Abstract
Myeloid cells are the most prominent amongst cells capable of presenting tumor-derived antigens to T cells and thereby maintaining the latter in an activated state. Myeloid populations of the tumor microenvironment prominently include monocytes and neutrophils (sometimes loosely grouped as myeloid-derived suppressor cells), macrophages and dendritic cells. While intratumoral myeloid populations, as a whole, have long been considered non-stimulatory or suppressive, it has only recently been appreciated that not all tumor-infiltrating myeloid cells are made equal. Because of advances in high-dimensional flow cytometry as well as more robust transcriptional profiling, we now also understand that the subsets of the tumor-myeloid compartment are far more diverse and notably even contain a rare population of stimulatory dendritic cells. As all of these myeloid populations represent major T-cell interacting partners for incoming tumor-reactive cytotoxic T lymphocytes, understanding the distinctions in their lineage and function reveals and guides numerous therapeutic avenues targeting these antigen-presenting cells. In this Cancer Immunology at the Crossroads overview, we review the recent progress in this rapidly evolving field and advance the hypothesis that the antigen-presenting compartment within tumor microenvironments may contain significant numbers of potent allies to be leveraged for immune-based tumor clearance.
Introduction
While tumor inflammation and tumor-mediated immune evasion have only recently been accepted as ‘Hallmarks of Cancer’ the relationship between inflammatory infiltrates and malignancy has been longstanding (1). In particular, myeloid cell expansion and extramedullary hematopoiesis have been observed as a characteristic of cancer progression since the early 1900’s (2). Furthermore, modulation of immune-cell function for therapeutic benefit dates back at least to ‘Coley’s toxins’ in the 1890’s (3). Today, it is evident that immunity plays critical roles in preventing tumor outgrowth; yet, tumor-mediated immunosuppressive mechanisms also promote malignant tumor survival (1). Understanding the balance between tumor elimination and tumor escape relies on a clear comprehension of the differential roles inflammatory infiltrates play in the tumor microenvironment (TME).
Of the many tumor-infiltrating immune-cell populations, myeloid cells constitute a major proportion. While a heterogeneous mix, these can be subdivided as granulocytes (especially neutrophils but occasionally and less numerous basophils and mast cells), monocytes, macrophages, and dendritic cells (DC) (4). In normal tissues, many of these cells are essential for proper functioning of both innate and adaptive immunity and notably for wound-repair. However in the setting of cancer, a significant excess of macrophages and dysfunctional or skewed populations of these and other cell types are commonly described. Macrophages in particular are known to be important, even outside the immune spectrum, insofar as tumor-associated macrophages (TAM) have been shown to promote tumorigenesis by multiple mechanisms including the release of angiogenic factors and matrix metalloproteases (MMP) (5). When considered as an aggregate population defined by single markers such as CD68 or CD163, ‘macrophage’ infiltration is correlated with worse outcomes in patients across multiple tumor types (6–9).
The precursors to many of the tumor-myeloid populations, including macrophages, are typically blood monocytes. Upon entry into a tumor, they undergo initially limited differentiation and may reside as immature or partially mature monocytes (10). Partially matured monocytes are found in other tissues and may play distinct roles in this state or may serve as a rapidly mobilized reservoir for macrophages and inflammatory dendritic cells (11,12). In mouse tumor biology, a heterogeneous population of monocytes and neutrophils of varying degrees of differentiation have been frequently grouped together and termed myeloid-derived suppressor cells (MDSC). This general categorization is coarsely defined by a positive stain with the broad myeloid marker CD11b and with the Gr-1 antibody clone (RB6-8C5), which binds to both the Ly6c and Ly6g antigens. These latter two markers, when co-expressed with CD11b, are better understood in hematology to simply define monocytes and neutrophils. In some cases, investigators have used Ly6c versus Ly6g to delineate MDSCs as either “monocytic MDSCs” or “neutrophilic MDSCs” although any unique characteristics of these compared to monocytes and neutrophils in other settings remains unclear (13). This collection of cells is commonly studied from spleens of tumor-bearing mice; when isolated and mixed in vitro with T cells and various cytokine mixtures to ‘mature’ them, they demonstrate the ability to suppress T-cell responses. Despite forward progress in this arena few studies have truly addressed how the MDSC populations differ from their steady-state immature myeloid brethren and more research will be necessary to elucidate this difference. Molecular studies have proposed various candidates including the expression of arginase I (Arg1), inducible nitric oxide synthase (iNOS), reactive oxygen species (ROS), and peroxynitrite as the suppressor mechanisms utilized by these cells (14). While the MDSC moniker was initially a useful paradigm for explaining the absence of T-cell reactivity in the TME, it remains to be determined if these are a cell type with in vivo functionality.
