Abstract
Nucleotide repeat expansions underlie numerous human neurological disorders. Repeats can trigger toxicity through multiple pathogenic mechanisms, including RNA gain-of-function, protein gain-of-function, and protein loss-of-function pathways. Traditionally, inference of the underlying pathogenic mechanism derives from the repeat location, with dominantly inherited repeats within transcribed noncoding sequences eliciting toxicity predominantly as RNA via sequestration of specific RNA binding proteins. However, recent findings question this assumption and suggest that repeats outside of annotated open reading frames may also trigger toxicity through a novel form of protein translational initiation known as repeat-associated non-AUG (RAN) translation. To date, RAN translation has been implicated in 4 nucleotide repeat expansion disorders: spinocerebellar ataxia type 8; myotonic dystrophy type 1 with CTG•CAG repeats; C9orf72 amyotrophic lateral sclerosis/frontotemporal dementia with GGGGCC•GGCCCC repeats; and fragile X-associated tremor/ataxia syndrome with CGG repeats. RAN translation contributes to hallmark pathological characteristics in these disorders by producing homopolymeric or dipeptide repeat proteins. Here, we review what is known about RAN translation, with an emphasis on how differences in both repeat sequence and context may confer different requirements for unconventional initiation. We then discuss how this new mechanism of translational initiation might function in normal physiology and lay out a roadmap for addressing the numerous questions that remain.
Electronic supplementary material
The online version of this article (doi:10.1007/s13311-014-0292-z) contains supplementary material, which is available to authorized users.
Keywords: Fragile X, Polyglutamine, C9orf72, Translation initiation, Myotonic dystrophy, ALS
Introduction
Nucleotide repeat expansions underlie over a dozen human neurological diseases, ranging broadly in severity, symptoms, sites of pathology, and prevalence [1, 2]. Repeat expansions are thought to elicit toxicity via 3 nonexclusive mechanisms. Repeats can alter transcription in cis, leading to suppressed RNA and protein expression from the gene in which they reside [3, 4]. Alternatively, transcribed repeats as RNA can bind to and sequester RNA binding proteins and prevent them from performing their normal functions [5, 6]. Lastly, translated repeats can alter the normal functions of the proteins in which they reside while also directly eliciting toxicity via alterations in proteostasis [5, 7].
The dominant mechanisms by which a given repeat acts to elicit toxicity is dependent on numerous variables, including the length of the repeat, its sequence context, and the native functions of the protein-coding gene with which it is associated. For example, large CGG repeat expansions (>200 repeats in length, i.e., “full mutations”) in the 5’ untranslated region (UTR) of FMR1 elicit transcriptional and translational silencing of the FMR1 locus (loss-of-function) resulting in absent expression of the fragile X protein (FMRP) [8]. The absence of FMRP leads to fragile X syndrome, a common cause of intellectual disability and autism. In contrast, moderate CGG expansions in FMR1 (50–200 repeats, i.e., “premutations”) are actively transcribed, allowing the CGG repeat as RNA to bind to a number of key RNA-binding proteins and alter their functions [9–12]. For example, the microRNA biogenesis-associated proteins DGCR8 and DROSHA are sequestered by the expanded CGG repeat, subsequently eliciting alterations in microRNA abundance [12]. Similarly, expanded CUG repeats in the 3’UTR of DMPK mRNA bind to muscleblind-like protein splicing factors, leading to alterations in alternative splicing of muscleblind-like protein target transcripts [5, 13–21]. For trinucleotide repeat expansions located within canonical open reading frames (ORFs), as occurs with CAG repeats in HTT, the repeat is translated into the native protein. In Huntington’s disease, this leads to an expanded polyglutamine repeat within the Huntingtin protein that aggregates in affected tissues and that elicits toxicity via alterations in both proteostasis and in the native functions of the protein [22–26].
However, recent data from multiple laboratories suggest that nucleotide repeat expansions are capable of eliciting translational initiation in the absence of a normal ORF or AUG start codon [27–30]. This process, referred to as repeat-associated non-AUG (RAN) translation, has the potential to significantly alter our understanding of nucleotide repeat disorder pathogenesis. In this review, we describe the discovery of RAN translation and its association with neurodegenerative diseases. We then discuss how RAN translation might occur, with specific insights into the roles of sequence and repeat context, and the potential for similar processes to expand the functional proteome. Lastly, we highlight specific questions that the field must address going forward.
Eukaryotic Translation Initiation
Upon transcription and RNA processing (e.g., capping, splicing, and polyadenylation), eukaryotic mRNAs are exported from the nucleus into the cytoplasm to serve as templates for translation. The majority of eukaryotic nuclear-encoded mRNAs rely on cap-dependent translational machinery for initiation. In this process, the 5’ m7GpppG cap acts as a critical cis-element that recruits the cap-binding protein eukaryotic initiation factor (eIF) eIF4E into a complex known as eIF4F to allow unwinding of the mRNA and loading of the 43S preinitiation complex (PIC; comprised of the small 40S ribosomal subunit and the eIF2 ternary complex) on the 5’ end of the mRNA (reviewed elsewhere [31–33]). This PIC then scans along the mRNA in a 5’ to 3’ direction with the help of RNA helicases until it reaches an AUG start codon in an optimal context (e.g., Kozak sequence) [34–40], at which point the 60S ribosomal subunit is recruited and translation begins.
Alternatively, some mRNAs utilize a cap-independent mechanism for translation initiation that allow for internal ribosome entry of the 43S PIC onto the mRNA at highly structured elements known as internal ribosome entry sites (IRES) [41–44]. IRES elements can also sometimes directly recruit a subset of eIFs to allow for unconventional translational initiation [43–46]. In one extreme example, the cricket paralysis virus-IRES initiates translation independent of any eIFs or initiator tRNA [47–50]. Although initially described only in viral RNAs, there are now numerous examples of eukaryotic cellular IRES in transcripts such as c-myc, p53 [51], FGF2 [52], eIF4G/eIF4G1 [53, 54], and Apaf-1 [55].
Additionally, while initiation typically begins at the first AUG codon in an appropriate sequence context, there are numerous documented examples where near-cognate start codons (differ from AUG by one nucleotide, e.g., CUG or GUG) are utilized [56]. Recent ribosome profiling studies, which map the position of ribosomes on mRNAs across the transcriptome, suggest that such near-cognate initiator codon usage may be quite prevalent [57–59]. These alternative initiation events play important regulatory roles in protein production from their associated transcripts, implying that a lack of stringency may be built into the system [60].
