Abstract
αβ T lymphocytes sense perturbations in host cellular body components induced by infectious pathogens, oncogenic transformation or chemical or physical damage. Millions-billions of these lymphocytes are generated through T-lineage development in the thymus, each endowed with a clonally-restricted surface T-cell receptor (TCR). An individual TCR has the capacity to recognize a distinct “foreign” peptide among the myriad of antigens that the mammalian host must be capable of detecting. TCRs explicitly distinguish foreign from self peptides bound to major histocompatibility complex (MHC) molecules. This is a daunting challenge, given that the MHC-linked peptidome consists of thousands of distinct peptides with a relevant non-self target antigen often embedded at low number, among orders of magnitude higher frequency self-peptides. In this Masters of Immunology article, I shall review how TCR structure and attendant mechanobiology involving non-linear responses impact sensitivity as well as specificity to meet this requirement. Assessment of human tumor-cell display using state of the art mass spectrometry physical detection methods that quantify epitope copy number can help inform as to requisite T-cell functional avidity affording protection and/or therapeutic immunity. Future rational CD8 cytotoxic T cell-based vaccines may follow, targeting virally-induced cancers, other non-viral immunogenic tumors, and potentially even non-immunogenic tumors whose peptide display can be purposely altered by MHC-binding drugs to stimulate immune attack.
Introduction
Adaptive immunity endows mammals and other jawed vertebrates with precursors of T (thymus-derived) and B (bone marrow-derived) lymphocytes able to generate a repertoire of clonotypic antigen receptors (TCR and BCR) of immense diversity from somatic rearrangements of variable gene segments (VDJ and VJ recombination). Spatio-temporally controlled differentiation and selection processes of those cells shape two complementary lineages of the immune system, offering protection with exquisite specificity, sensitivity and long-term memory.
Key discoveries during the last 50 years have unraveled the cellular and molecular nature of adaptive immunity. In the 1960s, T and B lymphocytes were identified and their interactions shown to be essential for antibody production (1, 2). The basic paradigm of immunoglobulin (Ig) gene rearrangements that generate antibody diversity was revealed in 1976 (3). The "dual" specificity of T cells for a foreign-peptide and a self-major histocompatibility complex (MHC) molecule by functional studies was discovered and clearly noted to be distinct from the "single" specificity of antibody recognition of foreign proteins (4, 5). This realization then led to an intense effort to understand the molecular puzzle represented by self versus non-self discrimination and the receptor and ancillary molecules on T cells responsible for this unusual recognition.
The discovery of how to expand T cells in vitro via IL2-dependent T-cell cloning (6), in conjunction with monoclonal antibody (7) and flow cytometry screening (8) technologies plus in vitro functional analyses were decisive in molecular identification for the long sought-after TCR. A key set of advances came in the early 1980s with the identification in human of a clonotypic disulfide-linked heterodimer, the Ti αβ TCR heterodimer, which together with CD3 molecules, were essential for the peptide-MHC (pMHC) recognition and cellular activation (9–14). Biochemical evidence showed that, similar to Ig molecules, both Ti α and β chains possessed variable and constant regions (9, 10). A comparable αβ Ti was also identified by Kappler and Marrack in the mouse with similar cognate immune recognition features (15, 16). Those murine studies supported the earlier conjecture by Allison and colleagues of a potential TCR-related molecule detected on a murine T-cell lymphoma (17). cDNAs for the TCR αβ genes were obtained from the cloning efforts of Davis and Mak (18–20) in mouse and human, respectively, identifying the β chain as shown by protein sequence (21). These studies showed that TCR combinatorial diversity was generated by the same type of site-specific gene recombination mechanisms as with Ig genes, but without somatic hypermutation and led to identification of a second type of TCR, the γδ TCR [reviewed in (3)].
CD4 and CD8, surface molecules identified during the same period, were recognized as co-receptors that optimize TCR recognition and T-cell activation via interaction with monomorphic segments of MHC class II and class I molecules, respectively (22, 23). The “dual recognition” puzzle was solved when it was shown that MHC class I and class II proteins bound foreign and self-peptides derived from degradation of intracellular or exogenous proteins and that such complexes could be recognized by the TCR (24). Three-dimensional structures of peptides complexed with MHC molecules or pMHC were defined (25, 26) as were structures of αβ TCR heterodimers in complex with pMHC ligands (27–30).
Self vs. non-self discrimination is at the core of T-lymphocyte recognition. αβ TCRs ligate foreign peptides bound to self-MHC resulting in specific T-cell activation, proliferation and effector function. In contrast, aside from homeostatic proliferation, self-peptide/self-MHC complexes are ignored and thus, inactivation-inert. This preoccupation with foreign epitopes by αβ T cells is generated in the thymus where self-reactive thymocytes are deleted by an apoptotic negative selection mechanism (31, and reviewed in ref. 32). Any residual self-reactive T cells that escape deletion are controlled by regulatory T cells in the peripheral lymphoid and tissue compartments (33).
If advances in molecular T-cell immunology can be rationally applied to tumor prevention and immunotherapy through vaccination, then precise identification and targeting of tumor antigens leading to destruction of cancer should be possible. The recent success of chimeric antigen receptors (CAR) using Ig-related ectodomains and TCR signaling cytoplasmic components to induce dramatic tumor remission in otherwise incurable patients supports the validity of T cell-based strategies (34). Eliminating toxicities and cost of personalized gene therapy is a significant advantage of an effective cytotoxic T cell (CTL)-based vaccine immunotherapy that can induce high avidity, tumor-specific CD8 T cells. Fulfilling this promise requires a deep understanding of adaptive T-cell recognition and the nuances of T-cell biology and molecular structure.
The HLA-peptidome challenge
In humans, the MHC molecules are called human leukocyte antigen (HLA) proteins. Different people express versions encoded by different gene variants (alleles). In general, peptides associate with HLA molecules by inserting parts of their amino-acid residues into a set of six binding pockets (termed A–F) in HLA. The structure of these pockets is highly allele-specific, thereby dictating peptide-binding preferences for each HLA molecule.