In addition to immature myeloid cells, most tumors in mouse and man are characterized by infiltration of mature populations of TAMs, frequently comprising more than 50% of CD45+ stroma. Different functions have been ascribed to macrophages located in different areas of tumors (e.g. at the tumor-stroma border, next to tumor blood vessels, and in hypoxic areas of tumors), suggesting that there are functionally-distinct subsets of these cells (15). Based largely on early in vitro studies skewing monocytes with IL4, it has also become common to delineate macrophages that express arginase and CD206 as ‘M2’ or ‘alternatively activated’ and the tissue equivalents of these are suggested to be involved in injury repair and skewing of T-cell responses to Th2 (IL4, IL10) production (16). Indeed, by the criterion of CD206 expression, these macrophages are very common in many tumors. A contrasting in vitro-derived lineage, differentiated with IFN γ and lipopolysaccharide (LPS), has been termed ‘M1’ or ‘classically activated’ (17). In most studies in tissues, these distinctions are not obviously applicable and macrophage polarization beyond a certain point may be better conceived as a continuous spectrum of activation states, variably able to access multiple effector pathways. Consequently, both designations, M1 and M2, may be attempting to binarize gene-expression into two bins where the diversity of gene-expression across myeloid cells means that this simply is not universally appropriate, at least using these particular markers (18). Regardless, TAMs in aggregate have been shown to be key players in promoting angiogenesis and facilitating tumor-cell invasion and metastasis and the M1/M2 distinction for TAMs, like the MDSC nomenclature for monocytes/neutrophils, is beginning to yield to a less simplistic explanation (15).
The careful phenotypic and functional subsetting of TAMs from the TME is complicated by the similarity of macrophages and DCs; this is a pervasive problem across immunobiology but may be most problematic in tumor biology, wherein multiple papers describe what are seemingly the same cells but by different names. A morphologic criterion has been often applied to the issue; one approach to try to differentiate DCs from macrophages was based on a more spikey or dendritic morphology for the former and more veiled or bulbous morphology for the latter (19). In one case, our laboratory used a similar criterion, along with the presence of the frequently used DC-associated marker CD11c, to define a population very closely associated with the borders of tumor as ‘marginating dendritic cells’ (20). In that work, we noted that others could equally call these cells macrophages on the basis of their expression of the frequently macrophage-associated marker F4/80 and indeed, following from careful phenotyping and RNA-sequencing, we now favor doing so for most of those particular cells (21). In general, given their similarities, it is often the case that DCs and macrophage assignments are best made by larger collections of lineage- and gene expression-based criteria.
Parsing of Tumor-Antigen Presenting Cells
Regardless of what actual name one uses, the reality is that there is diversity in the antigen-presenting compartment within tumors. This diversity is a critically important issue and is not simply semantics. T cells can tell the difference between different features of antigen-presenting cells (APC), and since it is understood that T cells are a major driver of tumor immunity, understanding the exact features of their cognate APC is quite important. Particularly in light of the recent advances in therapies specifically targeting T-cell activation in the TME, understanding the APC populations that interface with T cells and in what manner do they interact is crucial to achieving the most effective treatment (22).
The problems facing tumor biologist regarding this APC lineage is not a unique one. However, recent advances in the study of the mononuclear phagocytic system (MPS) in healthy tissues, notably those undertaken by the Immunological Genome Project (ImmGen), have helped to define surface markers and especially gene-expression profiles that more clearly delineate DC, macrophages, and monocytes (23,24). While CD11c, CD11b and Gr-1 markers broadly label tumor-myeloid populations and at one time were useful in tumor tissue-section staining, they unfortunately fail, at least by themselves, to fully distinguish one cell type from another. There are broadly five ways by which the antigen-presenting compartment can now be identified and parsed.
