Abstract
Histone mRNA levels are cell cycle regulated, and the major regulatory steps are at the posttranscriptional level. A major regulatory mechanism is S- phase restriction of Stem-loop binding protein (SLBP) which binds to the 3′ end of histone mRNA and participates in multiple steps of histone mRNA metabolism, including 3′ end processing, translation and regulation of mRNA stability. SLBP expression is cell cycle regulated without significant change in its mRNA level. SLBP expression is low in G1 until just before S phase where it functions and at the end of S phase SLBP is degraded by proteasome complex depending on phosphorylations on Thr 60 and Thr61. Here using synchronized HeLa cells we showed that SLBP production rate is low in early G1 and recovers back to S phase level somewhere between early and mid-G1. Further, we showed that SLBP is unstable in G1 due to proteasome mediated degradation as a novel mechanism to keep SLBP low in G1. Finally, the S/G2 stable mutant form of SLBP is degraded by proteasome in G1, indicating that the SLBP degradation mechanism in G1 is independent of the previously identified S/G2 degradation mechanism. In conclusion, as a mechanism to limit histone production to S phase, SLBP is kept low in G1 phase due to cooperative action of translation regulation and proteasome mediated degradation which is independent of previously known S/G2 degradation.
Keywords: Histone synthesis, SLBP, histone mRNA, replication dependent histone genes
Introduction
During each S phase, bulk amount of histone proteins are expressed in order to package the newly replicated DNA. In metazoans, canonical histone mRNAs lack polyA tails but end in a conserved stem-loop structure. These histone mRNAs are encoded by so called replication dependent histone genes which lack introns, and the only processing reaction to produce mature histone mRNA is an endonucleolytic cleavage after the conserved stem-loop structure at their 3′ end (Marzluff et al., 2008; Millevoi and Vagner, 2010; Rattray and Muller, 2012).
Expression of metazoan histone mRNAs is limited to the S phase due to both transcriptional and posttranscriptional mechanisms (Marzluff et al., 2008). At late G1, while the transcription of replication dependent histone genes increases 3–5 fold (DeLisle et al., 1983; Heintz et al., 1983), the histone mRNA processing efficiency increases 10 fold (Harris et al., 1991; Luscher and Schumperli, 1987; Stauber and Schumperli, 1988). At the end of S phase the stability of histone mRNAs decreases along with the processing efficiency to shut down the histone production (Marzluff et al., 2008). Stem-loop binding protein which binds to stem-loop structure of histone mRNAs is a major factor that coordinates the histone biosynthesis and cell cycle (Marzluff and Duronio, 2002; Marzluff et al., 2008; Zheng et al., 2003). SLBP is required for histone pre-mRNA processing (Dominski et al., 1999; Martin et al., 1997; Wang et al., 1996; Zhao et al., 2004), remains with mature mRNA and is involved in all other aspects of histone mRNA metabolism including nuclear export, translation and mRNA stability (Cakmakci et al., 2008; Gallie et al., 1996; Marzluff, 2009; Marzluff et al., 2008; Sanchez and Marzluff, 2002). SLBP expression is cell cycle regulated and is limited to S phase without significant change in its mRNA level (Whitfield et al., 2000). In G1, SLBP expression is low as a major mechanism to limit histone mRNA processing and mature histone mRNA production (Whitfield et al., 2000; Zheng et al., 2003). At late G1, SLBP expression increases significantly and this is proposed to be the main reason for the increase in histone mRNA processing efficiency towards S phase (Whitfield et al., 2000; Zheng et al., 2003). Beside the relatively modest increase in transcription of replication dependent histone genes, this increase in histone mRNA processing efficiency appears as the major reason for rapid accumulation of histone mRNAs in S phase (Harris et al., 1991; Marzluff et al., 2008). At the end of S phase, SLBP is rapidly degraded by proteasome (Whitfield et al., 2000). Cyclin A/Cdk1 which emerges as an important regulator of late S-G2 transition (Enders, 2012; Katsuno et al., 2009; Koseoglu et al., 2010a; Merrick and Fisher, 2012), phosphorylates SLBP at Thr 61 and triggers its degradation (Koseoglu et al., 2008). Phosphorylation of Thr 61 by cyclin A/Cdk1 primes the phosphorylation of Thr60 by CK2 and double phosphorylated SLBP is marked for degradation by proteasome to shut down histone mRNA processing (Koseoglu et al., 2008; Koseoglu et al., 2010b).
Previously, Whitfield et al. demonstrated that SLBP mRNA translation rate is low in G1 and proposed poor production efficiency as the reason for low level of SLBP in G1 phase (Whitfield et al., 2000). Here, we report that low production rate of SLBP is limited to early G1 phase. By labeling the newly produced proteins with 35S methionine, we showed that somewhere between early and mid-G1 SLBP translation reaches to the S phase level. As a novel mechanism to keep SLBP expression low during G1 we found that SLBP is unstable in G1 due to proteasome mediated degradation. Finally, using stably expressed S/G2 stable mutant version of SLBP (Zheng et al., 2003) we showed that SLBP degradation in G1 is independent of the previously identified S/G2 degradation mechanism.