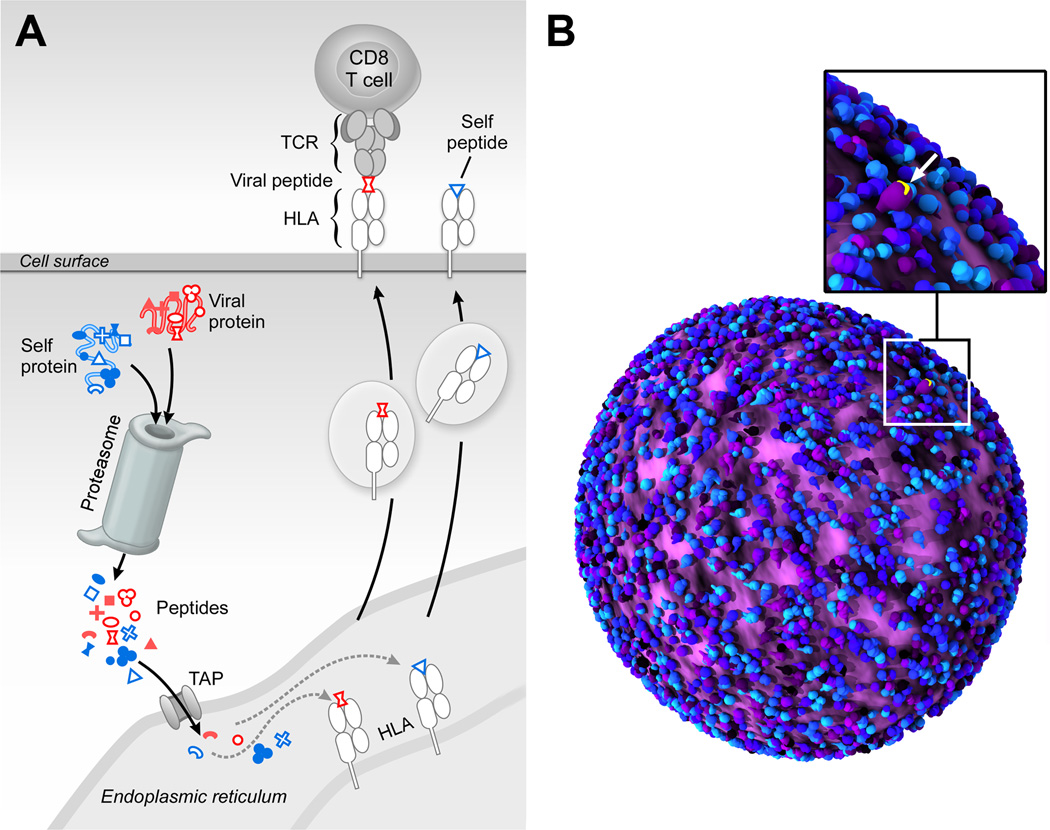
As shown in Fig. 1A, in the cytoplasm of most human cells, self proteins as well as foreign proteins, such as those from a virus that has infected a cell, are cleaved into multiple peptides by a complex called the proteasome. A subset of these peptides is then carried by transporters associated with antigen-processing (TAP) proteins to the endoplasmic reticulum. Here, an even more limited set of peptides are loaded onto HLA molecules, which are transported to the cell surface. These surface-displayed HLA-peptide complexes are recognized by TCRs on the surface of T cells. Specific CD8+ T cells can recognize the infected cells and induce a lytic program that kills these virally-infected targets.
Fig. 1. Processing and presentation of HLA-bound self- and foreign-peptides.
A) Peptides are generated through proteolysis in the proteosome, transported to the endoplasmic reticulum by transporters associated with antigen processing (TAP), associated with HLA molecules therein and then exported and cell surface displayed. B) Artistic rendition of 50,000 to 100,000 peptide-MHC complex (pMHC) molecules on a cell surface (blue/purple) with a single tumor antigen (yellow) among the MHC-bound peptidome, emphasizing the daunting challenge of TCR-based recognition. This figure was rendered by Steve Moskowitz of Advanced Medical Graphics, Boston, MA.
The MHC-bound peptides recognized by T cells are typically 8–11 amino acids in length with single amino acid substitutions readily sensed and discriminated by a TCR. The entire array of MHC-linked peptides is referred to as the HLA peptidome in humans. During immune surveillance, an individual high avidity T cell has the capacity to detect one to several copies of a specific pMHC on the antigen-presenting cell (APC) that expresses 100,000 chemically similar pMHC molecules (35). Discrimination of foreign vs. myriad self-peptides is manifested with precision; if this were not the case, then either autoimmunity or immunodeficiency would result. Fig. 1B shows a tumor-cell surface display with only one tumor antigen surface-expressed among a huge number of self-pMHC molecules. How TCRs are capable of mediating such exquisite specificity and sensitivity was previously a mystery, especially in view of the fact that unlike with B-cell receptors, there is no somatic hypermutation of TCR genes. Monomeric TCR-pMHC affinities are orders of magnitude weaker than high affinity antibody-antigen interactions (36). As described below, however, the elucidation of the TCR as a mechanotransducer with dynamic, non-linear responses explains this behavior.
TCR structure
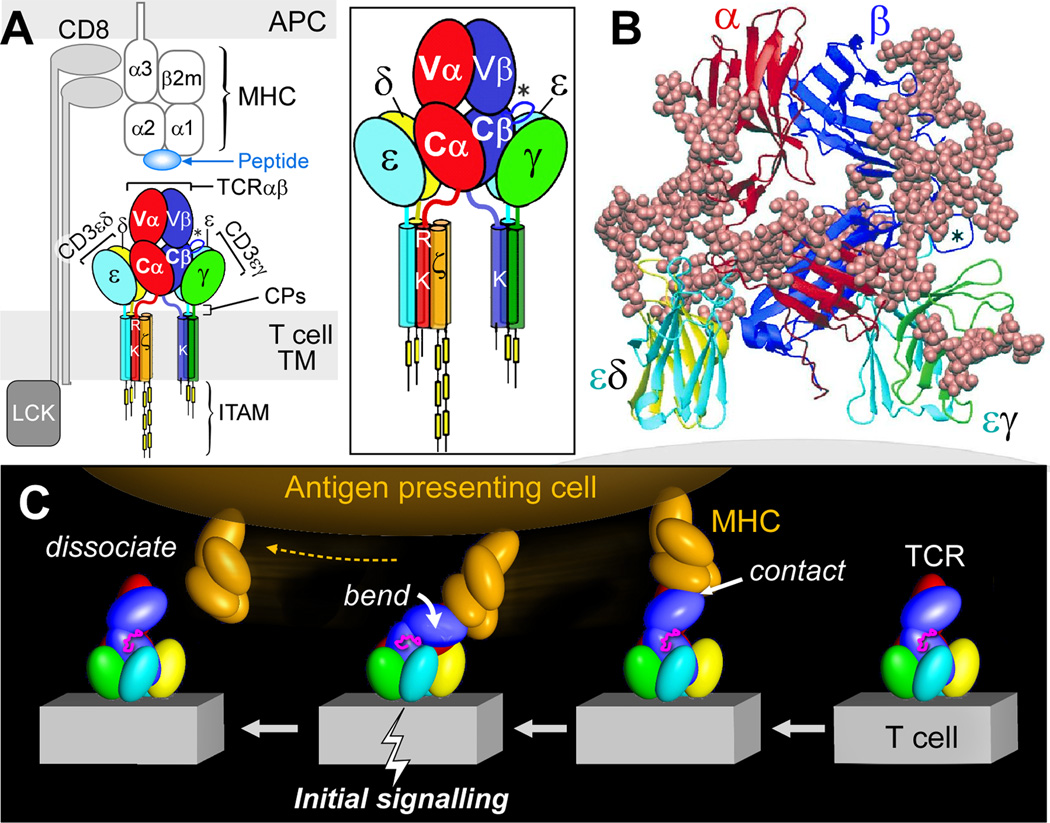
Fig. 2 offers a model of the TCR complex of the αβ heterodimer consisting of V (variable) and C (constant) modules, the CD3εγ and CD3εδ ectodomains (37), defines a plausible subunit topology, and emphasizes its glycan richness. The multiple N-linked glycan adducts of the TCR complex (Fig. 2B) help guide pMHC ligands to the TCR recognition surface, reducing entropic penalties by directing binding to the exposed, glycan-free complementarity-determining region (CDR) loops at the top of the structure. Given that CD3ζ has only a 9 amino acid long ectosegment, it is omitted from Fig. 2 as are the connecting peptides (CP). This rendering incorporates the consequences of several known TCR characteristics: i) putative transmembrane charge pairs involving TCR subunit chain association (Fig. 2A) with CD3ε-CD3δ-TCRα-CD3ζ-CD3ζ as one cluster and CD3ε-CD3γ-TCRβ as a second cluster (38, 39), ii) extracellular domain associations involving other in vitro chain association data (40–42), and iii) proximity of one CD3ε subunit to the TCR Cβ FG loop (designated by an asterisk * in Fig. 2) (43). Evident in Fig 2A-B is the central position of the TCRαβ heterodimer with a vertical dimension of 80Å projecting from the cell membrane, flanked on either side by the shorter (40Å) CD3 heterodimers, CD3εδ on the "left" TCRα side and CD3εγ on the "right" TCRβ side. The width of the CD3εδ and CD3εγ components, 50Å and 55Å, respectively, are comparable in size to that of the TCRαβ heterodimer (58Å), and together (excluding glycans) span ~160Å. These flanking CD3 ectodomain components will likely impede lateral movement of the TCRαβ heterodimer upon pMHC binding. The 5–10 amino acid squat and rigid CD3 CP segments (31, 44) contrast sharply with the long (19–26aa) and flexible TCR α and β CP linking their respective constant domains to the transmembrane (TM) segments (Fig. 2A).
Fig. 2. TCR complex interaction with peptide-MHC complex (pMHC).
