Abstract
The timing of flowering, and in particular the degree to which it is responsive to the environment, is a key factor in the adaptation of a given species to various eco-geographic locations and agricultural practices. Flowering time variation has been documented in many crop legumes, and selection for specific variants has permitted significant expansion and improvement in cultivation, from prehistoric times to the present day. Recent advances in legume genomics have accelerated the process of gene identification and functional analysis, and opened up new prospects for a molecular understanding of flowering time adaptation in this important crop group. Within the legumes, two species have been prominent in flowering time studies; the vernalization-responsive long-day species pea (Pisum sativum) and the warm-season short-day plant soybean (Glycine max). Analysis of flowering in these species is now being complemented by reverse genetics capabilities in the model legumes Medicago truncatula and Lotus japonicus, and the emergence of genome-scale resources in a range of other legumes. This review will outline the insights gained from detailed forward genetic analysis of flowering time in pea and soybean, highlighting the importance of light perception, the circadian clock and the FT family of flowering integrators. It discusses the current state of knowledge on genetic mechanisms for photoperiod and vernalization response, and concludes with a broader discussion of flowering time adaptation across legumes generally.
Keywords: legume, flowering, photoperiod, pea, soybean
Introduction
Many aspects of plant growth and development are highly attuned to the environment, and this is particularly true of the transition from vegetative growth to the flowering state. Genetic variation that affects the timing of flowering and its regulation by environmental factors has clear significance for the performance of crop species. Many species have a requirement for exposure to specific photoperiods and/or temperatures in order to flower, and flowering may be significantly delayed or prevented if these requirements are not met. Genetic changes that relax or eliminate these constraints have enabled expansion to a wider latitudinal and climatic range and allowed greater flexibility in seasonal cropping practices. Conversely, delayed flowering may be an advantage in other situations, as it may reduce damage from certain abiotic stresses and lead to greater yield through increased biomass accumulation prior to flowering. Knowledge of how individual genetic variants combine to provide adaptation in specific situations can be valuable for breeding purposes, as it can accelerate introgression of new traits into adapted backgrounds and may allow the directed modification of phenology for specific target environments.
The major crop legumes fall within two sister clades that are included in the larger group of papilionoid legumes, often referred to as the galegoid and phaseoloid clades (Cronk et al., 2006). In general, species within the galegoid clade (e.g., pea, lentil, chickpea, faba bean) are from temperate regions and with respect to flowering time control are vernalization-responsive long-day plants (LDPs), whereas those in the phaseoloid clade (e.g., soybean, cowpea, pigeonpea, common bean) generally originate from lower latitudes and are short-day plants (SDPs; Summerfield and Roberts, 1985b). Two other important crop legumes, peanut (Arachis hypogaea) and lupin (Lupinus spp.) are basal to both of these clades. Not surprisingly, in view of their respective origins at low and high latitudes, peanut has characteristics of a SDP (Bagnall and King, 1991) and the commonly grown lupin species are vernalization-reponsive LDP (Summerfield and Roberts, 1985c).
While the defining feature of the vegetative-to-reproductive transition is the conversion of meristems to produce flowers rather than vegetative shoots, in a natural environment this is also accompanied by significant changes to a wide range of other developmental traits, including stem elongation, apical dominance, lateral branching, resource allocation, maturity and yield. In some species, genes and environmental factors that affect the initial flowering transition can also continue to have a significant influence on post-flowering processes affecting fertility and pod development. Thus, although flowering time can be seen from one perspective as a relatively simple trait, in reality it is linked to fundamental decisions made by the plant about when and how to allocate resources, and thus participates in a complex network of two-way interactions with other developmental processes.
Over recent years there has been a dramatic increase in the number of legume species with genome sequences and/or significant genomic resources (Young and Bharti, 2012). However, in the study of flowering time, as for other developmental processes, no one legume species has emerged as the predominant model. Most work on flowering time has focused on the two species pea (Pisum sativum) and soybean (Glycine max), which respectively represent the temperate LDP and warm-season SDP. This is partly due to the long history in the use of both of these species for flowering research and the accumulation of knowledge on its genetic control (Murfet, 1985; Summerfield and Roberts, 1985a; Weller et al., 1997b, 2009; Watanabe et al., 2012). Interestingly, the two legume model species Medicago truncatula (barrel medic) and Lotus japonicus, which have been prominent in studies of nitrogen fixation and other fundamental processes, and have genome sequences available, have so far not been deeply exploited for investigation of flowering time control.
Flowering Genes
The availability of extensive genomic resources for several legume species and well-documented synteny has enabled a comprehensive inventory of genes potentially relevant for flowering time control. This has been useful both in identification of candidate genes for flowering loci, and in exploring the molecular physiology of flowering through gene expression studies and reverse genetics. It has also dramatically improved the prospects for discovering legume-specific genes through purely positional approaches. Well over 100 genes that contribute to control of flowering time have been identified in Arabidopsis and rice, and the functions and interactions of these genes are regularly reviewed (e.g., Kim et al., 2009; Amasino and Michaels, 2010; Andres and Coupland, 2012; Pin and Nilsson, 2012; Brambilla and Fornara, 2013; Song et al., 2013). In legumes, several compendia of flowering gene homologs are already available (Hecht et al., 2005; Kim et al., 2012; Watanabe et al., 2012). While the basic genes and gene families central to pathways controlling flowering time in Arabidopsis appear to be largely conserved in legumes, there are numerous examples of gene duplication and loss. This is likely to reflect the history and consequences of genome duplications after the divergence of the Arabidopsis and legume lineages (Young and Bharti, 2012).
Photoreceptors provide primary information about the light environment that enables detection of daylength. Legumes have a standard complement of only three phytochromes (phyA, phyB, and phyE; Hecht et al., 2005), and lack a representative of the ancient phyC clade, although the phyA lineage has undergone a more recent duplication in the phasioloid legumes (Liu et al., 2008). The cryptochrome gene CRY1 has also been duplicated in the phasioloid legumes, while an older duplication of the CRY2 gene is common to all legumes (Platten et al., 2005b). In contrast, only two LOV-domain flavoprotein photoreceptors in the FKF1/ZTL family are present in legumes. The circadian clock is also important for photoperiod measurement, and while all major Arabidopsis clock genes are represented, the galegoid legumes appear to have only a single gene orthologous to the circadian-clock related MYB transcription factor genes CCA1 and LHY (Hecht et al., 2007), whereas the TOC1, GI, and ELF3 genes have variously undergone duplication in the two legume groups. The CONSTANS protein, which integrates light and circadian signaling for photoperiod-specific induction in Arabidopsis, is represented by two co-orthologs in the SD legumes, but only one in the LD legumes (Wong et al., 2014).
Another feature of legumes is the expansion of the FT/TFL1 gene family. In Arabidopsis, these genes integrate environmental signaling for induction of flowering, and guide the fate of meristems during inflorescence development. Both major crop legume groups have multiple TFL1 genes and in the galegoid legumes two of these sequences are distinctively divergent from Arabidopsis TFL1 (Foucher et al., 2003; Cronk et al., 2006; Hecht et al., 2011). Legumes also have three distinct subclades of FT genes; FTa, FTb, and FTc (Hecht et al., 2011). Within the large family of MADS domain genes, legumes have additional SVP and SOC1 genes (Hecht et al., 2005; Jaudal et al., 2014), while the FLC clade, which has an important role in vernalization response in Arabidopsis, appears to be absent in the galegoid legumes and at most vestigial in soybean (Hecht et al., 2005; Ruelens et al., 2013).
Genetic Analysis of Flowering in Legumes
Pea (Pisum sativum L.)
Over 20 loci related to flowering time and inflorescence development have been identified in pea (Table 1). Initial work on genetic control of flowering resolved several loci from existing variation among various cultivars of garden and field pea, while other loci were subsequently identified through characterization of induced mutants and specific mutant screens (Murfet, 1985; Weller et al., 1997a, 2009).
Table 1.
Comparison of flowering loci in pea and soybean.
| Developmental role | Pea locus | Reference for molecular identity | Soybean locus | Reference for molecular identity | Molecular identity | ||
|---|---|---|---|---|---|---|---|
| Light perception/signaling | FUN1 | + | Weller et al. (2004) | E3 | - | Watanabe et al. (2009) | PHYA |
| E4 | - | Liu et al. (2008) | PHYA | ||||
| LV | - | Weller et al. (2001) | PHYB | ||||
| LIP1 | - | Sullivan and Gray (2000) | COP1 | ||||
| Circadian clock | SN | - | Liew et al. (2014) | LUX | |||
| DNE | - | Liew et al. (2009) | ELF4 | ||||
| PPD | - | ? | |||||
| HR | - | Weller et al. (2012) | ELF3 | ||||
| Photoperiod response | E | + | ? | ||||
| LATE1 | + | Hecht et al. (2007) | E2 | - | Watanabe et al. (2011) | GIGANTEA | |
| LATE2 | - | ||||||
| E1 | - | Xia et al. (2012) | B3-domain TF | ||||
| E7 | - | ? | |||||
| E8 | - | ? | |||||
| Signal integration and inflorescence development | GIGAS | + | Hecht et al. (2007) | FT (FTa1) | |||
| LF | - | Foucher et al. (2003) | TFL1 (TFL1c) | ||||
| VEG1 | + | Berbel et al. (2012) | Dt2 | Ping et al. (2014) | MADS box TF | ||
| VEG2 | + | Sussmilch et al. (2015) | FD | ||||
| LATE5 | + | ||||||
| DET | Foucher et al. (2003) | Dt1 | Liu et al. (2010) | TFL1 (TFL1a) | |||
| UNI | Hofer et al. (1997) | LFY | |||||
| Other/unknown | E5 | - | ? | ||||
| E6 | + | ? | |||||
| E9 | + | ? | |||||
| J | + | ? | |||||
| LATE3 | + | ? | |||||
| LATE4 | + | ? | |||||
| LW | + | ? | |||||
| AERO1 | - | ? |
References establishing the molecular identity of each locus are cited. The effect (or inferred effect) on flowering of the functional (wild-type) allele is indicated as either “+” (promoting flowering) or “-” (inhibiting flowering). The molecular identity refers to the Arabidopsis ortholog or the type of protein encoded.
