature in microtubule protofilaments and subsequent dissociation. We wanted to explore the depolymerization process induced by MCAK, which required producing a stabilized form of the curved state. This was achieved by capturing the protofilaments on preassembled microtubules through a spiral complex that capitalizes on the secondary tubulin binding sites presented by MCAK (Tan et al., 2006). This equilibrium state generates two forms of tubulin assembly, and may represent an intermediate in the depolymerization cycle. Data independent HX-MS2 was applied to free microtubules, a functional neck-motor construct of MCAK, and the captured state. The construct exceeds 1 giga-Dalton in size, presenting over 150 kDa of unique sequence. When applying the conventional HX-MS method, we detected 281 usable peptides for α/β-tubulin and 119 for the MCAK construct. HX-MS2 detected the same set of peptides, with an additional 27% tubulin peptides and 26% MCAK peptides detectable uniquely in fragment space (Figure 5A). The validity of these fragments was confirmed using a complete library of peptide identifications, collected from exhaustive proteomics analyses of each protein digest. With improved coverage, we observed that MCAK is strongly stabilized when captured in an intermediate state of depolymerization. This is consistent with a two-site tubulin binding mode previously reported (Zhang et al., 2013). Our analysis also supports an outwardly curving protofilament geometry: the tubulin surface normally within the lumen of a microtubule is now exposed (Figure 5B). The data-independent HX-MS2 experiment type presents a much simpler workflow. SWATH-based HX-MS2 avoids all the initial sequencing steps of the conventional experiment, because all peptide fragments are collected in every experiment.
Figure 5. SWATH-Based HX-MS2 Provides Increased Spectral Capacity for Analysis of the MCAK-Microtubule Interaction.
The addition of tandem MS data to the workflow increases the number of useable peptides for (A) MCAK (HX-MS, 119 peptides [green]; HX-MS2, 150 peptides [red]) and (B) α/β-tubulin (HX-MS, 281 peptides [green]; HX-MS2, 358 peptides [red]). (C) HX-MS2 perturbation data mapped to a model of the MCAK-tubulin depolymerized state, captured on an intact microtubule.
(D) Expanded element of the captured state. Blue is a protection and red is a deprotection upon assembly. Proteins are shown with a base color of dark gray (α-tubulin), light gray (β-tubulin), and light green (MCAK).
Covalent Label Analysis Package
HX methods support the investigation of backbone dynamics and solvent accessibility, but other covalent chemistries can provide complementary data through side-chain labeling. The covalent labeling package in the Studio permits the analysis of side-chain labeling chemistries of any type. It supports a bottom-up strategy in a targeted MS2 mode. Label distribution is calculated based upon the detection of peptides and their labeled counterparts (Figure 6). The labeled species in the distribution tend to separate during chromatography, which adds complexity to the analysis. However, all peaks in a distribution are found automatically and the distributions can be validated using a combination of accurate mass and fragment data. Labels can be quantified at the MS2 level to increase the resolution of the analysis. As with the HX-MS modes, data can be imported into the statistics package for assessment.
Figure 6. The Covalent Labeling Package Supports the Quantitation of any Covalent Labeling Chemistry for Footprinting Analysis.
(A) The processing path is similar to the high resolution HX mode, as peptides with binding-altered labeling can be reanalyzed by MS/MS for site-specific determinations.
(B) The example shows a peptide (HVMTNLGEK) from a tryptic digest of calmodulin labeled with a photoactivatable diazirine (azibutanol). Although one label is inserted, there are two distinct observable features seen in the chromatogram, which can indicate at least two label insertion sites.
(C) Fragment-level data is displayed in a generic format that compares two states assembled in the statistics package (here labeled minus unlabeled). Significant labeling changes are presented as histogram bars, normalized to the largest value (expressed as a percentage labeling). Direction of change is color-coded: red (increased labeling), blue (decreased labeling), and black (no change). Although different ion series from an ETD fragmentation of this peptide (c and z) were measured, the label insertion points are localized to M and E on the basis of z-type fragments.