A) TCR components (ectodomains, stalk connecting peptides [CP; depicted as red, blue, chartreuse, and turquoise lines], transmembrane [TM] segments and cytoplasmic tails) are labeled and shown in distinct colors. An enlarged view is shown in the box. Note that the single acid residue in each CD3 subunit is omitted here for clarity. The pMHC on the APC and the interacting CD8αβ co-receptor are not colored. In each panel, the Cβ FG loop is depicted by an asterisk (*). B) Lateral view of TCR receptor components in ribbon form (PDB:IFND, 1XM and 1JBJ) oriented above the T-cell membrane (grey). Adducted sugars are depicted in beige in space-filling (CPK) representation. CD3ζζ is absent since it lacks ecotodomains. C) Force on TCR-pMHC interaction initiates signaling (pMHC, orange; Cβ FG loop, magenta; and TCR complex, other colors). This figure was rendered by Steve Moskowitz of Advanced Medical Graphics, Boston, MA. See accompanying supplemental movies 1 and 2.
This contrasting array suggested that the αβ TCR heterodimer may bend and extend relative to the squat and rigid CD3 heterodimers. For example, if as shown in Fig. 2C (from right to left), the pMHC on the APC is first ligated by a specific TCR, then as the T cell continues to move prior to a stop movement signal mediated through inside-out integrin affinity up-regulation, pMHC functions as a force transducing handle to pull on the TCRαβ heterodimer. This force is amplified and exerted on CD3εγ by the lever arm aided by the Cβ FG loop (depicted in magenta) where the TCRβ TM acts as a fulcrum. Supplemental movies 1 and 2 reveal the large forces that result from T-cell immune surveillance of epithelial surfaces and the likely impact of this movement on the TCR complex quaternary conformational changes. Signal transduction involving common structural subunit conformational rearrangement rather than alterations within clonotypic αβ heterodimers per se offers a basic activation mechanism common to all TCRs.
Mechanotransduction
The simple notion that the TCR is a mechanosensor has been codified by several independent studies. Kim and colleagues provided the first direct evidence of the influence of mechanical force in TCR activation (44). Using an optically trapped bead coated with pMHC or anti-CD3 monoclonal antibody for engaging the TCR, T cells were mechanically triggered by applying an oscillating tangential force to the cell surface while monitoring their activation via intracellular calcium flux. Importantly, piconewton (pN) force application with cognate pMHC but not irrelevant pMHC triggered activation. Additional mechanosensor evidence was provided in studies by Li and colleagues using a micropipette to demonstrate shear force associated with activation (45) and Husson and colleagues employing a biomembrane force probe (BFP) to reveal pushing and pulling associated with T-cell triggering (46). Triggering was also shown by Judokusumo and colleagues to depend on substrate stiffness (47). Prediction that a nonlinear mechanical response such as catch-bond formation might facilitate TCR-based recognition (44) was elegantly confirmed by Liu and coworkers using a BFP (48).
Physical forces are generated through T-lymphocyte movement during immune surveillance as well as by cytoskeletal rearrangements at the immunological synapse following cessation of cell migration [(49) and references therein]. Nevertheless, the mechanistic explanation for how TCRs distinguish between foreign- and self-peptides bound to a given MHC molecule has been unclear: peptide residues themselves comprise few of the TCR contacts on the pMHC and pathogen-derived peptides are scant among myriad self-peptides bound to the same MHC class arrayed on infected cells as noted above. Using optical tweezers and DNA tether spacer technology which permit pN force application and nm scale precision, Das and colleagues (49) have determined how bioforces relate to self versus non-self discrimination. Single-molecule analyses involving isolated αβ heterodimers as well as complete TCR complexes on T lymphocytes reveal that the FG loop in the β subunit constant domain (Fig. 2; depicted in magenta) allosterically controls both the variable domain module’s catch-bond lifetime and peptide discrimination via force-driven conformational transition. In contrast to slip-bonds that release under physical load, catch-bonds become stronger so that TCR-pMHC single-bond lifetimes extend. For a representative CD8-derived αβ TCR heterodimer like N15 that binds a rabies family vesicular stomatitis peptide, the bond lifetime goes from 0.3 seconds at zero force to >3 seconds at 15 pN. Such low forces are readily achievable in biological systems.