Naturally Occurring Variation
Two major loci are known that delay flowering under non-inductive SD. Recessive alleles at the HIGH RESPONSE (HR) locus cause early flowering in SD and reduce, but do not eliminate, the photoperiod response, whereas recessive alleles at the STERILE NODES (SN) locus confer complete daylength insensitivity (Murfet, 1985). Only a single, naturally occurring mutant hr allele has been identified, but for SN both naturally occurring and induced mutant alleles have been described (Liew et al., 2014). Recent work has established HR and SN as pea orthologs of Arabidopsis circadian clock genes ELF3 and LUX, respectively (Weller et al., 2012; Liew et al., 2014), and analysis of sequence diversity suggests a wide distribution of the hr allele across domesticated pea germplasm and an important and ancient role for this mutation in the spring-flowering habit (Weller et al., 2012). In contrast, the main naturally occurring sn allele has a more restricted distribution and occurs only within the subset of lines carrying hr, implying a more recent origin (Liew et al., 2014).
The third locus, LATE FLOWERING (LF), inhibits flowering in both long and short days, and was identified over 10 years ago as a divergent homolog of TFL1 (Foucher et al., 2003). Numerous allelic variants of LF are known, including both naturally occurring and induced mutant alleles. Accessions in which the LF gene is deleted or inactivated by nonsense mutation show extremely early, photoperiod-insensitive initiation of flowering (Murfet, 1985; Foucher et al., 2003), but remain responsive to photoperiod in several other respects, suggesting that LF is not involved directly in the photoperiod response mechanism.
The fourth locus, EARLY (E), is the least well-understood of the naturally variant loci. Dominant alleles of E confer early initiation of flowering in some genetic backgrounds, but this effect shows complex interactions with other loci and incomplete penetrance. Recent identification of a major-effect quantitative trait loci (QTL) for flowering time in a chromosomal location similar to E (Lejeune-Hénaut et al., 2008; Weller et al., 2012) may help in its future molecular characterization.
Allelic differences at the HR, SN, LF, and E loci interact to specify an extremely wide range of flowering times in plants in non-inductive conditions. This range extends from the genotype lf sn which may flower as early as node 7 and is completely insensitive to photoperiod, to genotype LF SN HR e which flowers relatively late under LD and may not flower at all under SD (Murfet, 1985; Weller et al., 2012). Interestingly, most mutagenesis programs in pea have been conducted in spring-flowering (hr) cultivars, and in some cases in lines that also carry sn or lf alleles, and many are also likely to carry derived alleles at the E locus. Mutants isolated from these programs therefore carry at least one additional mutation affecting flowering time, and potentially as many as four.
Induced Mutants – Photoperiod Response
In addition to the SN and HR loci, several other photoperiod response loci have been identified through analysis of induced mutants. Two of these, DIE NEUTRALIS (DNE) and PHOTOPERIOD (PPD) have a role similar to SN, as recessive dne and ppd mutants show early flowering in SD. In the presence of hr, these mutations confer complete photoperiod insensitivity for flowering and other traits (King and Murfet, 1985; Arumingtyas and Murfet, 1994). DNE is the pea ortholog of another circadian clock gene, ELF4 (Liew et al., 2009), but PPD has not yet been identified. In Arabidopsis, the proteins encoded by ELF3, ELF4, and LUX genes participate in the so-called evening loop of the circadian clock and work together in a complex termed the evening complex (EC; Nagel and Kay, 2012), which may provide a mechanistic explanation for the fact that mutants for HR/PsELF3, DNE/PsELF4, and SN/PsLUX have similar phenotypes.
Loci involved in promoting flowering in pea under inductive (LD) conditions have also been identified. Mutants for the phyA photoreceptor were first identified in screens for seedling photomorphogenesis and subsequently shown to have a LD-specific late-flowering phenotype (Weller et al., 1997a). The phyA mutants are largely insensitive to LD, although day extensions with artificial light rich in blue or far-red wavelengths can result in earlier flowering, implying a contribution from other photoreceptors (Weller et al., 1997a; Platten et al., 2005a). While phyB and cry1 might seem plausible candidates for this activity, evidence from phyB and cry1 mutants suggests that neither is fundamentally involved in promotion of flowering (Weller et al., 2001; Platten et al., 2005a). The importance of PHYA is underlined by the dominant early flowering photoperiod-insensitive phyA-3D mutant, which has a higher level of phyA protein due to increased protein stability (Weller et al., 2004). Other loci LATE1 and LATE2 have been identified in specific mutant screens (Hecht et al., 2007). LATE2 has not yet been identified, but LATE1 is an ortholog of Arabidopsis circadian-clock-related gene GIGANTEA (GI; Hecht et al., 2007). The late1 mutants are similar to phyA mutants with respect to their effect on flowering and photoperiod responsiveness, but have only mild photomorphogenic defects. Similar to Arabidopsis GI, LATE1 shows strongly rhythmic expression, and late1 mutants affect the expression rhythms of key circadian clock genes, confirming that LATE1 has a role in clock function (Hecht et al., 2007; Kim et al., 2012; Weller et al., 2012; Liew et al., 2014).
Mutations at other pea loci affect the flowering transition without significantly interfering with the overall ability of the plant to respond to daylength. Under LD, gigas and vegetative1 (veg1) and veg2 mutants do not produce flowers, but instead show a profuse outgrowth of aerial vegetative branches (Murfet, 1985; Hecht et al., 2011; Berbel et al., 2012; Sussmilch et al., 2015). This phenotype appears to represent a failure to specify the identity of secondary inflorescences and to induce the expression of the floral meristem identity gene PROLIFERATING INFLORESCENCE MERISTEM (PIM), a co-ortholog of AP1 (Taylor et al., 2002; Berbel et al., 2012). GIGAS and VEG1 have been respectively identified as an FT homolog and as a divergent member of the AP1 clade of MADS-domain genes (Hecht et al., 2011; Berbel et al., 2012), and VEG2 was recently shown to be the pea ortholog of FD, an BZIP transcription factor that is an important signaling partner of FT proteins (Sussmilch et al., 2015).
Soybean [Glycine max (L.) Merr.]
At least 10 loci that affect flowering-related characteristics in the SDP soybean have now been described (Table 1). Variation at these loci is responsible for a major proportion of the latitudinal adaptation in soybean, which is grown from tropical regions to 50°N. Cultivars grown at lower latitudes experience a longer growing season and are relatively late to mature, whereas expansion to higher latitudes and completion of the growth cycle within the short summer growing season have required a reduction in sensitivity to the inhibitory effects of LD.
The well-known E series of maturity loci (E1 to E9) confer early flowering and maturity, particularly under non-inductive (LD) conditions (Cober and Morrison, 2010; Watanabe et al., 2012; Kong et al., 2014). With the exception of E6 and the recently described E9 locus, the early flowering alleles at the E loci are recessive (Watanabe et al., 2012; Kong et al., 2014). Addition of early flowering alleles at these loci results in incrementally earlier flowering under LD and improved adaptation to short summers at high latitudes. In contrast, “delayed juvenile” soybean lines are conspicuously later to flower under inductive SD conditions, a trait useful for adaptation to low latitudes generally and spring sowings at middle latitudes (Tomkins and Shipe, 1997). Genetic control of the delayed juvenile trait is not currently clear. One study proposed a single locus (J), with recessive alleles conferring late flowering in SD (Ray et al., 1995), whereas other analyses point to a more complex genetic control involving up to two other genes, one of which may be E6 (Cober, 2011). Flowering time in soybean has also been analyzed as a quantitative trait, and several QTL have been identified, many of which are likely to correspond to known maturity loci (Watanabe et al., 2012). The E1, E2, and E3 loci in particular appear to have been detected in a number of QTL studies and have also been referred to as Flowering Time 1 (FT1), FT2, and FT3, respectively (Yamanaka et al., 2000).
Several of the E loci have been characterized physiologically. As in pea, the effects of individual soybean loci have mostly been examined in genetic backgrounds already carrying hypomorphic alleles at other loci, making it difficult to gain a full picture of the action and interactions of any given locus. Nevertheless, five of the eight loci (E1, E3, E4, E7, E8) appear to specifically affect photoperiod responsiveness (Cober et al., 1996a, 2001, 2010; Cober and Voldeng, 2001). In particular, differential sensitivity of E3 and E4 loci to light quality of an artificial LD implicated them in the phytochrome system (Cober et al., 1996b), and both genes have subsequently been shown to encode phyA-type photoreceptors (Liu et al., 2008; Watanabe et al., 2009). Soybean contains four PHYA genes that consist of two pairs of homeologs, with E3 and E4 representing different homeolog pairs. The homeolog of E4, PHYA1, is apparently functional, whereas the homeolog of E3 carries a deletion and is probably a pseudogene (Watanabe et al., 2009). In most plant systems, phyA is important for de-etiolation under continuous far-red (FR) light. Interestingly, loss of E4 function reduced but did not abolish de-etiolation under FR, whereas loss of E3 function had no effect on this response, even in the absence of E4 (Liu et al., 2008). This suggests that the FR-sensing role during de-etiolation has become subfunctionalized to the E4/PHYA1 pair, and implies that phyA1 may also contribute to photomorphogenic responses. The presence of phyA1 may also explain why the e3 e4 mutant still shows delayed flowering in response to photoperiod extensions rich in far-red light. However, it is clear that three other loci, E1, E7, and E8 also contribute to this response (Cober and Voldeng, 2001; Cober et al., 2010).