Structure Building, a Pipeline to High Ambiguity Driven Biomolecular Docking
A principal driver for the development of the Studio was the integration of MS data into structure-building activities. The Studio supports a package designed to extract candidate interfacial data from HX and covalent labeling output. The data are combined with high ambiguity driven biomolecular docking (HADDOCK) (de Vries et al., 2010; Dominguez et al., 2003) in order to generate ambiguous interaction restraints (AIRs) for residues presented as part of the interface, or specific unambiguous restraints in the case of crosslinking data. HADDOCK supports docking by combining experimentally measured interface data with approaches based on energetics and shape complementarity. It has demonstrated high performance in multiple rounds of critical assessment of prediction of interactions (Rodrigues et al., 2013) and is well suited to handle the ambiguity presented by labeling chemistries. HX and covalent labeling can map interfaces, but neither identifies interacting residues across binding interfaces, nor can they readily discriminate between binding sites or allosteric effects. For each element in a complex, HADDOCK addresses ambiguity by using data to specify active residues (those known to make contact in a complex) and passive residues (those that may make contact), and it generates an interaction restraint between an active residue in a protein and the active and passive residues in the partner molecule (de Vries et al., 2010). It remains, then, to devise ways in which to identify active and passive residues from labeling data and prioritizing their use in generating interaction restraints.
The current version of the Studio provides ways to generate sets of putative active residues for iterative docking, using the statistical output from the labeling experiments. The statistics package was structured to process data for any combination of protein states necessary for the modeling activities, limited only by the requirements of HADDOCK (i.e., simultaneous docking of up to six molecules from any combination of proteins, nucleic acids, peptides, or drug-like molecules). The output from the statistical analysis of the protein states is recalled by the HADDOCK package in the Studio. In this package, the user can define criteria for the generation of active residues using any parameter in the data set, such as the magnitude of induced changes in label values from a binding partner, or the confidence level in these changes. Active residues identified in other experiment types (e.g., mutational analysis) can be entered here as well. An interface to the HADDOCK server allows the user to submit the list of active residues and monitor jobs (see Supplemental Experimental Procedures and Figure S8). The HADDOCK server automatically filters the active residues based on solvent accessibility and automatically identifies passive residues (see Supplemental Experimental Procedures).
In many cases, labeling data may not fully map a binding site, yet could also be clouded by allosteric effects, which may falsely mimic binding site data. We approach this as an optimization problem. The label data are sorted and used to generate multiple HADDOCK runs, one for each sort. The HADDOCK routine generates large number samplings for each run. Each run is assessed a score and clustered on the basis of structural similarity. Each cluster is assigned a representative docked structure. An effective optimization strategy assumes that the best subset of labeling data will generate a maximal cluster size with acceptable HADDOCK scores. There is one such strategy that involves the generation of data subsets using progressively lower thresholds applied to the labeling data (see Supplemental Experimental Procedures and Figure S11).
To demonstrate an application of HX-guided docking in HADDOCK, we processed HX-MS data from a novel drug binding site on the microtubule lattice. Laulimalide is a microtubule stabilizer and a promising new scaffold for the development of next-generation antimitotic cancer therapeutics (Risinger et al., 2009). An HX-based site mapping was performed where allosteric effects were suppressed, leaving only the HX data for the binding site. Together with biased docking, molecular dynamics, and mutational analysis, we demonstrated that laulimalide interacts with β-tubulin at a location remote from the well known taxol binding site (Bennett et al., 2010). This primary site has been recently confirmed by X-ray diffraction of a specially stabilized single protofilament segment (Prota et al., 2014).
