Abstract
Polyglutamine diseases are a class of neurodegenerative diseases that share an expansion of a glutamine-encoding CAG tract in the respective disease genes as a central hallmark. In all of these diseases there is progressive degeneration in a select subset of neurons, and the mechanisms behind this degeneration remain unclear. Emerging evidence from animal models of disease has identified abnormalities in synaptic signaling and intrinsic excitability in affected neurons, which coincide with the onset of symptoms and precede apparent neuropathology. The appearance of these early changes suggests that altered neuronal activity might be an important component of network dysfunction and that these alterations in network physiology could contribute to symptoms of disease. Here we review abnormalities in neuronal function that have been identified in both animal models and patients, and highlight ways in which these changes in neuronal activity may contribute to disease symptoms. We then review the literature supporting an emerging role for abnormalities in neuronal activity as a driver of neurodegeneration. Finally, we identify common themes that emerge from studies of neuronal dysfunction in polyglutamine disease.
Electronic supplementary material
The online version of this article (doi:10.1007/s13311-014-0289-7) contains supplementary material, which is available to authorized users.
Key Words: Polyglutamine disease, cerebellar ataxia, spinocerebellar ataxia, Huntington disease, synaptic transmission, intrinsic excitability, calcium
Introduction
The polyglutamine (polyQ) diseases are a class of neurodegenerative disorders characterized by expansion of a glutamine-encoding CAG tract within the coding region of the gene. To date, 9 polyQ diseases have been identified: spinal-bulbar muscular atrophy (SBMA) [1], Huntington disease (HD) [2], spinocerebellar ataxia (SCA) type 1 (SCA1) [3], SCA type 3 (SCA3) [4], dentatorubral-pallidoluysian atrophy (DRPLA) [5], SCA type 2 (SCA2) [6], SCA type 7 (SCA7) [7], SCA type 6 (SCA6) [8], and SCA type 17 (SCA17) [9]. All of these diseases share autosomal dominant inheritance, with the exception of SBMA, which shows X-linked inheritance.
The genes that are mutated in each of these conditions are widely or in some cases ubiquitously expressed. Despite their widespread expression, mutations do not result in global functional impairment but rather progressive focal neurologic impairment. The nature of impairment varies by disease, although there is overlap; for example, in the SCAs functional impairment is related to pathology in the cerebellum and its associated pathways. Postmortem analysis has revealed that each condition also shows a unique pattern of histopathology, with different brain structures demonstrating degenerative macroscopic changes (i.e., brain structure volume reduction), as well as microscopic changes (progressive cell loss in specific neuronal populations and the appearance of protein aggregates). The cerebellum and its associated pathways are involved in the polyQ ataxias, SCAs 1–3, SCA6, SCA7, and SCA17, and DRPLA; the striatum and cortex are the primary sites of pathology in HD; and the anterior horn cells in the spinal cord are the most severely affected cells in SBMA.
The discovery that brain areas with marked cell loss are responsible for controlling those functions that are affected in patients has provided the field with a common and overarching hypothesis that views degeneration and cell loss in affected populations of cells as a driver of symptom progression. In this view, the symptoms that are observed reflect “drop out” of neurons that normally participate in the networks which serve the affected neurologic function. The progressive nature of symptoms would therefore reflect a reduction in the participation of certain populations of neurons within the network.
PolyQ expansion disease research has benefited tremendously from the use of genetically modified mice as models of disease, as these models have recapitulated both the neurodegenerative features and progressive symptomatic impairment that is observed in patients. Studies of these models have yielded a surprising insight: the onset of apparent symptomatic impairment often precedes detectable neuropathology [10–17]. These results suggest that the cell loss observed in patient samples is representative of more advanced disease, and that reduced participation in an affected network due to neuronal loss may not be the only contributor to symptoms observed in patients.
In the search for the earliest abnormalities that are associated with the onset of motor symptoms, a surprising number of mouse models of polyQ disease have identified abnormalities in neuronal activity preceding overt neuronal loss. These changes in activity, broadly speaking, fall into 2 partially-overlapping categories: alterations in synaptic transmission and alterations in intrinsic excitability. In the context of these findings, then, one might think about progressive symptoms a little differently. Rather than reflecting a simple loss of network participation, progressive symptoms may, in fact, reflect aberrant activity of cells that are integrated within the network, resulting, in turn, in network dysfunction and the subsequent development of symptoms. This would obviously not be independent of cell loss, which would also be expected to contribute to symptomatic impairment. Abnormal neuronal activity in and of itself may therefore represent an important component of progressive impairment.
The aim of this review is to outline what is currently known about abnormalities in neuronal activity and their contribution to the development and progression of symptoms in polyQ disorders. It should be noted that there are many hypotheses regarding mechanisms of polyQ disease pathogenesis that are beyond the scope of this review, and are covered quite well elsewhere [18–21]. The review presents evidence from a variety of animal models and also from humans with disease, and outlines ways in which aberrant neuronal activity appears to contribute to progressive symptomatic impairment. We begin by outlining the literature that demonstrates changes in synaptic function and intrinsic excitability in each disorder. Subsequently, we explore the emerging link between alterations in activity and degenerative changes in affected neurons. Finally, we outline common themes that emerge from studies of altered neuronal activity in polyQ disease, and suggest multiple shared mechanisms by which altered activity contributes to neuropathology and how these changes may be targeted for the treatment of these conditions.
Evidence for Altered Neuronal Activity in polyQ Diseases
Studies from mouse models of polyQ diseases have demonstrated that there are changes in neuronal activity that are associated with the onset of symptoms. Subsequent to symptom onset, changes in neuronal activity have been found to correlate with symptom progression. Below we present data that support a link between altered neuronal activity and functional impairment in DRPLA, SCA1, SCA2, SCA3, SCA6, and HD (outlined in Fig. 1). It should be noted that although such a link may also exist in SCA7 and SCA17, these conditions have not been included because there are currently no data examining changes in neuronal function in these diseases.
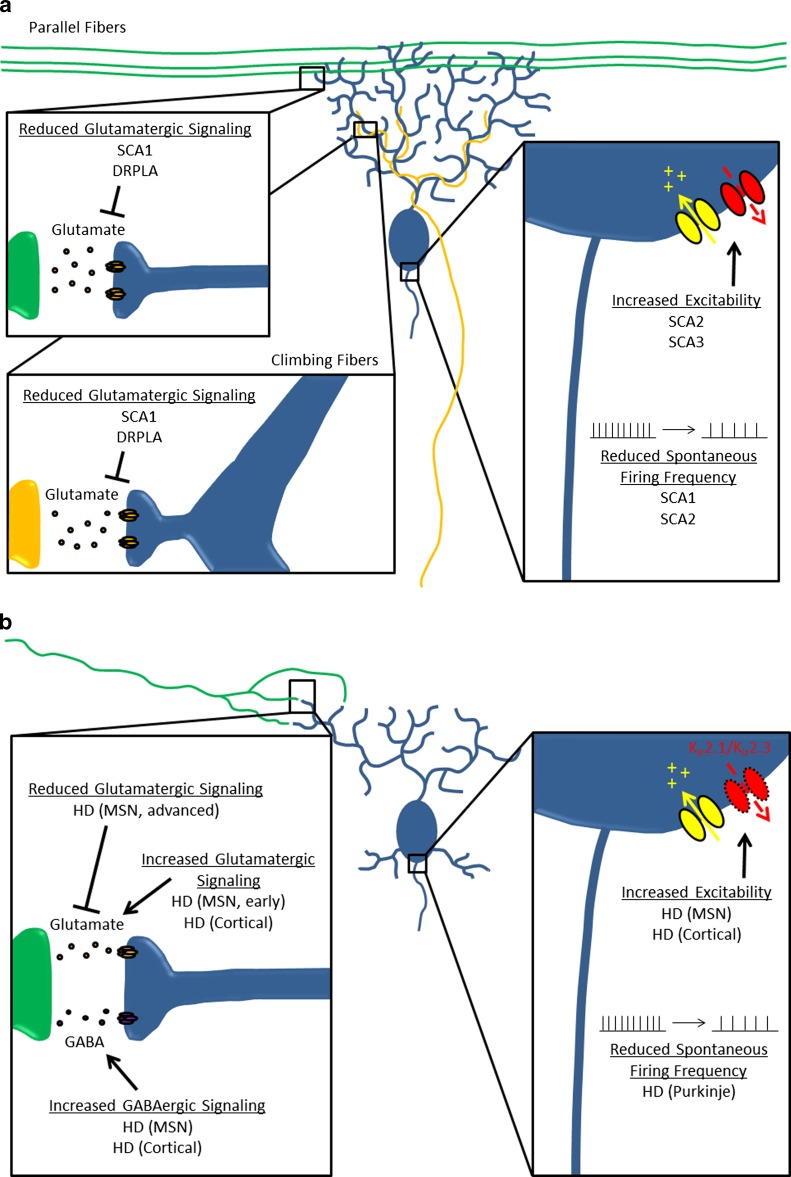
Fig. 1.