Like the pea LATE1 locus, soybean E2/FT2 was also identified as an ortholog of GIGANTEA, through a strategy involving fine mapping, candidate gene analysis and reverse genetics (Watanabe et al., 2011). Although recessive e2 alleles can promote flowering under both LD and SD, E2 is also reported to enhance the photoperiod response and clearly contributes to early flowering and latitudinal adaptation (Jiang et al., 2014). However, one of the most significant developments to emerge from analysis of soybean flowering loci has been the recent molecular characterization of the E1 gene. E1 has a major role in natural variation for flowering in soybean and has the largest effect among the E loci (Yamanaka et al., 2000). Positional cloning of E1 revealed that it possesses a region of weak similarity to the plant-specific B3 domain, in addition to a helix-turn-helix domain and a nuclear localization signal, all suggesting a probable role as a transcription factor (Xia et al., 2012). E1 essentially appears to define a legume-specific gene family, but it is distantly related to the RAV subfamily of B3 domain proteins, which includes the Arabidopsis TEMPRANILLO genes that are also transcriptional repressors of FT (Castillejo and Pelaz, 2008).
Flowering Pathways in Legumes
The FT Gene Family
FT genes are of particular interest for understanding flowering time control, in view of their well-documented roles in integration of environmental signals for flowering and in signaling from the site of photoperiod detection in the leaf to the site of flower formation at the shoot apex (Pin and Nilsson, 2012). The FT gene family has been studied in detail in pea, Medicago, Lotus, and soybean (Kong et al., 2010; Laurie et al., 2011; Yamashino et al., 2013; Zhai et al., 2014). Most species have at least five FT-like genes that comprise three distinct clades unique to legumes; FTa, FTb, and FTc (Hecht et al., 2011). Genes in the FTc group are the most divergent, and are distinguished from Arabidopsis FT and most other FT genes by substitution of several conserved residues.
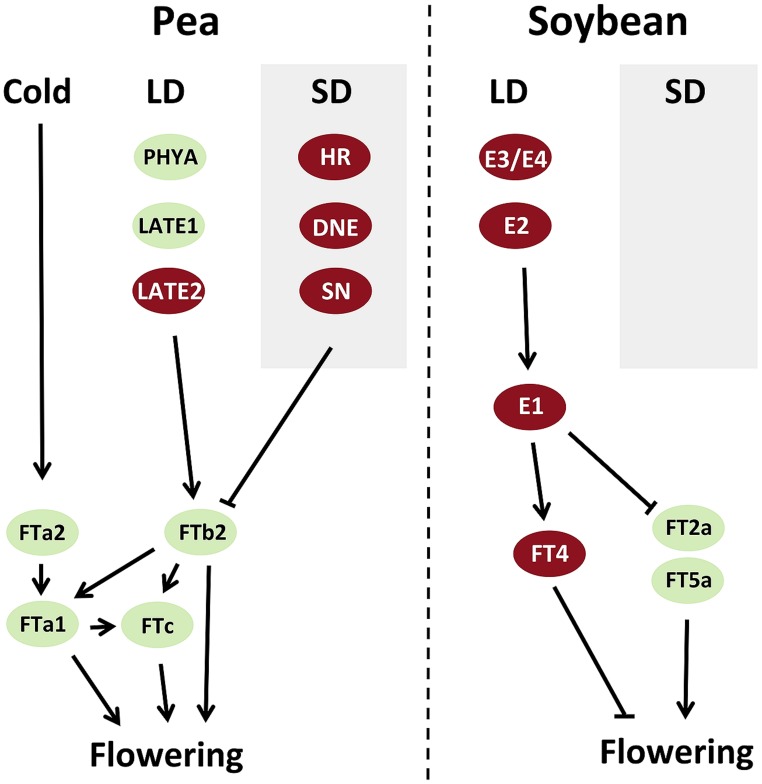
In pea and Medicago, study of expression patterns and mutant phenotypes suggest that FTa and FTb genes are expressed in leaves and may be important for targets of vernalization and photoperiod responses respectively, whereas FTc genes are expressed only in the shoot apex and may contribute to the integration signals from leaf-expressed FT genes (Hecht et al., 2011; Laurie et al., 2011). All pea FT genes can promote flowering to some extent when expressed in transgenic Arabidopsis (Hecht et al., 2011) but one in particular, FTb2, meets the characteristics of the classical “florigen.” It is expressed specifically in leaves under LD, and its upregulation correlates closely with the physiological induction of flowering and precedes the induction of all other FT genes. In addition, its expression correlates strongly with production of a graft-transmissible flowering stimulus (Hecht et al., 2011). Another pea FT gene, FTa1/GIGAS, is also expressed in leaves, and grafting experiments suggest that it may also generate a mobile flowering signal (Beveridge and Murfet, 1996; Hecht et al., 2011). However, the timing of FTa1 induction is delayed relative to FTb2, and gigas mutants have inflorescence identity defects but respond strongly to daylength. This suggests that the two main FT genes expressed in leaves in pea may both signal to the shoot apex but have different developmental roles. In addition, it appears that pea FT genes may regulate each other (Hecht et al., 2011; Figure 1).
FIGURE 1.
Models summarizing interactions between flowering genes in control of flowering time in pea and soybean. Genes that promote flowering are shown in green, and those that inhibit flowering are shown in red. LDs, long days; SDs, short days.
In soybean, two FT genes have been singled out as important promoters of flowering; FT2a (an FTa gene) and FT5a (an FTc gene). Expression of both genes is induced in leaves under inductive (SD) photoperiods, and both promote flowering when overexpressed in either Arabidopsis or soybean itself (Kong et al., 2010; Nan et al., 2014). Whereas FT2a appears to be qualitatively regulated by photoperiod, FT5a is expressed to some extent even in LD suggesting its role may not be restricted to photoperiod response (Kong et al., 2010). Two other FTa genes (FT3a and FT3b) also show significant expression in leaves in SD suggesting they may also participate in promotion of flowering (Kong et al., 2010). Surprisingly, one of the four soybean FTb genes, FT4, shows an opposite pattern of regulation and appears to act as a repressor of flowering. FT4 is induced in leaves in LD, and is able to delay flowering when overexpressed in Arabidopsis (Zhai et al., 2014). Interestingly, FT4 carries a substitution of a highly conserved glycine (G133R) in an important functional region, and the significance of this is supported by the fact that the same residue is also substituted in another FT known to repress flowering; FT1 in sugar beet (Pin et al., 2010). However, within the legumes the existence of a repressive FT may be unique to soybean, as all other legume FTb-type sequences known to date carry the canonical glycine in this position.
Response to Photoperiod
In species as diverse as Arabidopsis and rice, the photoperiod-dependent induction of FT genes relies on interactions between light perception and the circadian clock (Andres and Coupland, 2012; Brambilla and Fornara, 2013; Song et al., 2013). It is likely that this is also the case in legumes, in view of the fact that orthologs of PHYA (a photoreceptor) and GI (a gene affecting clock function) are important regulators of photoperiodic flowering in both pea and soybean (Weller et al., 1997a; Hecht et al., 2007; Liu et al., 2008; Watanabe et al., 2009, 2011). The importance of the clock for legume photoperiod responsiveness is further reinforced by the fact that the pea HR, DNE, and SN genes are all orthologs of clock genes and influence clock function (Liew et al., 2009, 2014; Weller et al., 2012). In both soybean and pea, functional variation in PHYA and GI orthologs is clearly associated with differences in the expression of FT genes. Expression of soybean FT2a is elevated by e2, e3, and e4 alleles under long days (Kong et al., 2010; Watanabe et al., 2011) indicating that all three of these E loci participate in a pathway for photoperiod response that converges on FT2a. In the pea late1 and phyA mutants, expression of FTb2 is not detected and other FT genes are more weakly expressed (Hecht et al., 2011), whereas expression of multiple FT genes is elevated in dne and sn mutants (Liew et al., 2009, 2014). These interactions are summarized in Figure 1.
Unfortunately, we do not yet have a clear picture of how these clock and photoreceptor inputs are integrated to provide photoperiod-specific regulation of FT genes. The paradigm for integration of light and clock signals in photoperiod measurement has been established in Arabidopsis and centers on the B-box transcription factor CONSTANS (CO), which is a direct transcriptional activator of FT (Tiwari et al., 2010). LD-specific induction of FT is achieved through transcriptional and post-translational regulation of CO. GI associates with the blue-light photoreceptor FKF1 to provide light-dependent promotion of CO transcription via degradation of the CDF family of CO transcriptional repressors (Andres and Coupland, 2012; Song et al., 2013). In contrast, phyA acts at the post-translational level by stabilizing CO protein under FR light, possibly through antagonism of CO degradation by the COP1 ubiquitin ligase complex (Andres and Coupland, 2012; Song et al., 2013) Although the GI-CO-FT regulatory relationship is also functionally significant in the SDP rice (Brambilla and Fornara, 2013), evidence for the more general conservation of this mechanism across flowering plants is limited (Ballerini and Kramer, 2011), which raises the question of whether it may operate in legumes.