Methods to Identify and Classify Intratumoral APCs
Flow Cytometry
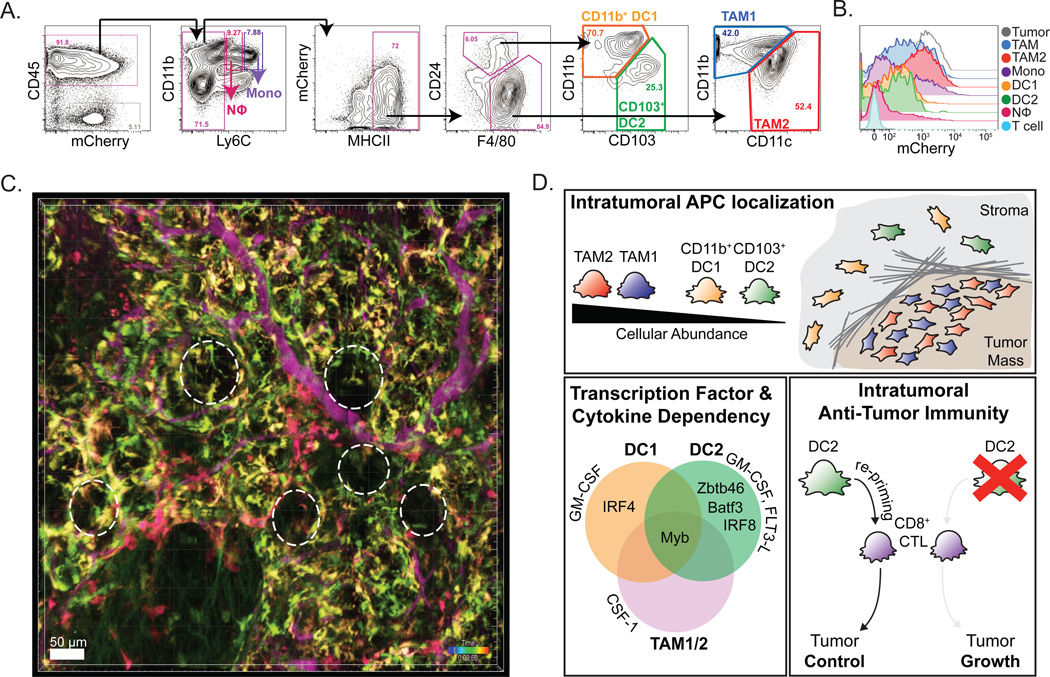
Using a 12-color flow panel of conventional surface markers we now routinely separate tumor-infiltrating myeloid cells across multiple mouse tumor models. By such analysis, neutrophils, monocytes, two subsets of infiltrating macrophages and two rare subsets of conventional tissue-resident DCs can be identified in multiple tumors. Importantly, while many have used CD11c alone as a marker of DCs, we show the need for multiple cell-surface markers including CD24, CD11b and CD103, to fully separate the DC lineage from the abundant tumor macrophages, which similarly express high levels of CD11c. Critically, for full myeloid population delineation, 9 conventional surface markers are required: CD45, CD11b, CD11c, Ly6C, CD24, CD90.2/1, MHCII, F4/80, and CD103. By this approach, two subsets of TAMs, defined as MHCII+ F4/80+, can be identified by modest but consistent differential CD11c, CD11b, and MHCII expression (Figure 1A). We refer to CD11clow CD11bhigh cells as TAM1 and CD11chigh CD11blow as TAM2, whereas others have suggested that TAM2s might be more tumor-specific and propose to call TAM1s tissue macrophages (25). However, it remains to be determined whether these macrophage populations are in fact tumor-specific, and indeed to determine how tumor macrophages deviate from normal macrophages. This flow panel also distinguishes two conventional DC (cDC) subsets, which are MHCII+ CD24+ F4/80− but can be split by the expression of CD11b and CD103. Similarly, infiltrating neutrophils and monocytes can be identified by differential CD11b, Ly6C and CD24 expression. This pattern of markers parses the previous CD11b+ Gr-1+ compartment into 6 discrete myeloid subsets across multiple mouse tumor models.
Figure 1. Parsing the Tumor Antigen-Presenting Cell Compartment.
(A) Flow cytometry and gating of tumor APC populations from digested and CD45-enriched PyMTchOVA tumor. Use of 9 different fluorophores to parse the myeloid compartment, revealing Monocytes (purple), Neutrophils (pink), CD11b+ DC (orange), CD103+ DC (green), TAM1 (blue) and TAM2 (red) populations (B) Histogram of tumor-derived mCherry fluorescence by tumor- infiltrating immune cells in a PyMTchOVA tumor, as a measure of endogenous tumor-antigen uptake. (C) Intravital 2-photon representative still image of an early carcinoma lesion from a PyMTchOVA × Cx3cr1-eGFP × Cd11c-mCherry reporter. Tumoral lesions indicated with dashed lines. Blood vessels are labeled (purple) with Evan’s Blue. Red (mCherry only cells = CD11b+ and CD103+ DC), yellow (mCherry and GFP double- positive cells = TAM2), and green (GFP only cells= TAM1) are shown. Scale bar = 50µm. (D) Summary cartoon of the tumor APC populations, their lineage factors and importance in CTL re-priming. Panels A and B were adapted from portions of figure 1 in ref. 28.
Importantly, a similar diversity of TAM and DC populations can also be found using a similar array of markers in human tumors, including in metastatic melanoma, breast tumors, and head neck squamous cell carcinomas. Here the corollary TAM subsets can be defined as CD45+ HLA-DR+ CD14+ CD11b+ CD11c+, while the cDC subsets are CD45+ HLA-DR+ CD14−, CD11c+ and either BDCA1+ (CD1c) for the CD11b+ DC counterpart, or BDCA3+ (CD141) for the CD103+ DC counterpart. These latter cells frequently bear the chemokine receptor XCR1, providing an alternative marker for their identification. Here, the precedence for using these specific markers comes in part from studies of normal tissue where these human DC markers in particular have been shown to isolate populations matching those in the mouse (26).
T-cell Stimulation
As revealed by intravital imaging of the TME using methods which we will discuss below, many of these myeloid subsets are active T-cell partners—namely T cells engage them in short- or long-lived contacts. As such, it becomes critical to understand the differential capacity of each population to induce, and sustain T-cell responses (20,27). This is important as the dogma of an “immunosuppressive tumor microenvironment” too easily suggests that all T cell-APC reactions are equally suppressed. Importantly, when separated from the group, the rare CD103+ DC subset found at the tumor are in fact robust T-cell stimulators, and when observed in situ by imaging are able to make prolonged synapses with T cells (28). This is an issue we will discuss in greater detail below but needless to say, an APC that stimulates intratumoral T cells is much desired when trying to boost T cell-mediated tumor killing.