Material and Methods
Cell culture and synchronization
HeLa cells were grown in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum and penicillin-streptomycin. All cultures were maintained in a humidified incubator at 37C and 5% CO2. Cells were synchronized by double-thymidine block method. Hela cells were plated at density of 2.5*105 cells per 10 cm plate the day before initiation of double-thymidine block. Cells were blocked for 19 hrs with 2 mM thymidine, released in to fresh media for 9 hrs after washing with phosphate-buffered saline (PBS), and then blocked again with 2 mM thymidine for 16 hrs to arrest all the cells at the beginning of S phase. The cells were released in to fresh media after washing out the thymidine by PBS, and collected at indicated time points and cell cycle phases. The cells progress through the cell cycle phases S, G2, M and enter next G1 synchronously. The cell cycle distribution of the cells was determined by propodium iodide staining and flow cytometry analysis in either University of North Carolina (Chapel Hill, NC) or Bogazici University (Istanbul, Turkey) flow cytometry facility.
Transfection and generation of stable cells
HeLa cells were seeded at 90% confluency on 6-well plate and transfected with 2 ug DNA and Lipofectamine2000 (Invitrogen) according to manufacturer’s protocol. 48 hrs after transfection 1/3 of transfected cells were collected for verification of exogenous protein expression by western blotting and 2/3 were seeded again for generation of stable cells. For selection of stable cells, cells were treated with 400 ug/ml of Geneticin (Gibco) for 3–4 weeks until separate colonies were detected by eye on the plates and these stably transfected cells were pooled together. To maintain the stably transfected cells, 200 μg of G418 per ml was kept in the medium and was removed just prior to synchronization.
Lysate preparation and western blot analysis
Cells were collected, washed in PBS and lysed in NP-40 lysis buffer (0.5 % NP-40, 150mM NaCl, 50mM Tris-HCl (pH 8), 1mM dithiothreitol, 1mM phenylmethylsulfonyl fluoride, 1X protease inhibitor mixture (Roche)) by 20 minutes of rocking in 4°C. The insoluble material was pelleted by 10 mins. centrifugation at 16000 g by a microcentrifuge at 4°C. Western blots were performed according to standard protocols. For western blot analysis typically 50 microg of total cell protein was resolved in 12% SDS PAGE and tranferred to nitrocellulose blot.
Analysis of SLBP synthesis rate
Synchronized cells at indicated cell cycle phases were preincubated in DMEM without methionine (ICN Pharmeceuticals, CA) supplemented with 10% dialyzed fetal bovine serum, for 30 mins prior to labeling to deplete intracellular stores of methionine. Next, cells were pulse labeled with 1 mCi of [35S] methionine (NEN Life Sciences, MA) for 15 min and collected. Cells were lysed in NP-40 lysis buffer as above. The insoluble materials were removed by ultracentrifugation. Equal protein amounts of lysates were precleared by protein A agarose beads (GE Healthcare) and were incubated with affinity purified SLBP antibody. The SLBP–antibody complexes were recovered by binding to protein A agarose beads (GE Healthcare) and were washed extensively with NP-40 lysis buffer. The bound proteins were eluted in SDS loading buffer and resolved by SDS PAGE for detection by autoradiography.
Antibodies and inhibitors
SLBP antibody is raised against the C-terminal 13 aminoacids of the protein (Wang et al., 1996). Cyclin A antibody (sc-569) was purchased from SantaCruz (Santa Cruz, CA). Cycloheximide and Mg132 were purchased from Sigma (St Louis, MO).
Results
Low synthesis rate of SLBP is limited to early G1
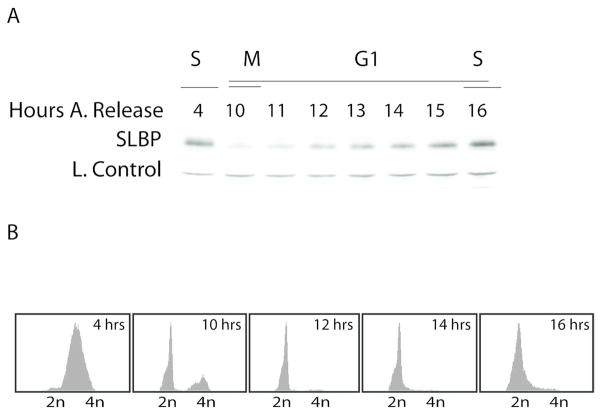
After the degradation of SLBP at S/G2, the SLBP level remains low until prior to the next S phase (Fig 1). As cells approach S phase, SLBP level increases significantly, without a significant change in SLBP mRNA level (Whitfield et al., 2000). The G1 stage of the cell cycle is often the longest period of mammalian somatic cell cycle, and varies greatly between different cell types and growth conditions. In rapidly cycling HeLa cells, it lasts up to 6–8 hours. In this study, using the cells synchronized by double thymidine method, we defined early G1 as first 2 hours after M/G1 phase and it takes roughly another 4 hours for SLBP to recover back to the S phase level at G1/S (Fig 1).