Soybean contains four genes orthologous to Arabidopsis CO (Wong et al., 2014) and all four are reported to promote flowering in transgenic Arabidopsis (Wu et al., 2014), but their endogenous role has not yet been determined. In addition, although the GmCOL1a and GmCOL1b genes showed higher expression levels under SD (Wu et al., 2014) and show some overlap with FT genes in their diurnal expression pattern, it is still unclear whether these genes are transcriptionally regulated by any of the E loci or whether they in turn regulate any of the FT genes. The situation in the galegoid legumes is simpler, with only a single CO ortholog (COLa) known to be present (Hecht et al., 2005; Yamashino et al., 2013; Wong et al., 2014). While the function of this gene has not been directly investigated in pea, its expression is not misregulated in either late1 (Psgi) or dne (Pself4) mutants, despite strong defects in FT regulation, flowering and photoperiod responsiveness in both mutants (Hecht et al., 2007, 2011; Liew et al., 2009). In addition, expression analyses, transgenic studies and characterization of specific mutants in M. truncatula also indicate that COLa does not have any substantial role in flowering time regulation, and suggest that the same is probably true for other COL genes (Wong et al., 2014).
The available evidence so far thus indicates that legume CO-like genes may not have a major role in integration of responses to photoperiod in the temperate legume group, and suggests that some alternative mechanism must be operating. One possibility is that the E1 gene may participate in this role. Analysis of soybean e1 mutants and E1 overexpression lines clearly show that E1 regulates expression of FT genes, including repression of FT2a and FT5a (Thakare et al., 2011; Xia et al., 2012) and induction of the repressive FT4 (Zhai et al., 2014; Figure 1). E1 is specifically expressed under LD, where it shows a strong diurnal expression rhythm, and is also transcriptionally regulated by E4 (Xia et al., 2012), suggesting that regulation by light and the circadian clock may be important for its function. In future, it will be interesting to learn whether E1 is also regulated by E2 and E3, and whether E1 orthologs also regulate flowering in the temperate legumes. However, even if E1 does participate as a key integrator in the soybean photoperiod response mechanism, the fact that it is a repressor of FT suggests that there still may be undiscovered components required for upregulation of FT genes in inductive conditions.
One possible scenario for CO-independent FT induction in legumes is provided by a recent study in Arabidopsis which showed that GI can bind directly to the FT promoter to activate FT transcription in a CO-independent manner (Sawa and Kay, 2011), and it can also interact physically with TEM proteins, which are direct transcriptional repressors of FT distantly related to E1 (Castillejo and Pelaz, 2008; Sawa and Kay, 2011). It is possible that legume E1 and GI orthologs might interact in a similar way. Another scenario may be that the DNA binding and protein-interacting properties of CO may be partitioned into two separate proteins, as has recently been suggested in sugarbeet, where a CCT-domain pseudo-response regulator (PRR) protein (similar to the C-terminal domain of CO) and a B-box zinc finger protein (similar to the N-terminal domain of CO) may interact to confer CO function (Dally et al., 2014).
In addition, the fact that PHYA and GI orthologs are central components of the photoperiod response mechanism in both LDP pea and SDP soybean, but show opposite effects on flowering and FT expression in these two species, implies that the SD/LD difference results from a reversed regulatory interaction at some point downstream of both genes. The relatively close taxonomic relationship between these two species make them an attractive model for understanding the evolution of this difference in photoperiod response mode.
Response to Vernalization
Regulation of flowering by vernalization is a phenomenon widespread across annual species from temperate regions, but is thought to have evolved independently in different plant lineages. As a result, the genes and genetic mechanisms conferring vernalization responsiveness are likely to differ across different groups (Kim et al., 2009). In legumes, the first insight into the genetic control of vernalization response has come from work in M. truncatula, where a survey of Medicago FT genes showed that two tandemly arranged FT genes (FTa1 and FTa2) are induced by vernalization but have different temporal patterns of response (Laurie et al., 2011). FTa2 is induced during exposure to cold, but FTa1 is only expressed following return to warm conditions. Loss-of-function FTa1 insertion mutants are insensitive to vernalization but retain sensitivity to photoperiod, indicating that FTa1 is necessary for response to vernalization, and may be the key target in the legume vernalization pathway (Figure 1; Laurie et al., 2011). Interestingly, lines carrying insertions close to but not within FTa1 coding regions show dominant inheritance of early, vernalization-independent flowering, which suggests that FTa1 is normally subject to repression by adjacent regulatory regions and that this repression is normally overcome by vernalization (Jaudal et al., 2013).
Control of Flowering in Other Legumes
Flowering time control is a key issue in adaptation of many other crop legumes (Summerfield and Roberts, 1985b; Nelson et al., 2010), and loci controlling flowering time and other flowering-related traits have now been identified in most crop and model legumes (Table 2). The majority of these loci have been detected as QTL, but in some cases they have been amenable to classical genetic analyses and Mendelian inheritance has been defined. It is likely that some of the same loci have been detected in different studies but it is difficult to assess this due to the lack of markers in common and/or sequence information.
Table 2.
List of flowering time QTL in legumes, indicating the linkage group (LG) on which they occur.
For chickpea, QTL on the same LG identified in different studies are distinguished by the LG number followed by a letter. More detail on these QTL is shown in Figure 2.
While all of this variation is naturally arising and therefore likely to be adaptive, several loci can be singled out for a particularly significant contribution to adaptation and range expansion. One example is the SDP common bean (Phaseolus vulgaris), where, like soybean, expansion to higher latitudes has been accompanied by earlier flowering under LD and a reduction in photoperiod responsiveness (Gepts and Debouck, 1991). A significant proportion of this variation can be attributed to PPD, a mendelized locus on LG1, where recessive alleles confer reduced photoperiod response and early flowering under LD (Koinange et al., 1996). A second example is narrow-leafed lupin (Lupinus angustifolius), where the acquisition of early, vernalization insensitive flowering conditioned by dominant alleles at the the Ku locus has been integral to deployment of this crop for Mediterranean climates with mild winters in which a vernalization requirement would not be met (Nelson et al., 2010). A third example is lentil (Lens culinaris), where an early flowering variant at the Sn locus has had an important role in developing early flowering cultivars for water-limited environments and broadening the genetic base of lentil in south Asia (Sarker and Erskine, 2006).
There is significant potential for translation of insights from the pea and soybean systems to achieve a better understanding of other legume species. This potential reflects advances in two areas. First, functional and phylogenetic analyses of flowering genes and gene families in pea and soybean can improve the identification of plausible candidate genes for particular loci. Second, availability of sequenced genomes and gene-based genetic maps have improved the technical ability to identify and evaluate candidate genes under QTL, and to identify those QTL that may have conserved locations across several species. In addition the fact that certain desirable traits such as early, photoperiod-insensitive flowering or determinate growth can result from simple monogenic loss-of-function mutations means that in species where such variants do not already exist it may be feasible to generate them through mutagenesis.
Within the legumes the value of a comparative approach is most clearly shown by recent findings that in several species, determinate inflorescence architecture is conferred by mutation of specific TFL1 genes [described by Benlloch et al. (2015) in this Research Topic]. A second example is the recent identification of the lentil SN locus as the ortholog of pea HR (Weller et al., 2012), suggesting that the nature of flowering time adaptation may be widely conserved. In addition to these well-established examples, it is now also possible to identify other potentially conserved QTL and to begin to speculate about their nature (Figure 2).
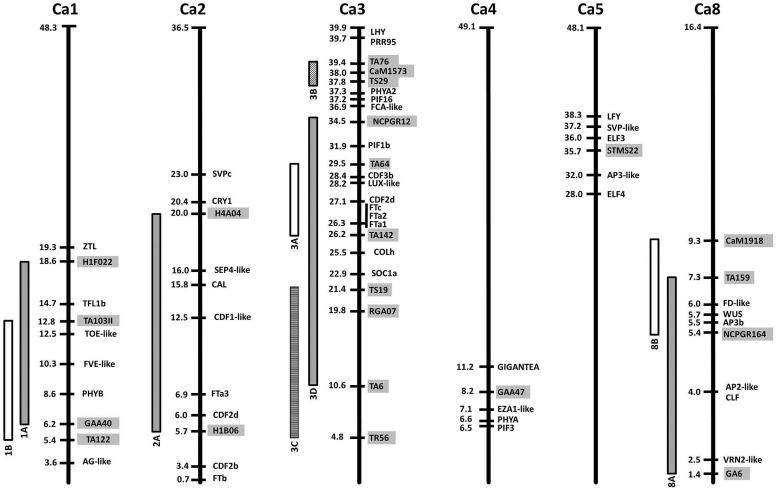
FIGURE 2.
Relative locations of flowering time gene homologs and quantitative trait loci (QTL) in the chickpea genome. Chromosomes are represented as bold vertical lines with marker/gene positions indicated in Mbp. QTL are shown as vertical bars, and the markers delimiting them are indicated by gray boxes. References for each QTL can be found in Table 2.