RAN Translation in Neurodegenerative Disease
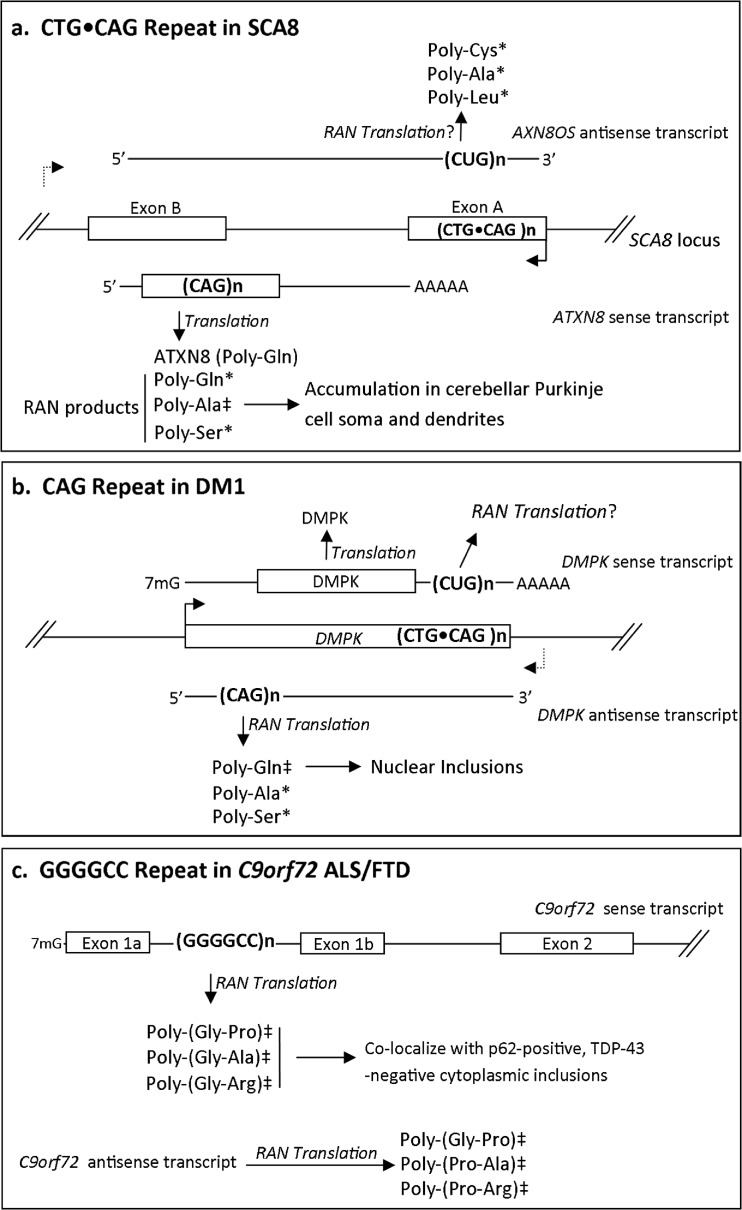
Thus far, RAN translation has been linked to CTG•CAG, GGGGCC•GGCCCC, and CGG microsatellite expansions (Figs. 1 and 2). RAN translation was initially reported by Zu et al. [27] in association with spinocerebellar ataxia type 8 (SCA8). SCA8 is a dominant, slowly progressive neurodegenerative disorder caused by a CAG•CTG repeat expansion within the coding sequence of the ATXN8/ATXN8OS gene [61–63]. Zu et al. [27] found that removal of the only ATG start codon from a SCA8 minigene construct failed to prevent production of a poly-Gln protein. Using a series of elegant epitope-tagged constructs, mass spectrometry, and tritium-labeling experiments, Zu et al. [27] demonstrated that translation of CAG repeats could occur without an AUG start codon in all 3 reading frames to produce poly-Gln, poly-Ala, and poly-Ser homopolymeric proteins. Immunofluorescence-based experiments demonstrated accumulation of all 3 RAN products within a single transfected cell, suggesting that these processes can occur in parallel. For SCA8 RAN-translated poly-Ala, mass spectroscopy measurements identified a series of peptides lacking an N-terminal methionine but with differing lengths of alanine peptides, suggesting initiation occurring throughout the GCA repeat itself. Evidence for similar translation in all 3 reading frames across CUG repeats from the ATXN8OS transcript was also observed, as was initiation in alternative ORFs when the repeats were placed within an AUG-initiated ORF. Zu et al. [27] found that antibodies targeting the predicted poly-Ala product from the ATXN8 transcript selectively recognized a protein in cerebellum in human SCA8 cases and SCA8 model mice. A similar approach provided in vivo evidence of a poly-Gln RAN translation product from an antisense CAG repeat containing transcript associated with the DMPK locus in myotonic dystrophy type 1 (DM1) (Fig. 1) [27, 64].
Fig. 1.
Repeat-associated non-AUG (RAN) translation at CAG, CUG, and GGGGCC repeats. (a) Spinocerebellar ataxia type 8 (SCA8) results from a CAG•CTG expansion in exon A of the ATXN8 gene, with an expanded CAG repeat in the sense transcript and CUG repeat in the antisense transcript, ATXN8OS. RAN translation produces poly-Gln-, poly-Ala-, and poly-Ser-containing proteins from ATXN8 and potentially poly-Ala-, poly-Cys-, and poly-Leu-containing proteins from ATXN8OS. The box in the ATXN8 sense transcript represents the open reading frame (ORF). The CAG repeat comprises the entire ORF. (b) Myotonic dystrophy type 1 (DM1) results from a CTG•CAG expansion in the 3’ untranslated region (UTR) of DMPK, where the CUG repeat resides in the 3’UTR and the CAG repeat is part of an antisense transcript of unknown function. In vivo antibody-based evidence suggests that a poly-Gln RAN product is produced in a DM1 mouse model and human tissues. (c) RAN translation of the expanded GGGGCC repeat located in intron 1 of the C9orf72 transcript results in 3 dipeptide repeat proteins: poly-(Gly-Pro), poly-(Gly-Ala), and poly-(Gly-Arg). In addition, an antisense transcript (containing GGCCCC repeats) also acts as a substrate for RAN translation, producing poly-(Gly-Pro), poly-(Pro-Ala), and poly-(Pro-Arg) dipeptide repeat proteins. *Evidence of production in vitro. ‡Evidence of production in vivo in patient tissues. ? = possible RAN products
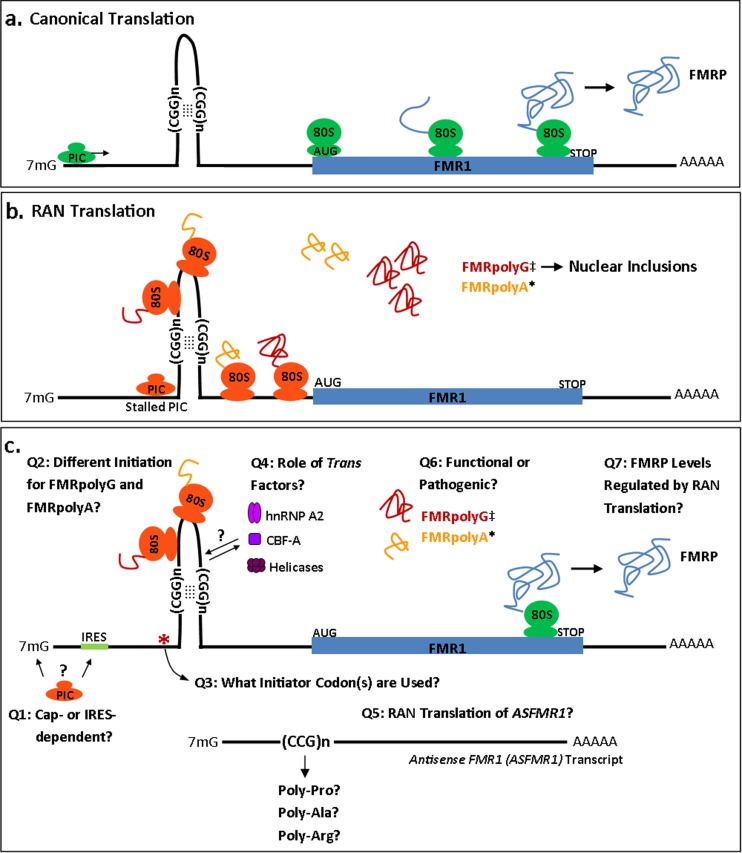
Fig. 2.
