SUMMARY
Gametogenesis is dependent on the expression of germline-specific genes. However, it remains unknown how the germline epigenome is distinctly established from that of somatic lineages. Here we show that genes commonly expressed in somatic lineages and spermatogenesis-progenitor cells undergo repression in a genome-wide manner in late stages of the male germline and identify underlying mechanisms. SCML2, a germline-specific subunit of a Polycomb repressive complex 1 (PRC1), establishes the unique epigenome of the male germline through two distinct antithetical mechanisms. SCML2 works with PRC1 and promotes RNF2-dependent ubiquitination of H2A, thereby marking somatic/progenitor genes on autosomes for repression. Paradoxically, SCML2 also prevents RNF2-dependent ubiquitination of H2A on sex chromosomes during meiosis, thereby enabling unique epigenetic programming of sex chromosomes for male reproduction. Our results reveal divergent mechanisms involving a shared regulator by which the male germline epigenome is distinguished from that of the soma and progenitor cells.
INTRODUCTION
The germline is the only heritable lineage across generations, and it ensures continuity of life. Although biological strategies to specify the germline are diverse among species, suppression of somatic transcriptional programs is a common feature in the germline (Nakamura et al., 2010). In mammals, primordial germ cells are specified during early embryonic development after gastrulation and actively migrate into developing gonads, where they differentiate into spermatozoa or oocytes according to the presence or absence of Y chromosomes (Svingen and Koopman, 2013). During this developmental period, the germline un-dergoes unique epigenetic programing distinct from that in somatic lineages and is naturally reprogrammed in the next generation (Gill et al., 2012; Kota and Feil, 2010; Saitou et al., 2012; Sasaki and Matsui, 2008). However, it remains elusive how the germline epigenome is distinctly established from that of somatic lineages.
A potential candidate is a Polycomb group (PcG)-based mechanism, which regulates epigenetic gene repression and is responsible for stem cell renewal, differentiation, and development (Aloia et al., 2013; Simon and Kingston, 2013). Recent evidence has demonstrated that multiple Polycomb subunits are exchanged to acquire specific functions for different biological contexts (Gao et al., 2012; Tavares et al., 2012). Therefore, if a germline-specific PcG protein exists, it could mediate germ-line-specific functions. In this context, it should be noted that germline genes are repressed by a PcG protein, L(3)mbt, in somatic cells of the fly. Importantly, tumors with a deletion of L(3)mbt exhibit soma-to-germline transformation (Janic et al., 2010). This study suggests that suppression of germline genes is important for tumor suppression in somatic cells, as also proposed in humans (Simpson et al., 2005). It further suggests that a PcG-based mechanism is a critical determinant between soma versus germline transcriptomes. However, it is unknown whether a germline-specific epigenetic silencer exists that suppresses the common features of the somatic program to define the unique epigenome of the germline.
One of the PcG complexes in mammals, Polycomb repressive complex 1 (PRC1), plays a central role during development by suppressing large numbers of genes through mono-ubiquitination of H2A at lysine 119 (H2AK119ub). This is mediated by a PRC1 core subunit, RNF2 (also known as RING1B) (Wang et al., 2004). In female primordial germ cells, PRC1 prevents precocious entry into meiosis and coordinates the timing of sex differentiation (Yokobayashi et al., 2013). It is unknown whether any specific PcG subunits have critical roles during spermatogenesis, the process in which spermatogonia undergo self-renewal, enter meiosis, and differentiate into sperm.
In this study, our unbiased proteomics screen unexpectedly identified Sex comb on midleg-like 2 (SCML2), a homolog of the Drosophila PRC1 subunit Sex comb on midleg (Scm). We demonstrate that SCML2 is a specific and essential epigenetic modifier in the male germline. SCML2 is one of the malignant brain tumor (MBT) domain-containing proteins (Montini et al., 1999), which often function together with PRC1. Two other MBT domain-containing proteins, SCMH1 and SFMBT1, have been suggested to be involved in spermatogenesis (Takada et al., 2007; Zhang et al., 2013). However, Sfmbt1 knockout mice are fertile (Qin et al., 2012), and subfertility of Scmh1 knockout mice is completely rescued by the additional deletion of another PRC1 subunit, PHC2 (Takada et al., 2007). In addition, the deletion of another MBT domain protein, L3MBTL1, does not affect fertility in mice (Qin et al., 2010). These results indicate that SFMBT1, SCMH1, PHC2, and L3MBTL1 are not essential in the male germline. Unlike these proteins, here we show that SCML2 is essential for spermatogenesis and that SCML2 suppresses a large group of genes that are commonly expressed in somatic lineages and in spermatogenesis progenitor cells. Additionally, we demonstrate that, in meiosis, SCML2 is essential for epigenetic programming of sex chromosomes, where unsynapsed sex chromosomes are epigenetically silenced by the action of DNA damage response (DDR) proteins through a process known as meiotic sex chromosome inactivation (MSCI) (Ichijima et al., 2012; Turner, 2007). Based on these two specific functions, SCML2 uniquely defines the specific epigenome of the male germline.
RESULTS
The Somatic/Progenitor Program Is Largely Silenced in the Male Germline
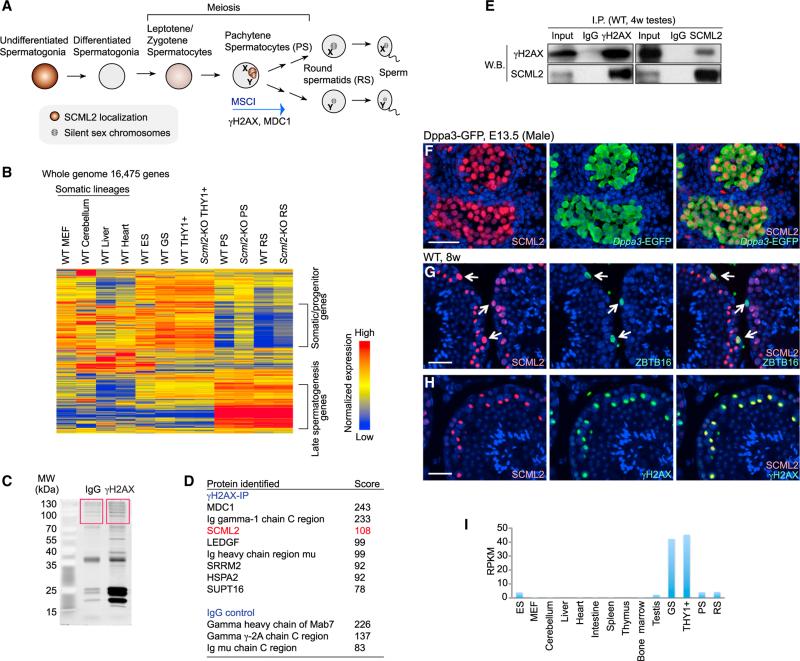
To elucidate unique features of the male germline, we compared the transcriptomes of the spermatogenic cells at different stages and somatic lineages in mice. Specifically, we compared RNA sequencing (RNA-seq) data of germline stem (GS) cells that represent mitotically active undifferentiated spermatogonia, the THY1+ undifferentiated spermatogonia that enrich the stem cell phase, the meiotic spermatocyte at the pachytene stage (pachytene spermatocytes [PS]), and post-meiotic round spermatids (RS) (described in Figure 1A) with those of other cell types by generating a heatmap that included all of the expressed RefSeq genes in the genome of these samples (16,475 genes) (Figure 1B). Importantly, we found that whole-genome transcriptomes in PS and RS were largely different from those of somatic lineages, embryonic stem (ES) cells, and the mitotic phase of germ cells including GS cells and THY1+ cells (Figure 1B). A large group of genes was expressed in somatic lineages, ES cells, GS cells, and the THY1+ fraction, but this group was specifically repressed in PS and RS. We defined this group of genes as somatic/progenitor genes because this group is commonly active in somatic lineages and mitotic phases of spermatogenesis-progenitor cells. In contrast, a large group of late spermatogenesis genes that are associated with male reproduction was specifically expressed in PS and RS. In accordance with this finding, previous reports have demonstrated that germ cells undergo a dynamic transcriptome change during the late stages of spermatogenesis (Chalmel et al., 2007; Khil et al., 2004; Namekawa et al., 2006; Shima et al., 2004). Our present study demonstrates that suppression of the somatic/progenitor program is a unique feature of the late stages of the male germline and also that a specific set of late spermatogenesis genes is activated at these stages.
Figure 1. A Global Transcriptome Change during Late Spermatogenesis and Identification of SCML2.
(A) Schematic of spermatogenesis and summary of SCML2 localization.
(B) A heatmap showing gene expression patterns among several germ cells versus somatic cells. MEF, mouse embryonic fibroblasts. All 16,475 genes that showed more than 3 RPKM in at least one stage are shown.