Tumor-Antigen Loading
As many APCs are actively phagocytic, another approach in their identification and delineation in the TME is differential levels of phagocytic uptake of tumor antigens. This can be measured either by in vivo or ex vivo methods. By genetic labeling of tumor cells with pH-stable fluorescent proteins (FP) such as mCherry, fluorescent tumor-antigen uptake can be tracked to the phagocytosing myeloid compartments, a method we first demonstrated for islet-draining lymph nodes in insulin-GFP mice (20,29). While this method certainly contains the caveat of differential processing and half-life of FPs, it allows for an in vivo snapshot of the populations actively taking up and retaining significant tumor antigens in the TME. By this method, the macrophage, DC, and monocyte subsets can be compared based on tumor-antigen processing. While both TAM subsets contain large amounts of fluorescent tumor materials, cDC subsets are dimmer for this, only a small subset of monocytes phagocytose and no neutrophils take up any tumor antigen (Figure 1B). While, as discussed, this measurement represents both materials ingested and retained, ex vivo dextran uptake assays on the isolated populations reveal similar results, suggesting that macrophages are the best phagocytes, followed by DCs. It remains to be examined whether those monocytes that phagocytose antigens are still immature or may represent a partially differentiated cell, perhaps on its way to become a TAM (11). Of note, only the CD103+ DCs appear to hold the ingested tumor antigen into a neutral-pH intracellular compartment, which is a phenotype associated with cells that are able to cross-present their internalized materials to stimulate CD8 T cells (28,30).
Direct Imaging, Using Lineage-Based Expression of FPs
Following from flow data that initially segregated APC compartments, we took advantage of the observation that TAM1 and TAM2 cells highly express CX3CR1, whereas cDCs do so only weakly or not at all. Thus in a Cx3cr1-GFP mouse, TAM1 and TAM2 are both bright for GFP. In contrast, TAM2 and the DC subsets are positive for CD11c and so are ‘red’ fluorescent in a Cd11c-mCherry genetic strain. As indicated in Figure 1C, this permits TAM1s to be observed as green-only, TAM2 as green-red (yellow) and the two DC subsets to be seen as red-only. Future adoption of Xcr1-Venus reporters or antibodies should allow complete spatial un-mixing of these cells within mouse tumor tissue. Similar, multi-spectral antibody labeling is likely also possible for human patient biopsies and may assist in better understanding the spatial distribution of these cells within tumors.
Lineage-Specific Cytokines and Transcription Factors for Intratumoral APCs
In many healthy tissue sites, specific transcription factors (TF) and cytokines are now also understood to drive different myeloid subsets. Myeloid differentiation is largely controlled by hematopoietic growth factors including CSF-1 (M-CSF), CSF-2 (GM-CSF), CSF-3 (G-CSF) and FLT3-L, and differential reliance on cytokines can similarly distinguish tumor-myeloid populations (31,32). We and others have variously used the understanding of the specificity of these growth factors for specific lineages as another way to parse myeloid lineages within the TME, with results that are in accord with flow-cytometry-based nomenclature (28,33,34). While cytokine antibody blockade studies demonstrated that TAM subsets uniquely require CSF-1, cytokine receptor knockout models revealed intratumoral neutrophils strictly require G-CSF, and intratumoral cDCs, especially the CD11b+ subset, need GM-CSF to populate the tumor. Tumor models overexpressing cytokines such as GM-CSF or FLT3-L have revealed the sufficiency of these cytokines to support and enhance the rare conventional DC subsets, CD11b+ and CD103+ DCs, respectively, at the tumor (28).
Similarly, discrete transcription factors can segregate these populations and distinguish DC and macrophage subsets (24). Specifically, lymphoid tissue CD8α+ DCs and peripheral tissue CD103+ DC lineages in healthy tissue have been defined by reliance on BATF3 and IRF8, while peripheral CD11b+ cDC lineages, especially in the skin have been shown to require IRF4 (35–38). Additionally, classical DC and their progenitors have been defined by their reliance on the TF ZBTB46 (39,40). Much has also been elucidated regarding macrophage subsets in healthy tissue, defining differential requirement of the TF c-Myb which separates yolk-sac-derived tissue-resident macrophages from bone marrow hematopoietic stem cell (HSC)-derived macrophages (41). First, expression of these different TFs in populations either by transcript as measured by qRT-PCR, or protein as measured by intracellular flow cytometry, can begin to parse lineage differences between populations. This reveals high expression of c-Myb by all tumor-myeloid populations, confirming HSC derivation. Conversely, while both cDC subsets express TF ZBTB46, only CD103+ DCs express IRF8 and CD11b+ DCs uniquely express high levels of IRF4. Genetic ablation models used in conjunction with various tumor models reveal that these TFs are in fact required for their respective population. Such analysis confirms a reliance on BATF3 and IRF8 for CD103+ DC; the CD11b+ DC subset partially requires IRF4 (Figure 1D). The incomplete reliance of the tumoral CD11b+ DC subset on IRF4 suggests even further heterogeneity within this lineage. These tumoral CD11b+ DCs may in fact be split among the PD-L2+ IRF4-dependent CD11b+ DCs and inflammatory DC-SIGN+ monocyte-derived CD11b+ DC lineages, however this remains to be determined (36,42).
Thus, through the methods of flow cytometry, T-cell activation, phagocytic capacity and lineage requirements, not only can the tumor-myeloid compartment be extensively parsed, but discrete functional roles can be tied to this identity revealing unique and rare populations of previously unappreciated myeloid subsets at the tumor (Figure 1D).