The most prominent case is the existence of a conserved major flowering time QTL in a region syntenic with a section of Medicago chromosome 7 containing a tandem array of FTa and FTc genes. This region is now implicated in control of flowering time in numerous members of the temperate legume clade, including M. truncatula itself (Pierre et al., 2008), faba bean (Cruz-Izquierdo et al., 2012), chickpea (Cobos et al., 2009; Aryamanesh et al., 2010), narrow-leafed lupin (Nelson et al., 2006), L. japonicus (Gondo et al., 2007) and alfalfa (Robins et al., 2007). As described above, functional studies in Medicago and pea have demonstrated the importance of FT genes in this cluster for vernalization responsiveness and other aspects of flowering (Laurie et al., 2011), and this implies that disruption to one or more of these genes is the most likely molecular basis for these QTL. Evidence from Medicago and pea indicates that FTa and FTc genes promote flowering, and a simple loss-of-function mutation would therefore be expected to be recessive and late-flowering. However, for several of these QTL the “derived” form appears to be early flowering, as the later-flowering variant is much more widespread and likely to be ancestral (faba bean, chickpea, lupin), and at least in lupin and chickpea the early flowering trait has been reported to show dominant inheritance (Aryamanesh et al., 2010; Nelson et al., 2010). These observations suggest that if genes in the FTa/c cluster are the basis for these QTL, the genetic changes are likely to be gain-of-function mutations, and it might also be expected that one or more of the FT genes in the cluster would show high and/or de-regulated expression. This also suggests that the causal genetic changes could be varied and complex, potentially involving variation in copy number or alteration to promoter or other regulatory regions, as observed for gain-of-function mutations affecting flowering time genes in cereals (e.g., Beales et al., 2007; Nitcher et al., 2013).
A closer look at individual species provides additional illustrations of a comparative approach to candidate gene identification. The first example is common bean where, in addition to several flowering time QTL (Table 2), two loci controlling photoperiod response have also been characterized; Ppd and Hr (Gu et al., 1998). The Ppd locus has been mapped to within 5 cM of the FIN locus controlling shoot determinacy (Kwak et al., 2008), and the molecular identity of FIN as a TFL1 co-ortholog (Repinski et al., 2012) therefore indicates the approximate genomic region in which PPD is located. This region is syntenic with the region in soybean containing the E3/PHYA3 gene (McClean et al., 2010) and, as expected, contains the bean E3 ortholog, suggesting this as an attractive candidate for the PPD locus. A second bean locus contributing to photoperiod sensitivity, Hr, is less well-defined but is positioned toward the other end of the same linkage group (Gu et al., 1998), a region that contains homologs of ELF3 and the FTa/c cluster. A third locus identified as a QTL in LG9 is located near the bean ortholog of ZEITLUPE, a gene important for circadian clock regulation in Arabidopsis (Tar’an et al., 2002; Kwak et al., 2008).
The second example is chickpea, where classical genetic analyses have distinguished four Mendelian loci, named Early flowering 1 (Efl1) to Efl4 (Gaur et al., 2015). Recessive alleles at these loci confer early flowering and at least two are likely to be widespread within the chickpea germplasm and have a major impact on flowering time adaptation. In addition, many linkage studies have been performed in chickpea, and six flowering time QTL have been defined in LG1, 2, 3, 4, and 8 (Table 2). Some of these studies have involved parents known to carry efl1 or efl2, suggesting that QTL may correspond to major loci in some cases. This, together with the availability of the chickpea genome sequence (Jain et al., 2013; Varshney et al., 2013) enables an assessment of approximate co-location between flowering gene homologs and flowering time loci or QTL (Figure 2). For example, Efl1 was defined in a cross using the early flowering line ICCV2 (Kumar and van Rheenen, 2000), which has also been used as a common parent in several studies reporting a major QTL (Cho et al., 2002; Vadez et al., 2012; Jamalabadi et al., 2013). Although its genomic location is still uncertain (as markers near the QTL have been variously assigned to LG3, 4, 5, 6, and 8), the clearest and most recent indication is given by Jamalabadi et al. (2013) who placed a major QTL between markers TA117 and STMS22, which both map to LG5 (Radhika et al., 2007; Hiremath et al., 2012). The latter is physically located near genes responsible for flowering time variation in other species such as ELF3 or ELF4, but also LEAFY (LFY), which is involved in flower initiation and inflorescence development (Hofer et al., 1997). Recessive alleles at the photoperiod response locus Efl2 are present in the cultivar ICC5810 (Or et al., 1999), a line also used as a parent by Lichtenzveig et al. (2006), who found a major QTL in LG1 explaining 60% of the flowering time variation. This chromosomal region contains several genes with the potential to explain these differences, including orthologs of PHYB and TFL1.
For the remaining reported QTL, there is no clear relationship to major flowering loci, but some potential candidates can be identified, including PHYA and GI for the LG4 QTL, another member of the FT family (FTa3) for the LG2 QTL, and a FD-like gene for the LG8 QTL (Figure 2). The QTL in the central region of LG3 deserves special attention as it has been reported several times (Table 2). The presence of the FTa/c cluster as the most likely candidate has been already mentioned, but other genes within the interval are also noteworthy, including, two CYCLING DOF FACTOR (CDF) homologs, a LUX-like gene, and an ortholog of the CO-like gene COLh. This last gene has also been highlighted as a candidate near the chromosome 7 QTL in M. truncatula (Pierre et al., 2011). A second QTL close to the bottom of chickpea LG3 has also been described (Aryamanesh et al., 2010), in a region containing genes involved in circadian clock and light signaling, including PRR and phytochrome-interacting factor (PIF) homologs, a PHYA paralog, and the ortholog of LATE ELONGATED HYPOCOTYL (LHY).
Conclusion
Detailed genetic studies in model legumes are bringing rapid progress to our understanding of flowering time control, and have identified significant similarities and differences with other plant groups. At the same time, new genome sequences and dense, gene-based genetic maps are accelerating the translation of these insights to a range of other legume species, and giving new insight into how flowering time gene functions may be conserved or diversified within this important group of crops. It now seems reasonable to expect that genes underlying most of the major flowering time loci in legumes will be identified within the next 5–10 years, which will help improve our understanding of how they work together to provide adaptation to specific locations and agronomic constraints.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank F. Sussmilch and R. Macknight for helpful comments on the manuscript, and the Australian Research Council for financial support through Discovery Project and Future Fellowship grants (JLW).
References
- Amasino R. M., Michaels S. D. (2010). The timing of flowering. Plant Physiol. 154 516–520 10.1104/pp.110.161653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andargie M., Pasquet R. S., Gowda B. S., Muluvi G. M., Timko M. P. (2014). Molecular mapping of QTLs for domestication-related traits in cowpea (V. unguiculata (L.) Walp.). Euphytica 200 401–412 10.1007/s10681-014-1170-9 [DOI] [Google Scholar]
- Andargie M., Pasquet R. S., Muluvi G. M., Timko M. P. (2013). Quantitative trait loci analysis of flowering time related traits identified in recombinant inbred lines of cowpea (Vigna unguiculata). Genome 56 289–294 10.1139/gen-2013-0028 [DOI] [PubMed] [Google Scholar]
- Andres F., Coupland G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13 627–639 10.1038/nrg3291 [DOI] [PubMed] [Google Scholar]
- Arumingtyas E. L., Murfet I. C. (1994). Flowering in Pisum: a further gene controlling response to photoperiod. J. Heredity 85 12–17. [Google Scholar]
- Aryamanesh N., Nelson M. N., Yan G., Clarke H. J., Siddique K. H. M. (2010). Mapping a major gene for growth habit and QTLs for ascochyta blight resistance and flowering time in a population between chickpea and Cicer reticulatum. Euphytica 173 307–319 10.1007/s10681-009-0086-2 [DOI] [Google Scholar]
- Bagnall D. J., King R. W. (1991). Response of peanut (Arachis hypogaea) to temperature, photoperiod and irradiance.1. Effect on flowering. Field Crops Res. 26 263–277 10.1016/0378-4290(91)90004-F [DOI] [Google Scholar]
- Ballerini E. S., Kramer E. M. (2011). In the light of evolution: a reevaluation of conservation in the CO-FT regulon and its role in photoperiodic regulation of flowering time. Front. Plant Sci. 2:81 10.3389/fpls.2011.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beales J., Turner A., Griffiths S., Snape J. W., Laurie D. A. (2007). A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor. Appl. Genet. 115 721–733 10.1007/s00122-007-0603-4 [DOI] [PubMed] [Google Scholar]