(C) SYPRO Ruby stained gel of γH2AX containing nucleosomes immunoprecipitated using γH2AX antibody. The boxed areas were subjected to mass spec-trometry-based protein identification.
(D) γH2AX-associated proteins identified by mass spectrometry.
(E) Co-immunoprecipitation of SCML2 and γH2AX using 4-week-old wild-type testes. (F) Immunostaining of SCML2 in a testicular section from a male embryo harboring Dppa3-EGFP transgene at E13.5. Germ cells were detected by anti-EGFP antibody. Nuclei were counterstained with DAPI. The scale bar represents 40 μm.
(G and H) Immunostaining of SCML2, ZBTB16 (G), and γH2AX (H) in wild-type adult testicular sections. Arrows indicate undifferentiated spermatogonia. Nuclei were counterstained with DAPI. The scale bars represent 40 μm.
(I) Expression profiles of Scml2 in different tissues, based on RNA-seq (n = 2). RPKM values (y axis) of Scml2 are shown. See also Figure S1.
Identification of SCML2 as an Essential Factor in the Male Germline
The next key question is whether there are any factors involved in establishing the unique transcriptomes of the late stages of the male germline. Interestingly, a factor we independently identified as a component of meiotic sex chromosomes has a role in shaping transcriptomes in the male germline (described below). In our attempt to identify a germline-specific epigenetic modifier, we focused on MSCI, a unique germline-specific event that is essential to male meiosis, which involves activation of the DDR pathway and deposition of γH2AX on sex chromosomes. Although several DDR proteins are required for MSCI, we reasoned that a germline-specific protein could regulate this process. To test this possibility, we performed immunoprecipitation of nucleosomes containing γH2AX, an essential modification of silent sex chromosomes, combined with mass spectrometry and identified an X chromosome linked protein, SCML2 (Figures 1C and 1D). Consistent with our previous study indicating the interaction of γH2AX and MDC1 on meiotic sex chromosomes (Ichijima et al., 2011), the mass spectrometry score for MDC1 was the highest. The score for SCML2 was the second highest. Subsequent immunoprecipitation, combined with western blotting, confirmed that SCML2 interacts with γH2AX in testicular extracts (Figure 1E).
Immunostaining revealed that the expression of SCML2 commenced in embryonic germ cells after entering the gonad in both males and females (Figures 1F and S1). In adult testes, SCML2 highly accumulated in ZBTB16 (also known as PLZF)-positive undifferentiated spermatogonia, which include the stem cell population (Figure 1G). Later during meiosis, SCML2 slightly accumulated on entire nuclei from the leptotene to the early pachytene stage (Figure S1) and highly accumulated on the sex chromosomes from the early to mid-pachytene stage to the diplotene stage (Figure 1H), consistent with the fact that γH2AX has a role in transcriptionally silencing sex chromosomes and interacts with SCML2. Similar to SCML2 protein localization, Scml2 transcripts were specifically detected in the germline (Figure 1I). Scml2 expression was especially high in GS cells that represent undifferentiated spermatogonia. Paradoxically, SCML2 is transcriptionally silenced by MSCI because of its X linkage (Mueller et al., 2008), despite being localized on the sex chromosomes during meiosis.
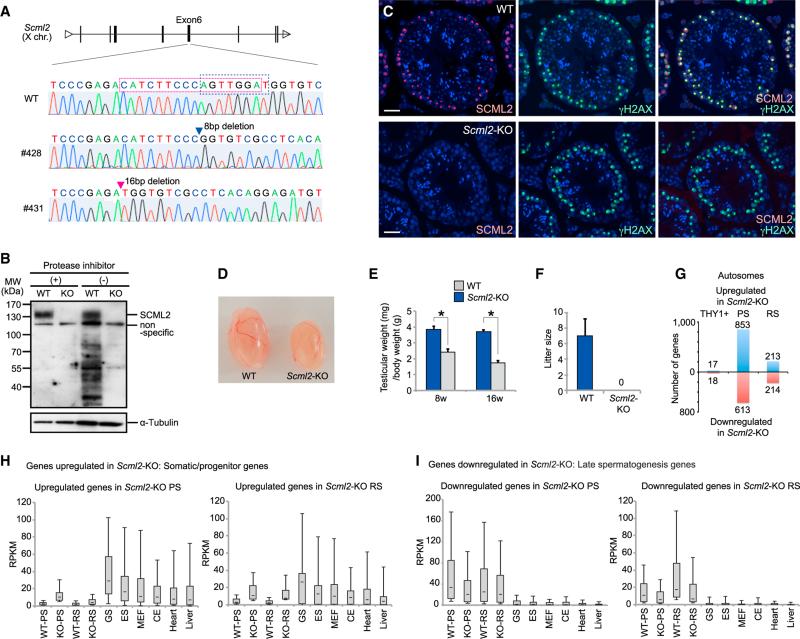
To better understand the function of SCML2, we generated Scml2-knockout (Scml2-KO) mice using zinc finger nuclease technology (Cui et al., 2011). We established two independent females harboring a heterozygous deletion at the X-linked Scml2 locus (Figure 2A). In male progeny possessing either of the mutations, SCML2 was not detectable by western blotting or immunostaining (Figures 2B and 2C), indicating that the mutations nullified SCML2 expression. Thus, we refer hereafter to both mutant lines as Scml2-KO mice. SCML2 was detected at 140 kDa in testicular lysate, although SCML2 is subject to protein degradation if a protease inhibitor is not added (Figure 2B). Scml2-KO males were born at Mendelian ratios and looked healthy (data not shown). However, their testes were smaller than littermate control testes, and Scml2-KO males were not able to impregnate female mice (Figures 2D–2F). Therefore, we conclude that Scml2-KO males are infertile and that SCML2 is essential in spermatogenesis.
Figure 2. SCML2 Regulates Global Silencing of Somatic/Progenitor Genes in the Male Germline.
(A) Genomic sequencing of male Scml2-KO mice, line #428 and line #431.
(B) Western blotting of testicular lysate obtained from wild-type and Scml2-KO mice.
(C) Immunostaining of SCML2 and γH2AX in adult testes. Nuclei were counterstained with DAPI. The scale bar represents 40 μm.
(D) Picture of wild-type and Scml2-KO testes at 8 weeks old.
(E) Testicular weight per body weight. Data are represented as mean ± SD (n = 4).
(F) Litter sizes when wild-type or Scml2-KO males were mated with wild-type females. Data are represented as mean ± SD (n = 6 for wild-type, n = 9 for Scml2-KO).
(G) The number of differentially expressed autosomal genes detected by RNA-seq in THY1+, PS, and RS between wild-type and Scml2-KO cells.
(H and I) Expression profiles of upregulated genes (H) and downregulated genes (I) in Scml2-KO spermatogenic cells across tissues. Expression data were obtained from RNA-seq (n = 2). CE, cerebellum. Distribution of RPKM values (y axis) is shown. The central dot is the median, the boxes encompass 50% of data points, and the error bars indicate 90% of data points.
See also Figure S2.
SCML2 Is Essential for the Global Silencing of Somatic/Progenitor Genes and for Activation of Late-Spermatogenesis Specific Genes
To elucidate gene expression changes in meiotic and post-meiotic stages resulting from SCML2 deletion, we performed RNA-seq using purified THY1+ fraction, PS, and RS from Scml2-KO mice. Because there was no germ cell arrest at a particular stage in Scml2-KO mice despite the occurrence of progressive apoptosis during differentiation in spermatogenesis (as shown below), purified germ cells enabled us to evaluate the gene expression change caused by SCML2 depletion without the secondary effect of developmental delay. Remarkably, the expression patterns of PS and RS were globally changed compared with those of wild-type mice; notably, somatic/progenitor genes were largely derepressed in Scml2-KO PS and RS, although gene expression in the THY1+ fraction was not largely altered (Figure 1B). Hereafter the autosomes and sex chromosomes were analyzed separately because the sex chromosomes are subject to MSCI. To identify specific targets regulated by SCML2, we applied stringent criteria to our RNA-seq data set (padj < 0.05, >2-fold difference, reads per kilobase per million (RPKM) ≥ 5 for higher-expression genes). We identified 853 and 613 auto-somal genes that were significantly up- and downregulated, respectively, in Scml2-KO PS (Figure 2G; gene lists are included in Table S1). In RS, 213 and 214 autosomal genes were significantly up- and downregulated, respectively. On the other hand, only 17 and 28 autosomal genes were up- and downregulated in Scml2-KO THY1+ cells. The genes upregulated in Scml2-KO PS and RS were mainly expressed in somatic lineages and spermatogenesis progenitor cells and strongly suppressed in wild-type PS and RS (Figures 2H and S2). By contrast, downregulated genes in Scml2-KO PS and RS were specifically expressed in PS and RS (Figures 2I and S2). Gene ontology (GO) enrichment analysis further confirmed that the set of genes downregulated in Scml2-KO cells showed enrichment for genes involved in the late stages of spermatogenesis (Figure S2). Taken together, SCML2 is essential for the suppression of somatic/progenitor genes and the activation of late-spermatogenesis-specific genes in PS and RS.