Ligation of the relevant TCRαβ heterodimer initiates a cascade of T-cell signaling events following exposure of the immunoreceptor tyrosine-based activation motif (ITAM) elements in the cytoplasmic tail of the non-covalently associated subunits (CD3εγ, CD3εδ and CD3ζζ) comprising the TCR complex in 1:1:1:1 dimer stoichiometry. This accessibility allows the active kinase, Lck, to bind and phosphorylate ITAMs followed by recruitment and activation of a second tyrosine kinase, ZAP-70 (50–53). In turn, multiple downstream pathways are engaged including transcriptional regulators controlling activation and differentiation of T cells (54, 55). Thymocyte development is also regulated by the TCR-pMHC interaction as it relates to repertoire selection (reviewed in ref. 32). The extended bond lifetime under force will foster greater subunit conformational alterations and membrane-lipid perturbation and facilitate exposure of sequestered CD8 cytoplasmic tail ITAMs.
Kinetic proofreading has been described as a mechanism permitting biological systems to finely discriminate between ligands which show small differences in their affinity (56, 57). Clearly, the non-equilibrium force-driven TCR-pMHC bond lifetime characteristics noted above allow considerable discrimination between self-pMHC versus foreign-pMHC complexes. The differential ligation kinetics and attendant conformational changes are sufficient to afford major signaling differences between stimulatory and non-stimulatory pMHC ligands. This discrimination is made even more robust in the formation of the signalosome at the immunological synapse. There in the immunological synapse, signaling is influenced by TCR ligation in both time and space, further enhancing discrimination between foreign-pMHC vs. self-pMHC interaction. Explicitly, T-cell triggering might occur as a result of one foreign-pMHC ligand interacting with several TCRs through serial engagement, or alternatively, more than one copy of the same foreign-pMHC interacting with several TCRs in proximity. Given the specificity of the TCR mechanosensing mechanism under force, it is highly improbably that repeated "mistakes" would follow and result in false activation. In conclusion to this section, it is clear that the TCR signaling behavior cannot be readily rationalized in the absence of force. Solution-binding may be strong between a TCR αβ heterodimer and pMHC but cellular activation may be nevertheless absent (58). The way in which a TCR binds to pMHC and the latter is co-ligated by the co-receptor (CD8 in the example shown in Fig. 2A) places restrictions on the permissible TCR-binding orientations to pMHC. TCR and co-receptor bind to the same pMHC, a so-called bidentate interaction, positioning lck associated with the co-receptor to phosphorylate ITAMs of the CD3 cytoplasmic tails
Functional avidity
Given the TCR mechanosensor properties noted above, CTL-targeting need not be dependent on display of a large number of epitopes by single tumor cell or infected cell. The key for protective T-cell effector function is the requisite match between the T cell's functional avidity and the relevant epitope copy number per target cell. Functional avidity refers to the sensitivity of a particular T cell to be triggered by pMHC on an APC (or target) to mediate its function. High avidity T cells are capable of recognizing a very small number of pMHC/target cells whereas low avidity T cells may require hundreds or even thousands (59–61) of such recognitions. Without such relevant avidity, effective cytolysis will not follow. Avidity is dependent on the TCR-pMHC interaction, CD8 co-receptor expression, intracellular signaling molecules and other factors (62). With respect to the TCR, the Vβ and Vα gene repertoires as well as the nature of the antigen itself contribute. Unfeatured pMHCI molecule surfaces, like influenza A M158-66 bound to HLA-A*0201, strongly elicit T cells which, although plentiful, comprise a low avidity immunodominant response (63, 64). As such, display requirements on infected epithelium for activation of those CTLs may be at too high a copy number to be achieved during natural infection. Consequently, this CTL response would be non-protective. Imagine that a high avidity T cell recognizes its target with a 20/20 vision while the low avidity T cell has 20/200, i.e. is legally blind. Identifying correct targets and deploying useful CTLs at a tumor site or nidas of infection is critical. In contrast, if ineffective T cells move into the site, it is counterproductive as T-cell infiltration is of finite magnitude within tissues and impedes deployment of protective CTLs. Moreover, high functional avidity T-cell interactions confer a cellular state refractory to inhibitors like TGFβ (65).
Physical detection and quantification of HLA-bound peptides
Until very recently, the primary approach for peptide identification has been to isolate antigen-experienced T cells from some tissue compartment and demonstrate that these cells functionally "recognize" specific pMHC using an activation-readout such as cytokine production. Beyond technical issues delineated elsewhere (66), this "reverse immunology" identifies only antigenic peptides and not those non-antigenic foreign peptides that are displayed as surface pMHC. However, non-antigenic peptides may be stealth as a consequence of immunodominance of other segments (i.e. M158-66 noted above) but such peptides may be readily capable of generating protective immune responses if they are identified and then used in vaccine formulations. A way around this limitation is the use of physical identification methods, such as mass spectrometry (MS), that have overcome analytical challenges posed by the complex set of peptides bound to MHC as derived from protein metabolism and displayed at the cell surface [(67–69) and reviewed in (66)]. The dynamic range of detection, defined as the target’s fraction of the total ion flux, has been directly estimated and is on the order of 105. The sensitivity of detection compares well with the most sensitive T-cell clone we have generated. A high avidity T cell cannot exceed a limit lower than one target pMHC among 100,000 irrelevant pMHC per target cell. The theory of the method and its use in identifying HPV-16 antigens has been published (67–69). Rapid electronic data capture, high-resolution mass spectrometers and information-rich precursor and ion fragment beams allow deep-targeted interrogation and re-interrogation of precious samples after the sample is acquired and data archived.
An objective for a therapeutic antitumor vaccine is to focus CTLs on HLA-bound peptides restricted to the cancer cell and deploy high avidity CTLs at the tumor site to foster elimination of the cancer. The cellular scale and sensitivity of the aforementioned MS detection methods permits such identification. An example is the set of cancers caused by human papillomavirus 16 (HPV-16). Of note, in cases in which HPV-16 infection has induced epithelial transformation and cervical cancer in HLA-A*0201 hosts, only a single epitope from the E7 oncogene product, E711-19, is naturally processed and presented by this allele on those tumor cells. Pointedly, although E711-20 is capable of binding equivalently to this same HLA allele, the 10-mer peptide, unlike the 9-mer, is not displayed on the primary epithelial cells. In contrast, when a large fragment of E7 as a synthetic peptide is exogenously added to HLA-A*0201 professional APCs, the E711-20 is displayed to a very large extent (67). Since T cells are not strongly crossreactive and are particularly specified to a single peptide length (70, 71), this discordance misguides the immune response. It also explains, in significant part, why the 10-mer vaccine was without clinical effect in a therapeutic HPV-16 cancer trial (72). These data also caution that what is directly presented by tumor cells vs. cross-presented by dendritic cells are not necessarily the same. Quantifying copy number of pMHC complexes per cell has also been implemented with this approach (68), providing insight into CTL functional avidity requirements.