Our site mapping identified two separate “islands” of perturbed deuteration kinetics on β-tubulin, well removed from each other. Initial structure-building activities considered only the primary island, as it represented the largest surface perturbation. We ran a docking exercise through the Studio using the data from this experiment to evaluate the impact of this secondary site, which sits across from the primary site in one protofilament on an adjacent β-tubulin subunit on a second protofilament (Bennett et al., 2012). Analysis of the results generated by the HADDOCK server highlighted a docked molecule in the top-ranked cluster in the correct site (Figure 7). The docked model aligns the two β-tubulin subunits with the protofilament axes in the lattice, with laulimalide occupying the interprotofilament groove. A key feature of the docked model agrees with our original study and the recent structural analysis, where the dehydropyran side chain penetrates a cleft in the primary binding site. The HADDOCK model presents a laulimalide conformation similar to that of the crystal structure, but with the macrocycle positioned closer to the secondary site. These results suggest that any pharmacophore model designed to exploit this novel microtubule stabilizing domain should consider both components of the binding site.
Figure 7. Structural Modeling of the Laulimalide-Microtubule Binding Site Based on HX-MS and Data-Directed Flexible Docking, as Directed by the Studio.
AIRs were extracted from the reductions in HX labeling induced by laulimalide binding to microtubules, and uploaded through the Studio to HADDOCK, along with a segment of microtubule structure (parallel β-tubulin monomers from PDB 2XRP). A representative pose (bottom-left) of the top-ranked cluster (red boxed region) is shown, oriented between two protofilaments. The reductions in HX labeling are shown in blue. Clustering of the results is based on ligand interfacial rmsd (Å) referenced to the best-scored model. A zoomed view of the representative pose is shown (A) and visually compared to the computational study using only single protofilament data (B), (Bennett et al., 2010) and a recent X-ray crystallographic structure derived from a stabilized single protofilament (C) (Prota et al., 2014).
DISCUSSION
The Studio is a freely available platform geared toward integrative structural biology. It supports the mining, integration, and application of restraint data for structural modeling of protein complexes and represents a complete rebuild and extension of the Hydra software for HX-MS. It generates restraints for data-directed structural modeling using the HADDOCK flexible docking program. We highlight a series of packages in the Studio that can process and integrate data from any labeling chemistry, not only HX. The Studio provides a strong design framework, which will simplify the addition of advanced algorithms for any aspect of the workflow, from MS data processing to structure visualization and restraint definition. There are two noteworthy components built into the Studio framework that include ultrafast processing of multidimensional LC-MS/MS data, and a utility for rapid validation of the raw data using a powerful interactive graph control. A full set of tutorials and test data will facilitate efficient training and use of the packages available in the Studio.
Analysis packages in the Studio support conventional HX-MS, targeted HX-MS2, and data-independent HX-MS2. The latter mode of operation is an important new addition to the field. It provides access to another dimension of data usually outside of HX-MS applications, and we show that it returns a significant amount of useable data for structural analysis, normally lost when combining multiple proteins in the conventional bottom-up approach to HX-MS. Conventional HX-MS methods suffer attrition in peptides useful for deuteration measurements. Data-independent acquisition methods such as SWATH return a significant fraction of these peptides by using fragments as surrogates, and additionally provide an overall simplification to the HX-MS experiment. This data-independent HX-MS2 concept leverages a well defined database of candidate peptides, derived from prior knowledge of the proteins involved in the complex. It is conceivable that data-independent implementations of HX-MS will permit the structural analysis of complexes isolated directly from cellular sources (e.g., IP’s), foregoing the need for recombinant protein production and the labor involved in reconstituting protein complexes.
The number of stable covalent chemistries employed for structural analysis is increasing, particularly through the use of novel oxidative methods and photochemistries. The Studio supports the extraction of labeling data in both the MS and MS2 dimensions, requiring only the specification of label mass, and it can integrate any number of labeling processes in one directed docking experiment. It provides the opportunity for combining results from HX experiments with those from covalent labeling experiments, plus data from any external source such as mutational analysis.
We anticipate that advances in MS-based structure-building, supported by utilities like the Studio, will widen the scope of rational drug design activities. The list of targets should grow to include unconventional systems such as multiprotein machines, as we illustrate with a refined modeling of a drug targeting an allosteric site for microtubule polymerization control. The mapping of binding sites at the highest structural resolution possible should support high quality structure-building activities for any class of interaction amenable to MS analysis.