Abnormal neuronal activity in polyglutamine (polyQ) disorders. This summarizes the abnormalities in intrinsic excitability and synaptic dysfunction that have been identified in models of polyQ disorders. The inset(s) to the left specifically highlights synaptic dysfunction, while the inset to the right highlights abnormalities of intrinsic excitability. (A) Abnormal Purkinje neuron activity in spinocerebellar ataxia (SCA)1, SCA2, and SCA3, and dentatorubral-pallidoluysian atrophy (DRPLA). (B) Abnormal activity of multiple neuronal subtypes in Huntington disease (HD). MSN = medium spiny neuron; GABA = gamma-aminobutyric acid
DRPLA
DRPLA is a rare autosomal dominant syndrome caused by pathogenic expansion of the CAG repeat in the atrophin-1 gene. There is considerable clinical heterogeneity amongst patients with DRPLA, and patients with DRPLA may present with an array of symptoms that includes cerebellar ataxia, choreoathetosis, myoclonus, epilepsy, dementia, and psychiatric symptoms [22]. Despite the tremendous clinical heterogeneity observed in DRPLA, postmortem examination of brains from patients with DRPLA exhibit a common pattern of degeneration: involvement of the dentatorubral and pallidoluysian systems [23, 24], as well as cerebral white matter abnormalities [25].
In a mouse model of DRPLA with 129 glutamine repeats in ATN1, alterations in neuronal activity have been found to precede overt cell loss. Cerebellar Purkinje neurons are a site of significant pathology in both this mouse model and in human patients. Purkinje neurons act as the principal output of the cerebellar cortex, and they receive 2 major types of modulatory synaptic input: climbing fibers and parallel fibers. In the mouse model of DRPLA at 12 weeks of age, a time point when there was already marked Purkinje neuron dendritic atrophy but no cell death, there is a reduction in paired-pulse ratio at both the climbing fiber-to-Purkinje neuron synapse and the parallel fiber-to-Purkinje neuron synapse [17]. The reduction in the paired-pulse ratios at both the climbing fiber and parallel fiber synapses suggests that glutamate released at these 2 synapses has reduced efficacy in its ability to activate postsynaptic receptors on Purkinje neurons. The fact that there is such a reduction in spite of dendritic atrophy, which would be expected to increase synaptic signal strength by increasing input resistance, suggests that the reduced paired-pulse ratios reflect a loss of molecules important for synaptic signaling.
SCA1
SCA1 is an inherited cerebellar ataxia with autosomal dominant inheritance. The disease arises as a result of a CAG triplet repeat in the N-terminal coding region of the ATXN1 gene on chromosome 6p23. The most common presentation of SCA1 is ataxia in addition to variable slowing of saccades, ophthalmoplegia, extrapyramidal feaures, dementia, and/or peripheral neuropathy [26]. In SCA1, as with other autosomal dominant ataxias, the Purkinje neurons of the cerebellum are severely affected, showing prominent atrophy and cell loss [27].
In work using a transgenic mouse model of SCA1 with 82 glutamine repeats in the gene, both climbing fiber and parallel fiber synapses on Purkinje neurons show profound abnormalities. The alterations in physiology correspond to symptom onset, and the degree of abnormalities correlate with the progression of symptoms. There is some evidence to suggest that parallel fiber synapses onto Purkinje neurons are abnormal, although the data are inconsistent. The earliest change is a reduction in the strength of the parallel fiber-to-Purkinje cell synapse in acute cerebellar slices several weeks before the onset of motor dysfunction [28]. In vivo optical imaging demonstrates no apparent change in parallel fiber synapse efficacy at this early time point, although there is reduced parallel fiber-to-Purkinje neuron signaling apparent at a later point in the disease [29].
There is very strong evidence to suggest that climbing fiber synapses onto Purkinje neurons show significant alterations in SCA1. Abnormalities are first apparent at the onset of motor symptoms, when Purkinje neuron responsiveness to climbing fiber input is dramatically reduced [29]. At this stage, there are also abnormalities in the structure of climbing fiber synaptic contacts, namely a loss of synapses in the distal dendritic tree and persistence of climbing fiber contacts on the soma that are normally pruned during development [30]. With disease progression, Purkinje neurons continue to show reduced responsiveness at climbing fiber synapses and a reduction in the volume of climbing fiber afferents onto Purkinje neurons [31]. All of these data, taken together, suggest that in SCA1 there is reduced Purkinje neuron responsiveness to both climbing fiber and parallel fiber synapses.
In addition to changes in synaptic physiology, Purkinje neurons have demonstrable changes in their intrinsic firing properties. Studies of the SCA1 mouse model show a reduction in basal pacemaker firing rate of Purkinje neurons from 2–3-week-old mice. Changes are evident before there are detectable changes in dendritic spine morphology, suggesting that they precede any degenerative structural changes in Purkinje neurons. These changes are also apparent before the onset of motor dysfunction [28].
SCA2
SCA2 is an autosomal dominant disorder caused by polyQ expansion of the ATXN2 gene on the long arm of chromosome 12. In typical patients the gene has the sequence (CAG)8(CAA)1(CAG)4(CAA)1(CAG)8, coding for 22 consecutive glutamine residues near the N-terminal end of the protein, but in affected individuals the length of the repeat generally surpasses 32 repeats [32]. Neuronal loss is seen in a variety of structures in SCA2, notably the cerebellum [33], thalamus [34], pons [35], medulla [36], midbrain, and cerebral cortex [37].
In SCA2, Purkinje neurons have demonstrable changes in their intrinsic firing properties, which correlate with the onset of symptoms. A progressive decrease in spontaneous pacemaker firing frequency is observed with age in Purkinje cells from a transgenic mouse model of SCA2 harboring 127 glutamine repeats in the gene. The progressive reduction in firing frequency begins before there is deficient motor performance or Purkinje cell degeneration, suggesting that it is an early event that might contribute to pathology in this condition [38].
SCA3 (Machado–Joseph Disease)
SCA3 (Machado–Joseph disease) is a dominantly inherited cerebellar ataxia that arises owing to CAG triplet repeat expansion in the ATXN3 gene. Patients typically present first with progressive gait imbalance and speech difficulties, eventually developing occulomotor difficulties, spasticity, dystonia, and, ultimately, severe dysarthria and dysphagia [39]. Pathology in SCA3 is fairly widespread, and cell loss is apparent in the cerebellum [40], cerebral cortex [37], thalamus [41], basal ganglia, midbrain, pons, and medulla [42].
In a transgenic mouse model of SCA3 harboring 84 glutamine repeats in the mutant gene, abnormal Purkinje neuron excitability has been found to be associated with the onset of motor impairment. Recordings from acute cerebellar slices from this model show that SCA3 Purkinje neurons have increased intrinsic excitability that causes them to undergo depolarization block and rendered them unable to sustain repetitive firing. Recordings were made at an age where animals were symptomatic but lacked any visible signs of neurodegeneration, suggesting that the abnormal neuronal activity is responsible for the motor dysfunction. The abnormal Purkinje neuron excitability can be improved in cerebellar slices by SKA-31, an activator of calcium-activated potassium channels. The role for abnormal neuronal excitability in motor dysfunction is further supported by an improvement in motor impairment following administration of SKA-31 in vivo [12].
SCA6
SCA6 is a dominantly inherited cerebellar ataxia that arises as a result of CAG triplet expansion in CACNA1A, the gene encoding the P/Q-type calcium channel Cav2.1. Patients with SCA6 typically present with a pure cerebellar ataxia, and multisystem involvement is generally uncommon [43]. This relatively pure cerebellar ataxia parallels the cellular histology, which shows a disproportionate loss of Purkinje neurons over other cerebellar and brainstem neurons [44].
Because this disorder is caused by mutations in an ion channel, one might anticipate that there are clear abnormalities in ion channel function that contribute to neuronal dysfunction and might even potentially underlie the observed pathology. However, the evidence regarding abnormal function of polyQ-expanded Cav2.1 is inconsistent. Studies in recombinant systems expressing CACNA1A cDNA from SCA6 patients suggest that the expanded CAG tract induces a hyperpolarizing shift in channel activation that could produce increased calcium load [45–47]. However, studies in Purkinje cells from the SCA6 knock-in mouse model have demonstrated no abnormalities in the intrinsic electrophysiologic properties of mutant CaV2.1 [13, 48], although it should be noted that these data might not be able to reflect accurately abnormalities in dendritic calcium currents because of technical limitations associated with whole-cell somatic patch clamp recordings.