Repeat-associated non-AUG (RAN) translation at CGG repeats in fragile X-associated tremor/ataxia syndrome. (a) Cap-dependent translational initiation at a canonical AUG start codon in FMR1 mRNA is likely required for production of fragile X protein (FMRP) in the setting of both normal and premutation repeat expansions. (b) Working model of RAN translation at CGG repeats. Premutation length repeats (50–200 repeats) form large thermostable hairpins or G-quadruplexes in FMR1 mRNA that stall scanning preinitiation complexes (PICs) [74]. Mutational analysis in the sequence upstream of the repeat suggests that initiation occurs at a non-AUG start codon just upstream of the repeat to produce FMRpolyG [30]. However, initiation in a different reading frame is not affected by placement of a stop codon just upstream of the repeat, suggesting that FMRpolyA initiation occurs within the repeat itself in a process that exhibits different repeat size requirements [30]. Of note, neither of these RAN products are fused to FMRP, as they exist in different open reading frames. (c) The mechanism of CGG RAN translation remains to be determined. Many open questions remain: 1) Is RAN translation cap-dependent or does it involve a previously defined internal ribosome entry site (IRES)?; 2) How does the initiation of FMRpolyG and FMRpolyA translational differ?; 3) What initiation codons are used to generate each RAN product?; 4) What role do trans-acting factors, such as G-quadruplex destabilizing proteins hnRNP A2 and CArG-box binding factor A (CBF), and other RNA helicases, play in RAN translation?; 5) Is the antisense transcript ASFMR1 through the repeat subject to RAN translation?; 6) Are FMRpolyA and FMRpolyG functional or pathogenic?; 7) What impact does RAN translation have on the production of FMRP? Transcript is not drawn to scale. *Evidence of production in vitro. ‡Evidence of production in vivo in patient tissues. ? = possible RAN product
More recently, 2 independent laboratories have reported pathologic evidence for RAN translation of a GGGGCC hexanucleotide repeat expansion in intron 1 of C9orf72 [28, 29]. This repeat, which is normally <23 repeats in controls and is often expanded into the hundreds of repeats in patients, is the most common known genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) [65, 66]. Using antibodies specific for poly-(Gly-Ala), poly-(Gly-Pro), and poly-(Gly-Arg)—putative dipeptide repeat RAN products from GGGGCC—Mori et al. [29] selectively detected proteins in postmortem brain samples with the C9orf72 expansion mutations by slot-blot analysis and immunohistochemistry. Similarly, Ash et al. [28] detected GGGGCC RAN-positive proteins in C9orf72 mutated samples, but not control tissues via immunohistochemistry [28]. Both groups found that poly-(Gly-Ala), poly-(Gly-Pro), and poly-(Gly-Arg) co-localize with p62-positive, TDP-43-negative cytoplasmic inclusions—a hallmark in C9orf72-related FTD. In both cases, the proteins appeared to be either too large or too insoluble to enter into polyacrylamide gels, making sizing analysis and quantitation challenging. One of the groups provided at least a hint at how an intronic repeat might get translated by demonstrating an increase in intronic retention in some cases [29]. Moreover, as occurred with CAG repeats [27], in transfected cells GGGGCC RAN translation in each reading frame exhibited different repeat size requirements before they became detectable by Western blot [29].
Since these initial reports, multiple laboratories have investigated how GGGGCC RAN translation affects toxicity and pathogenesis. Using similar strategies that defined GGGGCC RAN proteins from the C9orf72 sense transcript, multiple independent reports have recently confirmed dipeptide repeat RAN proteins in vivo—including poly-(Gly-Pro), poly-(Pro-Ala), and poly-(Pro-Arg)—originating from the C9orf72 antisense transcript [67–69]. Mori et al. [68] and Zu et al. [69] demonstrated that RAN products from both the sense and antisense transcript can be found in individual hippocampal neurons in C9orf72 patients. Interestingly, while GGGGCC RAN proteins are produced from expanded repeats, Mackenzie et al. [70] showed that poly-(Gly-Ala) aggregate distribution within the central nervous system of C9orf72 FTD/ALS patients is similar across clinical phenotypes and is anticorrelated with both cytoplasmic TDP-43 aggregate formation and neuronal loss. One possible explanation for this finding is that GGGGCC RAN proteins, or at least poly-(Gly-Ala), do not play an active role in pathogenesis. Alternatively, the formation of detectable aggregates may play a protective role, with “surviving” neurons being marked by their ability to efficiently form inclusions of these potentially toxic proteins, as has been reported for polyglutamine proteins [71]. Consistent with this concept, cell culture-based experiments in HEK293T cells show a strong correlation between cellular toxicity and increased production of GGGGCC RAN products from both the sense and antisense transcript, specifically poly-(Gly-Pro) and poly-(Pro-Arg) [69]. Conversely, studies using RNA based knockdown of C9orf72 in patient-derived induced pluripotent stem cells primarily support a RNA gain-of-function toxicity mechanism associated with the expanded GGGGCC allele, although these studies did not control for potential differences between soluble and insoluble RAN protein toxicity [72, 73]. The more difficult experiments aimed at testing the relative toxicity of GGGGCC•GGCCCC RAN proteins in animal models at physiologic concentrations in the absence of repeat RNA await.
RAN Translation at CGG Repeats: Insights into Mechanism
Recent work form our laboratory described RAN translation at expanded CGG repeats in the 5’UTR of FMR1, which underlie the neurodegenerative disorder fragile X-associated tremor/ataxia syndrome (FXTAS) [30] (Fig. 2). Experiments in both flies and mammalian cells show RAN translation in 2 reading frames: GGC (+1, poly-Gly) and GCG (+2, poly-Ala). Using antibody-based analyses akin to those described above, we demonstrated that the predicted poly-Gly containing protein, which we termed FMRpolyG, was present in neuronal intranuclear inclusions in human FXTAS brains. Importantly, FMRpolyG production elicited intranuclear inclusions in transfected cells, and modulating its production altered CGG repeat-associated toxicity in both cell culture and fly models of disease, providing direct in vivo evidence that RAN translation products can elicit toxicity [30].