EXPERIMENTAL PROCEDURES
Software Design
The Studio is a software package that provides a framework for efficient integration of components, geared toward fast and intuitive analysis of MS data. Software extensibility allows decoupling the application and its components, so that components can be easily added, modified, or removed without affecting the usability of the main application. The decoupling is achieved by only allowing components to communicate with each other through high-level interfaces, rather than requiring source code access. The core library contains generic interfaces and tools that provide a common platform for building analysis packages. An analysis package is a collection of components that apply to a specific MS-related task. Additional architectural details and analysis package functionality can be found in the Supplemental Experimental Procedures.
The Studio has several features that enhance usability and efficiency for processing, data storage, and user interaction. It takes advantage of multicore systems for computation in a distributed fashion, with merging of results. Data storage invokes a compressed file format that allows for fast MS data processing, including extracted ion chromatogram (XIC) extraction and spectrum summing. Data file compression is performed once at the start of each project. These improvements lead to a high degree of computing efficiency, benchmarked at 26 ms/peptide (XIC extraction, XIC smoothing, spectrum summing, data reduction, peak detection, and label quantification). The Studio also contains a new MS graph visualization tool. Most user interaction is focused around the graph control, such that data manipulation or validation can be performed directly on the graph. A set of tutorials is provided in Supplemental Experimental Procedures, and the executable can be found at http://www.structurems.ucalgary.ca/software.
Hydrogen/deuterium Exchange-Mass Spectrometry Experiments: Mitotic Centromere Associated Kinesin Nucleotide Exchange
Sample preparations for MCAK-based interactions have been described (Burns et al., 2014). Briefly, His-tagged EGFP-MCAK (neck-motor, 182–583) was expressed using a baculovirus expression system previously described (Maney et al., 1998), and purified. EGFP-MCAK was diluted in microtubule assembly buffer (10 mM K-PIPES, 100 mM KCl, 1 mM EGTA, 1 mM MgCl2, pH 6.8) containing 2 mM nucleotide (ATP or ADP) and held on ice until analysis. Deuterium labeling was initiated in by addition of deuterated assembly buffer + nucleotide, to 30% D2O at 25°C (labeling for 10, 30, 100, 300, and 1,000 s). The samples were quenched and digested at 10°C for 2.5 min using nepenthesin (Rey et al., 2013, pH 2.4. The sample was injected into an automated quadrupole time-of-flight-based HX-MS system previously described, employing an AB Sciex 5600 TripleTOF in MS mode (Burns et al., 2013) for mass analysis (positive ion mode, m/z 300–1,250). Samples were run in quadruplicate for each nucleotide state and time-point.
Targeted Hydrogen/deuterium Exchange-Mass Spectrometry2 Experiments
Peptides of EGFP-MCAK (182–583) for which a higher resolution analysis were required were identified using the ETD scheduler by interacting with the statistical output of the MCAK-nucleotide analyses (300 s time-point). The scheduler was applied to a structural region in the C-terminal of the motor domain. A ranked list of peptides detected in this region was generated for inclusion in a product ion experiment, replacing the TripleTOF with an LTQ Orbitrap-Velos ETD system, and adjusting transmission parameters to promote residue-level deuteration analysis with minimal scrambling. Additional details can be found in the Supplemental Experimental Procedures.