Recent work suggests that SCA6 may not arise from abnormalities or dysfunction associated with the CaV2.1 calcium channel, but rather from abnormalities in a second protein product that is generated from the same transcript. It has been demonstrated that CACNA1A is a bicistronic gene that produces 2 separate proteins products from a single mRNA transcript [49]. The first product is the pore-forming α1A subunit of the P/Q-type calcium channel, which is produced through canonical cap-dependent translation, and the second is a novel 75-kDa protein formed from the C terminus of the α1A subunit (termed α1-ACT), which arises from noncanonical translation at a cryptic internal ribosome entry site. Further characterization of this protein product has suggested that it is a nuclear transcription factor that promotes expression of genes involved in Purkinje neuron dendrite development and synapse formation.
SCA6 CAG tract expansions in CACNA1A fall within the coding sequence for α1-ACT, and SCA6 mutations appear to promote disease through both loss of function and toxic gain-of-function in α1-ACT [49, 50]. A number of consequences are postulated to arise from expression of this mutated transcription factor, but it is worth highlighting the fact that α1-ACT loss of function has a major impact on the integrity of parallel fiber synapses onto Purkinje neurons. This finding suggests that irrespective of whether or not P/Q-type channel function is normal in SCA6, there are still important consequences of SCA6 CACNA1A CAG tract expansion on Purkinje neuron activity that likely contribute to motor symptoms.
HD
HD is a fatal, progressive neurodegenerative disease that is characterized by cognitive dysfunction, psychiatric disturbance, and impairment of motor coordination. HD arises as a result of expansion of the CAG repeat in exon 1 of the huntingtin (HTT) gene [51]. In HD, the most striking pathology is observed in the striatum and cortex [52]. Importantly, a number of other areas of the brain have been noted to be affected in this condition, including the globus pallidus, hypothalamus [53], thalamus, and cerebellum [54].
Emerging evidence from several mouse models of HD has suggested that a number of different neuronal populations show abnormal activity that likely contributes to the motor and cognitive impairment seen in HD. The areas where this has been most effectively investigated are the medium spiny neurons (MSNs) of the striatum, the layer II/III pyramidal neurons of the cortex, and the Purkinje neurons of the cerebellum. Below, we present the current literature regarding abnormal neuronal activity in mouse models of HD. For the sake of clarity, the research that has been done examining dysfunction in each of these cell types is presented separately, and special attention is paid to establishing the timing of these changes and their relationship to the development of symptoms. This work has been derived primarily from 3 of the many rodent models of HD (reviewed in [55]), namely the R6/2 model, which expresses a fragment of the HTT gene containing the promoter, exon 1, and intron 1 [16]; the YAC128 model, which expresses full-length HTT within a yeast artificial chromosome [56]; and the HdhQ200 model, which contains a 200-repeat expansion of the endogenous CAG tract in the mouse Htt gene [57].
MSN Dysfunction in HD Models
One the most prominently affected cell types in HD are the MSNs of the striatum. There is substantial evidence in multiple HD models that early changes in multiple neurotransmitter systems that converge onto the MSN neurons correspond with the onset of motor symptoms. Glutamatergic inputs from the cortex appear to show abnormal increases in activity at the time when symptoms are first apparent, as evidenced by studies showing increased α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid responsiveness in MSNs from YAC128 HD mice [58], and increased large amplitude synaptic events in MSNs from the R6/2 model [59]. gamma-aminobutyric acid-ergic interneuron populations within the striatum also show abnormalities at this time, with MSNs from the R6/2 model demonstrating increases in the frequency of spontaneous inhibitory postsynaptic currents (IPSCs) that appear to originate from these interneuron populations rather than the MSNs themselves [60, 61]. Importantly, these changes have consequences for striatal network function, including a reduction in correlated firing of MSNs [62] and the emergence of glutamate-dependent MSN hyperactivity [63]. Taken together, these data suggest that the development of early motor symptoms associated with striatal dysfunction might reflect aberrant signaling through corticostriatal and intrastriatal inputs across multiple transmitter systems that influence MSN activity.
Once animals are fully symptomatic, there are additional changes in MSN synaptic function. Paradoxically, the changes seen in MSNs from symptomatic HD mice all reflect a reduction in synaptic excitability in MSNs. The corticostriatal glutamatergic inputs show a reduced capacity to excite MSNs, and there are multiple changes that indicate reduced efficacy at these synapses such as a reduction in spontaneous events [59], evoked synaptic events [58], and mini-EPSCs [59]. These reductions correlate with alterations in synaptic architecture and loss of dendritic spines [59, 64], suggesting that in more advanced disease the MSNs become disconnected from glutamatergic corticostriatal inputs. Meanwhile, the gamma-aminobutyric acid-ergic interneurons that synapse onto MSNs continue to exert an increased inhibitory effect [60]. One would anticipate that the net effect of these two coincident changes would be a dramatic suppression of MSN function, presumably resulting in more profound motor dysfunction.
In addition to changes in synaptic transmission onto MSNs, there are also changes in the intrinsic excitability of the MSNs that appear to reflect changes in intrinsic rather than synaptic conductances. The observable changes in MSN intrinsic excitability are profound, including a depolarized membrane potential [65] and altered interspike interval [66]. Some of these changes are linked to changes in specific channels, notably reduced inwardly-rectifying potassium current resulting from loss of Kir2.1 and Kir2.3 [67], and decreased voltage-gated Ca2+ currents [68]. It is unclear whether these changes reflect a primary consequence of mutant huntingtin expression or are a secondary consequence of neuronal atrophy or synaptic dysfunction. Nevertheless all of these changes would be characterized as increasing the excitability of MSNs, suggesting that increased MSN intrinsic excitability contributes to network dysfunction in HD.
An exciting novel finding is that astrocytes may play a role in increased MSN excitability observed in HD mouse models. Reduced activity of the astrocyte potassium channel Kir4.1 in HD mice was associated with increased extracellular potassium concentrations and a depolarized MSN membrane potential [69]. These data suggest that astrocytes might represent an important target for normalizing MSN function, in addition to the MSNs themselves.
Cortical Pyramidal Neuron Dysfunction in HD Models
In the cortex, pyramidal neurons of layers II, III, and V are the most prominently affected cells. The few studies investigating cortical pyramidal neuron activity in mouse models of HD suggest that pyramidal neuron dysfunction is not apparent until the onset of symptoms. Once motor and cognitive dysfunction becomes apparent, there are clear changes in both synaptic transmission and intrinsic excitability in cortical neurons.
A study using multiple HD models has demonstrated a common pattern of synaptic dysfunction in cortical neurons from symptomatic animals that is different in several respects from what is seen in MSNs from symptomatic HD animals. Pyramidal neurons from these models show an increase in both excitatory synaptic influence [increased spontaneous excitatory postsynaptic current (EPSC) frequency and evoked EPSC amplitude] and inhibitory synaptic influence (increased miniature IPSC amplitude and frequency, as well increased evoked IPSC amplitude) [70]. These data highlight the fact that despite the ubiquitous expression of mutant huntingtin, there are different functional consequences associated with expression of this mutant protein in different cell types, presumably related to the normal complement of synaptic proteins and ion channels that these cells express.
Analysis of intrinsic excitability in cortical neurons has largely been limited to the R6/2 model. Layer II/III neurons from symptomatic R6/2 mice undergo many of the same passive membrane changes that were observed in medium spiny neurons, notably a depolarized resting membrane potential and increased input resistance [70]. Given that the neurons at this age have already undergone substantial atrophy, it is difficult to distinguish whether these changes reflect changes in the expression or function of membrane ion channels, or whether those changes simply reflect the effect of small cell size on input resistance and membrane potential. However, it is worth noting that there is evidence for changes in active conductances expressed in pyramidal neurons, notably voltage-gated calcium currents, which show an increase in late symptomatic R6/2 animals [71].
Purkinje Neuron Dysfunction in HD Models
The cerebellum, like other non-neostriatal structures, has been traditionally viewed as largely unaffected in HD, but recent evidence from human autopsy samples suggests that there is a greater degree of cerebellar pathology in HD than had previously been appreciated, including Purkinje neuron loss, cerebellar nuclear neuron loss, and reactive astrogliosis within the cerebellum [72]. Recent evidence from several models of HD suggests that dysfunctional pacemaking in Purkinje neurons contributes to motor dysfunction and can precede the onset of obvious neuronal pathology. In the R6/2 model of HD, there is a striking reduction in Purkinje neuron firing frequency by postnatal day 28. It should be noted that there are no huntingtin inclusions in the Purkinje neurons until 12 weeks [73]. In the HdhQ200 model of HD, recordings were performed at a time where there was prominent Purkinje neuron loss, and loose patch electrophysiology of remaining Purkinje neurons identified a similar reduction in Purkinje neuron firing frequency in the HD mice compared with control littermates [74].