SCML2 Regulates Differentiation of Spermatogenic Cells
To further investigate the role of SCML2, we performed histological analyses. In Scml2-KO testes, massive loss of differentiated germ cells occurred (Figure 3A) and polynucleated cells were observed (Figure 3B). Additionally, we found that elongated spermatids in the Scml2-KO failed to condense (Figures 3C and 3D), and spermatozoa were rarely seen in epididymides (Figure 3E).
Figure 3. SCML2 Regulates Differentiation of Spermatogenic Cells.
(A–C) Histological testicular sections stained with H&E. Polynucleated cells and elongated spermatids and are shown with arrows in (B) and (C), respectively. The scale bars represent 60 μm. Quantitative data of the percentage of tubules containing polynucleated cells are shown on the right in (B). Data are represented as mean ± SD (n = 5).
(D) DAPI staining of elongated spermatids in germ cell slides that maintain chromatin morphology. Because abnormal chromatin was consistently observed in the Scml2-KO elongated spermatids, the quantification was not included. The scale bars represent 10 μm.
(E) Histological section of epididymis stained with H&E. The scale bars represent 60 μm.
(F–K) Immunostaining of ZBTB16, STRA8, and SYCP3 in 8 weeks testes. The scale bars represent 40 μm. Quantitative data are shown in (G), (I), and (K). Data are represented as mean ± SD (n = 4).
(L and M) TUNEL staining using testicular sections. The scale bars represent 40 μm. Quantitative data are shown in (M). Data are represented as mean ± SD (n = 4).
(N) Immunostaining of cleaved caspase-3, γH2AX, and H1T in adult testes. Apoptotic cells are shown with arrows. The scale bars represent 40 μm. *p < 0.05, unpaired t test.
Next, to investigate which cell types are affected by SCML2 deficiency, stage-specific spermatogenic cells were counted. The number of undifferentiated spermatogonia was unchanged (Figures 3F and 3G), suggesting that SCML2 is apparently not involved in the maintenance of spermatogonial stem cells, consistent with minor gene expression change in this population. In contrast, the number of STRA8-positive cells, which includes differentiating spermatogonia and pre-leptotene spermatocytes, as well as SYCP3-positive meiotic spermatocytes, was significantly decreased in the Scml2-KO (Figures 3H–3K). An increase of spermatocytes and spermatids undergoing apoptosis in Scml2-KO testes suggest that the reduction of differentiating cells is caused by apoptosis (Figures 3L–3N). In Scml2-KO testes, apoptosis occurred progressively in each stage following spermatogonia: H1T-negative spermatocytes (before the midpachytene stage), H1T-positive spermatocytes (during and after the mid-pachytene stage), and RS (Figure 3N). Although the decrease of SYCP3-positive meiotic spermatocytes could be due in part to the decrease of differentiated spermatogonia, apoptosis mainly occurred in pre-leptotene spermatocytes (the majority of STRA8-positve cells) and in later stages. Therefore, consistent with the global disruption of transcriptomes of Scml2-KO PS and RS, we conclude that SCML2 regulates the differentiation of spermatogenic cells.
SCML2 Is a Germline-Specific Subunit of PRC1
To elucidate the molecular function of SCML2, we investigated whether SCML2 is a part of PRC1 in spermatogenic cells because a previous proteomics study identified SCML2 as a PRC1 component (Gao et al., 2012). RNF2, a catalytic core component of PRC1, and RNF2-mediated H2AK119ub, which was detected by a rabbit monoclonal antibody (D27C4), were both detected in cells progressing from undifferentiated spermatogonia to spermatocytes (Figure S3). Consistent with the histological data, co-immunoprecipitation experiments revealed that SCML2 interacts with RNF2 in testicular extracts (Figure 4A). Furthermore, chromatin immunoprecipitation (ChIP) sequencing (ChIP-seq) using GS cells, where SCML2 is abundantly expressed, demonstrated that SCML2 frequently bound to PRC1 target genes, such as the Hoxd cluster (Woo et al., 2010) and Htra1 locus, and shared peaks with other PRC1 subunits, RNF2 and BMI1 (Figures 4B and S3). More than three-quarters (75.4% [n = 2,808]) of all SCML2-positive transcription start sites (TSSs; n = 3,723) overlapped with those of RNF2 (Figures 4C and S3), and 44.2% (n = 3,237) of all SCML2-positive islands (n = 7,326) were associated with RNF2-positive islands and enrichment of H2AK119ub throughout the entire genome (Figure 4D). Moreover, similar to other PcG proteins, SCML2 was also enriched at CpG islands, where gene-regulatory elements including promoters and enhancers reside (Ku et al., 2008) (Figure 4D). The ChIP-seq data were reproducible between native ChIP and cross-linking ChIP (Figure S4). Taken together, these results suggest that SCML2 forms a complex with PRC1 and binds to gene-regulatory regions in GS cells. Conversely, 13,771 RNF2-positive islands did not overlap with SCML2-positive islands (such as Tbx5 locus; Figure S3), suggesting that other PRC1 complexes, which lack SCML2, regulate these islands.
Figure 4. SCML2 Interacts with the PRC1 Complex in Germ Cells.
(A) Co-immunoprecipitation of SCML2 and RNF2 using 4-week-old wild-type testes.
(B) Binding peaks of SCML2, RNF2, BMI1, and H2AK119ub across the Hoxd cluster in GS cells.
(C) A Venn diagram showing the numbers of gene promoters bound by RNF2, SCML2, and BMI1 identified by ChIP-seq.
(D) A heatmap showing distribution of CpG, binding peaks of SCML2, RNF2, BMI1, and H2AK119ub in GS cells. Density around islands (±5 kb from the center of islands) is shown.
See also Figure S3.
SCML2 Regulates Establishment of H2AK119ub to Silence Somatic/Progenitor Genes on Autosomes
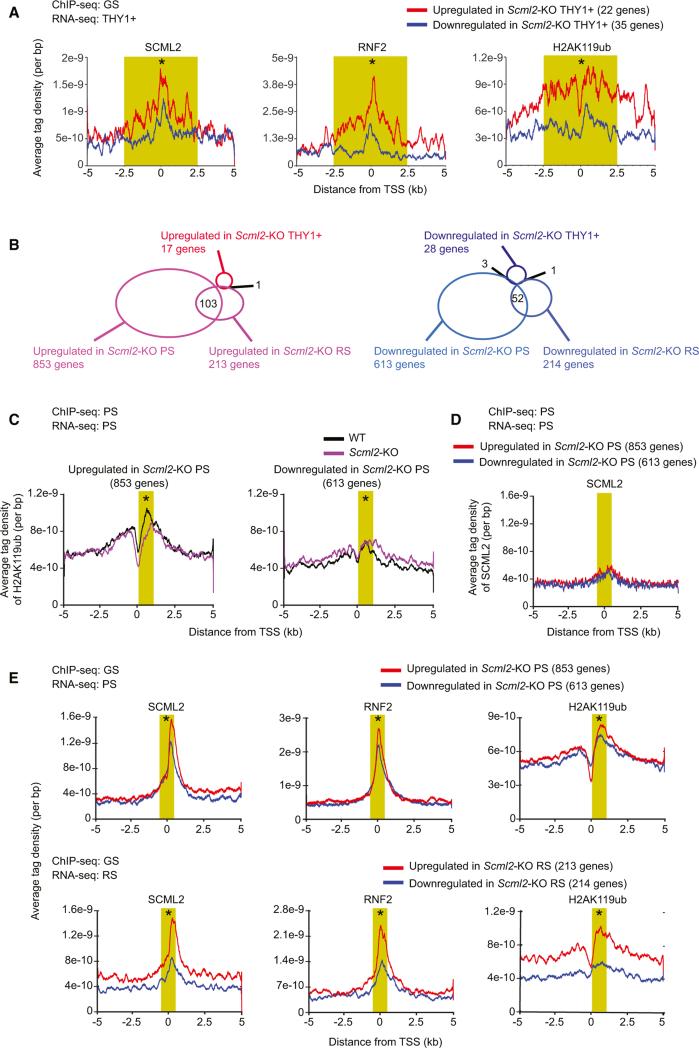
Because SCML2 forms a complex with RNF2 (Figure 4), we reasoned that SCML2 regulates target genes through H2AK119ub. To test this possibility, we first examined whether accumulation of H2AK119ub is associated with SCML2 in undifferentiated spermatogonia. Because we were not able to establish Scml2-KO GS cells, RNA-seq data from a THY1+ fraction of wild-type and Scml2-KO mice were used for this analysis. SCML2, RNF2, and H2AK119ub highly accumulated on the upregulated genes in Scml2-KO THY1+ cells compared with those downregulated (Figure 5A), suggesting that SCML2 regulates establishment of H2AK119ub for gene repression in undifferentiated spermatogonia.