Data Independent Hydrogen/deuterium Exchange-Mass Spectrometry2 Experiments: Mitotic Centromere Associated Kinesin-Microtubule Interaction
Recombinant truncated MCAK (neck-motor, 182–583) was produced using the same systems and methods described above. Purified bovine brain tubulin (Cytoskeleton) was reconstituted in ice-cold microtubule assembly buffer + 1 mM guanosine-5’-[(α,β)-methyleno]triphosphate (Jena Bioscience). Microtubules were recovered after polymerization cycling in assembly buffer, then stabilized with docetaxel (Sigma). Microtubules were diluted to 7 μM and kept at 37°C. Truncated MCAK was added to microtubules and incubated to allow depolymerization and protofilament capture. Complexes were pelleted and washed. The pellet was resuspended in 35% D2O in assembly buffer with 1 mM ATP for labeling. Samples were quenched and digested as above. The sample was injected into the HX-enabled TripleTOF system, operated in SWATH mode (one survey scan followed by 24 30Th bins, over an m/z range of 300–1,000). Samples were run in quadruplicate for each state: free MCAK, free microtubules, and complex. Additional details can be found in the Supplemental Experimental Procedures.
Covalent Labeling Experiments
Holo-calmodulin (calcium-bound) was labeled with 3,3-azibutanol, photolytically activated to generate the corresponding carbene. Photolysis was initiated with a single shot from an Nd:YAG laser (Quantel YG-980C, 355 nm at 50 mJ/shot). Quantitative analysis of label distribution was performed by tryptic digestion, followed by separation on an Easy-nLC-1000 for analysis on an LTQ Orbitrap Velos ETD, in both LC-MS and product ion LC-MS/MS modes. Additional details can be found in the Supplemental Experimental Procedures.
Data Dependent Acquisition for Peptide Feature List Generation
Peptide sequence identifications for each protein were generated by recursive data dependent acquisition by LC-MS/MS on the TripleTOF, involving multiple injections of each unlabeled protein digest under otherwise standard labeling conditions (LC gradient and temperature) until no new identifications were accrued. Data were searched against the known protein sequences using Mascot 2.3. and common modifications in the parameter space. Features were exported from Mascot files and assembled in a .CSV file for initial peptide feature lists as input to the Studio application packages.
HADDOCK
Models of the laulimalide-microtubule interaction were generated using a pipeline in the Studio to the HADDOCK web server (de Vries et al., 2010), using AIRs derived from previously published HX-MS data (Bennett et al., 2010). Briefly, active residues were generated by prefiltering peptide HX data for solvent accessibility, and passive residues were generated automatically in HADDOCK based on proximity to active residues (de Vries et al., 2010). The conformer of laulimalide used in docking was drawn from the X-ray crystallo-graphic structure of the molecule bound to a single protofilament (Prota et al., 2014), and its topology determined using PRODRG (Schüttelkopf and van Aalten, 2004). A total of 2,000 structures were sampled in the rigid body docking phase, and in the final iteration, the models were refined in explicit water and the 200 lowest-energy structures were clustered by ligand interface root-mean-square deviation (rmsd), using a 2.5A cutoff. See Supplemental Experimental Procedures for additional information on the use of HX-MS data in HADDOCK.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Natural Sciences and Engineering Research Council (DS) and the University of Calgary through grants to D.C.S. It was also supported by the Dutch Foundation for Scientific Research through a VICI grant (no. 700.56.442) to A.M.J.J.B. and by the WeNMR project (European FP7 e-Infrastructure grant, contract no. 261572, http://www.wenmr.eu). We present the “Woods plot” as a useful designation for a pairwise comparison of deuteration levels across linear sequence, in recognition of Virgil Woods, Jr., a pioneer in HX-MS who presented it, and who recently passed away.
Footnotes
AUTHOR CONTRIBUTIONS
All authors participated in the research. L.W. and A.M.J.J.B. assisted in research design, led by D.C.S. All authors participated in writing and editing the final manuscript.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, 13 figures, one table, software tutorials, and data and can be found with article online at http://dx.doi.org/10.1016/j.str.2014.08.013.