Figure 1. SLBP level stays low in G1 until S phase.
HeLa cells were synchronized by double thymidine block and collected at indicated times after the release. A) Western blot analysis was performed using anti-SLBP serum. Loading control is a cross-reacting band which is known to be stable throughout the cell cycle (Koseoglu et al., 2008). B) FACs analysis of the cells collected at indicated time points.
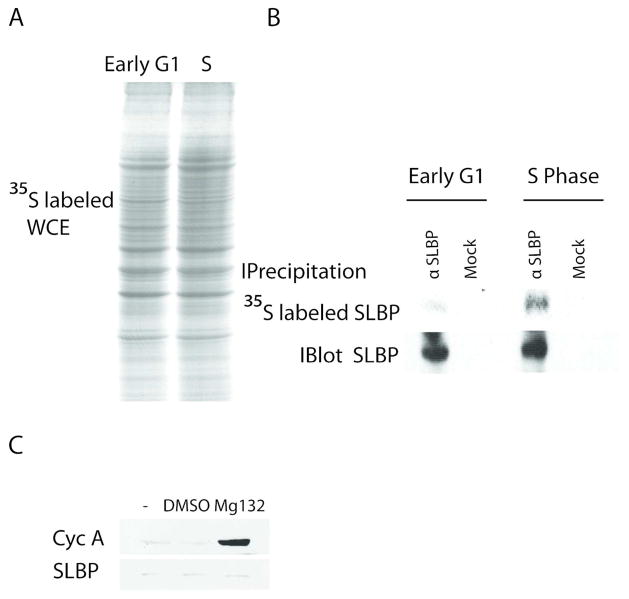
In parallel with previous findings by Whitfield et al., we found that in HeLa cells SLBP production rate is low at early G1(Fig 2). In order to compare production rate of SLBP in early G1 and S phases, we synchronized the cells with double thymidine method and pulse labeled the early G1 and S phase cells with media containing 35S methionine for 15 minutes. After labeling, first, we analyzed equal total protein amounts of radiolabeled extracts by SDS PAGE. We showed that the general level and pattern of newly produced radiolabeled proteins were comparable demonstrating that the synthesis rates of the most abundant cellular proteins are very similar in our early G1 and S phase cells (Fig 2A). From these lysates, we specifically immunoprecipitated SLBP and ran the samples on SDSPAGE to compare the levels of radiolabeled SLBP by autoradiography (Fig 2B, top panel). Since SLBP is low in G1, we also compared the total amount of SLBP that we can immunoprecipitate from early G1 and S phase cells, by performing similar SLBP immunoprecipitation experiment, but this time followed by western blot analysis (Fig. 2B, bottom panel). According to the western blot results (Fig. 2B, bottom panel) the levels of total SLBP immunoprecipitated form early G1 and S phase cell lysates were comparable. However, the amount of 35S labeled newly produced SLBP from early G1 cells was significantly lower than the amount of newly synthesized SLBP precipitated from the S-phase lysate (Fig. 2B) suggesting that the production rate in early G1 is much slower than the S phase.
Figure 2. SLBP production rate is low in early G1 cells.
HeLa cells were synchronized by double thymidine block method and labeled with 35S-methionine for 15 minutes at early G1 (1.5–2 hours after M/G1) or S phase (1.5 hours after release) after half hour incubation in methionine lacking media. A) Equal protein amounts of total lysates and B) immunoprecipitates with SLBP antibody or beads alone were run on SDS PAGE gel and detected with autoradiography (top panel). Cells at the same cell cycle points were lysed and SLBP was immunoprecipitated from equal protein amounts of total lysates. Immunoprecipitated SLBP was detected by western blot using SLBP antibody (bottom panel) C) Early G1 cells similarly synchronized by double thymidine method were collected after one hour Mg132 (50 μM), DMSO (carrier) or just media treatment and western blot analysis was performed using Cyclin A or SLBP antibody as indicated. Cell cycle phases were confirmed by FACs analysis.
Although we labeled cells for very short time, in order to check whether the low amount of the 35S labeled newly produced SLBP is due to very rapid degradation, we treated early G1 cells with the proteasome inhibitor MG132 for one hour. The level of SLBP protein remained low (Fig 2C), indicating that the main reason for the low amount of labeled SLBP is not rapid degradation but low production rate. On the other hand, the level of Cyclin A protein, which is known to be continuously produced and degraded in G1, increased significantly in response to treatment with proteasome inhibitor (Fig 2C).