Potential Contributions of Altered Neuronal Function to Neuronal Death
As discussed above, for many of the polyQ conditions there is evidence to suggest that neuronal dysfunction accompanies the development of symptoms and precedes the development of overt cellular pathology. This raises a question: Is there any link between neuronal dysfunction and cellular pathology? This hypothesis can potentially explain the cell-type specific toxicity of these conditions because it is the cell types that go on to develop selective neuropathology that show initial dysfunction in many of the models. Indeed, studies of both patients with polyQ expansion diseases and mouse models of these diseases have contributed to a growing body of evidence linking changes in function directly to neuropathology (outlined in Fig. 2).
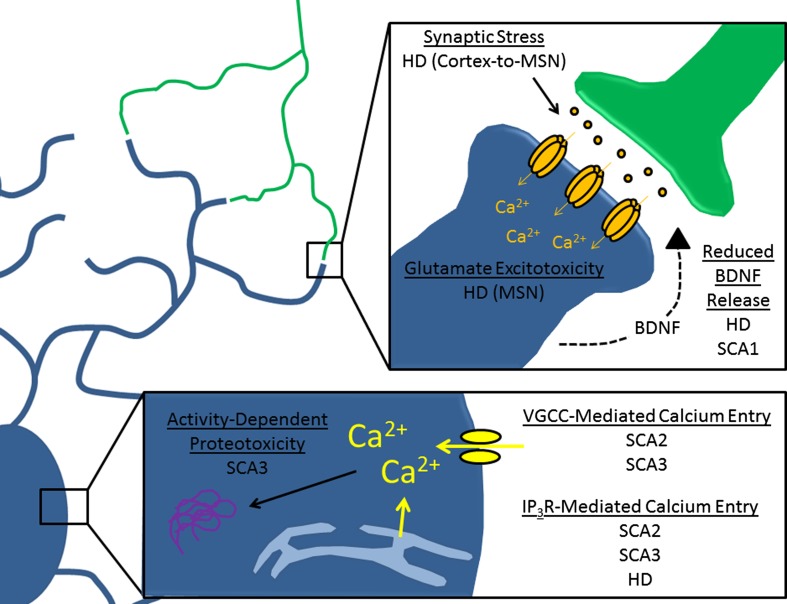
Fig. 2.
Mechanisms of toxicity linked to abnormal neuronal activity in polyglutamine (polyQ) disorders. This diagram highlights mechanisms by which abnormal neuronal activity might produce toxic injury in affected cells. The bottom inset summarizes changes associated with calcium handling, while the top inset summarizes changes associated with synaptic signaling. HD = Huntingdon disease; MSN = medium spiny neuron; BDNF = brain-derived neurotrophic factor; SCA = spinocerebellar ataxia; VGCC = voltage-gated calcium channels; IP3R = inositol-1,4,5 triphosphate receptor
In HD, there is mounting evidence highlighting a role for excitoxicity in the development of pathology. Excitoxicity is a process in which N-methyl-D-aspartate (NMDA) receptor hyperactivation results in toxic changes in a neuron that occur by a variety of mechanisms [75]. There is extensive evidence from both human and rodent studies to suggest that excitotoxic insult is a component of neurodegeneration in HD, particularly in the MSNs of the striatum. Relevant findings from rodents include the observation that kainic acid and other NMDA agonists can produce an HD-like syndrome and the observation that mouse models of HD show increased sensitivity to NMDA receptor agonists [76, 77]. There is also evidence for NMDA receptor-dependent toxicity in humans. Notably, NMDA receptor radioligand binding in the striatum is disproportionate to neuronal loss, arguing that those cells expressing high levels of NMDA receptor may be disproportionately affected in HD [78, 79]. It should be noted that excitoxicity likely represents the combined influence of altered synaptic signaling and altered intrinsic excitability. In HD, in particular, the evidence for early increases in both synaptic glutamatergic stimulation and intrinsic excitability in MSNs and cortical neurons suggests that these 2 types of changes might be acting in synchrony to promote excitotoxic damage.
There is evidence from mouse models to suggest that aberrant signaling through corticostriatal afferents may be particularly important in the establishment of striatal pathology. Perhaps the strongest evidence that these afferents contribute to pathology is the observation that lesions of the cortex [80] or selective removal of mutant huntingtin from cortical pyramidal neurons [81] can improve the behavioral and pathological HD phenotype in HD mouse models. In the context of these observations, it appears that synaptic stress at corticostriatal synapses might be a significant contributor to MSN pathology and might also explain the relationship between cortical and striatal pathology.
Studies of several dominant ataxias caused by polyQ tract expansion have not only demonstrated a role for altered excitability in pathology, but have also demonstrated that targeting excitability is a viable neuroprotective strategy in these conditions. In a mouse model of SCA1, as mentioned previously, Purkinje neurons show reduced firing frequency preceding the onset of neurodegenerative changes or motor symptoms. Treatment of SCA1 mice in vivo with aminopyridines, which corrects the abnormal firing frequency of Purkinje neurons, is able to limit the development of motor symptoms and also protects Purkinje cells from atrophy [28]. This effect was attributed to inhibition of Kv4 voltage-gated potassium channel family members, although the potential effect of aminopyridines in potentiating calcium-activated potassium channels was overlooked in this study [82]. Irrespective of the precise channel that mediates the neuroprotective effect, these data support a role for altered excitability in the development of cellular pathology and suggest that correcting aberrant membrane excitability is a viable therapeutic target in SCA1.
Similarly, a study in SCA2 has demonstrated that manipulating membrane excitability through modulating ion channel function serves to protect Purkinje neurons from neurodegenerative changes. Abnormal bursting behavior of Purkinje neurons has been identified in a mouse model of SCA2, and it was found that this bursting could be corrected with the application of a small conductance calcium-activated potassium (SK) channel activator. The same SK channel activator also alleviates behavioral and neuropathological changes in SCA2 mice [83]. Taken together, these data support a role for altered excitability in promoting pathology in SCA2.
A study in induced pluripotent stem cells (iPSC) from human SCA3 patients suggests a potential link between neuronal function and another known component of SCA3 pathology: aberrant protein processing. The formation of early aggregation intermediates is suggested to play an important role in polyQ diseases [84], and a study done in iPSC-derived neurons from subjects with SCA3 suggests that excitation of neurons might contribute to the formation of such intermediates [85]. These data demonstrate that L-glutamate-induced excitation of patient-specific iPSC-derived neurons results in ATXN3 cleavage and aggregation that is activity-dependent. It is speculated that this cleavage and aggregation is mediated by calcium entry through voltage-gated calcium channels (VGCC), which subsequently activates calpain family proteases that then cleave SCA3 to a more aggregate-prone form. These data demonstrate a direct connection between neuronal activity and cellular pathology. When this work is considered together with work characterizing elevated excitability of Purkinje neurons from a transgenic mouse model of SCA3 [12], a potential hypothesis of SCA3 pathogenesis could be considered, which views abnormalities in neuronal function as a potent modifier of mutant ATXN3 aggregation and toxicity.
Common Mechanisms of Disease
Across the polyQ diseases, there are common processes that are associated with changes in neuronal function. These processes might explain the mechanistic connection between expression of the expanded polyQ tract, altered neuronal activity, and degenerative changes in affected cells. Therefore, they not only inform our understanding of pathology, but also present themselves as useful targets for modifying cellular pathology influenced by altered neuronal function.
Transcriptional Dysregulation of Ion Channel Genes in PolyQ Diseases
It remains an open question as to how the CAG tract expansion induces changes in neuronal activity. This relationship is not very well studied, but there is emerging evidence in multiple polyQ diseases to suggest that the link may involve transcriptional dysregulation of ion channel genes and genes regulating synaptic components. As mentioned previously, studies in SCA6 have suggested that disease-causing mutations interfere with the normal function of a transcription factor that promotes stability of parallel fibers synapses onto Purkinje neurons [49]. In addition, gene ontology analysis of whole transcriptome sequencing data from patients with HD and from mouse models of SCA1 has revealed that ion channels are among the most prominently affected categories of genes in these conditions [86, 87]. Taken together, these data hint at mRNA production or processing as the site for pathogenic changes that result in pathogenic neuronal activity in polyQ diseases. Further studies will need to be done in order to assess the extent to which this is a general property of polyQ diseases, and to then assess whether transcriptional changes are sufficient to explain the abnormal activity that is observed.
Excitability, Altered Brain-derived Neurotrophic Factor Signaling, and Neurodegeneration
It is possible that normal neuronal activity provides trophic support that maintains appropriate morphology and survival. There is some evidence to suggest that alterations in activity-dependent secretion of neurotrophins, specifically the neurotrophin brain-derived neurotrophic factor (BDNF), are present in mouse models of disease. Because BDNF secretion in neurons is known to be regulated by neuronal activity [88–90], altered activity might be sufficient to produce pathologic changes in BDNF secretion. Indeed, there is emerging evidence for both HD and SCA1 that this may be the case.