REFERENCES
- Ahn J, Cao MJ, Yu YQ, Engen JR. Accessing the reproducibility and specificity of pepsin and other aspartic proteases. Biochim. Biophys. Acta. 2013;1834:1222–1229. doi: 10.1016/j.bbapap.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Barakat K, Huzil JT, Tuszynski J, Schriemer DC. Discovery and characterization of the laulimalide-microtubule binding mode by mass shift perturbation mapping. Chem. Biol. 2010;17:725–734. doi: 10.1016/j.chembiol.2010.05.019. [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Chan GK, Rattner JB, Schriemer DC. Low-dose laulimalide represents a novel molecular probe for investigating microtubule organization. Cell Cycle. 2012;11:3045–3054. doi: 10.4161/cc.21411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertsch A, Jung S, Zerck A, Pfeifer N, Nahnsen S, Henneges C, Nordheim A, Kohlbacher O. Optimal de novo design of MRM experiments for rapid assay development in targeted proteomics. J. Proteome Res. 2010;9:2696–2704. doi: 10.1021/pr1001803. [DOI] [PubMed] [Google Scholar]
- Burns KM, Rey M, Baker CA, Schriemer DC. Platform dependencies in bottom-up hydrogen/deuterium exchange mass spectrometry. Mol. Cell. Proteomics. 2013;12:539–548. doi: 10.1074/mcp.M112.023770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KM, Wagenbach M, Wordeman L, Schriemer DC. Nucleotide exchange in dimeric MCAK induces longitudinal and lateral stress at microtubule ends to support polymerization. Structure. 2014;22:1–11. doi: 10.1016/j.str.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JD, Goodlett DR, Masselon CD. Multiplexed and data-independent tandem mass spectrometry for global proteome profiling. Mass Spectrom. Rev. 2013 doi: 10.1002/mas.21400. Published online November 26, 2013. http://dx.doi.org/10.1002/mas.21400. [DOI] [PubMed]
- Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, De Luca JG, De Wulf P, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R, Cortens JP, Beavis RC. Open source system for analyzing, validating, and storing protein identification data. J. Proteome Res. 2004;3:1234–1242. doi: 10.1021/pr049882h. [DOI] [PubMed] [Google Scholar]
- de Vries SJ, van Dijk M, Bonvin AM. The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 2010;5:883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- Devarakonda S, Gupta K, Chalmers MJ, Hunt JF, Griffin PR, Van Duyne GD, Spiegelman BM. Disorder-to-order transition underlies the structural basis for the assembly of a transcriptionally active PGC-1α/ERRγ complex. Proc. Natl. Acad. Sci. USA. 2011;108:18678–18683. doi: 10.1073/pnas.1113813108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- Ems-McClung SC, Hainline SG, Devare J, Zong H, Cai S, Carnes SK, Shaw SL, Walczak CE. Aurora B inhibits MCAK activity through a phosphoconformational switch that reduces microtubule association. Curr. Biol. 2013;23:2491–2499. doi: 10.1016/j.cub.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber BJ, Boehringer D, Leitner A, Bieri P, Voigts-Hoffmann F, Erzberger JP, Leibundgut M, Aebersold R, Ban N. Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature. 2014;505:515–519. doi: 10.1038/nature12890. [DOI] [PubMed] [Google Scholar]
- Jørgensen TJ, Gårdsvoll H, Ploug M, Roepstorff P. Intramolecular migration of amide hydrogens in protonated peptides upon collisional activation. J. Am. Chem. Soc. 2005;127:2785–2793. doi: 10.1021/ja043789c. [DOI] [PubMed] [Google Scholar]
- Kahraman A, Herzog F, Leitner A, Rosenberger G, Aebersold R, Malmström L. Cross-link guided molecular modeling with ROSETTA. PLoS ONE. 2013;8:e73411. doi: 10.1371/journal.pone.0073411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaca E, Bonvin AM. Advances in integrative modeling of biomolecular complexes. Methods. 2013;59:372–381. doi: 10.1016/j.ymeth.2012.12.004. [DOI] [PubMed] [Google Scholar]