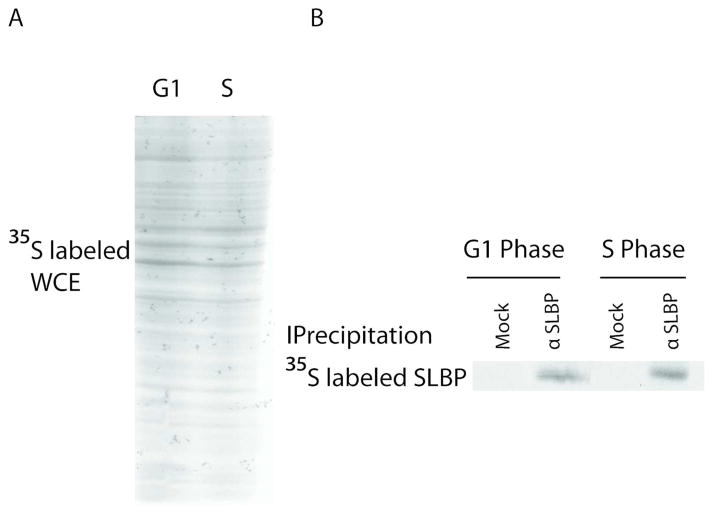
Next, in order to check whether the low level of SLBP production is the case for the entire G1 or only limited to early G1, we compared the production rate of SLBP at later points in G1 (2.5 hrs after M phase) with the rate in S phase cells by again 35S methionine labeling followed by SLBP immunoprecipitation experiments with cells at corresponding cell cycle stages. We showed that in contrast to early G1, later in G1 the level of newly produced SLBP is similar to S phase suggesting that translation efficiency recovers to the S phase level somewhere between early and mid-G1 (Fig 3). We obtained similar results in at least 3 different experiments.
Figure 3. Low SLBP production rate is limited to early G1.
HeLa cells were synchronized by double thymidine block method and labeled with 35S-methionine for 15 minutes at G1 (2.5–3 hours after M/G1) or S phases after half hour incubation in methionine lacking media. A) Equal protein amounts of total lysates and B) immunoprecipitates with SLBP antibody or beads alone were run on SDS PAGE gel and detected with autoradiography. Cell cycle phases were confirmed by FACs analysis.
SLBP is unstable in G1 due to proteasome mediated degradation
Although SLBP production rate increases to S phase level at somewhere between early and mid-G1, the amount of SLBP expression remains relatively low roughly for another 3–4 hours until late G1 or G1/S before recovering back to S phase level. This suggests additional mechanisms that keep SLBP expression limited in G1 until next S phase.
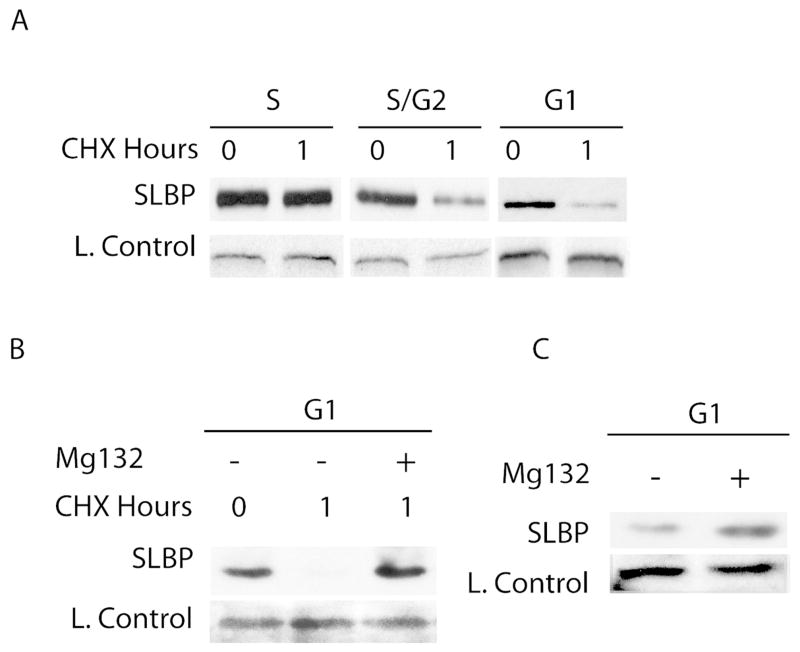
A possible mechanism to limit SLBP expression after the recovery of SLBP synthesis rate could be regulated degradation. This is the case in G1 for several other S phase related proteins like Cyclin A, Cdc6, Ams2 (Harper et al., 2002; Petersen, 2000; Trickey et al., 2013). To investigate this possibility, we compared the stability of SLBP in G1 after the recovery of the SLBP production rate, with different cell cycle phases by treating the cells with cycloheximide and checking the change in SLBP expression in one hour. After cycloheximide treatment SLBP level did not change much in S phase cells. On the other hand, in S/G2 and G1 cells, SLBP level significantly decreased after 1 hour cycloheximide treatment indicating that similar to S/G2, SLBP is unstable in G1 phase (Fig. 4A). Further when we treated these G1 cells with proteasome inhibitor along with cycloheximide, SLBP became stable suggesting that the instability we detect is due to proteasome mediated degradation (Fig. 4B). In early G1 cells, the SLBP level is already very low and it is difficult to make reliable conclusion about the stability of the SLBP with a similar experiment.