Tumors Themselves are Poor Stimulators for cytotoxic T lymphocytes (CTL): A Critical Role for Local APC
Tumor reactive CTLs face several barriers to effective killing of tumor cells. While many tumor cells produce tumor antigens, and can even present these antigens in the context of MHC class I/HLA, they are not sufficient APCs to induce CTL-mediated tumor-cell killing. In fact, most tumor cells at least partially down regulate surface MHC class I/HLA expression which, coupled with the selective pressure of ‘immuno-editing’ tumor antigens, and the absence of costimulatory molecule expression, puts them at a deficit for maintaining cytolytic function (43). This highlights the importance of having cross-priming and re-stimulating APCs in the TME for full CTL functionality. However, the most prominent tumor-infiltrating populations with APC potential, TAMs and monocytes, are equally poor T-cell partners, and contain little to no re-stimulation capacity, as discussed (10,20,28).
In contrast, among these immunosuppressive cells, the CD103+ DCs seem to be fully competent in a CTL re-priming role in vitro, and are in fact required for efficient repriming and effective tumor-cell killing by CTLs in vivo. This was demonstrated in a model of adoptive T-cell therapy, in which transferred OVA-specific CTLs eradicate established OVA-expressing tumors. The role of the CD103+ DCs was assessed in the context of the S1P1 receptor antagonist FTY-720, which blocks T-cell egress from the lymph node (LN), to eliminate any effect of LN priming. Using a Diphtheria Toxin Receptor ablation model (ZBTB46-DTR), which specifically ablates the CD103+DCs, in conjunction with T-cell therapy, revealed a critical role for this population in effective CTL-mediated tumor rejection (28). These results suggest that among the many poor stimulators for T cells in the TME, CD103+ DCs are instead robust T-cell partners for CTLs, promoting re-priming as a local mechanism of functional CTL tumor-cell killing. This result is consistent with previous studies of BATF3 knockout mice, lacking all cells of the CD103+DC lineage in all organs, which showed decreased spontaneous CTL activity and rejection of spontaneously regressing tumors (44,45). The interesting nuance of this finding is that it shows that the TME is not necessarily self-consistent; T-cell inhibitory APCs appear to coincide there with T-cell activating APCs.
Competition amongst APCs: Role of Stoichiometry and Location
Understanding the TME as a mixture of cues for T cells and figuring out which ones are available and in what abundance represents a next major challenge. While clearly available for T-cell priming, CD103+ DCs are both stoichiometrically and spatially at a disadvantage in the TME. In mouse models, total TAMs typically outnumber CD103+ DCs by approximately 10-fold and similar overabundance is seen in human tumor biopsies. This is probably important and helps to explain why the overall outcome is tolerizing because, based on ex vivo T-cell coupling assays, T cells are not very selective for forming synapses (T-APC couples) with the CD103+ DCs. So when all APCs are present at their normal densities, it may be that most tumor-specific T cells are captured in unproductive conjugates with the abundant TAM subsets. Furthermore, intravital imaging demonstrates that the abundant TAM populations are largely found marginating on tumor lobules and transformed tumor ducts, but the stimulatory DC subsets, instead lie distal to the tumoral lesions in collagen-rich areas (Figure 1C). When all CTLs are quantified during intravital imaging sessions, T-cell interactions with poorly stimulating TAMs dominate.
A working hypothesis emerges that the balance of myeloid APCs in the TME, as well as their location may be a critical factor for the effectiveness of antitumor T-cell responses, as ‘good’ and ‘bad’ APCs compete for T-cell contacts locally. While this has yet to be directly addressed, our group found a strong correlation with outcome in human patients when total tumor RNA from The Cancer Genome Atlas (TCGA) was assessed for transcripts expressed uniquely by ‘good’ CD103+ DCs (the human BDCA3+ DC equivalent), as a ratio to transcripts expressed uniquely by the ‘bad’ TAM and CD11b+ DC subsets (46,47). Remarkably, a significant improvement in hazard ratio was found across 12 different cancer types represented, showing that patients with a higher ratio of ‘good’ to ‘bad’ APC transcripts in tumor tissue, had significantly increased overall survival. Importantly, the ratio as a signature, and not the total expression of CD103+ genes alone, proved to be one of the strongest pro-immune survival signals for this data set (28). This supports a hypothesis that immuno-responsive tumors might be the ones with a relatively high density of this DC subset compared to TAMs in the TME, rather than the determining factor being an absolute density of CD103+ cells alone. Clearly more work needs to be done in this area and it is by no means clear whether this result holds consistent for every human tumor type.