In HD, reduced function and expression of striatal BDNF signaling is well documented in patient autopsy material and multiple animal models [91]. Studies have highlighted a direct role for the regulation of BDNF expression by wild-type huntingtin [92], and have suggested that the reduced BDNF expression observed in HD reflects a loss of this normal transcriptional function [93, 94]. Interestingly, some studies have shown a link between neuronal activity and transcriptional BDNF disruption. A cell culture model of cortical neuron microcircuits showed that blocking BDNF signaling could disrupt neuronal activity in HD neurons, but also showed that HD neurons themselves had reduced activity-dependent secretion of BDNF [95]. Taken together, these data point to a downward spiral effect, where initial changes in BDNF and changes in function might exacerbate one another.
In SCA1, preliminary evidence implicates activity-dependent changes in BDNF signaling in Purkinje neuron degeneration. Purkinje neurons show activity-dependent BDNF/TrkB signaling, with reduced activity tending to result in reduced signaling [90]. As described previously, a mouse model of SCA1 shows reduced firing frequency that could be corrected through aminopyridine treatment. Interestingly, the same interventions that are capable of restoring normal neuronal activity in Purkinje neurons also produce an increase in BDNF protein expression and a reduction in atrophy. Although these data are only correlational, they suggest that normalizing neuronal activity might rescue atrophy through BDNF. These findings also raise the possibility that the development of atrophy might reflect loss of activity-dependent BDNF trophic support [28].
Pathologic Calcium Handling in PolyQ Disorders
There is evidence to suggest that VGCC signaling is linked to huntingtin toxicity. One study found that mutant huntingtin may bind directly to the α2/δ auxiliary subunit (CACNA2D1) of VGCCs, although the effect of this binding on calcium channel function was not explored [96]. In another study, N-terminal fragments of exon 1-containing huntingtin were found to bind to the CaV2.2 pore-forming subunit of the N-type Ca2+ channel and block its regulation by syntaxin 1A, thereby causing these channels to allow for greater Ca2+ influx [97]. These data are in line with the hypothesis that expression of mutant huntingtin can increase calcium influx in a cell-autonomous manner. One would predict that this increase in calcium entry might have consequences for neuronal health, and a corollary to this prediction is that reducing calcium entry might be neuroprotective. In fact, it has been demonstrated that removal of the Drosophila L-type Ca2+ channel pore-forming subunit (Dmca1D) suppresses the eye degeneration phenotype of an HD fly model [98].
In addition to altered VGCC function, there is evidence to suggest that HD pathogenesis involves increased activity of the inositol-1,4,5 triphosphate (IP3) receptor, a receptor that mediates release of calcium from the endoplasmic reticulum. Mutant huntingtin has been identified to bind to the type 1 IP3 receptor and to increase its activity in MSNs [99]. This increase in activity was linked directly to MSN degeneration when several studies suggested that blocking IP3 receptor activity or preventing interactions between mutant huntingtin and type 1 IP3 receptor could protect MSN neurons from the YAC128 model of HD both in vitro and in vivo [100, 101]. These studies complement studies of VGCC in HD, and point to both as potential calcium sources for promoting cell degeneration. Future studies will be necessary to investigate whether both calcium sources participate in neuronal pathology simply by allowing for cytosolic calcium entry, or whether there are also unique roles for calcium entering through VGCCs or IP3 receptors.
Altered calcium handling also appears to play an important role in the pathogenesis of several of the autosomal dominant polyQ expansion ataxias. Purkinje neurons in the SCAs undergo dramatic simplification of their otherwise complex dendritic arbor. Interestingly, several mouse models with mutations in the calcium channel genes Trpc3 and Grid2 (in moonwalker and lurcher mice, respectively) result in increased dendritic calcium load and are associated with similar Purkinje neuron dendritic atrophy [102, 103].
For SCA1, the evidence supporting a role for calcium in Purkinje neurodegeneration is correlative, linked specifically to changes in the expression of the cytosolic calcium buffering factor calbindin. These studies from SCA1 mouse models show that changes in calcium buffering capacity correlate with the onset of neuropathology [104] and modify the progression of SCA1 motor dysfunction [105]. Taken together, these data suggest that altered calcium buffering capacity might contribute to atrophy and death in SCA1, and highlight atrophy as a potential driver of pathology.
For SCA2, it also appears that abnormal calcium handling contributes to pathology. In cerebellar Purkinje neurons, calcium-activated potassium channels serve as regulators of calcium entry by passing a repolarizing potassium current in response to increased cytosolic calcium concentrations. Interestingly, modulating the activity of calcium-activated potassium channels can rescue cellular pathology in a mouse model of SCA2, as described previously. Given the established role for these channels in regulating calcium entry, these data suggest that mitigating the influence of pathologic calcium handling by the cell could be neuroprotective [83].
Just as with HD, there is evidence that the endoplasmic reticulum calcium release mediated by the IP3 receptor may play a role in neuropathology for several of the SCAs. In both SCA2 and SCA3, the polyQ-expanded form of the protein binds to the IP3 receptor and increases IP3-mediated calcium release [106, 107]. Furthermore, chronic suppression of IP3 signaling in vivo reduces neuronal dysfunction and cell death in a transgenic mouse model of SCA2 [108] and SCA3 [106]. Taken together, these data support a role for increased IP3-mediated calcium release in the pathogenesis of SCA2 and SCA3.
Connecting Altered Neuronal Activity With Circuit Dysfunction
In all of the polyQ disease models mentioned in this review, there are 2 distinct phenomena that likely contribute to symptoms: early neuronal dysfunction and subsequent cell loss. This begs the question of whether abnormal neuronal activity contributes to circuit dysfunction in the same manner as neuronal loss, but to a lesser extent. We would argue that this is in fact the case, because the changes taking place in synaptic transmission or intrinsic excitability tend to have the aggregate effect of degrading the information encoded by a particular neuron. Through that framework, one can view early symptoms as partial “dropout” of neurons from the network before cell death completely eliminates them as participants.
Conclusions
Research in polyQ diseases is progressing rapidly, and the body of work highlighted in this review demonstrates that abnormal neuronal activity likely contributes to the development of symptoms and may also participate in the development of neuronal loss. Many unanswered questions still remain regarding the relationship between neuronal dysfunction, progression of symptoms, and, ultimately, neuronal death. Understanding the role of aberrant neuronal activity in disease pathogenesis is likely to yield more insights into the development of neuroprotective or symptomatic treatments, or both.
Electronic supplementary material
(PDF 1224 kb)
Acknowledgments
This work was supported by National Institutes of Health grants K08NS072158 and R01NS085054 (VGS) and through the University of Michigan Medical Scientist Training Program–National Institutes of Health grant T32GM007863 (RC).