Figure 5. SCML2 Regulates Establishment of H2AK119ub to Suppress Somatic/Progenitor Genes in Spermatogenesis.
(A) Occupancy of H2AK119ub around TSS in GS of the wild-type. ChIP-seq data of GS are shown for indicated genes based on RNA-seq analysis of THY1+ fraction.
(B) Diagrams showing the numbers of genes regulated based on RNA-seq analysis of THY1+ fraction, PS, and RS. Numbers of overlapping genes between two groups are shown in the diagram.
(C) Occupancy of H2AK119ub around TSS in PS of the wild-type and Scml2-KO. ChIP-seq data of PS (wild-type or Scml2-KO) are shown for indicated genes based on RNA-seq analysis of PS.
(D) Occupancy of SCML2 around TSS in wild-type PS. ChIP-seq data of wild-type PS are shown for indicated genes based on RNA-seq analysis of PS.
(E) Occupancy of SCML2, RNF2, and H2AK119ub around TSS in wild-type GS cells. ChIP-seq data of wild-type GS cells are shown for differentially expressed genes based on RNA-seq analysis of PS (top) and RS (bottom). The average tag density within 5 kb from TSS is shown. A Wilcoxon rank sum test was performed for read counts in the highlighted area (A): SCML2, RNF2, H2AK119ub: −2,500 to +2,500 bp from TSS; (C–E): SCML2 and RNF2: −500 to +500 bp from TSS; H2AK119ub: +200 to +1,200 bp from TSS (*p < 0.05).
See also Figure S4.
Next, we sought to determine whether suppression of somatic/progenitor genes that occurs in PS and RS are also mediated through the regulation of H2AK119ub. Consistent with the active transcription of somatic/progenitor genes in the THY1+ fraction, SCML2-regulated genes in PS and RS are largely distinct from those in the THY1+ fraction (Figure 5B). To examine the enrichment of H2AK119ub, we performed ChIP-seq of H2AK119ub in purified PS from the wild-type and the Scml2-KO (Figure 5C). Focusing on somatic/progenitor genes that are upregulated in Scml2-KO PS, H2AK119ub was relatively high around TSSs of such genes in the wild-type, compared with the Scml2-KO (Figure 5C). In contrast, when focusing on genes found downregulated in the Scml2-KO, accumulation of H2AK119ub was not observed around TSSs in either the wild-type or the Scml2-KO (Figure 5C). This result suggests that SCML2 promotes establishment of H2AK119ub for repression of somatic/progenitor genes in late spermatogenesis.
Because SCML2 was highly accumulated on entire nuclei in undifferentiated spermatogonia, but was not on the autosome regions during meiosis (Figures 1G, 1H, and S1), we hypothesized that SCML2 establishes H2AK119ub prior to spermatogenic differentiation. Consistent with our cytological data, in PS, SCML2 is no longer associated with TSSs of somatic/ progenitor genes found upregulated in Scml2-KO PS (Figure 5D). Moreover, in GS cells, SCML2, RNF2, and H2AK119ub were relatively high at TSSs of somatic/progenitor genes that were found to be repressed by SCML2 in the later stages (genes up-regulated in the Scml2-KO PS and RS: Figures 5E and S4). This tendency is evident in genes upregulated in the Scml2-KO RS. These results suggest that, prior to spermatogenic differentiation, SCML2 functions with RNF2 and regulates establishment of H2AK119ub, and that H2AK119ub persists into later stages for repression of somatic/progenitor genes after the removal of SCML2 from their TSSs. Because SCML2 was not especially high around TSSs of genes that were found downregulated in the Scml2-KO, activation of late spermatogenesis genes may be indirectly mediated through SCML2.
SCML2 Suppresses H2AK119ub on Meiotic Sex Chromosomes
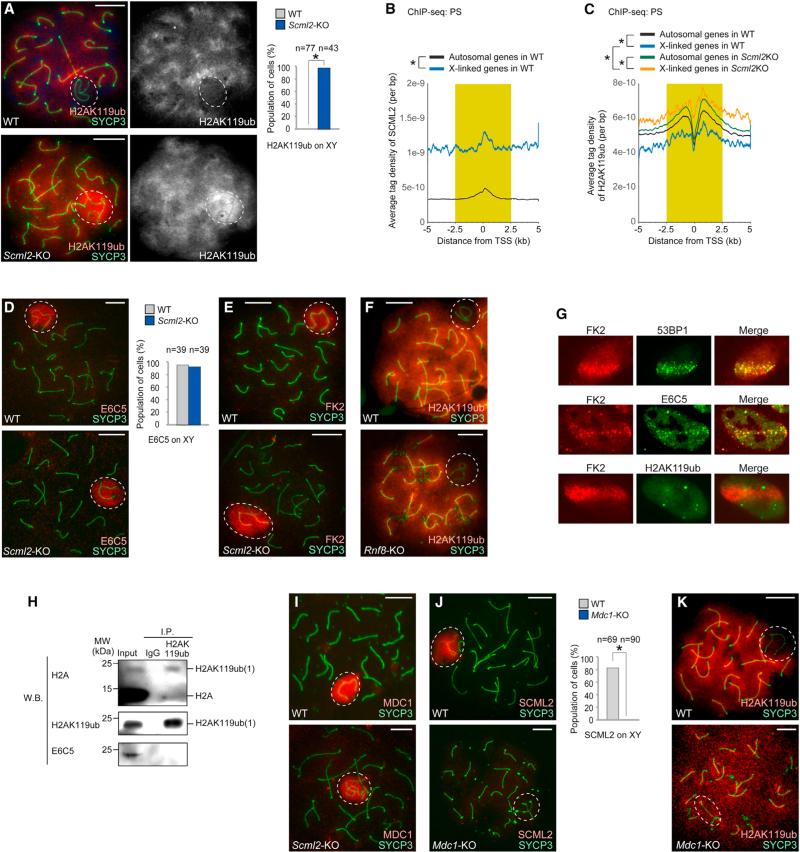
Because SCML2 was identified through its interaction with γH2AX and intensely localized on meiotic sex chromosomes (Figure 1), we next examined the function of SCML2 in the regulation of the sex chromosomes during meiosis. Surprisingly, H2AK119ub was decreased on the area of sex chromosomes in wild-type cells, but intensely accumulated there in Scml2-KO cells (Figures 6A and S5). Consistent with this, ChIP-seq data confirmed the enrichment of SCML2 on the X chromosomes in wild-type PS compared with that on autosomes (Figure 6B) and higher accumulation of H2AK119ub on the X chromosomes in Scml2-KO PS compared with that on the X chromosome in wild-type PS (Figure 6C).
Figure 6. SCML2 Suppresses H2AK119ub on Meiotic Sex Chromosomes.
(A, D–F, and I–K) Immunostaining using meiotic chromosome spreads of PS with the antibodies shown in panels. The sex chromosomes are circled with a white dashed line. The percentage of PS with positive staining on the sex chromosomes is shown on the right in (A), (D), and (J). *p < 0.05, unpaired t test. The scale bars represent 10 μm.
(B and C) The average tag density of all autosome genes and all X-linked genes within 5 kb from TSS is shown. A Wilcoxon rank sum test was performed for read counts in the highlighted area (−2,500 to +2,500 bp from TSS; *p < 0.05). Occupancy of SCML2 in PS (B). Occupancy of H2AK119ub in PS (C).
(G) Immunostaining of FK2, 53BP1, E6C5, and H2AK119ub in U2OS cells after laser stripe-induction of DNA DSBs. 53BP1 is a marker of DSBs. FK2 detects polyubiquitin conjugates. (H) The monoclonal antibody D27C4 immunoprecipitated H2AK119ub, while E6C5 did not detect H2AK119ub in adult testes.
See also Figure S5.