A path for creating CTL-targeting vaccines
Upwards of 20% of tumors worldwide are caused by viruses, with high-risk human papillomaviruses alone responsible for >5% of all cancers (73). A CTL-based approach should prove useful for combating infectious diseases in general and those 20% of cancers caused by viruses in particular. In this regard, there are four components required to create an effective CD8-based T-cell vaccine pipeline: 1) facile bioinformatic prediction of conserved protein segments of pathogens that include potential T-cell epitopes from variable viral strains; 2) physical detection MS methodologies to determine which of the predicted epitopes are actually arrayed on infected cells and APCs for T-cell recognition by TCRs; 3) nanovaccine technology to deliver conserved T-cell epitope payloads including adjuvants to APCs for stimulating epitope display in appropriate lymphoid tissue at optimal density and duration; and 4) insightful memory T-cell biology arising from transcriptomics, proteomics and other molecular analyses of CD8 subsets defining their development and elucidating the rules of physical deployment into tissue compartments. Collectively, these technologies and knowledge will create vaccines that elicit potent CD8 memory T cells with effector function that reside at sites of potential viral attack. Such resident-memory T cells (TRM) are positioned for immediate action and are in turn reinforced by subsequent recruitment of T effector (TEM) and T central memory (TCM) cells from blood and secondary lymphoid tissues (74, 75). In this manner, a prompt immune response is engendered that minimizes viral replication with attendant pathologic consequences.
Of note, in addition to the above MS technologies, computational methods are already available. These offer bioinformatic tools and databases to focus on peptide epitopes conserved among diverse strains of a given virus, predict peptide binding to multiple HLA alleles, and estimate population coverage based on HLA frequencies (76, 77) and references therein]. Vaccine technology using synthetic materials to target organs, tissues or cells and deliver concurrently epitopes and immunomodulatory payloads also exist (78). The above principles can be applied to immunogenic tumors as well as non-immunogenic tumors as described in the section below.
Regulation of immune responses
The effectiveness of adaptive immunity is predicated not only on the nature of cognate antigen recognition via TCR-pMHC interaction but, in addition, the various pathways that suppress T-cell activation. These anti-inflammatory mechanisms involve inhibitory small molecules like adenosine and carbon monoxide, proteins such as CTLA-4 and PD-1 and cells including regulatory T cells [reviewed in (79–81)]. It seems most probable, therefore, that a combination of focused CD8-based vaccine administration in conjunction with immunotherapy to block inhibitory pathways will elicit the greatest antitumor response. Because CD4 T-cell help is important for optimal T-cell function (see an overview in ref. 82), an epitope activating CD4 T cells should also be provided in the vaccination protocol.
Conclusions and future applications
The above MS technology will permit identification of relevant viral epitopes derived from acute infectious pathogens as well as virally-induced cancers. In turn, molecular TCR cloning methods can be used to identify those TCRs on naturally-derived and vaccine-elicited T cells to determine if necessary avidity thresholds have been achieved, either by assessing functional avidity of TCR transfectants and cytokine production readouts or alternatively, single-molecule quantification of TCR-pMHC interactions (49).
For non-viral tumors that are immunogenic, whole exome and/or transcriptome sequencing of individual tumors in conjunction with MS can identify mutant peptides for vaccine formulation in an individualized patient approach. The feasibility of such a strategy has been recently shown in a mouse model (83).
Lastly, for non-immunogenic tumors, induction of the expression of multiple neopeptide epitopes could target a polyclonal CTL attack against a cancer. In this regard, the anti-viral HIV drug abacavir (Ziagen; ViiV Healthcare) binds to one HLA molecule and alters its peptide-binding characteristics, causing the HLA molecule to load and display a new range of peptides on the cell surface. Because this repertoire includes self-peptides that were not displayed during T-cell development, the immune system contains T cells that can recognize these antigens and launch an immune attack against the cell (84). The abacavir interaction is specific to the HLA-B*57:01 allele's F peptide binding pocket but other drugs targeting products of other HLA alleles exist. In principle, these chemicals and medicinal compounds specific for various alleles could be directed in a restricted fashion to tumor cells by monoclonal antibody or other means to alter the tumor peptidome and to target it for CTL destruction. Such specificity would mitigate systemic allergic reactions, focusing only on the tumor. The use of the power and specificity of the immune system is being exploited for tumor immunotherapy with some exciting results. As more details about the adaptive T-cell immune system and its regulation are uncovered, this strategy is likely to rise exponentially.
Supplementary Material
Acknowledgments
EL. Reinherz supported by an SU2C-Farrah Fawcett Foundation Human Papillomavirus (HPV) Translational Cancer Research Team Grant (Grant Number SU2C-AACR-DT13-14). Stand Up To Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research. EL Reinherz also is supported by the National Institutes of Health (grant no. NIH UO1 AI90043).
The author is grateful to all members of the Laboratory of Immunobiology for their efforts and thoughtful insights.
Footnotes
Disclosure:
The CME staff disclosure is as stated.