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
References
- 1.La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 2.MacDonald ME, Ambrose CM, Duyao MP, et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 3.Orr HT, Chung MY, Banfi S, et al. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat Genet. 1993;4:221–226. doi: 10.1038/ng0793-221. [DOI] [PubMed] [Google Scholar]
- 4.Kawaguchi Y, Okamoto T, Taniwaki M, et al. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat Genet. 1994;8:221–228. doi: 10.1038/ng1194-221. [DOI] [PubMed] [Google Scholar]
- 5.Koide R, Ikeuchi T, Onodera O, et al. Unstable expansion of CAG repeat in hereditary dentatorubral-pallidoluysian atrophy (DRPLA) Nat Genet. 1994;6:9–13. doi: 10.1038/ng0194-9. [DOI] [PubMed] [Google Scholar]
- 6.Pulst SM, Nechiporuk A, Nechiporuk T, et al. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat Genet. 1996;14:269–276. doi: 10.1038/ng1196-269. [DOI] [PubMed] [Google Scholar]
- 7.Lindblad K, Savontaus ML, Stevanin G, et al. An expanded CAG repeat sequence in spinocerebellar ataxia type 7. Genome Res. 1996;6:965–971. doi: 10.1101/gr.6.10.965. [DOI] [PubMed] [Google Scholar]
- 8.Zhuchenko O, Bailey J, Bonnen P, et al. Autosomal dominant cerebellar ataxia (SCA6) associated with small polyglutamine expansions in the alpha 1A-voltage-dependent calcium channel. Nat Genet. 1997;15:62–69. doi: 10.1038/ng0197-62. [DOI] [PubMed] [Google Scholar]
- 9.Koide R, Kobayashi S, Shimohata T, et al. A neurological disease caused by an expanded CAG trinucleotide repeat in the TATA-binding protein gene: a new polyglutamine disease? Hum Mol Genet. 1999;8:2047–2053. doi: 10.1093/hmg/8.11.2047. [DOI] [PubMed] [Google Scholar]
- 10.Burright EN, Clark HB, Servadio A, et al. SCA1 transgenic mice: a model for neurodegeneration caused by an expanded CAG trinucleotide repeat. Cell. 1995;82:937–948. doi: 10.1016/0092-8674(95)90273-2. [DOI] [PubMed] [Google Scholar]
- 11.Huynh DP, Figueroa K, Hoang N, Pulst SM. Nuclear localization or inclusion body formation of ataxin-2 are not necessary for SCA2 pathogenesis in mouse or human. Nat Genet. 2000;26:44–50. doi: 10.1038/79162. [DOI] [PubMed] [Google Scholar]
- 12.Shakkottai VG. do Carmo Costa M, Dell’Orco JM, Sankaranarayanan A, Wulff H, Paulson HL. Early changes in cerebellar physiology accompany motor dysfunction in the polyglutamine disease spinocerebellar ataxia type 3. J Neurosci. 2011;31:13002–13014. doi: 10.1523/JNEUROSCI.2789-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watase K, Barrett CF, Miyazaki T, et al. Spinocerebellar ataxia type 6 knockin mice develop a progressive neuronal dysfunction with age-dependent accumulation of mutant CaV2.1 channels. Proc Natl Acad Sci U S A. 2008;105:11987–11992. doi: 10.1073/pnas.0804350105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yvert G, Lindenberg KS, Picaud S, Landwehrmeyer GB, Sahel JA, Mandel JL. Expanded polyglutamines induce neurodegeneration and trans-neuronal alterations in cerebellum and retina of SCA7 transgenic mice. Hum Mol Genet. 2000;9:2491–2506. doi: 10.1093/hmg/9.17.2491. [DOI] [PubMed] [Google Scholar]
- 15.Kelp A, Koeppen AH, Petrasch-Parwez E, et al. A novel transgenic rat model for spinocerebellar ataxia type 17 recapitulates neuropathological changes and supplies in vivo imaging biomarkers. J Neurosci. 2013;33:9068–9081. doi: 10.1523/JNEUROSCI.5622-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mangiarini L, Sathasivam K, Seller M, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 17.Sato T, Miura M, Yamada M, et al. Severe neurological phenotypes of Q129 DRPLA transgenic mice serendipitously created by en masse expansion of CAG repeats in Q76 DRPLA mice. Hum Mol Genet. 2009;18:723–736. doi: 10.1093/hmg/ddn403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams AJ, Paulson HL. Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci. 2008;31:521–528. doi: 10.1016/j.tins.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shao J, Diamond MI. Polyglutamine diseases: emerging concepts in pathogenesis and therapy. Hum Mol Genet. 2007;16:R115–R123. doi: 10.1093/hmg/ddm213. [DOI] [PubMed] [Google Scholar]
- 20.Zuccato C, Valenza M, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol Rev. 2010;90:905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- 21.Orr HT. Polyglutamine neurodegeneration: expanded glutamines enhance native functions. Curr Opin Genet Dev. 2012;22:251–255. doi: 10.1016/j.gde.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsuji S. Dentatorubral-pallidoluysian atrophy. Handb Clin Neurol. 2012;103:587–594. doi: 10.1016/B978-0-444-51892-7.00041-3. [DOI] [PubMed] [Google Scholar]
- 23.Nagafuchi S, Yanagisawa H, Sato K, et al. Dentatorubral and pallidoluysian atrophy expansion of an unstable CAG trinucleotide on chromosome 12p. Nat Genet. 1994;6:14–18. doi: 10.1038/ng0194-14. [DOI] [PubMed] [Google Scholar]
- 24.Naito H, Oyanagi S. Familial myoclonus epilepsy and choreoathetosis: hereditary dentatorubral-pallidoluysian atrophy. Neurology. 1982;32:798–807. doi: 10.1212/wnl.32.8.798. [DOI] [PubMed] [Google Scholar]
- 25.Yagishita S, Inoue M. Clinicopathology of spinocerebellar degeneration: its correlation to the unstable CAG repeat of the affected gene. Pathol Int. 1997;47:1–15. doi: 10.1111/j.1440-1827.1997.tb04429.x. [DOI] [PubMed] [Google Scholar]
- 26.Donato SD, Mariotti C, Taroni F. Spinocerebellar ataxia type 1. Handb Clin Neurol. 2012;103:399–421. doi: 10.1016/B978-0-444-51892-7.00025-5. [DOI] [PubMed] [Google Scholar]
- 27.Ferrer I, Genis D, Davalos A, Bernado L, Sant F, Serrano T. The Purkinje cell in olivopontocerebellar atrophy. A Golgi and immunocytochemical study. Neuropathol Appl Neurobiol. 1994;20:38–46. doi: 10.1111/j.1365-2990.1994.tb00955.x. [DOI] [PubMed] [Google Scholar]
- 28.Hourez R, Servais L, Orduz D, et al. Aminopyridines correct early dysfunction and delay neurodegeneration in a mouse model of spinocerebellar ataxia type 1. J Neurosci. 2011;31:11795–11807. doi: 10.1523/JNEUROSCI.0905-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barnes JA, Ebner BA, Duvick LA, et al. Abnormalities in the climbing fiber-Purkinje cell circuitry contribute to neuronal dysfunction in ATXN1[82Q] mice. J Neurosci. 2011;31:12778–12789. doi: 10.1523/JNEUROSCI.2579-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ebner BA, Ingram MA, Barnes JA, et al. Purkinje cell ataxin-1 modulates climbing fiber synaptic input in developing and adult mouse cerebellum. J Neurosci. 2013;33:5806–5820. doi: 10.1523/JNEUROSCI.6311-11.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duvick L, Barnes J, Ebner B, et al. SCA1-like disease in mice expressing wild-type ataxin-1 with a serine to aspartic acid replacement at residue 776. Neuron. 2010;67:929–935. doi: 10.1016/j.neuron.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Auburger GW. Spinocerebellar ataxia type 2. Handb Clin Neurol. 2012;103:423–436. doi: 10.1016/B978-0-444-51892-7.00026-7. [DOI] [PubMed] [Google Scholar]
- 33.Orozco G, Estrada R, Perry TL, et al. Dominantly inherited olivopontocerebellar atrophy from eastern Cuba. Clinical, neuropathological, and biochemical findings. J Neurol Sci. 1989;93:37–50. doi: 10.1016/0022-510x(89)90159-7. [DOI] [PubMed] [Google Scholar]
- 34.Rub U, Del Turco D, Del Tredici K, et al. Thalamic involvement in a spinocerebellar ataxia type 2 (SCA2) and a spinocerebellar ataxia type 3 (SCA3) patient, and its clinical relevance. Brain. 2003;126:2257–2272. doi: 10.1093/brain/awg234. [DOI] [PubMed] [Google Scholar]
- 35.Rub U, Burk K, Schols L, et al. Damage to the reticulotegmental nucleus of the pons in spinocerebellar ataxia type 1, 2, and 3. Neurology. 2004;63:1258–1263. doi: 10.1212/01.wnl.0000140498.24112.8c. [DOI] [PubMed] [Google Scholar]
- 36.Gierga K, Burk K, Bauer M, et al. Involvement of the cranial nerves and their nuclei in spinocerebellar ataxia type 2 (SCA2) Acta Neuropathol. 2005;109:617–631. doi: 10.1007/s00401-005-1014-8. [DOI] [PubMed] [Google Scholar]
- 37.Rub U, Seidel K, Ozerden I, et al. Consistent affection of the central somatosensory system in spinocerebellar ataxia type 2 and type 3 and its significance for clinical symptoms and rehabilitative therapy. Brain Res Rev. 2007;53:235–249. doi: 10.1016/j.brainresrev.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 38.Hansen ST, Meera P, Otis TS, Pulst SM. Changes in Purkinje cell firing and gene expression precede behavioral pathology in a mouse model of SCA2. Hum Mol Genet. 2013;22:271–283. doi: 10.1093/hmg/dds427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paulson H. Machado-Joseph disease/spinocerebellar ataxia type 3. Handb Clin Neurol. 2012;103:437–449. doi: 10.1016/B978-0-444-51892-7.00027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scherzed W, Brunt ER, Heinsen H, et al. Pathoanatomy of cerebellar degeneration in spinocerebellar ataxia type 2 (SCA2) and type 3 (SCA3) Cerebellum. 2012;11:749–760. doi: 10.1007/s12311-011-0340-8. [DOI] [PubMed] [Google Scholar]