Studies in cell culture provide some mechanistic insights into what cis factors influence when RAN translation occurs at CGG repeats. In the +1 (GGC, Gly) frame, initiation appears to occur predominantly outside of the repeat itself [30]. Placing a stop codon 12 nucleotides, but not 21 nucleotides, upstream of the repeat blocked detectable FMRpolyG production. As a stop codon would not halt or interfere with the scanning PIC, but world terminate translation if initiation was already established, we reasoned that RAN translation in this reading frame must initiate upstream of the repeat. Yet serial mutation of near-cognate start codons or removal of this entire region (the 48 nucleotides just upstream of the repeat) hindered, but did not abolish, RAN translation. However, importantly, at least 1 in-frame near-cognate initiation codon was always required [30]. Thus, FMRpolyG translation is not strictly dependent on the sequence at the site of initiation, but does require something that looks like an AUG start codon and a secondary structure in the CGG repeat that is predicted to stall a scanning ribosomal PIC [74, 75]. A similar observation was seen with stalling of the PIC by a stable hairpin and initiation at non-AUG codon or an AUG codon in a nonoptimal context [35, 74, 75]. This model is further supported by ribosome profiling studies [57], which demonstrate ribosome-protected fragments over numerous near cognate start codons just upstream of the CGG repeat in both human cell lines and mouse embryonic stem cells [30].
However, these rules for one form of RAN translation effectively do not apply to the other CGG repeat reading frames. For example, while RAN translation in the +1 (Gly) frame occurs even at shorter CGG repeat sizes (25–30 repeats) within the “normal” range in humans, RAN translation in the +2 (GCG, Ala) frame was observed only at larger (>70 CGGs) repeat sizes. Moreover, placement of stop codons just prior to the repeat in this +2 (Ala) frame had no impact on translation, suggesting initiation within the repeat [27]. Further, even at larger repeat sizes (100 CGGs), no products were detectable in the +0 (CGG, Arg) reading frame. Consistent with this, there is no evidence for an N-terminal extension on FMRP in patients with large, unmethylated repeat expansions, and FMRP is not found in the ubiquitinated inclusions in patients [76], both of which would be predicted if such a product were made. Thus, the mechanisms by which translation initiation occurs at each repeat and, indeed, each reading frame of each repeat, may be different and interdependent on numerous factors, including the surrounding sequence context, the amino acid produced, and the length of the repeat expansion.
What other cis factors are required for RAN translation at different repeats remains unexplored. The FMR1 locus may provide some early insights. While in vitro studies suggest that the primary mode of FMR1 mRNA translational initiation is 5’-cap-dependent [77], the FMR1 5’UTR does contain a functional IRES upstream of the CGG repeat [78, 79]. Interestingly, Ludwig et al. [77] also showed that replacement of the CGG repeat with a stable hairpin blocked translation in a synthetic 5’UTR, but not in FMR1 5’UTR containing mRNA. This lack of blockade resulted from translation initiation at a near-AUG start codon (GUG) within the synthetic hairpin itself [77], suggesting the secondary structure within the FMR1 5’UTR is somehow permissive of nonconical translation initiation. Other components of the FMR1 5’UTR may also modulate FMRP translation. Premutation length repeats confer a propensity towards a different transcription start site than that typically used at normal length repeats [80, 81]. Use of this alternative transcription start site is associated with less efficient FMRP translation independent of repeat length [77]. These findings suggest that the FMR1 5’UTR may contain independent cis-elements that favor translational initiation at non-AUG start codons and RAN translational events, even at normal repeat sizes. However, the functional roles that these specific alternative transcriptional initiation sites and IRES sequences play in CGG RAN translation remain untested.
Trans-acting factors, such as specific initiation factors, RNA helicases, and other RNA-binding proteins, may be critical for RAN translation and may also differ between repeats. Interestingly, a few trans-acting factors, including hnRNPA2/B1 and CArG-box binding factor A, bind to CGG repeats (as either G-quadruplex [82–86] or hairpin structures [87, 88]) and augment translation of reporters placed downstream of the FMR1 5’UTR both in vitro and in cell culture systems [85, 89]. Although production of the +1 CGG RAN product and initiation at the downstream canonical AUG start codon appear to track together in cell-based systems [30], it is unknown whether the interaction of these RNA binding proteins might enhance or impair CGG RAN translation. Given that hnRNPA2/B1 overexpression suppresses CGG repeat-associated toxicity in Drosophila [10], these relationships deserve further exploration.
Impact on the Proteome and Translation Regulation
RAN translation has the potential to significantly contribute to proteome diversity by increasing the number of potential proteins generated from each transcript. The human genome harbors more repetitive elements than previously predicted [90], including microsatellites, and is pervasively transcribed [91–94]. Furthermore, there is evidence from many emerging techniques that generation of alternative protein products from noncanonical ORFs is common, even in some annotated “noncoding” RNAs [57, 59, 95, 96]. Ribosomal profiling studies suggest a significant underestimation of non-AUG ORFs and upstream ORFs (uORFs) in previous genome annotation [57, 97], and the use of these alternative uORFs appears to be a regulated event, given that alterations in internal states or environmental conditions can lead to shifts in ribosomal positioning on mRNAs in a transcription-independent manner [58]. If a portion of these uORFs initiate via a RAN translation-like mechanism, then understanding how they work may have a broader impact on our comprehension of the genome’s coding potential.
Additionally, RAN translation may play specific regulatory roles in certain sequence contexts [98]. In FMR1, translation through the repeat (the uORF) may assist in destabilizing the inhibitory CGG repeat RNA structure, allowing for normal scanning of subsequent ribosomes and proper initiation of the canonical ORF (Fig. 2) [30]. This is a particularly intriguing mechanism to consider for 3 reasons. First, uORFs within FMR1 appear to be conserved, given that the FMR1 5’UTR (outside of the repeat) is surprisingly invariant in humans [99], that ribosomal profiling peaks reveal multiple uORFs in mice just proximal to the CGG repeat, and that noncanonical upstream initiation occurs in association with dfxr, the Drosophila homolog of FMR1 [30, 100]. Second, the inhibitory effects of the FMR1 5’UTR on translation are not as significant as would be predicted based on RNA secondary structure modeling. Specifically, the GC rich repeat and surrounding sequence, even at “normal” repeat sizes, has a predicted minimal free energy in excess of hairpins that completely impair ribosomal scanning [35, 74, 101, 102]. This suggests that something (perhaps RAN translation?) must assist in unwinding this hairpin to allow FMRP production. Third, FMRP translation is itself a highly regulated event that is critical for appropriate spatiotemporal regulation of translation within neuronal dendrites [103], where it acts as a suppressor of translation [104]. In response to activation of metabotropic glutamate receptors, it is rapidly phosphorylated and degraded, allowing translation of the transcripts with which it is associated [105, 106]. Additionally, FMRP itself is rapidly synthesized by mGluR activation, and this appears critical for maintenance of normal synaptic responses [107–109]. However, how FMR1 translation is initiated and regulated remains poorly defined. CGG RAN translation may offer a mechanism by which the repeat and 5’UTR could allow for such regulation, and similar mechanisms may exist for other neuronal or cellular transcripts to provide a previously unappreciated level of translational regulation.
A Roadmap Forward in RAN Translation
Our understanding of how RAN translation occurs and what roles it plays in neurodegeneration and normal physiology is still in its infancy. Already, it is clear that there may well be multiple types of RAN translation, with different initiation requirements and pathologic consequences for each repeat—or, more specifically, for each reading frame associated with each transcript (sense and antisense) produced from each repeat [110–112]. For all of these, it will be important to address the following 3 questions.