Figure 4. SLBP is degraded by proteasome in G1.
HeLa cells were synchronized by double thymidine block method and treated with A) Cycloheximide alone (100 μM), B) Cycloheximide and Mg132 C) Mg132 alone for 1 hour at indicated cell cycle phases. Western blot analysis is performed using anti-SLBP serum. The cross-reacting band recognized by anti-SLBP serum serves as loading control. Cell cycle phases were confirmed by FACs analysis.
Based on our findings, after the recovery of SLBP synthesis rate, SLBP is simultaneously produced and degraded in G1. If this is the case, we expected to see increase in SLBP level when the cells at this point of cell cycle are treated with just proteasome inhibitor. When we treated cells with proteasome inhibitor alone, SLBP level increased significantly confirming that at this point of G1 SLBP is produced efficiently but kept low by proteasome mediated degradation (Fig 4C).
G1 and S/G2 degradations of SLBP are regulated by different mechanisms
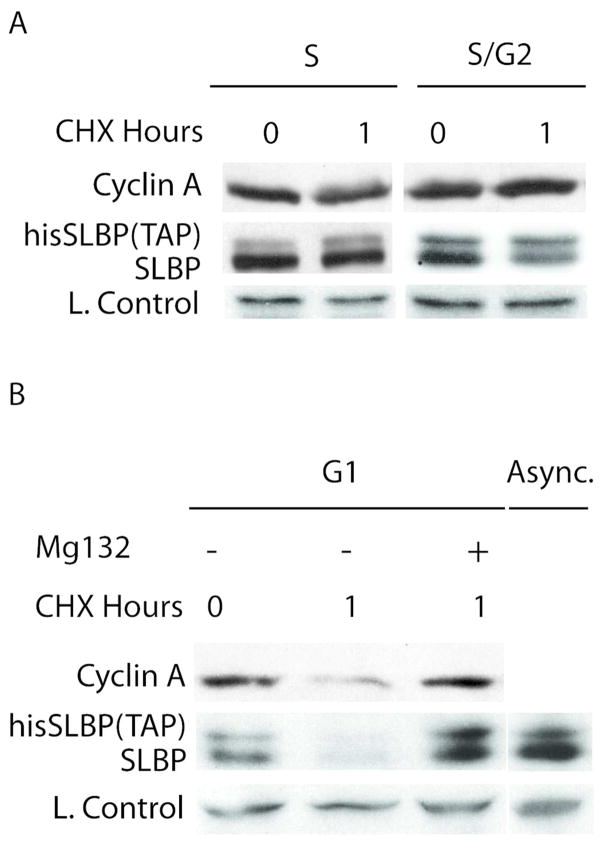
Finally, in order to check whether SLBP degradation mechanism at G1, is different than previously identified S/G2 degradation, we produced HeLa cells stably expressing mutant his-tagged SLBP (Threonine 61 is mutated to Alanine) which is known to be stable at S/G2 transition. When we treated those cells with cycloheximide for 1 hour, S/G2 stable mutant SLBP level did not change much in S or S/G2 phases whereas in G1 cells parallel to endogenous wild type SLBP, it decreased significantly (Fig. 5). When we treated those G1 cells with proteasome inhibitor along with cycloheximide, like endogenous SLBP, mutant SLBP became stable suggesting that the instability we detect is due to proteasome mediated degradation (Fig. 5B). Based on these experiments, we showed that S/G2 stable mutant version of SLBP is degraded in G1 indicating that SLBP degradation at G1 is regulated by different site(s) on SLBP.
Figure 5. S/G2 stable mutant SLBP is degraded by proteasome in G1.
HeLa cells stably expressing his-tagged mutant SLBP were synchronized by double thymidine method and treated with A) Cycloheximide alone B) Cycloheximide and Mg132 for 1 hour at indicated cell cycle phases. Cells were collected before and after treatment and western blot analysis is performed using anti- SLBP serum. Cell cycle phases were confirmed by FACs analysis.