- 41.Rub U, de Vos RA, Brunt ER, et al. Spinocerebellar ataxia type 3 (SCA3): thalamic neurodegeneration occurs independently from thalamic ataxin-3 immunopositive neuronal intranuclear inclusions. Brain Pathol. 2006;16:218–227. doi: 10.1111/j.1750-3639.2006.00022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seidel K, Siswanto S, Brunt ER, den Dunnen W, Korf HW, Rub U. Brain pathology of spinocerebellar ataxias. Acta Neuropathol. 2012;124:1–21. doi: 10.1007/s00401-012-1000-x. [DOI] [PubMed] [Google Scholar]
- 43.Matsumura R, Futamura N, Fujimoto Y, et al. Spinocerebellar ataxia type 6. Molecular and clinical features of 35 Japanese patients including one homozygous for the CAG repeat expansion. Neurology. 1997;49:1238–1243. doi: 10.1212/wnl.49.5.1238. [DOI] [PubMed] [Google Scholar]
- 44.Gierga K, Schelhaas HJ, Brunt ER, et al. Spinocerebellar ataxia type 6 (SCA6): neurodegeneration goes beyond the known brain predilection sites. Neuropathol Appl Neurobiol. 2009;35:515–527. doi: 10.1111/j.1365-2990.2009.01015.x. [DOI] [PubMed] [Google Scholar]
- 45.Restituito S, Thompson RM, Eliet J, et al. The polyglutamine expansion in spinocerebellar ataxia type 6 causes a beta subunit-specific enhanced activation of P/Q-type calcium channels in Xenopus oocytes. J Neurosci. 2000;20:6394–6403. doi: 10.1523/JNEUROSCI.20-17-06394.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toru S, Murakoshi T, Ishikawa K, et al. Spinocerebellar ataxia type 6 mutation alters P-type calcium channel function. J Biol Chem. 2000;275:10893–10898. doi: 10.1074/jbc.275.15.10893. [DOI] [PubMed] [Google Scholar]
- 47.Matsuyama Z, Wakamori M, Mori Y, Kawakami H, Nakamura S, Imoto K. Direct alteration of the P/Q-type Ca2+ channel property by polyglutamine expansion in spinocerebellar ataxia 6. J Neurosci. 1999;19:Rc14. doi: 10.1523/JNEUROSCI.19-12-j0004.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saegusa H, Wakamori M, Matsuda Y, et al. Properties of human Cav2.1 channel with a spinocerebellar ataxia type 6 mutation expressed in Purkinje cells. Mol Cell Neurosci. 2007;34:261–270. doi: 10.1016/j.mcn.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 49.Du X, Wang J, Zhu H, et al. Second cistron in CACNA1A gene encodes a transcription factor mediating cerebellar development and SCA6. Cell. 2013;154:118–133. doi: 10.1016/j.cell.2013.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kordasiewicz HB, Thompson RM, Clark HB, Gomez CM. C-termini of P/Q-type Ca2+ channel alpha1A subunits translocate to nuclei and promote polyglutamine-mediated toxicity. Hum Mol Genet. 2006;15:1587–1599. doi: 10.1093/hmg/ddl080. [DOI] [PubMed] [Google Scholar]
- 51.Pouladi MA, Morton AJ, Hayden MR. Choosing an animal model for the study of Huntington’s disease. Nat Rev Neurosci. 2013;14:708–721. doi: 10.1038/nrn3570. [DOI] [PubMed] [Google Scholar]
- 52.Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- 53.Gabery S, Murphy K, Schultz K, et al. Changes in key hypothalamic neuropeptide populations in Huntington disease revealed by neuropathological analyses. Acta Neuropathol. 2010;120:777–788. doi: 10.1007/s00401-010-0742-6. [DOI] [PubMed] [Google Scholar]
- 54.Fennema-Notestine C, Archibald SL, Jacobson MW, et al. In vivo evidence of cerebellar atrophy and cerebral white matter loss in Huntington disease. Neurology. 2004;63:989–995. doi: 10.1212/01.wnl.0000138434.68093.67. [DOI] [PubMed] [Google Scholar]
- 55.Figiel M, Szlachcic WJ, Switonski PM, Gabka A, Krzyzosiak WJ. Mouse models of polyglutamine diseases: review and data table. Part I. Mol Neurobiol. 2012;46:393–429. doi: 10.1007/s12035-012-8315-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Slow EJ, van Raamsdonk J, Rogers D, et al. Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum Mol Genet. 2003;12:1555–1567. doi: 10.1093/hmg/ddg169. [DOI] [PubMed] [Google Scholar]
- 57.Heng MY, Duong DK, Albin RL, et al. Early autophagic response in a novel knock-in model of Huntington disease. Hum Mol Genet. 2010;19:3702–3720. doi: 10.1093/hmg/ddq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Joshi PR, Wu NP, Andre VM, et al. Age-dependent alterations of corticostriatal activity in the YAC128 mouse model of Huntington disease. J Neurosci. 2009;29:2414–2427. doi: 10.1523/JNEUROSCI.5687-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cepeda C, Hurst RS, Calvert CR, et al. Transient and progressive electrophysiological alterations in the corticostriatal pathway in a mouse model of Huntington’s disease. J Neurosci. 2003;23:961–969. doi: 10.1523/JNEUROSCI.23-03-00961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cepeda C, Starling AJ, Wu N, et al. Increased GABAergic function in mouse models of Huntington’s disease: reversal by BDNF. J Neurosci Res. 2004;78:855–867. doi: 10.1002/jnr.20344. [DOI] [PubMed] [Google Scholar]
- 61.Cepeda C, Galvan L, Holley SM, et al. Multiple sources of striatal inhibition are differentially affected in Huntington’s disease mouse models. J Neurosci. 2013;33:7393–7406. doi: 10.1523/JNEUROSCI.2137-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miller BR, Walker AG, Shah AS, Barton SJ, Rebec GV. Dysregulated information processing by medium spiny neurons in striatum of freely behaving mouse models of Huntington’s disease. J Neurophysiol. 2008;100:2205–2216. doi: 10.1152/jn.90606.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rebec GV, Conroy SK, Barton SJ. Hyperactive striatal neurons in symptomatic Huntington R6/2 mice: variations with behavioral state and repeated ascorbate treatment. Neuroscience. 2006;137:327–336. doi: 10.1016/j.neuroscience.2005.08.062. [DOI] [PubMed] [Google Scholar]
- 64.Singaraja RR, Huang K, Sanders SS, et al. Altered palmitoylation and neuropathological deficits in mice lacking HIP14. Hum Mol Genet. 2011;20:3899–3909. doi: 10.1093/hmg/ddr308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Levine MS, Klapstein GJ, Koppel A, et al. Enhanced sensitivity to N-methyl-D-aspartate receptor activation in transgenic and knockin mouse models of Huntington’s disease. J Neurosci Res. 1999;58:515–532. [PubMed] [Google Scholar]
- 66.Klapstein GJ, Fisher RS, Zanjani H, et al. Electrophysiological and morphological changes in striatal spiny neurons in R6/2 Huntington’s disease transgenic mice. J Neurophysiol. 2001;86:2667–2677. doi: 10.1152/jn.2001.86.6.2667. [DOI] [PubMed] [Google Scholar]
- 67.Ariano MA, Cepeda C, Calvert CR, et al. Striatal potassium channel dysfunction in Huntington’s disease transgenic mice. J Neurophysiol. 2005;93:2565–2574. doi: 10.1152/jn.00791.2004. [DOI] [PubMed] [Google Scholar]
- 68.Cepeda C, Ariano MA, Calvert CR, et al. NMDA receptor function in mouse models of Huntington disease. J Neurosci Res. 2001;66:525–539. doi: 10.1002/jnr.1244. [DOI] [PubMed] [Google Scholar]
- 69.Tong X, Ao Y, Faas GC, et al. Astrocyte Kir4.1 ion channel deficits contribute to neuronal dysfunction in Huntington’s disease model mice. Nat Neurosci. 2014;17:694–703. doi: 10.1038/nn.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cummings DM, Andre VM, Uzgil BO, et al. Alterations in cortical excitation and inhibition in genetic mouse models of Huntington’s disease. J Neurosci. 2009;29:10371–10386. doi: 10.1523/JNEUROSCI.1592-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Andre VM, Cepeda C, Venegas A, Gomez Y, Levine MS. Altered cortical glutamate receptor function in the R6/2 model of Huntington’s disease. J Neurophysiol. 2006;95:2108–2119. doi: 10.1152/jn.01118.2005. [DOI] [PubMed] [Google Scholar]
- 72.Rub U, Hoche F, Brunt ER, et al. Degeneration of the cerebellum in Huntington’s disease (HD): possible relevance for the clinical picture and potential gateway to pathological mechanisms of the disease process. Brain Pathol. 2013;23:165–177. doi: 10.1111/j.1750-3639.2012.00629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dougherty SE, Reeves JL, Lucas EK, Gamble KL, Lesort M, Cowell RM. Disruption of Purkinje cell function prior to huntingtin accumulation and cell loss in an animal model of Huntington disease. Exp Neurol. 2012;236:171–178. doi: 10.1016/j.expneurol.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dougherty SE, Reeves JL, Lesort M, Detloff PJ, Cowell RM. Purkinje cell dysfunction and loss in a knock-in mouse model of Huntington disease. Exp Neurol. 2013;240:96–102. doi: 10.1016/j.expneurol.2012.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang Y, Qin ZH. Molecular and cellular mechanisms of excitotoxic neuronal death. Apoptosis. 2010;15:1382–1402. doi: 10.1007/s10495-010-0481-0. [DOI] [PubMed] [Google Scholar]