First, what are the critical cis and trans factors that allow for RAN translational initiation to occur at repetitive RNA sequences? For each repeat in a setting as close as possible to its native context, we must evaluate what the key steps are in translational initiation, from whether the transcript must be capped and spliced to what impact neighboring sequence differences play, to what initiation factors are utilized to accomplish initiation of the translational event. Additionally, for certain repeat contexts, even more complicated questions must be answered. For example, the GGGGCC repeat-associated with C9orf72 is in an intron, which predicts that it should normally be rapidly targeted for degradation and excluded from the cytoplasm. Are introns retained in select mRNAs and exported from the nucleus, or is RAN translation a unique form of nuclear translation [113, 114]? Recent work from Haeusler et al. [115] demonstrates that both GGGGCC repeat DNA and RNA form G-quadruplex secondary structures, as well as RNA–DNA hybrids, which stimulate the accumulation of repeat-containing abortive transcripts. One intriguing possibility is that such transcripts might bypass typical intron degradation machinery, allowing for subsequent nuclear export and RAN translation [115].
One important question that has not been addressed is whether the canonical initiator tRNA (Met-tRNAMeti) is required, and, if so, how it is delivered to the PIC and non-AUG start codon. As part of the initiating ternary complex, Met-tRNAMeti is typically delivered to the 40S subunit by eIF2–guanosine triphosphate to form the canonical PIC. Importantly, the eIF2–guanosine triphosphate-bound PIC has very low selectively for non-AUG start codons [116–121], which may be inconsistent with the relatively high levels of RAN products seen in cell culture and fly models [27–30, 68, 69]. However, multiple translation factors—such as eIF2A [122, 123], eIF5B [124–127], and eIF2D [128, 129]—have been noted to bind and deliver Met-tRNAMeti to the 40S subunit, although still preferentially initiating at AUG start codons. Nevertheless, this raises the interesting possibility that atypical eIFs may have a unique role in RAN translation initiation. Alternative translation initiation approaches bypassing the need for Met-tRNAMeti have been reported, particularly for noncanonical initiation at the cricket paralysis virus–IRES. Not requiring Met-tRNAMeti or any known eIF [49], the cricket paralysis virus–IRES directly recruits the 40S subunit and allows for the first codon to be decoded by eEF1A-aminoacyl-tRNA at the A-site [130]. Whether similar mechanisms or translation factors are responsible for RAN translation remains to be determined.
Once a clearer picture of the minimal requirements for RAN translation emerges, we can begin to address questions related to the apparent differences between repeats [110–112]. Identifying the critical cis-elements and trans-factors that play roles in RAN translation may elucidate a core RAN translation machinery. In addition, determining what drives the nonuniformity in repeat length dependence between reading frames of a single transcript will provide crucial mechanistic insight.
Second, what are the normal physiologic functions, if any, for RAN translation? Although most RAN products appear at first glance to be purposeless, it is premature to assume that they lack normal functions. Characterization of their interacting partners and distribution within cells may provide insights into both their direct roles in toxicity and their native roles, if any [110–112]. Furthermore, it will also be important to define what roles RAN translation plays in regulating translation from other ORFs found in the same transcripts in which it occurs. A broader question is whether these same RAN translational mechanisms might mediate the use of non-AUG codons throughout the transcriptome on highly structured RNAs. While identifying such genes will not be trivial, characterizing the minimal cis elements that are required for RAN translation may allow for in silico identification of additional genes that harbor similar elements. Likewise, RAN translation-specific trans-acting factors could allow for identification of target transcripts from which RAN products might be produced. Together, such approaches may shed light on the prevalence of RAN translation and its potential roles in cellular homeostasis [110–112].
The repetitive nature of the RAN homopolymeric and dipeptide repeat proteins pose some specific challenges in terms of questions related to relative abundance, toxicity, stability, and cellular clearance. For example, degradation pathways such as the N-end rule pathway (reviewed in [131, 132]) can strongly influence the half-life of a given protein. Certain N-terminal amino acids, such as Arg and Gln, target proteins for rapid turnover [133, 134]. Thus the N-terminal amino acid utilized during RAN translational initiation may influence both the relative stability of the protein and its potential for cellular function. Such pathways could explain why, for example, no CGG RAN product is observed in the Arg (+0) frame [30], despite evidence for accumulation of RAN translation products in all other reading frames tested to date. Alternatively, some of the RAN proteins may be particularly resistant to endoproteolytic cleavage owing to a lack of enzymes that recognize the specific repetitive motifs they present. Detailed biochemical analysis of each RAN protein N-terminus and degradation pathways will likely be revealing.
Third, what is the pathological consequence of RAN translation and is its correction a meaningful therapeutic target? The accumulation of RAN translation products in aggregates in human disorders suggests that they may be pathologic, but the evidence for their direct roles in pathogenesis are largely lacking, especially in vivo in mammalian systems. To achieve this, it will be necessary to dissociate the toxicity associated with repeats as RNA from the potential toxicity of RAN products as protein. Given that many of these repeats exist in RNAs that are of low abundance (introns in pre-mRNAs and antisense transcripts typically are present at a fraction of the sense mRNAs with which they are associated), it will be important to study these processes at physiologic concentrations. Howe-ver, if they are highly toxic, even low-level production and accumulation may contribute meaningfully to pathogenesis.
As is true for many inherited neurodegenerative disorders, there is a decades-long delay between the initial expression of the toxic mutated genes and the subsequent development of clinical symptoms in FXTAS and ALS/FTD. If RAN proteins contribute meaningfully to neurodegeneration, then it will be important to determine if their delayed toxicity reflects a loss of a compensatory mechanism for dealing with toxic proteins or instead whether there are tissue-specific changes in the relative production of RAN proteins. For example, ribosome profiling of nonlog phase yeast demonstrates a dramatic increase in nonconical translation, suggesting that different cellular conditions might be more or less permissive of alternative translational initiation mechanisms [58]. Could the aging brain provide a similarly permissive environment for RAN translation?
If RAN products are important, then RAN translation may serve as a novel therapeutic target. The feasibility of such an approach will depend heavily on what native roles RAN translational initiation processes underlie and whether they are mechanistically separable from those involved in the translation of most mammalian proteins. However, it is tempting to speculate that if these translational events are truly aberrant, then identifying agents that can selectively block them is a potentially fertile area for drug development.
Conclusion
Discovery of RAN translation has shed light on a new facet of eukaryotic translation initiation and human disease. With further mining into the mechanism, a novel therapeutic target for multiple neurodegenerative disorders is plausible. It will be critical to determine how prevalent RAN translation is across the transcriptome and whether it contributes to normal cellular functions and human diseases.