Therapeutic Avenues and Conclusions
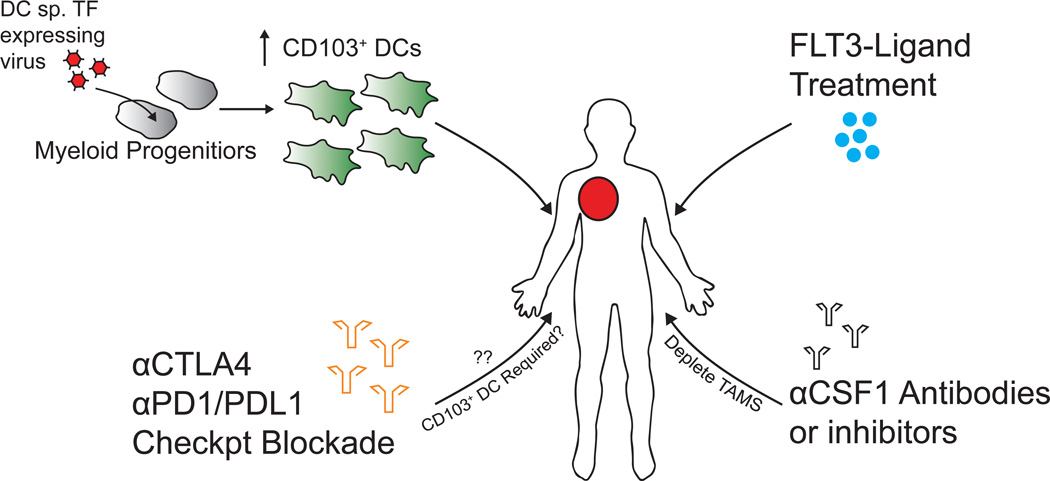
From the multiple studies that have characterized and functionally tested tumor-myeloid subsets, it is becoming exceedingly clear that not all tumor myeloid cells are pro-tumor or immune-inhibitory (25,48). Critically, the identification of the stimulatory CD103+ DCs in both mouse and human tumors has revealed a novel target whereby enhancing the tumoral load of these cells, while possibly limiting TAMs, offers a unique therapeutic axis (Figure 2). By understanding the lineage requirements of this population, several approaches can be used to increase the tumoral CD103/BDCA3 frequency. FLT3-L is a critical cytokine for the development of this lineage, and FLT3-L treatment in mice expands CD103+ DC at the tumor, as well as globally throughout the animal, suggesting that FLT3-L treatment may be beneficial in certain settings (28,49,50). In fact, there is currently a phase II clinical trial underway in melanoma, combining radiotherapy and targeted antigen delivery to DCs with Poly I.C. adjuvant either in the presence or absence of FLT3-L administration (51). Similarly, in mouse models, many groups have studied the effect of combining checkpoint blockade therapies with cancer vaccines such as G-VAX and F-VAX (52,53). Here, the efficacy of αCTLA-4 is markedly increased in melanoma models when combined with F-VAX (irradiated, FLT3-L-expressing tumor cells), which is likely acting on the CD103+ DC population at the tumor and suggests a role for these cells in immunotherapy responsiveness. Alternatively, CD103/BDCA3+ DC numbers could be enhanced at the tumor by forced retroviral expression of key transcription factors (e.g. IRF8, BATF3, ZBTB46) in myeloid progenitors to skew cells into this stimulatory lineage. More importantly might be to simultaneously boost CD103/BDCA3+ DC numbers while concomitantly knocking down TAM frequency at the tumor. In fact, the success of recent CSF-1 inhibitors in human trials, which specifically depletes TAMs, may be explained by the resulting ratio of ‘good’ to ‘bad’ myeloid cells, as BDCA3+ DCs are spared under such treatment (54).
Figure 2. Therapeutic Avenues to Increase Tumoral CD103+ DCs.
Schematic diagram of potential methods to increase the abundance of CD103/BDCA3+ DCs at the tumor in order to enhance antitumor T-cell immunity locally.
Ultimately, many new questions are now raised regarding the role of this population in antitumor immunity and in response to therapy. For example, what role does the CD103/BDCA3+ DC population play in checkpoint blockade therapies? Are patients with higher proportions of BDCA3+ DCs more likely to respond to cancer immunotherapies? How is this population changed with radio- and chemo-therapy? Can the tumoral abundance of BDCA3+ DC be used as a diagnostic biomarker during treatment? All of these are relevant question we should now be asking. In conclusion, careful parsing of the tumor-myeloid subsets, particularly the macrophage and DCs, at the tumor has allowed the discovery of rare and targetable myeloid populations. The CD103+ DCs in mouse or BDCA3+ DCs in human tumors display functional relevance for antitumor T-cell responses and patient survival, highlighting how tumor myeloid-cell subsetting has revealed a new cellular axis of immune therapeutic targeting.