Importantly, this staining pattern was distinct from that of ubiquitination mediated by RNF8, which establishes polyubiquitination of H2A in the context of the DDR in somatic cells (Feng and Chen, 2012; Huen et al., 2007; Kolas et al., 2007; Mai-land et al., 2007). RNF8-mediated ubiquitination, detected by a mouse monoclonal antibody against histone H2A ubiquitinated at Lys119 (clone E6C5), accumulated on meiotic sex chromosomes (Ichijima et al., 2011; Lu et al., 2010; Sin et al., 2012a) and this accumulation was not dependent on SCML2 (Figure 6D). On meiotic sex chromosomes, the signals detected with the E6C5 antibody also overlap with signals of an FK2 antibody (Ichijima et al., 2011; Lu et al., 2010; Sin et al., 2012a) that detects polyubiquitin conjugates at sites of double-strand breaks (DSBs) in somatic cells (Mailand et al., 2007; Polanowska et al., 2006; Sobhian et al., 2007).
Consistent with the pattern of E6C5, FK2 signals were unchanged on the meiotic sex chromosomes in Scml2-KO cells (Figure 6E). On the other hand, exclusion of H2AK119ub from the meiotic sex chromosomes was unchanged in Rnf8-KO PS (Figure 6F). Therefore, on the meiotic sex chromosomes, H2AK119ub and the signals detected with E6C5 are distinctly regulated. We conclude that SCML2 specifically suppresses accumulation of H2AK119ub, but not the signals detected with E6C5, on meiotic sex chromosomes.
We further clarified the difference between H2AK119ub and signals detected with E6C5 by testing how they respond to the induction of DNA DSBs in somatic cells. Consistent with previous reports (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007), signals detected with E6C5 were robustly recruited to laser-induced DSBs concomitantly with polyubiquitin conjugates detected by FK2. In contrast, H2AK119ub was not induced (Figure 6G). Therefore, H2AK119ub and the signals detected with E6C5 are distinctly regulated in the contexts of both meiotic sex chromosomes and somatic DSBs.
To clarify the difference between two distinct signals detected by two different antibodies, we performed biochemical assays to detect ubiquitinated proteins (Choo and Zhang, 2009). In this assay, testicular cells were treated with 2M SDS for solubilization and denaturing proteins prior to immunoprecipitation. With this condition, after the immunoprecipitation, covalently bound ubiquitination to immunoprecipitated targets can only be detected. The rabbit anti-H2AK119ub indeed immunoprecipitated and recognized mono-ubiquitinated H2AK119ub (Figure 6H). On the other hand, E6C5 and FK2 did not immunoprecipitate H2A but recognized unknown high-molecular weight proteins in testes (Figure S5). Furthermore, γH2AX did not contain H2AK119ub or epitopes for E6C5 (Figure S5), suggesting that γH2AX itself is not the target for ubiquitination in testes. This result, together with the ChIP-seq data (Figure 6C), excluded the possibility that, in the wild-type, SCML2 masks the epitope for H2AK119ub because of its binding to γH2AX. Altogether, we conclude that two different antibodies recognize distinct targets and SCML2 specifically suppresses H2AK119ub on the sex chromosome.
SCML2 Works Downstream of γH2AX-MDC1 Signaling on Meiotic Sex Chromosomes
We next examined how localization of SCML2 and H2AK119ub are regulated on meiotic sex chromosomes. Localization of γH2AX and MDC1, an interacting partner of γH2AX that mediates γH2AX spreading to the entire chromosome-wide domain of the sex chromosomes (Ichijima et al., 2011), remained unchanged in the Scml2-KO (Figures 2C and 6I). On the other hand, MDC1 is required for SCML2 localization on meiotic sex chromosomes (Figure 6J). Thus, during meiosis, SCML2 may be recruited on the sex chromosomes through its interaction with γH2AX (Figures 1C–1E), and SCML2 may function downstream of γH2AX-MDC1 signaling. Additionally, exclusion of H2AK119ub from meiotic sex chromosomes was not observed in the Mdc1-KO (Figure 6K), suggesting that the removal of the H2AK119ub by SCML2 is an active process that specifically occurs on the sex chromosomes downstream of MDC1.
Consistent with the normal γH2AX-MDC1 signaling, MSCI appeared to take place normally in Scml2-KO PS based on the staining on RNA polymerase II (Figure 7A). However in RNA-seq, transcription of the X chromosome was modestly dere-pressed in PS (Figure 7B). On the basis of the stringent criteria (padj < 0.05, >2-fold difference, RPKM R 5 for higher expression genes), 87 genes on the X chromosomes were upregulated in Scml2-KO PS (Figure 7C), suggesting partial disruption of MSCI. On the contrary, Scml2-KO RS exhibited specific downregulation of 17 genes on the X chromosomes (Figure 7C). This suggests that activation of a group of X-linked genes that escape from sex chromosome inactivation in RS (Sin et al., 2012a; Sin et al., 2012b) is partially affected in Scml2-KO RS.
Figure 7. SCML2 Is Required for Epigenetic Programming of Meiotic Sex Chromosomes.
(A) Immunostaining of RNA polymerase II and MDC1 using germ cell slides that maintain chromatin morphology. The sex chromosomes are circled with a white dashed line. The scale bars represent 10 μm.
(B) Heatmap showing the expression on the X chromosome based on RNA-seq. Five hundred twenty-four X-linked genes that showed more than 3 RPKM in at least one stage of THY1+ fraction, PS, or RS are shown.
(C) The number of differentially expressed genes encoded on the X chromosome, based on RNA-seq in Scml2-KO THY1+ fraction, PS, and RS.
(D–F and H–J) Immunostaining using meiotic chromosome spreads of PS with the antibodies shown in each panel. The sex chromosomes are circled with a white dashed line. The scale bars represent 10 μm. The percentage of PS with the described feature is shown on the right in (D) and (F). Arrowheads in (E) indicate MLH1 foci. The number of MLH1 foci per cell is shown on the right in (E).
(G) Co-immunoprecipitation of SCML2 using 4-week-old wild-type testes. USP7 was detected by western blotting.
(K) Model of epigenetic regulation on the meiotic sex chromosomes. SCML2 works downstream of the γH2AX-MDC1 signaling and suppresses H2AK119ub. On the other hand, RNF8 works downstream of the γH2AX-MDC1 signaling and establishes ubiquitination and active epigenetic modifications (Sin et al., 2012a).
(L) Model of two distinct antithetical functions of SCML2 in the establishment of the male germline epigenome. First, SCML2 regulates establishment of H2AK119ub prior to spermatogenic differentiation, and H2AK119ub persists into later stages to suppress the somatic/progenitor program during differentiation of spermatogenic cells. Second, SCML2 suppresses chromosome-wide accumulation of H2AK119ub on the sex chromosomes during meiosis, which is specifically required for proper epigenetic programming of the meiotic sex chromosomes.
See also Figures S6 and S7.
Additionally, we investigated whether SCML2 affects other critical steps of meiosis. Chromosome synapsis, MLH1 foci that represent sites of crossovers, and DDR signals on the sex chromosome axes, such as BRCA1, ATR, and TOPBP1, were not affected in the Scml2-KO (Figures 7D, 7E, and S6). However, in accordance with the existence of polynucleated cells (Figure 3B), we found reduced expression of Aurkb, Ccna1, and Boll in Scml2-KO PS (Figure S6), all of which are involved in completion of cytokinesis during meiosis (Fernández-Miranda et al., 2011; Nickerson et al., 2007; VanGompel and Xu, 2010). Accumulation of aurora kinase B and histone H3 phosphorylation, which is directly catalyzed by aurora kinase B, were decreased in Scml2-KO diplotene spermatocytes (Figure S6), suggesting that SCML2 regulates meiosis in part by activating Aurkb expression. These results, in turn, indicate that the function of SCML2 on the sex chromosome during meiotic prophase is uncoupled from these meiotic events.
SCML2 Works with USP7 and Is Required for Epigenetic Programming of Meiotic Sex Chromosomes
Next, we searched for a candidate that removes H2AK119ub from the meiotic sex chromosomes. We found that the deubiquitinase USP7, which functions with PRC1 and mediates deubiquitination of H2A in vitro (Maertens et al., 2010), accumulated on meiotic sex chromosomes in an SCML2-dependent manner (Figure 7F). We further confirmed that SCML2 interacts with USP7 in testicular extracts (Figure 7G). These results suggest that SCML2 works with USP7 deubiquitinase and removes H2AK119ub from meiotic sex chromosomes.
Previously, it was suggested that SCMH1, another homolog of Drosophila Scm, mediates exclusion of PcG proteins from meiotic sex chromosomes (Takada et al., 2007). However, in the Scml2-KO, we did not observe any change in the localization of PcG proteins (RNF2, BMI1, RYBP, MEL18) on the sex chromosomes or in the exclusion of H3K27me3, which is mediated by PRC2, from the sex chromosomes (Figure S6). These results indicate that the function of SCML2 is distinct from that of SCMH1.