Electronic supplementary material
(PDF 1225 kb)
Acknowledgments
We thank Hank Paulson for critical reading of this manuscript. Funding for this work was provided by the M-Cubed Initiative, the Department of Veterans Affairs (BLRD #11212652), the NIH (R01NS086810, K08NS069809 and P30-AG13283), and the Harris Professorship to PKT. Additional funding for this work was provided by the NIH (F32NS089124) to MGK.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
References
- 1.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 2.Almeida B, Fernandes S, Abreu IA, Macedo-Ribeiro S. Trinucleotide repeats: a structural perspective. Front Neurol. 2013;4:76. doi: 10.3389/fneur.2013.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang YH, Gellibolian R, Shimizu M, Wells RD, Griffith J. Long CCG triplet repeat blocks exclude nucleosomes: a possible mechanism for the nature of fragile sites in chromosomes. J Mol Biol. 1996;263:511–516. doi: 10.1006/jmbi.1996.0593. [DOI] [PubMed] [Google Scholar]
- 4.Mulvihill DJ, Nichol Edamura K, Hagerman KA, Pearson CE, Wang YH. Effect of CAT or AGG interruptions and CpG methylation on nucleosome assembly upon trinucleotide repeats on spinocerebellar ataxia, type 1 and fragile X syndrome. J Biol Chem. 2005;280:4498–4503. doi: 10.1074/jbc.M413239200. [DOI] [PubMed] [Google Scholar]
- 5.Renoux AJ, Todd PK. Neurodegeneration the RNA way. Prog Neurobiol. 2012;97:173–189. doi: 10.1016/j.pneurobio.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Rourke JR, Swanson MS. Mechanisms of RNA-mediated disease. J Biol Chem. 2009;284:7419–7423. doi: 10.1074/jbc.R800025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams AJ, Paulson HL. Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci. 2008;31:521–528. doi: 10.1016/j.tins.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng Y, Zhang FP, Lokey LK, et al. Translational Suppression by Trinucleotide Repeat Expansion at Fmr1. Science. 1995;268:731–734. doi: 10.1126/science.7732383. [DOI] [PubMed] [Google Scholar]
- 9.Jin P, Duan R, Qurashi A, et al. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55:556–564. doi: 10.1016/j.neuron.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sofola OA, Jin P, Qin Y, et al. RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron. 2007;55:565–571. doi: 10.1016/j.neuron.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sellier C, Rau F, Liu Y, et al. Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J. 2010;29:1248–1261. doi: 10.1038/emboj.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sellier C, Freyermuth F, Tabet R, et al. Sequestration of DROSHA and DGCR8 by expanded CGG RNA repeats alters microRNA processing in fragile X-associated tremor/ataxia syndrome. Cell Rep. 2013;3:869–880. doi: 10.1016/j.celrep.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller JW, Urbinati CR, Teng-Umnuay P, et al. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mankodi A, Logigian E, Callahan L, et al. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 15.Kanadia RN, Johnstone KA, Mankodi A, et al. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 16.Kanadia RN, Shin J, Yuan Y, et al. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc Natl Acad Sci U S A. 2006;103:11748–11753. doi: 10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osborne RJ, Lin X, Welle S, et al. Transcriptional and post-transcriptional impact of toxic RNA in myotonic dystrophy. Hum Mol Genet. 2009;18:1471–1481. doi: 10.1093/hmg/ddp058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du H, Cline MS, Osborne RJ, et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol. 2010;17:187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charizanis K, Lee KY, Batra R, et al. Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron. 2012;75:437–450. doi: 10.1016/j.neuron.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang ET, Cody NA, Jog S, et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The Huntington's Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 23.Scherzinger E, Lurz R, Turmaine M, et al. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto A, Lucas JJ, Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington's disease. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez I, Mahlke C, Yuan JY. Pivotal role of oligomerization in expanded polyglutamine neurodegenerative disorders. Nature. 2003;421:373–379. doi: 10.1038/nature01301. [DOI] [PubMed] [Google Scholar]
- 26.Finkbeiner S. Huntington's disease. Cold Spring Harb Perspect Biol 2011;3. [DOI] [PMC free article] [PubMed]
- 27.Zu T, Gibbens B, Doty NS, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ash PE, Bieniek KF, Gendron TF, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mori K, Weng SM, Arzberger T, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 30.Todd PK, Oh SY, Krans A, et al. CGG Repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron. 2013;78:440–455. doi: 10.1016/j.neuron.2013.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol. 2012;19:568–576. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- 33.Hinnebusch AG, Lorsch JR. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb Perspect Biol 2012;4. [DOI] [PMC free article] [PubMed]
- 34.Kozak M, Nathans D. Translation of the genome of a ribonucleic acid bacteriophage. Bacteriol Rev. 1972;36:109–134. doi: 10.1128/br.36.1.109-134.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci U S A. 1986;83:2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 37.Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozak M. Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol Cell Biol. 1989;9:5073–5080. doi: 10.1128/mcb.9.11.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kozak M. Features in the 5' non-coding sequences of rabbit alpha and beta-globin mRNAs that affect translational efficiency. J Mol Biol. 1994;235:95–110. doi: 10.1016/s0022-2836(05)80019-1. [DOI] [PubMed] [Google Scholar]
- 40.Kozak M. Adherence to the first-AUG rule when a second AUG codon follows closely upon the first. Proc Natl Acad Sci U S A. 1995;92:7134. doi: 10.1073/pnas.92.15.7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 42.Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5' nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pestova TV, Shatsky IN, Hellen CU. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pestova TV, Hellen CU, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reynolds JE, Kaminski A, Carroll AR, Clarke BE, Rowlands DJ, Jackson RJ. Internal initiation of translation of hepatitis C virus RNA: the ribosome entry site is at the authentic initiation codon. RNA. 1996;2:867–878. [PMC free article] [PubMed] [Google Scholar]
- 46.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Develop. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson JE, Pestova TV, Hellen CU, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 48.Jan E, Thompson SR, Wilson JE, Pestova TV, Hellen CU, Sarnow P. Initiator Met-tRNA-independent translation mediated by an internal ribosome entry site element in cricket paralysis virus-like insect viruses. Cold Spring Harbor Symp Quant Biol. 2001;66:285–292. doi: 10.1101/sqb.2001.66.285. [DOI] [PubMed] [Google Scholar]
- 49.Pestova TV, Hellen CU. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Develop. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pestova TV, Lomakin IB, Hellen CU. Position of the CrPV IRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly. EMBO Rep. 2004;5:906–913. doi: 10.1038/sj.embor.7400240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ray PS, Grover R, Das S. Two internal ribosome entry sites mediate the translation of p53 isoforms. EMBO Rep. 2006;7:404–410. doi: 10.1038/sj.embor.7400623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vagner S, Gensac MC, Maret A, et al. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol Cell Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gan WN, Rhoads RE. Internal initiation of translation directed by the 5'-untranslated region of the mRNA for eIF4G, a factor involved in the picornavirus-induced switch from cap-dependent to internal initiation. J Biol Chem. 1996;271:623–626. doi: 10.1074/jbc.271.2.623. [DOI] [PubMed] [Google Scholar]