Acknowledgements
We thank Mark Headley for critical reading of the manuscript. Supported by National Institutes of Health grants U54 CA163123 and U01 CA141451.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Talmadge JE, Gabrilovich DI. History of myeloid-derived suppressor cells. Nat Rev Cancer. 2013;13:739–752. doi: 10.1038/nrc3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiemann B, Starnes CO. Coley's toxins, tumor necrosis factor and cancer research: a historical perspective. Pharmacol Ther. 1994;64:529–564. doi: 10.1016/0163-7258(94)90023-x. [DOI] [PubMed] [Google Scholar]
- 4.Schmid MC, Varner JA. Myeloid cells in the tumor microenvironment: modulation of tumor angiogenesis and tumor inflammation. J Oncol. 2010;2010:201026. doi: 10.1155/2010/201026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 6.de Visser KE. Spontaneous immune responses to sporadic tumors: tumor-promoting, tumor-protective or both? Cancer Immunol Immunother. 2008;57:1531–1539. doi: 10.1007/s00262-008-0501-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanada T, Nakagawa M, Emoto A, Nomura T, Nasu N, Nomura Y. Prognostic value of tumor-associated macrophage count in human bladder cancer. Int J Urol. 2000;7:263–269. doi: 10.1046/j.1442-2042.2000.00190.x. [DOI] [PubMed] [Google Scholar]
- 8.Yao Y, Kubota T, Sato K, Kitai R. Macrophage infiltration-associated thymidine phosphorylase expression correlates with increased microvessel density and poor prognosis in astrocytic tumors. Clin Cancer Res. 2001;7:4021–4026. [PubMed] [Google Scholar]
- 9.Ruffell B, Au A, Rugo HS, Esserman LJ, Hwang ES, Coussens LM. Leukocyte composition of human breast cancer. Proc Natl Acad Sci U S A. 2012;109:2796–2801. doi: 10.1073/pnas.1104303108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallina G, Dolcetti L, Serafini P, De Santo C, Marigo I, Colombo MP, et al. Tumors induce a subset of inflammatory monocytes with immunosuppressive activity on CD8+ T cells. J Clin Invest. 2006;116:77–90. doi: 10.1172/JCI28828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jakubzick C, Gautier EL, Gibbings SL, Sojka DK, Schlitzer A, Johnson TE, et al. Minimal differentiation of classical monocytes as they survey steady-state tissues and transport antigen to lymph nodes. Immunity. 2013;39:599–610. doi: 10.1016/j.immuni.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zigmond E, Varol C, Farache J, Elmaliah E, Satpathy AT, Friedlander G, et al. Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity. 2012;37:1076–1090. doi: 10.1016/j.immuni.2012.08.026. [DOI] [PubMed] [Google Scholar]
- 13.Youn JI, Gabrilovich DI. The biology of myeloid-derived suppressor cells: the blessing and the curse of morphological and functional heterogeneity. Eur J Immunol. 2010;40:2969–2975. doi: 10.1002/eji.201040895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis CE, Pollard JW. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 2006;66:605–612. doi: 10.1158/0008-5472.CAN-05-4005. [DOI] [PubMed] [Google Scholar]
- 16.Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J Exp Med. 1992;176:287–292. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becker S, Daniel EG. Antagonistic and additive effects of IL-4 and interferon-gamma on human monocytes and macrophages: effects on Fc receptors, HLA-D antigens, and superoxide production. Cell Immunol. 1990;129:351–362. doi: 10.1016/0008-8749(90)90211-9. [DOI] [PubMed] [Google Scholar]
- 18.Xue J, Schmidt SV, Sander J, Draffehn A, Krebs W, Quester I, et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40:274–288. doi: 10.1016/j.immuni.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bell D, Chomarat P, Broyles D, Netto G, Harb GM, Lebecque S, et al. In Breast Carcinoma Tissue, Immature Dendritic Cells Reside within the Tumor, Whereas Mature Dendritic Cells Are Located in Peritumoral Areas. J Exp Med. 1999;190:1417–1426. doi: 10.1084/jem.190.10.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engelhardt JJ, Boldajipour B, Beemiller P, Pandurangi P, Sorensen C, Werb Z, et al. Marginating dendritic cells of the tumor microenvironment cross-present tumor antigens and stably engage tumor-specific T cells. Cancer cell. 2012;21:402–417. doi: 10.1016/j.ccr.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hume DA, Robinson AP, MacPherson GG, Gordon S. The mononuclear phagocyte system of the mouse defined by immunohistochemical localization of antigen F4/80. Relationship between macrophages, Langerhans cells, reticular cells, and dendritic cells in lymphoid and hematopoietic organs. J Exp Med. 1983;158:1522–1536. doi: 10.1084/jem.158.5.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller JC, Brown BD, Shay T, Gautier EL, Jojic V, Cohain A, et al. Deciphering the transcriptional network of the dendritic cell lineage. Nat Immunol. 2012;13:888–899. doi: 10.1038/ni.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franklin RA, Liao W, Sarkar A, Kim MV, Bivona MR, Liu K, et al. The cellular and molecular origin of tumor-associated macrophages. Science. 2014;344:921–925. doi: 10.1126/science.1252510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haniffa M, Shin A, Bigley V, McGovern N, Teo P, See P, et al. Human tissues contain CD141hi cross-presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity. 2012;37:60–73. doi: 10.1016/j.immuni.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boissonnas A, Licata F, Poupel L, Jacquelin S, Fetler L, Krumeich S, et al. CD8+ tumor-infiltrating T cells are trapped in the tumor-dendritic cell network. Neoplasia. 2013;15:85–94. doi: 10.1593/neo.121572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Broz M, Binnewies M, Boldajipour B, Nelson AE, Pollack JL, Erle DJ, et al. Dissecting the Tumor Myeloid Compartment Reveals A Rare Antigen Presenting Critical for T cell Immunity. Cancer Cell. 2014;26:638–652. doi: 10.1016/j.ccell.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang Q, Adams JY, Tooley AJ, Bi M, Fife BT, Serra P, et al. Visualizing regulatory T cell control of autoimmune responses in nonobese diabetic mice. Nat Immunol. 2006;7:83–92. doi: 10.1038/ni1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savina A, Jancic C, Hugues S, Guermonprez P, Vargas P, Moura IC, et al. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell. 2006;126:205–218. doi: 10.1016/j.cell.2006.05.035. [DOI] [PubMed] [Google Scholar]