- Benlloch R., Berbel A., Ali L., Gohari G., Millan T., Madueño F. (2015). Genetic control of inflorescence architecture in legumes. Front. Plant. Sci. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbel A., Ferrandiz C., Hecht V., Dalmais M., Lund O. S., Sussmilch F. C., et al. (2012). VEGETATIVE1 is essential for development of the compound inflorescence in pea. Nat. Commun. 3 797 10.1038/ncomms1801 [DOI] [PubMed] [Google Scholar]
- Beveridge C. A., Murfet I. C. (1996). The gigas mutant in pea is deficient in the floral stimulus. Physiol. Plant 96 637–645 10.1111/j.1399-3054.1996.tb00237.x [DOI] [Google Scholar]
- Blair M. W., Iriarte G., Beebe S. (2006). QTL analysis of yield traits in an advanced backcross population derived from a cultivated Andean x wild common bean (Phaseolus vulgaris L.) cross. Theor. Appl. Genet. 112 1149–1163 10.1007/s00122-006-0217-2 [DOI] [PubMed] [Google Scholar]
- Brambilla V., Fornara F. (2013). Molecular control of flowering in response to day length in rice. J. Integr. Plant Biol. 55 410–418 10.1111/jipb.12033 [DOI] [PubMed] [Google Scholar]
- Castillejo C., Pelaz S. (2008). The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 18 1–6 10.1016/j.cub.2008.07.075 [DOI] [PubMed] [Google Scholar]
- Chavarro M. C., Blair M. W. (2010). QTL Analysis and effect of the fin locus on tropical adaptation in an inter-genepool common bean population. Tropical. Plant Biol. 3 204–218 10.1007/s12042-010-9058-x [DOI] [Google Scholar]
- Cho S. H., Kumar J., Shultz J. L., Anupama K., Tefera F., Muehlbauer F. J. (2002). Mapping genes for double podding and other morphological traits in chickpea. Euphytica 128 285–292 10.1023/A:1020872009306 [DOI] [Google Scholar]
- Cober E. R. (2011). Long juvenile soybean flowering responses under very short photoperiods. Crop Sci. 51 140–145 10.2135/Cropsci2010.05.0262 [DOI] [Google Scholar]
- Cober E. R., Molnar S. J., Charette M., Voldeng H. D. (2010). A new locus for early maturity in soybean. Crop Sci. 50 524–527 10.2135/Cropsci2009.04.0174 [DOI] [Google Scholar]
- Cober E. R., Morrison M. J. (2010). Regulation of seed yield and agronomic characters by photoperiod sensitivity and growth habit genes in soybean. Theor. Appl. Genet. 120 1005–1012 10.1007/s00122-009-1228-6 [DOI] [PubMed] [Google Scholar]
- Cober E. R., Stewart D. W., Voldeng H. D. (2001). Photoperiod and temperature responses in early-maturing, near-isogenic soybean lines. Crop Sci. 41 721–727 10.2135/cropsci2001.413721x [DOI] [Google Scholar]
- Cober E. R., Tanner J. W., Voldeng H. D. (1996a). Genetic control of photoperiod response in early-maturing, near-isogenic soybean lines. Crop Sci. 36 601–605 10.2135/cropsci1996.0011183X003600030013x [DOI] [Google Scholar]
- Cober E. R., Tanner J. W., Voldeng H. D. (1996b). Soybean photoperiod-sensitivity loci respond differently to light quality. Crop Sci. 36 606–610 10.2135/cropsci1996.0011183X003600030014x [DOI] [Google Scholar]
- Cober E. R., Voldeng H. D. (2001). A new soybean maturity and photoperiod-sensitivity locus linked to E1 and T. Crop Sci. 41 698–701 10.2135/cropsci2001.413698x [DOI] [Google Scholar]
- Cobos M. J., Rubio J., Fernández-Romero M. D., Garza R., Moreno M. T., Millán T., et al. (2007). Genetic analysis of seed size, yield and days to flowering in a chickpea recombinant inbred line population derived from a Kabuli × Desi cross. Ann. Appl. Biol. 151 33–42 10.1111/j.1744-7348.2007.00152.x [DOI] [Google Scholar]
- Cobos M. J., Winter P., Kharrat M., Cubero J. I., Gil J., Milian T., et al. (2009). Genetic analysis of agronomic traits in a wide cross of chickpea. Field Crops Res. 111 130–136 10.1016/J.Fcr.2008.11.006 [DOI] [Google Scholar]
- Cronk Q., Ojeda I., Pennington R. T. (2006). Legume comparative genomics: progress in phylogenetics and phylogenomics. Curr. Opin. Plant Biol. 9 99–103 10.1016/j.pbi.2006.01.011 [DOI] [PubMed] [Google Scholar]
- Cruz-Izquierdo S., Avila C. M., Satovic Z., Palomino C., Gutierrez N., Ellwood S. R., et al. (2012). Comparative genomics to bridge Vicia faba with model and closely-related legume species: stability of QTLs for flowering and yield-related traits. Theor. Appl. Genet. 125 1767–1782 10.1007/S00122-012-1952-1 [DOI] [PubMed] [Google Scholar]
- Dally N., Xiao K., Holtgrawe D., Jung C. (2014). The B2 flowering time locus of beet encodes a zinc finger transcription factor. Proc. Natl. Acad. Sci. U.S.A. 111 10365–10370 10.1073/Pnas.1404829111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foucher F., Morin J., Courtiade J., Cadioux S., Ellis N., Banfield M. J., et al. (2003). DETERMINATE and LATE FLOWERING are two TERMINAL FLOWER1/CENTRORADIALIS homologs that control two distinct phases of flowering initiation and development in pea. Plant Cell 15 2742–2754 10.1105/tpc.015701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur P. M., Samineni S., Tripathi S., Varshney R. K., Gowda C. L. L. (2015). Allelic relationships of flowering time genes in chickpea. Euphytica 10.1007/s10681-014-1261-7 [DOI] [Google Scholar]
- Gepts P., Debouck D. G. (1991). “Origin, domestication and evolution of the common bean (Phaseolus vulgaris),” in Common Bean: Research for Crop Improvement, eds Schoonhoven A., Voysest O. (Oxford: CAB; ), 7–53. [Google Scholar]
- Gondo T., Sato S., Okumura K., Tabata S., Akashi R., Isobe S. (2007). Quantitative trait locus analysis of multiple agronomic traits in the model legume Lotus japonicus. Genome 50 627–637 10.1139/G07-040 [DOI] [PubMed] [Google Scholar]
- Gu W., Zhu J., Wallace D. H., Singh S. P., Weeden N. F. (1998). Analysis of genes controlling photoperiod sensitivity in common bean using DNA markers. Euphytica 102 125–132 10.1023/A:1018340514388 [DOI] [Google Scholar]
- Hecht V., Foucher F., Ferrandiz C., Macknight R., Navarro C., Morin J., et al. (2005). Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 137 1420–1434 10.1104/pp.104.057018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V., Knowles C. L., Vander Schoor J. K., Liew L. C., Jones S. E., Lambert M. J. M., et al. (2007). Pea LATE BLOOMER1 is a GIGANTEA ortholog with roles in photoperiodic flowering, deetiolation, and transcriptional regulation of circadian clock gene homologs. Plant Physiol. 144 648–661 10.1104/pp.107.096818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V., Laurie R. E., Vander Schoor J. K., Ridge S., Knowles C. L., Liew L. C., et al. (2011). The pea GIGAS gene is a FLOWERING LOCUS T homolog necessary for graft-transmissible specification of flowering but not for responsiveness to photoperiod. Plant Cell 23 147–161 10.1105/tpc.110.081042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiremath P. J., Kumar A., Penmetsa R. V., Farmer A., Schlueter J. A., Chamarthi S. K., et al. (2012). Large-scale development of cost-effective SNP marker assays for diversity assessment and genetic mapping in chickpea and comparative mapping in legumes. Plant Biotechnol. J. 10 716–732 10.1111/J.1467-7652.2012.00710.X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer J., Turner L., Hellens R., Ambrose M., Matthews P., Michael A., et al. (1997). UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr. Biol. 7 581–587 10.1016/S0960-9822(06)00257-0 [DOI] [PubMed] [Google Scholar]
- Hossain S., Ford R., McNeil D., Pittock C., Panozzo J. F. (2010). Development of a selection tool for seed shape and QTL analysis of seed shape with other morphological traits for selective breeding in chickpea (Cicer arietinum L.). Aust. J. Crop Sci. 4 278–288. [Google Scholar]
- Isemura T., Kaga A., Konishi S., Ando T., Tomooka N., Han O. K., et al. (2007). Genome dissection of traits related to domestication in azuki bean (Vigna angularis) and comparison with other warm-season legumes. Ann. Bot. 100 1053–1071 10.1093/aob/mcm155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isemura T., Kaga A., Tabata S., Somta P., Srinives P., Shimizu T., et al. (2012). Construction of a genetic linkage map and genetic analysis of domestication related traits in mungbean (Vigna radiata). PLoS ONE 7:e41304 10.1371/journal.pone.0041304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M., Misra G., Patel R. K., Priya P., Jhanwar S., Khan A. W., et al. (2013). A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.). Plant J. 74 715–729 10.1111/Tpj.12173 [DOI] [PubMed] [Google Scholar]
- Jamalabadi J. G., Saidi A., Karami E., Kharkesh M., Talebi R. (2013). Molecular Mapping and characterization of genes governing time to flowering, seed weight, and plant height in an intraspecific genetic linkage map of chickpea (Cicer arietinum). Biochem. Genet. 51 387–397 10.1007/S10528-013-9571-3 [DOI] [PubMed] [Google Scholar]
- Jaudal M., Monash J., Zhang L., Wen J., Mysore K. S., Macknight R., et al. (2014). Overexpression of Medicago SVP genes causes floral defects and delayed flowering in Arabidopsis but only affects floral development in Medicago. J. Exp. Bot. 65 429–442 10.1093/jxb/ert384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaudal M., Yeoh C. C., Zhang L., Stockum C., Mysore K. S., Ratet P., et al. (2013). Retroelement insertions at the Medicago FTa1 locus in spring mutants eliminate vernalisation but not long-day requirements for early flowering. Plant J. 76 580–591 10.1111/tpj.12315 [DOI] [PubMed] [Google Scholar]