- 54.Johannes G, Sarnow P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA. 1998;4:1500–1513. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coldwell MJ, Mitchell SA, Stoneley M, MacFarlane M, Willis AE. Initiation of Apaf-1 translation by internal ribosome entry. Oncogene. 2000;19:899–905. doi: 10.1038/sj.onc.1203407. [DOI] [PubMed] [Google Scholar]
- 56.Ivanov IP, Firth AE, Michel AM, Atkins JF, Baranov PV. Identification of evolutionarily conserved non-AUG-initiated N-terminal extensions in human coding sequences. Nucleic Acids Res. 2011;39:4220–4234. doi: 10.1093/nar/gkr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, Weissman JS. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2012;335:552–557. doi: 10.1126/science.1215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Michel AM, Choudhury KR, Firth AE, Ingolia NT, Atkins JF, Baranov PV. Observation of dually decoded regions of the human genome using ribosome profiling data. Genome Res. 2012;22:2219–2229. doi: 10.1101/gr.133249.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hinnebusch AG. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol Mol Biol Rev. 2011;75:434–467. doi: 10.1128/MMBR.00008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koob MD, Moseley ML, Schut LJ, et al. An untranslated CTG expansion causes a novel form of spinocerebellar ataxia (SCA8) Nat Genet. 1999;21:379–384. doi: 10.1038/7710. [DOI] [PubMed] [Google Scholar]
- 62.Daughters RS, Tuttle DL, Gao W, et al. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5:e1000600. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moseley ML, Zu T, Ikeda Y, et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat Genet. 2006;38:758–769. doi: 10.1038/ng1827. [DOI] [PubMed] [Google Scholar]
- 64.Cho DH, Thienes CP, Mahoney SE, Analau E, Filippova GN, Tapscott SJ. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell. 2005;20:483–489. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 65.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gendron TF, Bieniek KF, Zhang YJ, et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013;126:829–844. doi: 10.1007/s00401-013-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mori K, Arzberger T, Grasser FA, et al. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 2013;126:881–893. doi: 10.1007/s00401-013-1189-3. [DOI] [PubMed] [Google Scholar]
- 69.Zu T, Liu Y, Banez-Coronel M, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci U S A. 2013;110:E4968–E4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mackenzie IR, Arzberger T, Kremmer E, et al. Dipeptide repeat protein pathology in C9ORF72 mutation cases: clinico-pathological correlations. Acta Neuropathol. 2013;126:859–879. doi: 10.1007/s00401-013-1181-y. [DOI] [PubMed] [Google Scholar]
- 71.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 72.Donnelly CJ, Zhang PW, Pham JT, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sareen D, O'Rourke JG, Meera P, et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med. 2013;5:208ra149. doi: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol Cell Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kozak M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci U S A. 1990;87:8301–8305. doi: 10.1073/pnas.87.21.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Iwahashi CK, Yasui DH, An HJ, et al. Protein composition of the intranuclear inclusions of FXTAS. Brain. 2006;129:256–271. doi: 10.1093/brain/awh650. [DOI] [PubMed] [Google Scholar]
- 77.Ludwig AL, Hershey JW, Hagerman PJ. Initiation of translation of the FMR1 mRNA Occurs predominantly through 5'-end-dependent ribosomal scanning. J Mol Biol. 2011;407:21–34. doi: 10.1016/j.jmb.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chiang PW, Carpenter LE, Hagerman PJ. The 5'-untranslated region of the FMR1 message facilitates translation by internal ribosome entry. J Biol Chem. 2001;276:37916–37921. doi: 10.1074/jbc.M105584200. [DOI] [PubMed] [Google Scholar]
- 79.Dobson T, Kube E, Timmerman S, Krushel LA. Identifying intrinsic and extrinsic determinants that regulate internal initiation of translation mediated by the FMRI 5 ' leader. BMC Mol Biol 2008;9. [DOI] [PMC free article] [PubMed]
- 80.Beilina A, Tassone F, Schwartz PH, Sahota P, Hagerman PJ. Redistribution of transcription start sites within the FMR1 promoter region with expansion of the downstream CGG-repeat element. Hum Mol Genet. 2004;13:543–549. doi: 10.1093/hmg/ddh053. [DOI] [PubMed] [Google Scholar]
- 81.Tassone F, De Rubeis S, Carosi C, et al. Differential usage of transcriptional start sites and polyadenylation sites in FMR1 premutation alleles. Nucleic Acids Res. 2011;39:6172–6185. doi: 10.1093/nar/gkr100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Usdin K. NGG-triplet repeats form similar intrastrand structures: implications for the triplet expansion diseases. Nucleic Acids Res. 1998;26:4078–4085. doi: 10.1093/nar/26.17.4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weisman-Shomer P, Cohen E, Fry M. Distinct domains in the CArG-box binding factor A destabilize tetraplex forms of the fragile X expanded sequence d(CGG)(n) Nucleic Acids Res. 2002;30:3672–3681. doi: 10.1093/nar/gkf506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Khateb S, Weisman-Shomer P, Hershco I, Loeb LA, Fry M. Destabilization of tetraplex structures of the fragile X repeat sequence (CGG)n is mediated by homolog-conserved domains in three members of the hnRNP family. Nucleic Acids Res. 2004;32:4145–4154. doi: 10.1093/nar/gkh745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Khateb S, Weisman-Shomer P, Hershco-Shani I, Ludwig AL, Fry M. The tetraplex (CGG)n destabilizing proteins hnRNP A2 and CBF-A enhance the in vivo translation of fragile X premutation mRNA. Nucleic Acids Res. 2007;35:5775–5788. doi: 10.1093/nar/gkm636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bugaut A, Balasubramanian S. 5 '-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012;40:4727–4741. doi: 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Handa V, Saha T, Usdin K. The fragile X syndrome repeats form RNA hairpins that do not activate the interferon-inducible protein kinase, PKR, but are cut by Dicer. Nucleic Acids Res. 2003;31:6243–6248. doi: 10.1093/nar/gkg818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zumwalt M, Ludwig A, Hagerman PJ, Dieckmann T. Secondary structure and dynamics of the r(CGG) repeat in the mRNA of the fragile X mental retardation 1 (FMR1) gene. RNA Biol. 2007;4:93–100. doi: 10.4161/rna.4.2.5039. [DOI] [PubMed] [Google Scholar]
- 89.Ofer N, Weisman-Shomer P, Shklover J, Fry M. The quadruplex r(CGG)n destabilizing cationic porphyrin TMPyP4 cooperates with hnRNPs to increase the translation efficiency of fragile X premutation mRNA. Nucleic Acids Res. 2009;37:2712–2722. doi: 10.1093/nar/gkp130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1 % of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dunham I, Kundaje A, Aldred SF, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013;9:e1003569. doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kochetov AV. Alternative translation start sites and hidden coding potential of eukaryotic mRNAs. Bioessays. 2008;30:683–691. doi: 10.1002/bies.20771. [DOI] [PubMed] [Google Scholar]