Discussion
As a key player in histone mRNA metabolism, SLBP expression is tightly regulated during the cell cycle. Previously, we showed that Cyclin A/Cdk1 triggers rapid degradation of SLBP at S/G2 border (Koseoglu et al., 2008). In G1, SLBP level is kept low as major mechanism to keep the histone mRNA processing thus histone production closed (Whitfield et al., 2000; Zheng et al., 2003). Here, we found that in G1, along with the previously identified low translation efficiency, SLBP is kept low by proteasome mediated degradation. We showed that low production rate of SLBP is limited to early G1 and SLBP is kept low by simultaneous synthesis and degradation during rest of G1 until S phase. Finally, we found that the regulation mechanism of this newly found SLBP degradation in G1 is different than the previously identified degradation of SLBP at S/G2.
Regulation of SLBP translation
Previously, Whitfield et al. found that SLBP expression is low in G1 and towards the G1/S SLBP level increases without significant change in its mRNA level (Whitfield et al., 2000). They proposed low translation efficiency of SLBP mRNA as the reason for the low level of SLBP in G1. Using 35S methionine pulse labeled cells, they showed that SLBP production rate is low in G1 in comparison to S phase (Whitfield et al., 2000). Here, we repeated similar experiments with HeLa cells and confirmed that SLBP production rate is low in early G1 (Fig 2). As new finding, we showed that around two hours after the M phase somewhere between early and mid- G1, SLBP production recovers to the S phase level (Fig. 3).
Although the SLBP translation rate is low in early G1, the nature of this regulation is still unknown. It is still not clear whether SLBP translation is kept low in early G1 by active inhibition due to binding of specific protein or miRNA, which is simply removed as cells approach S-phase, or whether there is active triggering by activation or expression of specific factors that facilitate SLBP translation towards S phase.
Since cap-dependent general translation is globally shut down during M phase (Cormier et al., 2003; Pyronnet and Sonenberg, 2001), we checked whether global translation had recovered in our early G1 cells (Fig 2A). For this reason we compared the levels and pattern of newly synthesized radiolabeled proteins by running equal protein amount of extracts from our 35S methionine labeled early G1 and S phase cells on SDSPAGE. There is no detectable difference in the pattern and levels of the bands (Fig. 2A) suggesting that general translation had recovered from the M phase shut down for the most abundant proteins detected on the SDS PAGE.
Although we showed that translation for most proteins recovered from the M phase inhibition in our early G1 cells (Fig 2A), it is possible that the activity of translation machinery in early G1, is not sufficient to induce translation of specific set of mRNAs including SLBP mRNA. During M phase, cap-dependent translation is shut down majorly due to inhibition of active eIF4F complex assembly (Cormier et al., 2003; Pyronnet et al., 2001). From M to G1 cap-dependent translation recovers to active state. In G1, mTOR and MAPK pathways further boost the translation initiation machinery and are important for progression to S phase(Cormier et al., 2003; Mamane et al., 2006; Pyronnet and Sonenberg, 2001; Sivan and Elroy-Stein, 2008). It is known that translation of sizable fraction of mRNAs, including several cell cycle related protein coding ones, are more sensitive to the level of eIF4F complex and its helicase activity due to their special sequence properties such as presence of 5′ secondary structures (Gebauer and Hentze, 2004). It is possible that SLBP mRNA is one those mRNAs vulnerable to decrease in the eIF4F activity and although translation recovered for most of the mRNAs in our early G1 cells, SLBP mRNA may require some more time possibly for the complete removal of the M phase inhibition of eIF4F complex formation or even further boost like the one by mTOR pathway.
SLBP degradation in G1
We found that SLBP production rate recovers back to S phase level somewhere between early and mid-G1. But SLBP level is kept low for another 3–4 hours until next G1/S (Fig. 1). When we concentrated on this period of G1 where SLBP is produced as efficiently as S phase but kept low, we found that SLBP is unstable due to rapid degradation by proteasome (Fig. 4). Further, treatment of Mg132 alone accumulated SLBP significantly confirming that SLBP is synthesized and degraded simultaneously at this period of G1 (Fig 4C). This kind of simultaneous production and degradation is the case for several cell cycle proteins (Brandeis and Hunt, 1996; Petersen, 2000) so that when those proteins are needed cells can accumulate them in very short time by shutting down the degradation mechanism.
In G1, before the recovery of SLBP synthesis neither Whitfield et al. (Whitfield et al., 2000) nor we could see significant accumulation of SLBP in response to Mg132 alone (Fig 2C). Since SLBP synthesis is off and the level of SLBP is already very low (lowest during cell cycle) this data can not totally exclude the activity of proteosome mediated SLBP degradation mechanism at early G1. Although this data shows that in early G1 there is no simultaneous synthesis and proteosome mediated degradation of SLBP, it is difficult to make conclusion about whether SLBP degradation mechanism is active at this period of G1.