References
- 1.Miller JFAP, Mitchell GF. Cell to cell interaction in the immune response. I. Hemolysin-forming cells in neonatally thymectomized mice reconstituted with thymus or thoracic duct lymphocytes. J Exp Med. 1968;128:801–820. doi: 10.1084/jem.128.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitchell GF, Miller JFAP. Cell to cell interaction in the immune response. II. The source of hemolysin-forming cells in irradiated mice given bone marrow and thymus or thoracic duct lymphocytes. J Exp Med. 1968;128:821–837. doi: 10.1084/jem.128.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tonegawa S. In: Nobel Lectures: Physiology or Medicine 1981 – 1990. Frångsmyr T, Lindsten J, editors. Singapore: World Scientific Publishing; 1993. pp. 381–405. [Google Scholar]
- 4.Doherty PC. The nobel lectures in immunology. The Nobel prize for physiology or medicine, 1996; Cell mediated immunity in virus infections. Scand J Immunol. 1996;46:527–540. doi: 10.1046/j.1365-3083.1997.d01-170.x. [DOI] [PubMed] [Google Scholar]
- 5.Zinkernagel RM. The nobel lectures in immunology. The Nobel prize for physiology or medicine, 1996; Cellular immune recognition and the biological role of major transplantation antigens. Scand J Immunol. 1997;46:421–436. [PubMed] [Google Scholar]
- 6.Baker PE, Gillis S, Smith KA. Monoclonal cytolytic T-cell lines. J Exp Med. 1979;149:273–278. doi: 10.1084/jem.149.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milstein C. In: Nobel Lectures, Physiology or Medicine 1981 – 1990. Frängsmyr T, Lindsten J, editors. Singapore: World Scientific Publishing; 1993. pp. 248–270. [Google Scholar]
- 8.Julius MH, Masuda T, Herzenberg LA. Demonstration that antigen-binding cells are precursors of antibody-producing cells after purification with a fluorescence-activated cell sorter. Proc Natl Acad Sci USA. 1972;69:1934–1938. doi: 10.1073/pnas.69.7.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Acuto O, Hussey RE, Fitzgerald KA, Protentis JP, Meuer SC, Schlossman SF, et al. The human T cell receptor: appearance in ontogeny and biochemical relationship of alpha and beta subunits on IL-2 dependent clones and T cell tumors. Cell. 1983;34:717–726. doi: 10.1016/0092-8674(83)90528-7. [DOI] [PubMed] [Google Scholar]
- 10.Acuto O, Meuer SC, Hodgdon JC, Schlossman SF, Reinherz EL. Peptide variability exists within alpha and beta subunits of the T cell receptor for antigen. J Exp Med. 1983;158:1368–1273. doi: 10.1084/jem.158.4.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clevers H, Alarcon B, Wileman T, Terhorst C. The T cell receptor/CD3 complex: a dynamic protein ensemble. Annu Rev Immunol. 1988;6:629–662. doi: 10.1146/annurev.iy.06.040188.003213. [DOI] [PubMed] [Google Scholar]
- 12.Meuer SC, Acuto O, Hussey RE, Hodgdon JC, Fitzgerald KA, Schlossman SF, et al. Evidence for the T3-associated 90K heterodimer as the T-cell antigen receptor. Nature. 1983;303:808–810. doi: 10.1038/303808a0. [DOI] [PubMed] [Google Scholar]
- 13.Meuer SC, Fitzgerald KA, Hussey RE, Hodgdon JC, Schlossman SF, Reinherz EL. Clonotypic structures involved in antigen-specific human T cell function. Relationship to the T3 molecular complex. J Exp Med. 1983;157:705–719. doi: 10.1084/jem.157.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reinherz EL, Meuer S, Fitzgerald KA, Hussey RE, Levine H, Schlossman SF. Antigen recognition by human T lymphocytes is linked to surface expression of the T3 molecular complex. Cell. 1982;30:735–743. doi: 10.1016/0092-8674(82)90278-1. [DOI] [PubMed] [Google Scholar]
- 15.Haskins K, Kubo R, White J, Pigeon M, Kappler J, Marrack P. The major histocompatibility complex-restricted antigen receptor on T cells. I. Isolation with a monoclonal antibody. J Exp Med. 1983;157:1149–1169. doi: 10.1084/jem.157.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kappler J, Kubo R, Haskins K, Hannum C, Marrack P, Pigeon M, et al. The major histocompatibility complex-restricted antigen receptor on T cells in mouse and man: identification of constant and variable peptides. Cell. 1983;35:295–302. doi: 10.1016/0092-8674(83)90232-5. [DOI] [PubMed] [Google Scholar]
- 17.Allison JP, McIntyre BW, Bloch D. Tumor-specific antigen of murine T-lymphoma defined with monoclonal antibody. 1982. J Immunol. 2005;174:1144–1151. [PubMed] [Google Scholar]
- 18.Hedrick SM, Cohen DI, Nielsen EA, Davis MM. Isolation of cDNA clones encoding T cell-specific membrane-associated proteins. Nature. 1984;308:149–153. doi: 10.1038/308149a0. [DOI] [PubMed] [Google Scholar]
- 19.Hedrick SM, Nielsen EA, Kavaler J, Cohen DI, Davis MM. Sequence relationships between putative T-cell receptor polypeptides and immunoglobulins. Nature. 1984;308:153–158. doi: 10.1038/308153a0. [DOI] [PubMed] [Google Scholar]
- 20.Yanagi Y, Yoshikai Y, Leggett K, Clark SP, Aleksander I, Mak TW. A human T cell-specific cDNA clone encodes a protein having extensive homology to immunoglobulin chains. Nature. 1984;308:145–149. doi: 10.1038/308145a0. [DOI] [PubMed] [Google Scholar]
- 21.Acuto O, Fabbi M, Smart J, Poole CB, Protentis J, Royer HD, et al. Purification and NH2-terminal amino acid sequencing of the beta subunit of a human T-cell antigen receptor. Proc Natl Acad Sci USA. 1984;81:3851–3855. doi: 10.1073/pnas.81.12.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meuer SC, Schlossman SF, Reinherz EL. Clonal analysis of human cytotoxic T lymphocytes: T4+ and T8+ effector T cells recognize products of different major histocompatibility complex regions. Proc Natl Acad Sci U S A. 1982;79:4395–4399. doi: 10.1073/pnas.79.14.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reinherz EL, Meuer SC, Schlossman SF. The delineation of antigen receptors on human T lymphocytes. Immunol Today. 1983;4:5–8. doi: 10.1016/0167-5699(83)90094-4. [DOI] [PubMed] [Google Scholar]
- 24.Unanue ER. From antigen processing to peptide-MHC binding. Nat Immunol. 2006;7:1277–1279. doi: 10.1038/ni1206-1277. [DOI] [PubMed] [Google Scholar]
- 25.Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature. 1987;329:506–512. doi: 10.1038/329512a0. [DOI] [PubMed] [Google Scholar]
- 26.Brown JH, Jardetzky TS, Gorga JC, Stern LJ, Urban RG, Strominger JL, et al. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature. 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- 27.Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 28.Garcia KC, Degano M, Stanfield RL, Brunmark A, Jackson MR, Peterson PA, et al. An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science. 1996;274:209–219. doi: 10.1126/science.274.5285.209. [DOI] [PubMed] [Google Scholar]