- 96.Vanderperre B, Lucier JF, Bissonnette C, et al. Direct detection of alternative open reading frames translation products in human significantly expands the proteome. PloS One. 2013;8:e70698. doi: 10.1371/journal.pone.0070698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fritsch C, Herrmann A, Nothnagel M, et al. Genome-wide search for novel human uORFs and N-terminal protein extensions using ribosomal footprinting. Genome Res. 2012;22:2208–2218. doi: 10.1101/gr.139568.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chatterjee S, Pal JK. Role of 5'- and 3'-untranslated regions of mRNAs in human diseases. Biol Cell. 2009;101:251–262. doi: 10.1042/BC20080104. [DOI] [PubMed] [Google Scholar]
- 99.Collins SC, Bray SM, Suhl JA, et al. Identification of novel FMR1 variants by massively parallel sequencing in developmentally delayed males. Am J Med Genet A. 2010;152A:2512–2520. doi: 10.1002/ajmg.a.33626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Beerman RW, Jongens TA. A non-canonical start codon in the Drosophila fragile X gene yields two functional isoforms. Neuroscience. 2011;181:48–66. doi: 10.1016/j.neuroscience.2011.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pelletier J, Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5' noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell. 1985;40:515–526. doi: 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- 102.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 103.Bhakar AL, Dolen G, Bear MF. The pathophysiology of Fragile X (and what it teaches us about synapses) Annu Rev Neurosci. 2012;35:417–423. doi: 10.1146/annurev-neuro-060909-153138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Darnell JC, Van Driesche SJ, Zhang C, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nalavadi VC, Muddashetty RS, Gross C, Bassell GJ. Dephosphorylation-induced ubiquitination and degradation of FMRP in dendrites: a role in immediate early mGluR-stimulated translation. J Neurosci. 2012;32:2582–2587. doi: 10.1523/JNEUROSCI.5057-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hou L, Antion MD, Hu D, Spencer CM, Paylor R, Klann E. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron. 2006;51:441–454. doi: 10.1016/j.neuron.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 107.Weiler IJ, Irwin SA, Klintsova AY, et al. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc Natl Acad Sci U S A. 1997;94:5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Todd PK, Mack KJ, Malter JS. The fragile X mental retardation protein is required for type-I metabotropic glutamate receptor-dependent translation of PSD-95. Proc Natl Acad Sci U S A. 2003;100:14374–14378. doi: 10.1073/pnas.2336265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Iliff AJ, Renoux AJ, Krans A, Usdin K, Sutton MA, Todd PK. Impaired activity-dependent FMRP translation and enhanced mGluR-dependent LTD in Fragile X premutation mice. Hum Mol Genet. 2013;22:1180–1192. doi: 10.1093/hmg/dds525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pearson CE. Repeat-associated non-ATG translation initiation: one DNA, two transcripts, seven reading frames, potentially nine toxic entities! PLoS Genet 2011;7:e1002018. [DOI] [PMC free article] [PubMed]
- 111.Cleary JD, Ranum LPW. Repeat-associated non-ATG (RAN) translation in neurological disease. Hum Mol Genet. 2013;22:R45–R51. doi: 10.1093/hmg/ddt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gendron TF, Belzil VV, Zhang YJ, Petrucelli L. Mechanisms of toxicity in C9FTLD/ALS. Acta Neuropathol. 2014;127:359–376. doi: 10.1007/s00401-013-1237-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Maquat LE, Tarn WY, Isken O. The pioneer round of translation: features and functions. Cell. 2010;142:368–374. doi: 10.1016/j.cell.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.David A, Dolan BP, Hickman HD, et al. Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J Cell Biol. 2012;197:45–57. doi: 10.1083/jcb.201112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Haeusler AR, Donnelly CJ, Periz G, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Donahue TF, Cigan AM, Pabich EK, Valavicius BC. Mutations at a Zn(Ii) Finger motif in the yeast eLF-2-beta gene alter ribosomal start-site selection during the scanning process. Cell. 1988;54:621–632. doi: 10.1016/s0092-8674(88)80006-0. [DOI] [PubMed] [Google Scholar]
- 117.Cigan AM, Pabich EK, Feng L, Donahue TF. Yeast translation initiation suppressor sui2 encodes the alpha subunit of eukaryotic initiation factor 2 and shares sequence identity with the human alpha subunit. Proc Natl Acad Sci U S A. 1989;86:2784–2788. doi: 10.1073/pnas.86.8.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yoon HJ, Donahue TF. The Sui1 suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNA(iMet) recognition of the start codon. Mol Cell Biol. 1992;12:248–260. doi: 10.1128/mcb.12.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Huang HK, Yoon H, Hannig EM, Donahue TF. GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Develop. 1997;11:2396–2413. doi: 10.1101/gad.11.18.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Maduzia LL, Moreau A, Poullet N, Chaffre S, Zhang Y. The role of eIF1 in translation initiation codon selection in Caenorhabditis elegans. Genetics. 2010;186:1187–1196. doi: 10.1534/genetics.110.121541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang YH, Maduzia LL. Mutations in Caenorhabditis elegans eIF2 beta permit translation initiation from non-AUG start codons. Genetics. 2010;185:141–U245. doi: 10.1534/genetics.110.115485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Merrick WC, Anderson WF. Purification and characterization of homogeneous protein synthesis initiation factor M1 from rabbit reticulocytes. J Biol Chem. 1975;250:1197–1206. [PubMed] [Google Scholar]
- 123.Kim JH, Park SM, Park JH, Keum SJ, Jang SK. eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J. 2011;30:2454–2464. doi: 10.1038/emboj.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Choi SK, Lee JH, Zoll WL, Merrick WC, Dever TE. Promotion of met-tRNAiMet binding to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science. 1998;280:1757–1760. doi: 10.1126/science.280.5370.1757. [DOI] [PubMed] [Google Scholar]
- 125.Roll-Mecak A, Cao C, Dever TE, Burley SK. X-Ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding. Cell. 2000;103:781–792. doi: 10.1016/s0092-8674(00)00181-1. [DOI] [PubMed] [Google Scholar]
- 126.Pestova TV, de Breyne S, Pisarev AV, Abaeva IS, Hellen CU. eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: a common role of domain II. EMBO J. 2008;27:1060–1072. doi: 10.1038/emboj.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat Struct Mol Biol. 2008;15:836–841. doi: 10.1038/nsmb.1445. [DOI] [PubMed] [Google Scholar]
- 128.Dmitriev SE, Terenin IM, Andreev DE, et al. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J Biol Chem. 2010;285:26779–26787. doi: 10.1074/jbc.M110.119693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Skabkin MA, Skabkina OV, Dhote V, Komar AA, Hellen CU, Pestova TV. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Develop. 2010;24:1787–1801. doi: 10.1101/gad.1957510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fernandez IS, Bai XC, Murshudov G, Scheres SH, Ramakrishnan V. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell. 2014;157:823–831. doi: 10.1016/j.cell.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tasaki T, Sriram SM, Park KS, Kwon YT. The N-end rule pathway. Ann Rev Biochem. 2012;81:261–289. doi: 10.1146/annurev-biochem-051710-093308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dougan DA, Micevski D, Truscott KN. The N-end rule pathway: from recognition by N-recognins, to destruction by AAA + proteases. Biochim Biophys Acta. 1823;2012:83–91. doi: 10.1016/j.bbamcr.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 133.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 134.Gonda DK, Bachmair A, Wunning I, Tobias JW, Lane WS, Varshavsky A. Universality and structure of the N-end rule. J Biol Chem. 1989;264:16700–16712. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1225 kb)