SLBP is degraded by proteasome also at S/G2 (Whitfield et al., 2000). Previously, we found that cyclin A/Cdk1 phosphorylates Thr 61 on SLBP and triggers this degradation (Koseoglu et al., 2008). Since cyclin A is degraded in G1, we were expecting that the SLBP degradation in G1 will probably be mediated by different site on SLBP. In order to clarify this we checked whether S/G2 stable mutant (Thr61 to Alanine) SLBP is degraded in G1. As expected we found that S/G2 stable mutant SLBP is also unstable in G1 due to proteasome mediated degradation (Fig 5B). Based on these findings we concluded that G1 and S/G2 degradations of SLBP are mediated by different SLBP regions and most probably by different mechanisms
The E3 ligase responsible for the G1 degradation of SLBP is yet to be identified. Krishnan et al. recently suggested Ser20 and Ser23 as a new phosphodegron involved in regulation of SLBP stability (Krishnan et al., 2012). It is possible that there may be an phosphorylation dependent degradation based on these sites in G1.
In G1, several other S phase related proteins like Cyclin A, Cdc6, Ams2 (Petersen, 2000; Trickey et al., 2013) are similarly degraded. Anaphase promoting complex (APC/C) which is activated in M phase and remains active in the following G1 phase, ubiqitinates and triggers degradation of all of these proteins. It is possible that similar to these S phase related proteins, SLBP is ubiquitinated by APC/C for degradation.
In conclusion, here we found that as a major mechanism to keep histone production low in G1, SLBP is kept low by cooperative action of translation regulation and proteasome mediated degradation. Low production rate of SLBP is limited to early G1, and during rest of G1 SLBP is simultaneously produced and degraded, providing cells a system to increase SLBP expression in very short time by shutting down the degradation mechanism. Finally, we found that previously known S/G2 and newly found G1 degradation of SLBP are regulated by different mechanisms.
Acknowledgments
Contract grant sponsor: TUBITAK; Contract grant number:110T987 (To MMK)
Contract grant sponsor: NIH; Contract grant number: GM29832 (To WFM)
This work was supported by The Scientific and Technological Research Council of Turkey (TUBITAK) grant # 110T987 (to MMK), Fatih University Research Fund (to MMK) and NIH grant GM29832 (to WFM).
Special thanks to Dr. Qahru Yüksel and Dr. Nesrin Özören for allowing and helping us to use Flow cytometry facility at Bogazici University.
Footnotes
Authors have no conflict of interest.
References
- Brandeis M, Hunt T. The proteolysis of mitotic cyclins in mammalian cells persists from the end of mitosis until the onset of S phase. The EMBO journal. 1996;15:5280–5289. [PMC free article] [PubMed] [Google Scholar]
- Cakmakci NG, Lerner RS, Wagner EJ, Zheng L, Marzluff WF. SLIP1, a factor required for activation of histone mRNA translation by the stem-loop binding protein. Molecular and cellular biology. 2008;28:1182–1194. doi: 10.1128/MCB.01500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier P, Pyronnet S, Salaun P, Mulner-Lorillon O, Sonenberg N. Cap-dependent translation and control of the cell cycle. Progress in cell cycle research. 2003;5:469–475. [PubMed] [Google Scholar]
- DeLisle AJ, Graves RA, Marzluff WF, Johnson LF. Regulation of histone mRNA production and stability in serum-stimulated mouse 3T6 fibroblasts. Molecular and cellular biology. 1983;3:1920–1929. doi: 10.1128/mcb.3.11.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Zheng LX, Sanchez R, Marzluff WF. Stem-loop binding protein facilitates 3′-end formation by stabilizing U7 snRNP binding to histone pre-mRNA. Molecular and cellular biology. 1999;19:3561–3570. doi: 10.1128/mcb.19.5.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enders GH. Mammalian interphase cdks: dispensable master regulators of the cell cycle. Genes & cancer. 2012;3:614–618. doi: 10.1177/1947601913479799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR, Lewis NJ, Marzluff WF. The histone 3′-terminal stem-loop is necessary for translation in Chinese hamster ovary cells. Nucleic acids research. 1996;24:1954–1962. doi: 10.1093/nar/24.10.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nature reviews Molecular cell biology. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Burton JL, Solomon MJ. The anaphase-promoting complex: it’s not just for mitosis any more. Genes & development. 2002;16:2179–2206. doi: 10.1101/gad.1013102. [DOI] [PubMed] [Google Scholar]