- 31.Chow A, Brown BD, Merad M. Studying the mononuclear phagocyte system in the molecular age. Nat Rev Immunol. 2011;11:788–798. doi: 10.1038/nri3087. [DOI] [PubMed] [Google Scholar]
- 32.Lavin Y, Merad M. Macrophages: Gatekeepers of Tissue Integrity. Cancer Immunol Res. 2013;1:201–209. doi: 10.1158/2326-6066.CIR-13-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchem JB, Brennan DJ, Knolhoff BL, Belt BA, Zhu Y, Sanford DE, et al. Targeting tumor-infiltrating macrophages decreases tumor-initiating cells, relieves immunosuppression, and improves chemotherapeutic responses. Cancer Res. 2013;73:1128–1141. doi: 10.1158/0008-5472.CAN-12-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruffell B, Chang-Strachan D, Chan V, Rosenbusch A, Ho CM, Pryer N, et al. Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell. 2014;26:623–637. doi: 10.1016/j.ccell.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy KM. Transcriptional control of dendritic cell development. Adv Immunol. 2013;120:239–267. doi: 10.1016/B978-0-12-417028-5.00009-0. [DOI] [PubMed] [Google Scholar]
- 36.Gao Y, Nish SA, Jiang R, Hou L, Licona-Limón P, Weinstein JS, et al. Control of T helper 2 responses by transcription factor IRF4-dependent dendritic cells. Immunity. 2013;39:722–732. doi: 10.1016/j.immuni.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams JW, Tjota MY, Clay BS, Vander Lugt B, Bandukwala HS, Hrusch CL, et al. Transcription factor IRF4 drives dendritic cells to promote Th2 differentiation. Nat Commun. 2013;4:2990. doi: 10.1038/ncomms3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlitzer A, McGovern N, Teo P, Zelante T, Atarashi K, Low D, et al. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity. 2013;38:970–983. doi: 10.1016/j.immuni.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meredith MM, Liu K, Darrasse-Jeze G, Kamphorst AO, Schreiber HA, Guermonprez P, et al. Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J Exp Med. 2012;209:1153–1165. doi: 10.1084/jem.20112675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Satpathy AT, KC W, Albring JC, Edelson BT, Kretzer NM, Bhattacharya D, et al. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J Exp Med. 2012;209:1135–1152. doi: 10.1084/jem.20120030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 42.Cheong C, Matos I, Choi JH, Dandamudi DB, Shrestha E, Longhi MP, et al. Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209(+) dendritic cells for immune T cell areas. Cell. 2010;143:416–429. doi: 10.1016/j.cell.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krummel MF, Heath WR, Allison JP. Differential coupling of second signals for cytotoxicity and proliferation in CD8+ T cell effectors: Amplification of the lytic potential by B7. J Immunol. 1999;163:2999–3006. [PubMed] [Google Scholar]
- 44.Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fuertes MB, Kacha AK, Kline J, Woo SR, Kranz DM, Murphy KM, et al. Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8{alpha}+ dendritic cells. J Exp Med. 2011;208:2005–2016. doi: 10.1084/jem.20101159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, et al. Multiplatform Analysis of 12 Cancer Types Reveals Molecular Classification within and across Tissues of Origin. Cell. 2014;158:929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruffell B, Chang-Strachan D, Chan V, Rosenbusch A, Ho CM, Pryer N, et al. Macrophage IL-10 Blocks CD8(+) T Cell-Dependent Responses to Chemotherapy by Suppressing IL-12 Expression in Intratumoral Dendritic Cells. Cancer Cell. 2014;26:623–637. doi: 10.1016/j.ccell.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karsunky H, Merad M, Cozzio A, Weissman IL, Manz MG. Flt3 ligand regulates dendritic cell development from Flt3+ lymphoid and myeloid-committed progenitors to Flt3+ dendritic cells in vivo. J Exp Med. 2003;198:305–313. doi: 10.1084/jem.20030323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKenna HJ. Generating a T cell tumor-specific immune response in vivo: can flt3-ligand-generated dendritic cells tip the balance? Cancer Immunol Immunother. 1999;48:281–286. doi: 10.1007/s002620050576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brody J. ClinicalTrials.gov [Internet] Bethesda (MD): National Library of Medicine (US); In Situ Vaccine for Low-Grade Lymphoma: Combination of Intratumoral Flt3L and Poly-ICLC with Low Dose Radiotherpy. 2000-[Dec 2013]. Available from: https://clinicaltrials.gov/ct2/show/NCT01976585: NCT01976585. [Google Scholar]
- 52.Curran MA, Allison JP. Tumor vaccines expressing flt3 ligand synergize with ctla-4 blockade to reject preimplanted tumors. Cancer Res. 2009;69:7747–7755. doi: 10.1158/0008-5472.CAN-08-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Elsas A, Hurwitz AA, Allison JP. Combination immunotherapy of B16 melanoma using anti-cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) and granulocyte/macrophage colony-stimulating factor (GM-CSF)-producing vaccines induces rejection of subcutaneous and metastatic tumors accompanied by autoimmune depigmentation. J Exp Med. 1999;1890:355–366. doi: 10.1084/jem.190.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ries CH, Cannarile MA, Hoves S, Benz J, Wartha K, Runza V, Rey-Giraud F, et al. Targeting Tumor-Associated Macrophages with Anti-CSF-1R Antibody Reveals a Strategy for Cancer Therapy. Cancer Cell. 2014;25:846–859. doi: 10.1016/j.ccr.2014.05.016. [DOI] [PubMed] [Google Scholar]