Because meiotic sex chromosomes are subject to extensive epigenetic programming downstream of DDR proteins (Ichijima et al., 2011; Sin et al., 2012a), we further examined how SCML2 deficiency and abnormal accumulation of H2AK119ub affected the distribution of other modifications on meiotic sex chromosomes. In the Scml2-KO, mono-methylation of H3K9 was highly upregulated when compared with the wild-type (Figures 7H and S7), although di- or tri-methylation of H3K9 was not changed in the Scml2-KO (data not shown). Histone variant MacroH2A1 locally accumulates on unsynapsed axes in the early pachytene stage, and on the pericentric heterochromatin of the X and Y chromosomes and pseudo-autosomal region in the mid to late pachytene stages of wild-type (Hoyer-Fender et al., 2000). In the Scml2-KO, MacroH2A1 highly accumulated evenly on the entire region of the meiotic sex chromosomes throughout early to late pachytene stages in the Scml2-KO (Figures 7I and S7). Furthermore, localization of XMR, a classical marker of meiotic sex chromosomes (Escalier and Garchon, 2000) was SCML2 dependent (Figure 7J). Altogether, SCML2 regulates epigenetic programming of the sex chromosomes for both gene silencing in PS and gene activation in RS. Thus, SCML2 regulates activity of the sex chromosomes for male reproduction.
DISCUSSION
The germline is the only lineage that ensures the perpetuation of genetic and epigenetic information across generations. The male germline undergoes a unique differentiation program, including meiosis and post-meiotic maturation, which culminates in packaging of the heritable genome into sperm. Subsequent to highly specialized epigenetic programming in the male germline, reprogramming in the next generation is required for reacquisition of totipotency. In this study, we have identified unique features of the transcriptome of the late stages of the male germline and have discovered mechanisms by which the male germline epigenome is distinguished from that of the soma and spermatogenesis-progenitor cells.
We demonstrate two distinct antithetical functions of SCML2 in the establishment of the male germline epigenome (Figure 7L). To suppress the somatic/progenitor program during late spermatogenesis, SCML2 regulates establishment of H2AK119ub prior to spermatogenic differentiation, and H2AK119ub persists into later stages. This repression may, in turn, indirectly ensure activation of spermatogenesis-specific genes, which is essential for the production of mature spermatozoa. Therefore, our results suggest that epigenetic programming largely predetermines transcriptomes during the late stages of the male germline. However, the modest SCML2-dependent establishment of H2AK119ub suggests that not all SCML2-regulated genes are the direct targets of SCML2 and that there can also be indirect regulation of gene repression by SCML2. Furthermore, it is unclear what directly accounts for the apoptosis and, thereby, what specific function of SCML2 is responsible for maintenance or differentiation of undifferentiated spermatogonia. The direct causal relationship of SCML2 function needs to be investigated in future studies.
Additionally, SCML2 prevents chromosome-wide accumulation of H2AK119ub on the sex chromosomes during meiosis, which is specifically required for proper epigenetic programming of the meiotic sex chromosomes for male reproduction. Together with suppression of the somatic/progenitor program, these two functions of SCML2 are likely to be independently regulated. The establishment of H2AK119ub occurs prior to spermatogenic differentiation, while suppression of H2AK119ub on the sex chromosomes occurs during meiosis in a γH2AX MDC1-dependent manner. These results suggest that Scml2 positively regulates H2AK119ub on autosomes and negatively regulates H2AK119ub on meiotic sex chromosomes. Taken together, SCML2 establishes the male germline-specific epigenome through the regulation of H2AK119ub and defines a unique transcriptome in late spermatogenesis, which is essential to acquire fertility as male gametes.
The two distinct functions of SCML2 we describe could be mediated through independent complexes (Figure 7L). In GS cells, there are many RNF2 islands independent of SCML2 (Figure 4). Therefore, not all PRC1 complexes contain SCML2, and a subset of PRC1s containing SCML2 may have a critical function in the establishment of the unique epigenome of the male germ-line. On the other hand, SCML2 may not necessarily be in the PRC1 complex on meiotic sex chromosomes. This is based on two observations: (1) in our mass spectrometry analysis, which was performed using γH2AX-containing nucleosomes, we did not detect other PcG proteins, and (2) SCML2 and USP7 are the only Polycomb-related factors that highly accumulate on the sex chromosomes in the wild-type, and although RNF2 exists evenly in the nucleus of PS, RNF2 does not specifically accumulate on the meiotic sex chromosomes (Figure S6).
Furthermore, we identified an epigenetic pathway by which SCML2 regulates the meiotic sex chromosomes. Our previous study demonstrated that RNF8 mediates ubiquitination of the meiotic sex chromosomes, subsequently establishes active epigenetic modifications, and activates a group of X-linked genes that escape from sex chromosome inactivation in RS (Sin et al., 2012a). In the present study, we show that RNF8 targets are likely to be non-histone ubiquitinated proteins that are distinct from H2AK119ub. On the basis of the distinct regulation of H2AK119ub and RNF8 targets, our results suggest that the SCML2 and RNF8 pathways have distinct functions downstream of γH2AX-MDC1 signaling in regulating meiotic sex chromosomes (Figure 7K).
In mammals, SCML2 is an MBT domain-containing protein that is essential in the male germline. Our results suggest that germline-specific expression of SCML2 is related to its function in establishing germline-specific features. Although a previous study suggested that mammalian Scm homologs have redundant functions and compensate for the loss of Scmh1 (Takada et al., 2007), in this study we found that the phenotype of the Scml2-KO is distinct from that of the Scmh1-KO and that the function of SCML2 is not redundant with that of SCMH1. Thus, SCML2 has a unique function in the germline that cannot be compensated by other MBT domain-containing proteins. A recent study suggested that human SCML2 is expressed ubiquitously in somatic tissues (Bonasio et al., 2014), so it would be intriguing to examine the difference between human SCML2 and mouse SCML2.
Previous studies revealed functions of PRC1 in the female germline. PRC1 coordinates the timing of entry into meiosis during female embryonic development (Yokobayashi et al., 2013) and establishes developmental competence for the following generation by defining gene expression patterns during oogenesis (Posfai et al., 2012). Although we found that SCML2 was also expressed in female embryonic germ cells (Figure S1), we were not able to examine the role of SCML2 in the female germ-line, because the Scml2-KO is not available due to its X linkage (male Scml2-KO [Y/−] mice are infertile). Conditional deletion of Scml2 is needed to clarify the function of SCML2 in the female germline.
Our study reveals a PcG-based mechanism that defines the specific epigenome of the germline. In contrast, fly L(3)mbt, which also contains the MBT domain, is implicated in suppression of germline genes in somatic cells in the fly (Janic et al., 2010). Additionally, one of the Drosophila PcG components, enhancer of zeste, which mediates H3K27me3, suppresses the somatic program in male germ cells in a non-cell autonomous manner (Eun et al., 2014). Although the function of SCML2 is likely to be cell autonomous in mice, a PcG-based mechanism could be an evolutionarily conserved determinant between soma and germline programs.
The identification of SCML2 as the critical determinant of the germline epigenome also has important implications for the genomics and evolution of the male germline. Remarkably, the testis transcriptome and its evolutionary traits are highly diverged from those of other organs (Brawand et al., 2011). In particular, meiotic spermatocyte and post-meiotic spermatid transcriptomes have diverged from those in soma to enable unique gene expression programs. This may explain the higher transcriptome complexity of the testis as a whole (Soumillon et al., 2013). Because germline transcriptomes are suggested to mirror the evolutionary history of germline genes, both on autosomes and on sex chromosomes, in mammals (Brawand et al., 2011; Sin et al., 2012b; Zhang et al., 2010), it is tempting to speculate that the two distinct SCML2-based mechanisms we describe, one on the autosomes and one on the sex chromosomes, are an evolutionarily conserved driving force of the complexity of the germline program. Altogether, our study here reveals mechanisms by which the germline-specific epigenome is established during spermatogenesis and may thus contribute to understanding the etiology of male infertility.
EXPERIMENTAL PROCEDURES
Animals
Dppa3-EGFP, Mdc1-KO, and Rnf8-KO mice were previously reported (Lou et al., 2006; Minter-Dykhouse et al., 2008; Payer et al., 2006). Scml2-KO mice were produced by using ZFN technology (Sigma Aldrich) (Cui et al., 2011). All animals were maintained in accordance with CCHMC's guidelines.
Protein Identification by Mass Spectrometry
Protein bands or gel regions from control immunoglobulin G- and γH2AX-specific antibody capture were identified by tandem mass spectrometry as described previously (Ridsdale et al., 2011).
Antibodies
The antibody list used in this study is available in Supplemental Experimental Procedures. Briefly, RNF2-dependent ubiquitinated H2A (H2AK119ub) is detected by a rabbit monoclonal antibody against histone H2A ubiquitinated at Lys 119 (D27C4: Cell Signaling #8240). Rnf8-dependent ubiquitination is detected by a mouse monoclonal antibody against histone H2A ubiquitinated at Lys119 (clone E6C5: Millipore #05-678).