- Jiang B., Nan H., Gao Y., Tang L., Yue Y., Lu S., et al. (2014). Allelic combinations of soybean maturity loci E1, E2, E3 and E4 result in diversity of maturity and adaptation to different latitudes. PLoS ONE 9:e106042 10.1371/journal.pone.0106042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaga A., Isemura T., Tomooka N., Vaughan D. A. (2008). The genetics of domestication of the azuki bean (Vigna angularis). Genetics 178 1013–1036 10.1534/genetics.107.078451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajonphol T., Sangsiri C., Somta P., Toojinda T., Srinives P. (2012). SSR map construction and quantitative trait loci (QTL) identification of major agronomic traits in mungbean (Vigna radiata (L.) Wilczek). Sabrao J. Breed. Genet. 44 71–86. [Google Scholar]
- Kim D. H., Doyle M. R., Sung S., Amasino R. M. (2009). Vernalization: winter and the timing of flowering in plants. Annu. Rev. Cell Dev. Biol. 25 277–299 10.1146/annurev.cellbio.042308.113411 [DOI] [PubMed] [Google Scholar]
- Kim M. Y., Shin J. H., Kang Y. J., Shim S. R., Lee S. H. (2012). Divergence of flowering genes in soybean. J. Biosci. 37 857–870 10.1007/s12038-012-9252-0 [DOI] [PubMed] [Google Scholar]
- King W. M., Murfet I. C. (1985). Flowering in Pisum: a sixth locus, Dne. Ann. Bot. 56 835–846. [Google Scholar]
- Koinange E. M. K., Singh S. P., Gepts P. (1996). Genetic control of the domestication syndrome in common bean. Crop Sci. 36 1037–1045 10.2135/cropsci1996.0011183X003600040037x [DOI] [Google Scholar]
- Kong F., Liu B., Xia Z., Sato S., Kim B. M., Watanabe S., et al. (2010). Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol. 154 1220–1231 10.1104/pp.110.160796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F. J., Nan H. Y., Cao D., Li Y., Wu F. F., Wang J. L., et al. (2014). A new dominant gene E9 conditions early flowering and maturity in soybean. Crop Sci. 54 2529–2535 10.2135/Cropsci2014.03.0228 [DOI] [Google Scholar]
- Kongjaimun A., Kaga A., Tomooka N., Somta P., Vaughan D. A., Srinives P. (2012). The genetics of domestication of yardlong bean, Vigna unguiculata (L.) Walp. ssp. unguiculata cv.-gr. sesquipedalis. Ann. Bot. 109 1185–1200 10.1093/aob/mcs048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroc M., Koczyk G., Świkęcicki W., Kilian A., Nelson M. N. (2014). New evidence of ancestral polyploidy in the Genistoid legume Lupinus angustifolius L. (narrow-leafed lupin). Theor. Appl. Genet. 127 1237–1249 10.1007/s00122-014-2294-y [DOI] [PubMed] [Google Scholar]
- Kumar J., van Rheenen H. A. (2000). A major gene for time of flowering in chickpea. J. Hered. 91 67–68 10.1093/jhered/91.1.67 [DOI] [PubMed] [Google Scholar]
- Kumawat G., Raje R. S., Bhutani S., Pal J. K., Mithra A. S. V. C. R., Gaikwad K., et al. (2012). Molecular mapping of QTLs for plant type and earliness traits in pigeonpea (Cajanus cajan L. Millsp.). BMC Genet. 13:84 10.1186/1471-2156-13-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak M., Velasco D., Gepts P. (2008). Mapping homologous sequences for determinacy and photoperiod sensitivity in common bean (Phaseolus vulgaris). J. Hered. 99 283–291 10.1093/jhered/esn005 [DOI] [PubMed] [Google Scholar]
- Laurie R. E., Diwadkar P., Jaudal M., Zhang L., Hecht V., Wen J., et al. (2011). The Medicago FLOWERING LOCUS T homolog, MtFTa1, is a key regulator of flowering time. Plant Physiol. 156 2207–2224 10.1104/pp.111.180182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune-Hénaut I., Hanocq E., Béthencourt L., Fontaine V., Delbreil B., Morin J., et al. (2008). The flowering locus Hr colocalizes with a major QTL affecting winter frost tolerance in Pisum sativum L. Theor. Appl. Genet. 116 1105–1116 10.1007/s00122-008-0739-x [DOI] [PubMed] [Google Scholar]
- Lichtenzveig J., Bonfil D. J., Zhang H. B., Shtienberg D., Abbo S. (2006). Mapping quantitative trait loci in chickpea associated with time to flowering and resistance to Didymella rabiei the causal agent of Ascochyta blight. Theor. Appl. Genet. 113 1357–1369 10.1007/s00122-006-0390-3 [DOI] [PubMed] [Google Scholar]
- Liew L. C., Hecht V., Laurie R. E., Knowles C. L., Vander Schoor J. K., Macknight R. C., et al. (2009). DIE NEUTRALIS and LATE BLOOMER 1 contribute to regulation of the pea circadian clock. Plant Cell 21 3198–3211 10.1105/tpc.109.067223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew L. C., Hecht V., Sussmilch F. C., Weller J. L. (2014). The pea photoperiod response gene STERILE NODES is an ortholog of LUX ARRHYTHMO. Plant Physiol. 165 648–657 10.1104/pp.114.237008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Kanazawa A., Matsumura H., Takahashi R., Harada K., Abe J. (2008). Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome A gene. Genetics 180 995–1007 10.1534/genetics.108.092742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Watanabe S., Uchiyama T., Kong F., Kanazawa A., Xia Z., et al. (2010). The soybean stem growth habit gene Dt1 is an ortholog of Arabidopsis TERMINAL FLOWER1. Plant Physiol. 153 198–210 10.1104/pp.109.150607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClean P. E., Mamidi S., Mcconnell M., Chikara S., Lee R. (2010). Synteny mapping between common bean and soybean reveals extensive blocks of shared loci. BMC Genomics 11:184 10.1186/1471-2164-11-184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfet I. C. (1985). “Pisum sativum,” in CRC Handbook of Flowering, ed.Halevy A. H. (Boca Raton: CRC Press; ), 97–126. [Google Scholar]
- Nagel D. H., Kay S. A. (2012). Complexity in the wiring and regulation of plant circadian networks. Curr. Biol. 22 R648–R657 10.1016/j.cub.2012.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan H., Cao D., Zhang D., Li Y., Lu S., Tang L., et al. (2014). GmFT2a and GmFT5a redundantly and differentially regulate flowering through interaction with and upregulation of the bZIP transcription factor GmFDL19 in soybean. PLoS ONE 9:e97669 10.1371/journal.pone.0097669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M. N., Berger J. D., Erskine W. (2010). Flowering time control in annual legumes: prospects in a changing global climate. CAB Rev. Perspect. Agric. Veterinary Sci. Nutr. Nat. Res. 5 1–14 10.1079/PAVSNNR20105017 [DOI] [Google Scholar]
- Nelson M. N., Phan H. T., Ellwood S. R., Moolhuijzen P. M., Hane J., Williams A., et al. (2006). The first gene-based map of Lupinus angustifolius L.-location of domestication genes and conserved synteny with Medicago truncatula. Theor. Appl. Genet. 113 225–238 10.1007/s00122-006-0288-0 [DOI] [PubMed] [Google Scholar]
- Nitcher R., Distelfeld A., Tan C., Yan L. L., Dubcovsky J. (2013). Increased copy number at the HvFT1 locus is associated with accelerated flowering time in barley. Mol. Genet. Genomics 288 261–275 10.1007/S00438-013-0746-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Or E., Hovav R., Abbo S. (1999). A major gene for flowering time in chickpea. Crop Sci. 39 315–322. [Google Scholar]
- Pérez-Vega E., Pañeda A., Rodríguez-Suárez C., Campa A., Giraldez R., Ferreira J. J. (2010). Mapping of QTLs for morpho-agronomic and seed quality traits in a RIL population of common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 120 1367–1380 10.1007/s00122-010-1261-5 [DOI] [PubMed] [Google Scholar]
- Pierre J. B., Bogard M., Herrmann D., Huyghe C., Julier B. (2011). A CONSTANS-like gene candidate that could explain most of the genetic variation for flowering date in Medicago truncatula. Mol. Breed. 28 25–35 10.1007/S11032-010-9457-6 [DOI] [Google Scholar]
- Pierre J. B., Huguet T., Barre P., Huyghe C., Julier B. (2008). Detection of QTLs for flowering date in three mapping populations of the model legume species Medicago truncatula. Theor. Appl. Genet. 117 609–620 10.1007/S00122-008-0805-4 [DOI] [PubMed] [Google Scholar]
- Pin P. A., Benlloch R., Bonnet D., Wremerth-Weich E., Kraft T., Gielen J. J., et al. (2010). An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 330 1397–1400 10.1126/science.1197004 [DOI] [PubMed] [Google Scholar]
- Pin P. A., Nilsson O. (2012). The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ. 35 1742–1755 10.1111/j.1365-3040.2012.02558.x [DOI] [PubMed] [Google Scholar]
- Ping J., Liu Y., Sun L., Zhao M., Li Y., She M., et al. (2014). Dt2 is a gain-of-function MADS-domain factor gene that specifies semideterminacy in soybean. Plant Cell 26 2831–2842 10.1105/tpc.114.126938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platten J. D., Foo E., Elliott R. C., Hecht V., Reid J. B., Weller J. L. (2005a). Cryptochrome 1 contributes to blue-light sensing in pea. Plant Physiol. 139 1472–1482 10.1104/pp.105.067462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platten J. D., Foo E., Foucher F., Hecht V., Reid J. B., Weller J. L. (2005b). The cryptochrome gene family in pea includes two differentially expressed CRY2 genes. Plant Mol. Biol. 59 683–696 10.1007/s11103-005-0828-z [DOI] [PubMed] [Google Scholar]
- Radhika P., Gowda S. J. M., Kadoo N. Y., Mhase L. B., Jamadagni B. M., Sainani M. N., et al. (2007). Development of an integrated intraspecific map of chickpea (Cicer arietinum L.) using two recombinant inbred line populations. Theor. Appl. Genet. 115 209–216 10.1007/S00122-007-0556-7 [DOI] [PubMed] [Google Scholar]