- 76.Zeron MM, Hansson O, Chen N, et al. Increased sensitivity to N-methyl-D-aspartate receptor-mediated excitotoxicity in a mouse model of Huntington’s disease. Neuron. 2002;33:849–860. doi: 10.1016/s0896-6273(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 77.Graham RK, Pouladi MA, Joshi P, et al. Differential susceptibility to excitotoxic stress in YAC128 mouse models of Huntington disease between initiation and progression of disease. J Neurosci. 2009;29:2193–2204. doi: 10.1523/JNEUROSCI.5473-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Albin RL, Young AB, Penney JB, et al. Abnormalities of striatal projection neurons and N-methyl-D-aspartate receptors in presymptomatic Huntington’s disease. N Engl J Med. 1990;322:1293–1298. doi: 10.1056/NEJM199005033221807. [DOI] [PubMed] [Google Scholar]
- 79.Young AB, Greenamyre JT, Hollingsworth Z, et al. NMDA receptor losses in putamen from patients with Huntington’s disease. Science. 1988;241:981–983. doi: 10.1126/science.2841762. [DOI] [PubMed] [Google Scholar]
- 80.Stack EC, Dedeoglu A, Smith KM, et al. Neuroprotective effects of synaptic modulation in Huntington’s disease R6/2 mice. J Neurosci. 2007;27:12908–12915. doi: 10.1523/JNEUROSCI.4318-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gu X, Andre VM, Cepeda C, et al. Pathological cell-cell interactions are necessary for striatal pathogenesis in a conditional mouse model of Huntington’s disease. Mol Neurodegener. 2007;2:8. doi: 10.1186/1750-1326-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alvina K, Khodakhah K. The therapeutic mode of action of 4-aminopyridine in cerebellar ataxia. J Neurosci. 2010;30:7258–7268. doi: 10.1523/JNEUROSCI.3582-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kasumu AW, Hougaard C, Rode F, et al. Selective positive modulator of calcium-activated potassium channels exerts beneficial effects in a mouse model of spinocerebellar ataxia type 2. Chem Biol. 2012;19:1340–1353. doi: 10.1016/j.chembiol.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Takahashi T, Kikuchi S, Katada S, Nagai Y, Nishizawa M, Onodera O. Soluble polyglutamine oligomers formed prior to inclusion body formation are cytotoxic. Hum Mol Genet. 2008;17:345–356. doi: 10.1093/hmg/ddm311. [DOI] [PubMed] [Google Scholar]
- 85.Koch P, Breuer P, Peitz M, et al. Excitation-induced ataxin-3 aggregation in neurons from patients with Machado-Joseph disease. Nature. 2011;480:543–546. doi: 10.1038/nature10671. [DOI] [PubMed] [Google Scholar]
- 86.Hodges A, Strand AD, Aragaki AK, et al. Regional and cellular gene expression changes in human Huntington’s disease brain. Hum Mol Genet. 2006;15:965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- 87.Crespo-Barreto J, Fryer JD, Shaw CA, Orr HT, Zoghbi HY. Partial loss of ataxin-1 function contributes to transcriptional dysregulation in spinocerebellar ataxia type 1 pathogenesis. PLoS Genet. 2010;6:e1001021. doi: 10.1371/journal.pgen.1001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Balkowiec A, Katz DM. Cellular mechanisms regulating activity-dependent release of native brain-derived neurotrophic factor from hippocampal neurons. J Neurosci. 2002;22:10399–10407. doi: 10.1523/JNEUROSCI.22-23-10399.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ghiglieri V, Sgobio C, Patassini S, et al. TrkB/BDNF-dependent striatal plasticity and behavior in a genetic model of epilepsy: modulation by valproic acid. Neuropsychopharmacology. 2010;35:1531–1540. doi: 10.1038/npp.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Miki T, Hirai H, Takahashi T. Activity-dependent neurotrophin signaling underlies developmental switch of Ca2+ channel subtypes mediating neurotransmitter release. J Neurosci. 2013;33:18755–18763. doi: 10.1523/JNEUROSCI.3161-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zuccato C, Cattaneo E. Brain-derived neurotrophic factor in neurodegenerative diseases. Nat Rev Neurol. 2009;5:311–322. doi: 10.1038/nrneurol.2009.54. [DOI] [PubMed] [Google Scholar]
- 92.Zuccato C, Ciammola A, Rigamonti D, et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington’s disease. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 93.Zuccato C, Tartari M, Crotti A, et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- 94.Zuccato C, Belyaev N, Conforti P, et al. Widespread disruption of repressor element-1 silencing transcription factor/neuron-restrictive silencer factor occupancy at its target genes in Huntington’s disease. J Neurosci. 2007;27:6972–6983. doi: 10.1523/JNEUROSCI.4278-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gambazzi L, Gokce O, Seredenina T, et al. Diminished activity-dependent brain-derived neurotrophic factor expression underlies cortical neuron microcircuit hypoconnectivity resulting from exposure to mutant huntingtin fragments. J Pharmacol Exp Ther. 2010;335:13–22. doi: 10.1124/jpet.110.167551. [DOI] [PubMed] [Google Scholar]
- 96.Kaltenbach LS, Romero E, Becklin RR, et al. Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 2007;3:e82. doi: 10.1371/journal.pgen.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Swayne LA, Chen L, Hameed S, et al. Crosstalk between huntingtin and syntaxin 1A regulates N-type calcium channels. Mol Cell Neurosci. 2005;30:339–351. doi: 10.1016/j.mcn.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 98.Romero E, Cha GH, Verstreken P, et al. Suppression of neurodegeneration and increased neurotransmission caused by expanded full-length huntingtin accumulating in the cytoplasm. Neuron. 2008;57:27–40. doi: 10.1016/j.neuron.2007.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tang TS, Tu H, Chan EY, et al. Huntingtin and huntingtin-associated protein 1 influence neuronal calcium signaling mediated by inositol-(1,4,5) triphosphate receptor type 1. Neuron. 2003;39:227–239. doi: 10.1016/s0896-6273(03)00366-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tang TS, Slow E, Lupu V, et al. Disturbed Ca2+ signaling and apoptosis of medium spiny neurons in Huntington’s disease. Proc Natl Acad Sci U S A. 2005;102:2602–2607. doi: 10.1073/pnas.0409402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tang TS, Guo C, Wang H, Chen X, Bezprozvanny I. Neuroprotective effects of inositol 1,4,5-trisphosphate receptor C-terminal fragment in a Huntington’s disease mouse model. The J Neurosci. 2009;29:1257–1266. doi: 10.1523/JNEUROSCI.4411-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Becker EB, Oliver PL, Glitsch MD, et al. A point mutation in TRPC3 causes abnormal Purkinje cell development and cerebellar ataxia in moonwalker mice. Proc Natl Acad Sci U S A. 2009;106:6706–6711. doi: 10.1073/pnas.0810599106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zanjani HS, McFarland R, Cavelier P, et al. Death and survival of heterozygous Lurcher Purkinje cells in vitro. Develop Neurobiol. 2009;69:505–517. doi: 10.1002/dneu.20715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vig PJ, Subramony SH, Burright EN, et al. Reduced immunoreactivity to calcium-binding proteins in Purkinje cells precedes onset of ataxia in spinocerebellar ataxia-1 transgenic mice. Neurology. 1998;50:106–113. doi: 10.1212/wnl.50.1.106. [DOI] [PubMed] [Google Scholar]
- 105.Vig PJ, Wei J, Shao Q, Lopez ME, Halperin R, Gerber J. Suppression of calbindin-D28k expression exacerbates SCA1 phenotype in a disease mouse model. Cerebellum. 2012;11:718–732. doi: 10.1007/s12311-011-0323-9. [DOI] [PubMed] [Google Scholar]
- 106.Chen X, Tang TS, Tu H, et al. Deranged calcium signaling and neurodegeneration in spinocerebellar ataxia type 3. J Neurosci. 2008;28:12713–12724. doi: 10.1523/JNEUROSCI.3909-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu J, Tang TS, Tu H, et al. Deranged calcium signaling and neurodegeneration in spinocerebellar ataxia type 2. J Neurosci. 2009;29:9148–9162. doi: 10.1523/JNEUROSCI.0660-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kasumu AW, Liang X, Egorova P, Vorontsova D, Bezprozvanny I. Chronic suppression of inositol 1,4,5-triphosphate receptor-mediated calcium signaling in cerebellar purkinje cells alleviates pathological phenotype in spinocerebellar ataxia 2 mice. J Neurosci. 2012;32:12786–12796. doi: 10.1523/JNEUROSCI.1643-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1224 kb)