- 29.Reinherz EL, Tan K, Tang L, Kern P, Liu J, Xiong Y, et al. The crystal structure of a T cell receptor in complex with peptide and MHC class II. Science. 1999;286:1913–1921. doi: 10.1126/science.286.5446.1913. [DOI] [PubMed] [Google Scholar]
- 30.Teng MK, Smolyar A, Tse AG, Liu JH, Liu J, Hussey RE, et al. Identification of a common docking topology with substantial variation among different TCR-peptide-MHC complexes. Curr Biol. 1998;8:409–412. doi: 10.1016/s0960-9822(98)70160-5. [DOI] [PubMed] [Google Scholar]
- 31.Touma M, Sun ZY, Clayton LK, Marissen WE, Kruisbeek AM, Wagner G, et al. Importance of the CD3gamma ectodomain terminal beta-strand and membrane proximal stalk in thymic development and receptor assembly. J Immunol. 2007;178:3668–3679. doi: 10.4049/jimmunol.178.6.3668. [DOI] [PubMed] [Google Scholar]
- 32.von Boehmer H. The thymus in immunity and in malignancy. Cancer Immunol Res. 2014;2:592–597. doi: 10.1158/2326-6066.CIR-14-0070. [DOI] [PubMed] [Google Scholar]
- 33.Sakaguchi S. Naturally arising Foxp3-expressing CD25+CD4+ regulatory T cells in immunological tolerance to self and non-self. Nat Immunol. 2005;6:345–352. doi: 10.1038/ni1178. [DOI] [PubMed] [Google Scholar]
- 34.Maus MV, Grupp SA, Porter DL, June CH. Antibody-modified T cells: CARs take the front seat for hematologic malignancies. Blood. 2014;123:2625–2635. doi: 10.1182/blood-2013-11-492231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sykulev Y, Joo M, Vturina I, Tsomides TJ, Eisen HN. Evidence that a single peptide-MHC complex on a target cell can elicit a cytolytic T cell response. Immunity. 1996;4:565–571. doi: 10.1016/s1074-7613(00)80483-5. [DOI] [PubMed] [Google Scholar]
- 36.Kato L, Stanlie A, Begum NA, Kobayashi M, Aida M, Honjo T. An evolutionary view of the mechanism for immune and genome diversity. J Immunol. 2012;188:3559–3566. doi: 10.4049/jimmunol.1102397. [DOI] [PubMed] [Google Scholar]
- 37.Sun ZY, Kim ST, Kim IC, Fahmy A, Reinherz EL, Wagner G. Solution structure of the CD3epsilondelta ectodomain and comparison with CD3epsilongamma as a basis for modeling T cell receptor topology and signaling. Proc Natl Acad Sci U S A. 2004;101:16867–16872. doi: 10.1073/pnas.0407576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Call ME, Pyrdol J, Wiedmann M, Wucherpfennig KW. The organizing principle in the formation of the T cell receptor-CD3 complex. Cell. 2002;111:967–979. doi: 10.1016/s0092-8674(02)01194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Call ME, Pyrdol J, Wucherpfennig KW. Stoichiometry of the T-cell receptor-CD3 complex and key intermediates assembled in the endoplasmic reticulum. EMBO J. 2004;23:2348–2357. doi: 10.1038/sj.emboj.7600245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manolios N, Kemp O, Li ZG. The T cell antigen receptor alpha and beta chains interact via distinct regions with CD3 chains. Eur J Immunol. 1994;24:84–92. doi: 10.1002/eji.1830240114. [DOI] [PubMed] [Google Scholar]
- 41.Manolios N, Letourneur F, Bonifacino JS, Klausner RD. Pairwise, cooperative and inhibitory interactions describe the assembly and probable structure of the T-cell antigen receptor. EMBO J. 1991;10:1643–1651. doi: 10.1002/j.1460-2075.1991.tb07687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koning F, Maloy WL, Coligan JE. The implications of subunit interactions for the structure of the T cell receptor-CD3 complex. Eur J Immunol. 1990;20:299–305. doi: 10.1002/eji.1830200211. [DOI] [PubMed] [Google Scholar]
- 43.Ghendler Y, Smolyar A, Chang HC, Reinherz EL. One of the CD3epsilon subunits within a T cell receptor complex lies in close proximity to the Cbeta FG loop. J Exp Med. 1998;187:1529–1536. doi: 10.1084/jem.187.9.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim ST, Takeuchi K, Sun ZY, Touma M, Castro CE, Fahmy A, et al. The alphabeta T cell receptor is an anisotropic mechanosensor. J Biol Chem. 2009;284:31028–31037. doi: 10.1074/jbc.M109.052712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li YC, Chen BM, Wu PC, Cheng TL, Kao LS, Tao MH, et al. Cutting Edge: mechanical forces acting on T cells immobilized via the TCR complex can trigger TCR signaling. J Immunol. 2010;184:5959–5963. doi: 10.4049/jimmunol.0900775. [DOI] [PubMed] [Google Scholar]
- 46.Husson J, Chemin K, Bohineust A, Hivroz C, Henry N. Force generation upon T cell receptor engagement. PLoS One. 2011;6:e19680. doi: 10.1371/journal.pone.0019680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Judokusumo E, Tabdanov E, Kumari S, Dustin ML, Kam LC. Mechanosensing in T lymphocyte activation. Biophys J. 2012;102:L5–L7. doi: 10.1016/j.bpj.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu B, Chen W, Evavold BD, Zhu C. Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell. 2014;157:357–368. doi: 10.1016/j.cell.2014.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Das DK, Feng Y, Mallis RJ, Li X, Keskin DB, Hussey RE, et al. Force-dependent T cell receptor structural transitions regulate peptide detection. Proc Natl Acad Sci USA. 2015;112:1517–1522. doi: 10.1073/pnas.1424829112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aivazian D, Stern LJ. Phosphorylation of T cell receptor zeta is regulated by a lipid dependent folding transition. Nat Struct Biol. 2000;7:1023–1026. doi: 10.1038/80930. [DOI] [PubMed] [Google Scholar]
- 51.Au-Yeung BB, Deindl S, Hsu LY, Palacios EH, Levin SE, Kuriyan J, et al. The structure, regulation, and function of ZAP-70. Immunol Rev. 2009;228:41–57. doi: 10.1111/j.1600-065X.2008.00753.x. [DOI] [PubMed] [Google Scholar]
- 52.Nika K, Soldani C, Salek M, Paster W, Gray A, Etzensperger R, et al. Constitutively active Lck kinase in T cells drives antigen receptor signal transduction. Immunity. 2010;32:766–777. doi: 10.1016/j.immuni.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu C, Gagnon E, Call ME, Schnell JR, Schwieters CD, Carman CV, et al. Regulation of T cell receptor activation by dynamic membrane binding of the CD3epsilon cytoplasmic tyrosine-based motif. Cell. 2008;135:702–713. doi: 10.1016/j.cell.2008.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol. 2012;12:749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Man K, Miasari M, Shi W, Xin A, Henstridge DC, Preston S, et al. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat Immunol. 2013;14:1155–1165. doi: 10.1038/ni.2710. [DOI] [PubMed] [Google Scholar]