- Ray J. D., Hinson K., Mankono J. E. B., Malo M. F. (1995). Genetic control of a long-juvenile trait in soybean. Crop Sci. 35 1001–1006 10.2135/cropsci1995.0011183X003500040012x [DOI] [Google Scholar]
- Rehman A. U., Malhotra R. S., Bett K., Tar’an B., Bueckert R., Warkentin T. D. (2011). Mapping QTL associated with traits affecting grain yield in chickpea (Cicer arietinum L.) under terminal drought stress. Crop Sci. 51 450–463 10.2135/cropsci2010.03.0129 [DOI] [Google Scholar]
- Repinski S. L., Kwak M., Gepts P. (2012). The common bean growth habit gene PvTFL1y is a functional homolog of Arabidopsis TFL1. Theor. Appl. Genet. 124 1539–1547 10.1007/S00122-012-1808-8 [DOI] [PubMed] [Google Scholar]
- Robins J. G., Bauchan G. R., Brummer E. C. (2007). Genetic mapping forage yield, plant height, and regrowth at multiple harvests in tetraploid alfalfa (Medicago sativa L.). Crop Sci. 47 11–18 10.2135/Cropsci2006.07.0447 [DOI] [Google Scholar]
- Ruelens P., De Maagd R. A., Proost S., Theissen G., Geuten K., Kaufmann K. (2013). FLOWERING LOCUS C in monocots and the tandem origin of angiosperm-specific MADS-box genes. Nat. Commun. 4 2280 10.1038/ncomms3280 [DOI] [PubMed] [Google Scholar]
- Sarker A., Erskine W. (2006). Recent progress in the ancient lentil. J. Agric. Sci. 144 19–29 10.1017/s0021859605005800 [DOI] [Google Scholar]
- Sawa M., Kay S. A. (2011). GIGANTEA directly activates FLOWERING LOCUS T in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 108 11698–11703 10.1073/Pnas.1106771108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y. H., Ito S., Imaizumi T. (2013). Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 18 575–583 10.1016/j.tplants.2013.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan J. A., Gray J. C. (2000). The pea light-independent photomorphogenesis1 mutant results from partial duplication of COP1 generating an internal promoter and producing two distinct transcripts. Plant Cell 12 1927–1937 10.1105/tpc.12.10.1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerfield R. J., Roberts E. H. (1985a). “Glycine max,” in CRC Handbook of Flowering, ed.Halevy A. H. (Boca Raton: CRC press; ), 100–117. [Google Scholar]
- Summerfield R. J., Roberts E. H. (1985b). “Grain legume species of significant importance in world agriculture,” in CRC Handbook of Flowering, ed.Halevy A. H. (Boca Raton: CRC press; ), 61–73. [Google Scholar]
- Summerfield R. J., Roberts E. H. (1985c). “Lupinus,” in CRC Handbook of Flowering, ed.Halevy A. H. (Boca Raton: CRC press; ), 125–133. [Google Scholar]
- Sussmilch F. C., Berbel A., Hecht V., Vander Schoor J. K., Ferrándiz C., Madueño F., et al. (2015). Pea VEGETATIVE2 is an FD homolog that is essential for flowering and compound inflorescence Development. Plant Cell 10.1105/tpc.115.136150 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tar’an B., Michaels T. E., Pauls K. P. (2002). Genetic mapping of agronomic traits in common bean. Crop Sci. 42 544–556 10.2135/cropsci2002.0544 [DOI] [Google Scholar]
- Taylor S. A., Hofer J. M., Murfet I. C., Sollinger J. D., Singer S. R., Knox M. R., et al. (2002). PROLIFERATING INFLORESCENCE MERISTEM, a MADS-box gene that regulates floral meristem identity in pea. Plant Physiol. 129 1150–1159 10.1104/pp.001677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakare D., Kumudini S., Dinkins R. D. (2011). The alleles at the E1 locus impact the expression pattern of two soybean FT-like genes shown to induce flowering in Arabidopsis. Planta 234 933–943 10.1007/S00425-011-1450-8 [DOI] [PubMed] [Google Scholar]
- Tiwari S. B., Shen Y., Chang H. C., Hou Y., Harris A., Ma S. F., et al. (2010). The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 187 57–66 10.1111/j.1469-8137.2010.03251.x [DOI] [PubMed] [Google Scholar]
- Tomkins J. P., Shipe E. R. (1997). Environmental adaptation of long-juvenile soybean cultivars and elite strains. Agron. J. 89 257–262 10.2134/agronj1997.00021962008900020017x [DOI] [Google Scholar]
- Vadez V., Krishnamurthy L., Thudi M., Anuradha C., Colmer T. D., Turner N. C., et al. (2012). Assessment of ICCV 2 x JG 62 chickpea progenies shows sensitivity of reproduction to salt stress and reveals QTL for seed yield and yield components. Mol. Breed. 30 9–21 10.1007/S11032-011-9594-6 [DOI] [Google Scholar]
- Varshney R. K., Song C., Saxena R. K., Azam S., Yu S., Sharpe A. G., et al. (2013). Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat. Biotechnol. 31 240–246 10.1038/Nbt.2491 [DOI] [PubMed] [Google Scholar]
- Varshney R., Thudi M., Nayak S., Gaur P., Kashiwagi J., Krishnamurthy L., et al. (2014). Genetic dissection of drought tolerance in chickpea (Cicer arietinum L.) . Theor. Appl. Genet. 127 445–462 10.1007/s00122-013-2230-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Harada K., Abe J. (2012). Genetic and molecular bases of photoperiod responses of flowering in soybean. Breed. Sci. 61 531–543 10.1270/jsbbs.61.531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Hideshima R., Xia Z., Tsubokura Y., Sato S., Nakamoto Y., et al. (2009). Map-based cloning of the gene associated with the soybean maturity locus E3. Genetics 182 1251–1262 10.1534/genetics.108.098772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Xia Z., Hideshima R., Tsubokura Y., Sato S., Yamanaka N., et al. (2011). A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics 188 395–407 10.1534/genetics.110.125062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller J. L., Batge S. L., Smith J. J., Kerckhoffs L. H. J., Sineshchekov V. A., Murfet I. C., et al. (2004). A dominant mutation in the pea PHYA gene confers enhanced responses to light and impairs the light-dependent degradation of phytochrome A. Plant Physiol. 135 2186–2195 10.1104/pp.103.036103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller J. L., Beauchamp N., Kerckhoffs L. H. J., Platten J. D., Reid J. B. (2001). Interaction of phytochromes A and B in the control of de-etiolation and flowering in pea. Plant J. 26 283–294 10.1046/j.1365-313X.2001.01027.x [DOI] [PubMed] [Google Scholar]
- Weller J. L., Hecht V., Liew L. C., Sussmilch F. C., Wenden B., Knowles C. L., et al. (2009). Update on the genetic control of flowering in garden pea. J. Exp. Bot. 60 2493–2499 10.1093/jxb/erp120 [DOI] [PubMed] [Google Scholar]
- Weller J. L., Liew L. C., Hecht V. F., Rajandran V., Laurie R. E., Ridge S., et al. (2012). A conserved molecular basis for photoperiod adaptation in two temperate legumes. Proc. Natl. Acad. Sci. U.S.A. 109 21158–21163 10.1073/pnas.1207943110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller J. L., Murfet I. C., Reid J. B. (1997a). Pea mutants with reduced sensitivity to far-red light define an important role for phytochrome A in day-length detection. Plant Physiol. 114 1225–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller J. L., Reid J. B., Taylor S. A., Murfet I. C. (1997b). The genetic control of flowering in pea. Trends Plant Sci. 2 412–418 10.1016/S1360-1385(97)85580-X [DOI] [Google Scholar]
- Wong A. C., Hecht V. F., Picard K., Diwadkar P., Laurie R. E., Wen J., et al. (2014). Isolation and functional analysis of CONSTANS-LIKE genes suggests that a central role for CONSTANS in flowering time control is not evolutionarily conserved in Medicago truncatula. Front. Plant Sci. 5:486 10.3389/fpls.2014.00486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Price B. W., Haider W., Seufferheld G., Nelson R., Hanzawa Y. (2014). Functional and evolutionary characterization of the CONSTANS gene family in short-day photoperiodic flowering in soybean. PLoS ONE 9:e85754 10.1371/journal.pone.0085754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z., Watanabe S., Yamada T., Tsubokura Y., Nakashima H., Zhai H., et al. (2012). Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc. Natl. Acad. Sci. U.S.A. 109 E2155–E2164 10.1073/pnas.1117982109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Wu X., Wang B., Hu T., Lu Z., Liu Y., et al. (2013). QTL mapping and epistatic interaction analysis in asparagus bean for several characterized and novel horticulturally important traits. BMC Genet. 14:4 10.1186/1471-2156-14-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka N., Nagamura Y., Tsubokura Y., Yamamoto K., Takahashi R., Kouchi H., et al. (2000). Quantitative trait locus analysis of flowering time in soybean using a RFLP linkage map. Breed. Sci. 50 109–115 10.1270/jsbbs.50.109 [DOI] [Google Scholar]
- Yamashino T., Yamawaki S., Hagui E., Ueoka-Nakanishi H., Nakamichi N., Ito S., et al. (2013). Clock-controlled and FLOWERING LOCUS T (FT)-dependent photoperiodic pathway in Lotus japonicus I: verification of the flowering-associated function of an FT homolog. Biosci. Biotechnol. Biochem. 77 747–753 10.1271/bbb.120871 [DOI] [PubMed] [Google Scholar]
- Young N. D., Bharti A. K. (2012). Genome-enabled insights into legume biology. Annu. Rev. Plant Biol. 63 283–305 10.1146/annurev-arplant-042110-103754 [DOI] [PubMed] [Google Scholar]
- Zhai H., Lu S., Liang S., Wu H., Zhang X., Liu B., et al. (2014). GmFT4 a homolog of FLOWERING LOCUS T, is positively regulated by E1 and functions as a flowering repressor in soybean. PLoS ONE 9:e89030 10.1371/journal.pone.0089030 [DOI] [PMC free article] [PubMed] [Google Scholar]