Histological Analysis and Germ Cell Slide Preparation
For histological analysis, immunohistochemistry, and TUNEL staining, testes without tunica albuginea were fixed with 4% paraformaldehyde overnight. Testes were dehydrated and embedded in paraffin. Chromosome spreads were prepared using hypotonic treatment as described (Peters et al., 1997). In order to preserve the morphology of chromatin and the relative 3D nuclear structure in mouse testes, slides were prepared as described (Namekawa, 2014; Namekawa and Lee, 2011; Namekawa et al., 2006).
Germ Cell Fractionation
PS and RS were isolated with BSA gravity sedimentation as described previously (Bellvé, 1993). Purity was confirmed by nuclear staining with DAPI using fluorescence microscopy. More than 90% of purity was confirmed for each purification. Isolation of the THY1+ stem cell fraction was performed as described previously (Hammoud et al., 2014).
ChIP-Seq, RNA-Seq, and Data Analysis
Both native ChIP-seq and crosslinking ChIP-seq were performed for H2AK119ub, and crosslinking ChIP-seq was performed for SCML2, RNF2, and BMI1. For both ChIP-seq and RNA-seq, the DNA libraries were prepared with NEBNext ChIP-Seq Library Prep Master Mix Set for Illumina (NEB) and Agencourt AMPure XP (Beckman Coulter) or ThruPLEX kit (Rubicon Genomics). DNA libraries were adjusted to 5 nM in TE and sequenced with Illumina HiSeq 2000. Data analysis for both ChIP-seq and RNA-seq were performed in the Wardrobe Experiment Management System (https://code.google.com/p/genome-tools/; Kartashov and Barski, 2014). Briefly, reads were aligned to the mouse genome (mm10) with bowtie (version 1.0.0; Langmead et al., 2009), assigned to the refseq genes (or isoforms) using the Wardrobe algorithm, and displayed on a local mirror of UCSC Genome Browser as coverage. For ChIP-seq analysis, islands of SCML2, RNF2, and BMI1 enrichment were identified using MACS2 (version 2.0.10.20130712; Zhang et al., 2008). For ChIP-seq analysis, differentially expressed genes were identified using the following parameters: (1) ≥2-fold change; (2) DESeq (Anders and Huber, 2010), padj ≤ 0.05; and (3) RPKM ≥ 5 for higher expression genes. Subsequently, enriched gene ontology for differentially expressed genes between wild-type and Scml2-KO mice was characterized using the Database for Annotation, Visualization and Integrated Discovery version 6.7 (Huang et al., 2009).
Supplementary Material
Highlights.
SCML2 regulates global silencing of somatic/progenitor genes in the male germline
SCML2 is a germline-specific subunit of Polycomb repressive complex 1
SCML2 establishes H2A ubiquitination to silence somatic/ progenitor genes on autosomes
SCML2 prevents H2A ubiquitination on meiotic sex chromosomes
ACKNOWLEDGMENTS
We thank Yuya Ogawa, Gang Huang, Jérôme Déjardin, Rafael Kopan, Harinder Singh, Yueh-Chiang Hu, and members of the Namekawa lab for discussion and helpful comments regarding the manuscript; Azim Surani for providing Dppa3-EGFP mice; and Junjie Chen for providing the Mdc1-KO and Rnf8-KO mice. This work was supported by a Japan Society for the Promotion of Science postdoctoral fellowship for K.H., a Lalor Foundation postdoctoral fellowship for H.-S.S., the Developmental Fund at Cincinnati Children's Hospital Medical Center to S.H.N., research grant FY13-510 from the March of Dimes Foundation to S.H.N., and NIH grants HL085587 to P.R.A., HL098691 to A.B., and GM098605 to S.H.N.
Footnotes
AUTHOR CONTRIBUTIONS
K.H, P.R.A., A.B., and S.H.N. designed the experiments. K.H., H.-S.S., S.M., K.G.A., and W.C.B. performed ChIP-seq and RNA-seq experiments and prepared libraries. K.H., T.J.B., and K.G.A. examined meiotic phenotypes of Scml2-KO. A.V.K. and A.B. performed bioinformatics analyses with K.H., H.-S.S., and S.H.N.. S.M. performed biochemical assays to detect protein ubiquitination and purification of THY1+ fractions. Y.I. performed immunoprecipitation and initially identified SCML2. K.D.G. performed and interpreted the mass spectrometry data. F.Z. and P.R.A examined the somatic DDR. K.H., H.-S.S., S.M., T.J.B., P.R.A, A.B., and S.H.N. interpreted the results. K.H. and S.H.N. wrote the manuscript with all other authors.
ACCESSION NUMBERS
The Gene Expression Omnibus accession number for the ChIP-seq and gene expression data reported in this paper is GSE55060.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.devcel.2015.01.014.
REFERENCES
- Aloia L, Di Stefano B, Di Croce L. Polycomb complexes in stem cells and embryonic development. Development. 2013;140:2525–2534. doi: 10.1242/dev.091553. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellvé AR. Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol. 1993;225:84–113. doi: 10.1016/0076-6879(93)25009-q. [DOI] [PubMed] [Google Scholar]
- Bonasio R, Lecona E, Narendra V, Voigt P, Parisi F, Kluger Y, Reinberg D. Interactions with RNA direct the Polycomb group protein SCML2 to chromatin where it represses target genes. eLife. 2014;3:e02637. doi: 10.7554/eLife.02637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D, Soumillon M, Necsulea A, Julien P, Csárdi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- Chalmel F, Rolland AD, Niederhauser-Wiederkehr C, Chung SS, Demougin P, Gattiker A, Moore J, Patard JJ, Wolgemuth DJ, Jégou B, Primig M. The conserved transcriptome in human and rodent male gametogenesis. Proc. Natl. Acad. Sci. U S A. 2007;104:8346–8351. doi: 10.1073/pnas.0701883104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo YS, Zhang Z. Detection of protein ubiquitination. J. Vis. Exp. 2009;(30):1293. doi: 10.3791/1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Ji D, Fisher DA, Wu Y, Briner DM, Weinstein EJ. Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat. Biotechnol. 2011;29:64–67. doi: 10.1038/nbt.1731. [DOI] [PubMed] [Google Scholar]
- Escalier D, Garchon HJ. XMR is associated with the asynapsed segments of sex chromosomes in the XY body of mouse primary spermatocytes. Chromosoma. 2000;109:259–265. doi: 10.1007/s004120000075. [DOI] [PubMed] [Google Scholar]
- Eun SH, Shi Z, Cui K, Zhao K, Chen X. A non-cell autonomous role of E(z) to prevent germ cells from turning on a somatic cell marker. Science. 2014;343:1513–1516. doi: 10.1126/science.1246514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Chen J. The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat. Struct. Mol. Biol. 2012;19:201–206. doi: 10.1038/nsmb.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Miranda G, Trakala M, Martín J, Escobar B, González A, Ghyselinck NB, Ortega S, Cañamero M, Pérez de Castro I, Malumbres M. Genetic disruption of aurora B uncovers an essential role for aurora C during early mammalian development. Development. 2011;138:2661–2672. doi: 10.1242/dev.066381. [DOI] [PubMed] [Google Scholar]
- Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell. 2012;45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill ME, Erkek S, Peters AH. Parental epigenetic control of embryogenesis: a balance between inheritance and reprogramming? Curr. Opin. Cell Biol. 2012;24:387–396. doi: 10.1016/j.ceb.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Low DH, Yi C, Carrell DT, Guccione E, Cairns BR. Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell. 2014;15:239–253. doi: 10.1016/j.stem.2014.04.006. [DOI] [PubMed] [Google Scholar]
- Hoyer-Fender S, Costanzi C, Pehrson JR. Histone macroH2A1.2 is concentrated in the XY-body by the early pachytene stage of spermatogenesis. Exp. Cell Res. 2000;258:254–260. doi: 10.1006/excr.2000.4951. [DOI] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichijima Y, Ichijima M, Lou Z, Nussenzweig A, Camerini-Otero RD, Chen J, Andreassen PR, Namekawa SH. MDC1 directs chromosome-wide silencing of the sex chromosomes in male germ cells. Genes Dev. 2011;25:959–971. doi: 10.1101/gad.2030811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichijima Y, Sin HS, Namekawa SH. Sex chromosome inactivation in germ cells: emerging roles of DNA damage response pathways. Cell. Mol. Life Sci. 2012;69:2559–2572. doi: 10.1007/s00018-012-0941-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janic A, Mendizabal L, Llamazares S, Rossell D, Gonzalez C. Ectopic expression of germline genes drives malignant brain tumor growth in Drosophila. Science. 2010;330:1824–1827. doi: 10.1126/science.1195481. [DOI] [PubMed] [Google Scholar]