- 56.Hopfield JJ. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci USA. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKeithan TW. Kinetic proofreading in T-cell receptor signal transduction. Proc Natl Acad Sci USA. 1995;92:5042–5046. doi: 10.1073/pnas.92.11.5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adams JJ, Narayanan S, Liu B, Birnbaum ME, Kruse AC, Bowerman NA, et al. T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity. 2011;35:681–693. doi: 10.1016/j.immuni.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alexander-Miller MA, Leggatt GR, Berzofsky JA. Selective expansion of high- or low-avidity cytotoxic T lymphocytes and efficacy for adoptive immunotherapy. Proc Natl Acad Sci USA. 1996;93:4102–4107. doi: 10.1073/pnas.93.9.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sedlik C, Dadaglio G, Saron MF, Deriaud E, Rojas M, Casal SI, et al. In vivo induction of a high-avidity, high-frequency cytotoxic T-lymphocyte response is associated with antiviral protective immunity. J Virol. 2000;74:5769–5775. doi: 10.1128/jvi.74.13.5769-5775.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yee C, Savage PA, Lee PP, Davis MM, Greenberg PD. Isolation of high avidity melanoma-reactive CTL from heterogeneous populations using peptide-MHC tetramers. J Immunol. 1999;162:2227–2234. [PubMed] [Google Scholar]
- 62.von Essen MR, Kongsbak M, Geisler C. Mechanisms behind functional avidity maturation in T cells. Clin Dev Immunol. 2012;2012:163453. doi: 10.1155/2012/163453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tan AC, La Gruta NL, Zeng W, Jackson DC. Precursor frequency and competition dictate the HLA-A2-restricted CD8+ T cell responses to influenza A infection and vaccination in HLA-A2.1 transgenic mice. J Immunol. 2011;187:1895–1902. doi: 10.4049/jimmunol.1100664. [DOI] [PubMed] [Google Scholar]
- 64.Stewart-Jones GB, McMichael AJ, Bell JI, Stuart DI, Jones EY. A structural basis for immunodominant human T cell receptor recognition. Nat Immunol. 2003;4:657–663. doi: 10.1038/ni942. [DOI] [PubMed] [Google Scholar]
- 65.Giroux M, Delisle JS, O'Brien A, Hébert MJ, Perreault C. T cell activation leads to protein kinase C theta-dependent inhibition of TGF-beta signaling. J Immunol. 2010;185:1568–1576. doi: 10.4049/jimmunol.1000137. [DOI] [PubMed] [Google Scholar]
- 66.Reinherz EL, Keskin DB, Reinhold B. Forward vaccinology: CTL targeting based upon physical detection of HLA-bound peptides. Front Immunol. 2014;5:418. doi: 10.3389/fimmu.2014.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Keskin DB, Reinhold B, Lee SY, Zhang GL, Lank S, O'Conno DH, et al. Direct identification of an HPV-16 tumor antigen from cervical cancer biopsy specimens. Front Immunol. 2011;2:1–11. doi: 10.3389/fimmu.2011.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reinhold B, Keskin DB, Reinherz EL. Molecular detection of targeted major histocompatibility complex I-bound peptides using a probabilistic measure and nanospray MS(3) on a hybrid quadrupole-linear ion trap. Anal Chem. 2010;82:9090–9099. doi: 10.1021/ac102387t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Riemer A, Keskin DB, Zhang G, Handley M, Anderson KS, Brusic V, et al. A conserved E7-derived CTL epitope expressed on human papillomavirus-16 transformed HLA-A2+ human epithelial cancers. J Biol Chem. 2010;285:29608–29622. doi: 10.1074/jbc.M110.126722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ekeruche-Makinde J, Miles JJ, van den Berg HA, Skowera A, Cole DK, Dolton G, et al. Peptide length determines the outcome of TCR/peptide-MHCI engagement. Blood. 2013;121:1112–1123. doi: 10.1182/blood-2012-06-437202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Birnbaum ME, Mendoza JL, Sethi DK, Dong S, Glanville J, Dobbins J, et al. Deconstructing the peptide-MHC specificity of T cell recognition. Cell. 2014;157:1073–1087. doi: 10.1016/j.cell.2014.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Driel WJ, Ressing ME, Kenter GG, Brandt RM, Krul EJ, van Rossum AB, et al. Vaccination with HPV16 peptides of patients with advanced cervical carcinoma: clinical evaluation of a phase I-II trial. Eur J Cancer. 1999;35:946–952. doi: 10.1016/s0959-8049(99)00048-9. [DOI] [PubMed] [Google Scholar]
- 73.White MK, Pagano JS, Khalili K. Viruses and human cancers: a long road of discovery of molecular paradigms. Clin Microbiol Rev. 2014;27:463–481. doi: 10.1128/CMR.00124-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ariotti S, Beltman JB, Chodaczek G, Hoekstra ME, van Beek AE, Gomez-Eerland R, et al. Tissue-resident memory CD8+ T cells continuously patrol skin epithelia to quickly recognize local antigen. Proc Natl Acad Sci USA. 2012;109:19739–19744. doi: 10.1073/pnas.1208927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sheridan BS, Lefrancois L. Regional and mucosal memory T cells. Nat Immunol. 2011;12:485–491. doi: 10.1038/ni.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang GL, Deluca DS, Keskin DB, Chitkushev L, Zlateva T, Lund O, et al. MULTIPRED2: A computational system for large-scale identification of peptides predicted to bind to HLA supertypes and alleles. J Immunol Meth. 2011;374:53–61. doi: 10.1016/j.jim.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang GL, Riemer AB, Keskin DB, Chitkushev L, Reinherz EL, Brusic V. HPVdb: a data mining system for knowledge discovery in human papillomavirus with applications in T cell immunology and vaccinology. Database (Oxford) 2014 doi: 10.1093/database/bau031. 2014:bau031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Irvine DJ, Swartz MA, Szeto GL. Engineering synthetic vaccines using cues from natural immunity. Nat Mater. 2013;12:978–990. doi: 10.1038/nmat3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ohta A, Sitkovsky M. Extracellular adenosine-mediated modulation of regulatory T cells. Front Immunol. 2014;5:304. doi: 10.3389/fimmu.2014.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sitkovsky MV, Hatfield S, Abbott R, Belikoff B, Lukasjev D, Ohta A. Hostile, hypoxia-A2-adenosinergic tumor biology as the next barrier to overcome for tumor immunologists. Cancer Immunol Res. 2014;2:598–605. doi: 10.1158/2326-6066.CIR-14-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim H, Cantor H. The path to reactivation of antitumor immunity and checkpoint immunotherapy. Cancer Immunol Res. 2014;2:926–936. doi: 10.1158/2326-6066.CIR-14-0153. [DOI] [PubMed] [Google Scholar]
- 82.Kim HJ, Cantor H. CD4 T-cell subsets and tumor immunity: the helpful and the not-so-helpful. Cancer Immunol Res. 2041;2:1–8. doi: 10.1158/2326-6066.CIR-13-0216. [DOI] [PubMed] [Google Scholar]
- 83.Yadav M, Jhunjhunwala S, Phung QT, Lupardus P, Tanguay J, Bumbaca S, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515:572–576. doi: 10.1038/nature14001. [DOI] [PubMed] [Google Scholar]
- 84.Illing PT, Vivian JP, Dudek NL, Kostenko L, Chen Z, Bharadwaj M, et al. Immune self-reactivity triggered by drug-modified HLA-peptide repertoire. Nature. 2012;486:554–558. doi: 10.1038/nature11147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.