- Konermann L, Pan J, Liu YH. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem. Soc. Rev. 2011;40:1224–1234. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- Landgraf RR, Chalmers MJ, Griffin PR. Automated hydrogen/deuterium exchange electron transfer dissociation high resolution mass spectrometry measured at single-amide resolution. J. Am. Soc. Mass Spectrom. 2012;23:301–309. doi: 10.1007/s13361-011-0298-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasker K, Förster F, Bohn S, Walzthoeni T, Villa E, Unverdorben P, Beck F, Aebersold R, Sali A, Baumeister W. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc. Natl. Acad. Sci. USA. 2012;109:1380–1387. doi: 10.1073/pnas.1120559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau WC, Rubinstein JL. Subnanometre-resolution structure of the intact Thermus thermophilus H+-driven ATP synthase. Nature. 2012;481:214–218. doi: 10.1038/nature10699. [DOI] [PubMed] [Google Scholar]
- Lindner R, Lou X, Reinstein J, Shoeman RL, Hamprecht FA, Winkler A. Hexicon 2: automated processing of hydrogen-deuterium exchange mass spectrometry data with improved deuteration distribution estimation. J. Am. Soc. Mass Spectrom. 2014;25:1018–1028. doi: 10.1007/s13361-014-0850-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Easterling ML, Agar JN. Resolving isotopic fine structure to detect and quantify natural abundance- and hydrogen/deuterium exchange-derived isotopomers. Anal. Chem. 2014;86:820–825. doi: 10.1021/ac403365g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney T, Hunter AW, Wagenbach M, Wordeman L. Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J. Cell Biol. 1998;142:787–801. doi: 10.1083/jcb.142.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcsisin SR, Engen JR. Hydrogen exchange mass spectrometry: what is it and what can it tell us? Anal. Bioanal. Chem. 2010;397:967–972. doi: 10.1007/s00216-010-3556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melero R, Buchwald G, Castano R, Raabe M, Gil D, Lazaro M, Urlaub H, Conti E, Llorca O. The cryo-EM structure of the UPF-EJC complex shows UPF1 poised toward the RNA 3′ end. Nat Struct Mol Biol. 2012;19:498–505. S491–492. doi: 10.1038/nsmb.2287. [DOI] [PubMed] [Google Scholar]
- Mendoza VL, Vachet RW. Probing protein structure by amino acid-specific covalent labeling and mass spectrometry. Mass Spectrom. Rev. 2009;28:785–815. doi: 10.1002/mas.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkley ED, Rysavy S, Kahraman A, Hafen RP, Daggett V, Adkins JN. Distance restraints from crosslinking mass spectrometry: mining a molecular dynamics simulation database to evaluate lysine-lysine distances. Protein Sci. 2014;23:747–759. doi: 10.1002/pro.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Ruan X, Valvano MA, Konermann L. Validation of membrane protein topology models by oxidative labeling and mass spectrometry. J. Am. Soc. Mass Spectrom. 2012;23:889–898. doi: 10.1007/s13361-012-0342-x. [DOI] [PubMed] [Google Scholar]
- Pascal BD, Willis S, Lauer JL, Landgraf RR, West GM, Marciano D, Novick S, Goswami D, Chalmers MJ, Griffin PR. HDX workbench: software for the analysis of H/D exchange MS data. J. Am. Soc. Mass Spectrom. 2012;23:1512–1521. doi: 10.1007/s13361-012-0419-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percy AJ, Schriemer DC. MRM methods for high precision shift measurements in H/DX-MS. Int. J. Mass Spectrom. 2011;302:26–35. [Google Scholar]
- Politis A, Stengel F, Hall Z, Hernández H, Leitner A, Walzthoeni T, Robinson CV, Aebersold R. A mass spectrometry-based hybrid method for structural modeling of protein complexes. Nat. Methods. 2014;11:403–406. doi: 10.1038/nmeth.2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prota AE, Bargsten K, Northcote PT, Marsh M, Altmann KH, Miller JH, Díaz JF, Steinmetz MO. Structural basis of microtubule stabilization by laulimalide and peloruside A. Angew. Chem. Int. Ed. Engl. 2014;53:1621–1625. doi: 10.1002/anie.201307749. [DOI] [PubMed] [Google Scholar]
- Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- Rey M, Yang M, Burns KM, Yu Y, Lees-Miller SP, Schriemer DC. Nepenthesin from monkey cups for hydrogen/deuterium exchange mass spectrometry. Mol. Cell. Proteomics. 2013;12:464–472. doi: 10.1074/mcp.M112.025221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risinger AL, Giles FJ, Mooberry SL. Microtubule dynamics as a target in oncology. Cancer Treat. Rev. 2009;35:255–261. doi: 10.1016/j.ctrv.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts VA, Pique ME, Hsu S, Li S, Slupphaug G, Rambo RP, Jamison JW, Liu T, Lee JH, Tainer JA, et al. Combining H/D exchange mass spectroscopy and computational docking reveals extended DNA-binding surface on uracil-DNA glycosylase. Nucleic Acids Res. 2012;40:6070–6081. doi: 10.1093/nar/gks291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues JP, Melquiond AS, Karaca E, Trellet M, van Dijk M, van Zundert GC, Schmitz C, de Vries SJ, Bordogna A, Bonati L, et al. Defining the limits of homology modeling in information-driven protein docking. Proteins. 2013;81:2119–2128. doi: 10.1002/prot.24382. [DOI] [PubMed] [Google Scholar]
- Schraidt O, Marlovits TC. Three-dimensional model of Salmonella's needle complex at subnanometer resolution. Science. 2011;331:1192–1195. doi: 10.1126/science.1199358. [DOI] [PubMed] [Google Scholar]
- Schüttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- Slysz GW, Baker CA, Bozsa BM, Dang A, Percy AJ, Bennett M, Schriemer DC. Hydra: software for tailored processing of H/D exchange data from MS or tandem MS analyses. BMC Bioinformatics. 2009;10:162. doi: 10.1186/1471-2105-10-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D, Asenjo AB, Mennella V, Sharp DJ, Sosa H. Kinesin-13s form rings around microtubules. J. Cell Biol. 2006;175:25–31. doi: 10.1083/jcb.200605194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalassinos K, Pandurangan AP, Xu M, Alber F, Topf M. Conformational States of macromolecular assemblies explored by integrative structure calculation. Structure. 2013;21:1500–1508. doi: 10.1016/j.str.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topf M, Lasker K, Webb B, Wolfson H, Chiu W, Sali A. Protein structure fitting and refinement guided by cryo-EM density. Structure. 2008;16:295–307. doi: 10.1016/j.str.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wales TE, Eggertson MJ, Engen JR. Considerations in the analysis of hydrogen exchange mass spectrometry data. Methods Mol. Biol. 2013;1007:263–288. doi: 10.1007/978-1-62703-392-3_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walzthoeni T, Leitner A, Stengel F, Aebersold R. Mass spectrometry supported determination of protein complex structure. Curr. Opin. Struct. Biol. 2013;23:252–260. doi: 10.1016/j.sbi.2013.02.008. [DOI] [PubMed] [Google Scholar]
- Ward AB, Sali A, Wilson IA. Biochemistry. Integrative structural biology. Science. 2013;339:913–915. doi: 10.1126/science.1228565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L, Mitchison TJ. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J. Cell Biol. 1995;128:95–104. doi: 10.1083/jcb.128.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L, Wagenbach M, von Dassow G. MCAK facilitates chromosome movement by promoting kinetochore microtubule turnover. J. Cell Biol. 2007;179:869–879. doi: 10.1083/jcb.200707120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Willison LN, Tripathi P, Sathe SK, Roux KH, Emmett MR, Blakney GT, Zhang HM, Marshall AG. Epitope mapping of a 95 kDa antigen in complex with antibody by solution-phase amide backbone hydrogen/deuterium exchange monitored by Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 2011;83:7129–7136. doi: 10.1021/ac201501z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Asenjo AB, Greenbaum M, Xie L, Sharp DJ, Sosa H. A second tubulin binding site on the kinesin-13 motor head domain is important during mitosis. PLoS ONE. 2013;8:e73075. doi: 10.1371/journal.pone.0073075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.