- Kartashov AV, Barski A. Wardrobe - an integrated system for analysis of epigenomics and transcriptomics data. 2014 doi: 10.1186/s13059-015-0720-3. bioRxiv, http://biorxiv.org/content/early/2014/12/22/012799. [DOI] [PMC free article] [PubMed]
- Khil PP, Smirnova NA, Romanienko PJ, Camerini-Otero RD. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat. Genet. 2004;36:642–646. doi: 10.1038/ng1368. [DOI] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota SK, Feil R. Epigenetic transitions in germ cell development and meiosis. Dev. Cell. 2010;19:675–686. doi: 10.1016/j.devcel.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, Manis JP, van Deursen J, Nussenzweig A, Paull TT, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- Lu LY, Wu J, Ye L, Gavrilina GB, Saunders TL, Yu X. RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis. Dev. Cell. 2010;18:371–384. doi: 10.1016/j.devcel.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens GN, El Messaoudi-Aubert S, Elderkin S, Hiom K, Peters G. Ubiquitin-specific proteases 7 and 11 modulate Polycomb regulation of the INK4a tumour suppressor. EMBO J. 2010;29:2553–2565. doi: 10.1038/emboj.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- Minter-Dykhouse K, Ward I, Huen MS, Chen J, Lou Z. Distinct versus overlapping functions of MDC1 and 53BP1 in DNA damage response and tumorigenesis. J. Cell Biol. 2008;181:727–735. doi: 10.1083/jcb.200801083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montini E, Buchner G, Spalluto C, Andolfi G, Caruso A, den Dunnen JT, Trump D, Rocchi M, Ballabio A, Franco B. Identification of SCML2, a second human gene homologous to the Drosophila sex comb on midleg (Scm): a new gene cluster on Xp22. Genomics. 1999;58:65–72. doi: 10.1006/geno.1999.5755. [DOI] [PubMed] [Google Scholar]
- Mueller JL, Mahadevaiah SK, Park PJ, Warburton PE, Page DC, Turner JM. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat. Genet. 2008;40:794–799. doi: 10.1038/ng.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Shirae-Kurabayashi M, Hanyu-Nakamura K. Repression of early zygotic transcription in the germline. Curr. Opin. Cell Biol. 2010;22:709–714. doi: 10.1016/j.ceb.2010.08.012. [DOI] [PubMed] [Google Scholar]
- Namekawa SH. Slide preparation method to preserve three-dimensional chromatin architecture of testicular germ cells. J. Vis. Exp. 2014;(83):e50819. doi: 10.3791/50819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namekawa SH, Lee JT. Detection of nascent RNA, single-copy DNA and protein localization by immunoFISH in mouse germ cells and preim-plantation embryos. Nat. Protoc. 2011;6:270–284. doi: 10.1038/nprot.2010.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namekawa SH, Park PJ, Zhang LF, Shima JE, McCarrey JR, Griswold MD, Lee JT. Postmeiotic sex chromatin in the male germline of mice. Curr. Biol. 2006;16:660–667. doi: 10.1016/j.cub.2006.01.066. [DOI] [PubMed] [Google Scholar]
- Nickerson HD, Joshi A, Wolgemuth DJ. Cyclin A1-deficient mice lack histone H3 serine 10 phosphorylation and exhibit altered aurora B dynamics in late prophase of male meiosis. Dev. Biol. 2007;306:725–735. doi: 10.1016/j.ydbio.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B, Chuva de Sousa Lopes SM, Barton SC, Lee C, Saitou M, Surani MA. Generation of stella-GFP transgenic mice: a novel tool to study germ cell development. Genesis (New York, NY: 2000) 2006;44:75–83. doi: 10.1002/gene.20187. [DOI] [PubMed] [Google Scholar]
- Peters AH, Plug AW, van Vugt MJ, de Boer P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 1997;5:66–68. doi: 10.1023/a:1018445520117. [DOI] [PubMed] [Google Scholar]
- Polanowska J, Martin JS, Garcia-Muse T, Petalcorin MI, Boulton SJ. A conserved pathway to activate BRCA1-dependent ubiquitylation at DNA damage sites. EMBO J. 2006;25:2178–2188. doi: 10.1038/sj.emboj.7601102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posfai E, Kunzmann R, Brochard V, Salvaing J, Cabuy E, Roloff TC, Liu Z, Tardat M, van Lohuizen M, Vidal M, et al. Polycomb function during oogenesis is required for mouse embryonic development. Genes Dev. 2012;26:920–932. doi: 10.1101/gad.188094.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Van Buren D, Huang HS, Zhong L, Mostoslavsky R, Akbarian S, Hock H. Chromatin protein L3MBTL1 is dispensable for development and tumor suppression in mice. J. Biol. Chem. 2010;285:27767–27775. doi: 10.1074/jbc.M110.115410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Whyte WA, Anderssen E, Apostolou E, Chen HH, Akbarian S, Bronson RT, Hochedlinger K, Ramaswamy S, Young RA, Hock H. The Polycomb group protein L3mbtl2 assembles an atypical PRC1-family complex that is essential in pluripotent stem cells and early development. Cell Stem Cell. 2012;11:319–332. doi: 10.1016/j.stem.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridsdale R, Na CL, Xu Y, Greis KD, Weaver T. Comparative proteomic analysis of lung lamellar bodies and lysosome-related organelles. PLoS ONE. 2011;6:e16482. doi: 10.1371/journal.pone.0016482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M, Kagiwada S, Kurimoto K. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development. 2012;139:15–31. doi: 10.1242/dev.050849. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat. Rev. Genet. 2008;9:129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol. Reprod. 2004;71:319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell. 2013;49:808–824. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat. Rev. Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- Sin HS, Barski A, Zhang F, Kartashov AV, Nussenzweig A, Chen J, Andreassen PR, Namekawa SH. RNF8 regulates active epigenetic modifications and escape gene activation from inactive sex chromosomes in post-meiotic spermatids. Genes Dev. 2012a;26:2737–2748. doi: 10.1101/gad.202713.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin HS, Ichijima Y, Koh E, Namiki M, Namekawa SH. Human postmeiotic sex chromatin and its impact on sex chromosome evolution. Genome Res. 2012b;22:827–836. doi: 10.1101/gr.135046.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumillon M, Necsulea A, Weier M, Brawand D, Zhang X, Gu H, Barthès P, Kokkinaki M, Nef S, Gnirke A, et al. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep. 2013;3:2179–2190. doi: 10.1016/j.celrep.2013.05.031. [DOI] [PubMed] [Google Scholar]
- Svingen T, Koopman P. Building the mammalian testis: origins, differentiation, and assembly of the component cell populations. Genes Dev. 2013;27:2409–2426. doi: 10.1101/gad.228080.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada Y, Isono K, Shinga J, Turner JM, Kitamura H, Ohara O, Watanabe G, Singh PB, Kamijo T, Jenuwein T, et al. Mammalian Polycomb Scmh1 mediates exclusion of Polycomb complexes from the XY body in the pachytene spermatocytes. Development. 2007;134:579–590. doi: 10.1242/dev.02747. [DOI] [PubMed] [Google Scholar]
- Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell. 2012;148:664–678. doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JM. Meiotic sex chromosome inactivation. Development. 2007;134:1823–1831. doi: 10.1242/dev.000018. [DOI] [PubMed] [Google Scholar]
- VanGompel MJ, Xu EY. A novel requirement in mammalian spermatid differentiation for the DAZ-family protein Boule. Hum. Mol. Genet. 2010;19:2360–2369. doi: 10.1093/hmg/ddq109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. A region of the human HOXD cluster that confers Polycomb-group responsiveness. Cell. 2010;140:99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobayashi S, Liang CY, Kohler H, Nestorov P, Liu Z, Vidal M, van Lohuizen M, Roloff TC, Peters AH. PRC1 coordinates timing of sexual differentiation of female primordial germ cells. Nature. 2013;495:236–240. doi: 10.1038/nature11918. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YE, Vibranovski MD, Landback P, Marais GA, Long M. Chromosomal redistribution of male-biased genes in mammalian evolution with two bursts of gene gain on the X chromosome. PLoS Biol. 2010;8:8. doi: 10.1371/journal.pbio.1000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Bonasio R, Strino F, Kluger Y, Holloway JK, Modzelewski AJ, Cohen PE, Reinberg D. SFMBT1 functions with LSD1 to regulate expression of canonical histone genes and chromatin-related factors. Genes Dev. 2013;27:749–766. doi: 10.1101/gad.210963.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.