- Harris ME, Bohni R, Schneiderman MH, Ramamurthy L, Schumperli D, Marzluff WF. Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Molecular and cellular biology. 1991;11:2416–2424. doi: 10.1128/mcb.11.5.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N, Sive HL, Roeder RG. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Molecular and cellular biology. 1983;3:539–550. doi: 10.1128/mcb.3.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuno Y, Suzuki A, Sugimura K, Okumura K, Zineldeen DH, Shimada M, Niida H, Mizuno T, Hanaoka F, Nakanishi M. Cyclin A-Cdk1 regulates the origin firing program in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3184–3189. doi: 10.1073/pnas.0809350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koseoglu MM, Dong J, Marzluff WF. Coordinate regulation of histone mRNA metabolism and DNA replication: cyclin A/cdk1 is involved in inactivation of histone mRNA metabolism and DNA replication at the end of S phase. Cell cycle. 2010a;9:3857–3863. doi: 10.4161/cc.9.19.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koseoglu MM, Graves LM, Marzluff WF. Phosphorylation of threonine 61 by cyclin a/Cdk1 triggers degradation of stem-loop binding protein at the end of S phase. Molecular and cellular biology. 2008;28:4469–4479. doi: 10.1128/MCB.01416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koseoglu MM, Graves LM, Marzluff WF. Cyclin A/Cdk1 and CK2 cooperate to trigger degradation of the Stem-loop binding protein (SLBP) at the end of S phase inhibiting histone mRNA biosynthesis. Febs J. 2010b;277:143–143. [Google Scholar]
- Krishnan N, Lam TT, Fritz A, Rempinski D, O’Loughlin K, Minderman H, Berezney R, Marzluff WF, Thapar R. The prolyl isomerase Pin1 targets stem-loop binding protein (SLBP) to dissociate the SLBP-histone mRNA complex linking histone mRNA decay with SLBP ubiquitination. Molecular and cellular biology. 2012;32:4306–4322. doi: 10.1128/MCB.00382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscher B, Schumperli D. RNA 3′ processing regulates histone mRNA levels in a mammalian cell cycle mutant. A processing factor becomes limiting in G1-arrested cells. The EMBO journal. 1987;6:1721–1726. doi: 10.1002/j.1460-2075.1987.tb02423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamane Y, Petroulakis E, LeBacquer O, Sonenberg N. mTOR, translation initiation and cancer. Oncogene. 2006;25:6416–6422. doi: 10.1038/sj.onc.1209888. [DOI] [PubMed] [Google Scholar]
- Martin F, Schaller A, Eglite S, Schumperli D, Muller B. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. The EMBO journal. 1997;16:769–778. doi: 10.1093/emboj/16.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff W. A new way to initiate mRNA degradation. Nature structural & molecular biology. 2009;16:613–614. doi: 10.1038/nsmb0609-613. [DOI] [PubMed] [Google Scholar]
- Marzluff WF, Duronio RJ. Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Current opinion in cell biology. 2002;14:692–699. doi: 10.1016/s0955-0674(02)00387-3. [DOI] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nature reviews Genetics. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick KA, Fisher RP. Why minimal is not optimal: driving the mammalian cell cycle--and drug discovery--with a physiologic CDK control network. Cell cycle. 2012;11:2600–2605. doi: 10.4161/cc.20758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic acids research. 2010;38:2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen BO. Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes & development. 2000;14:2330–2343. doi: 10.1101/gad.832500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyronnet S, Dostie J, Sonenberg N. Suppression of cap-dependent translation in mitosis. Genes & development. 2001;15:2083–2093. doi: 10.1101/gad.889201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyronnet S, Sonenberg N. Cell-cycle-dependent translational control. Current opinion in genetics & development. 2001;11:13–18. doi: 10.1016/s0959-437x(00)00150-7. [DOI] [PubMed] [Google Scholar]
- Rattray AM, Muller B. The control of histone gene expression. Biochemical Society transactions. 2012;40:880–885. doi: 10.1042/BST20120065. [DOI] [PubMed] [Google Scholar]
- Sanchez R, Marzluff WF. The stem-loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Molecular and cellular biology. 2002;22:7093–7104. doi: 10.1128/MCB.22.20.7093-7104.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivan G, Elroy-Stein O. Regulation of mRNA Translation during cellular division. Cell cycle. 2008;7:741–744. doi: 10.4161/cc.7.6.5596. [DOI] [PubMed] [Google Scholar]
- Stauber C, Schumperli D. 3′ processing of pre-mRNA plays a major role in proliferation-dependent regulation of histone gene expression. Nucleic acids research. 1988;16:9399–9414. doi: 10.1093/nar/16.20.9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trickey M, Fujimitsu K, Yamano H. Anaphase-promoting complex/cyclosome-mediated proteolysis of Ams2 in the G1 phase ensures the coupling of histone gene expression to DNA replication in fission yeast. The Journal of biological chemistry. 2013;288:928–937. doi: 10.1074/jbc.M112.410241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZF, Whitfield ML, Ingledue TC, 3rd, Dominski Z, Marzluff WF. The protein that binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes & development. 1996;10:3028–3040. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- Whitfield ML, Zheng LX, Baldwin A, Ohta T, Hurt MM, Marzluff WF. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Molecular and cellular biology. 2000;20:4188–4198. doi: 10.1128/mcb.20.12.4188-4198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, McKillop-Smith S, Muller B. The human histone gene expression regulator HBP/SLBP is required for histone and DNA synthesis, cell cycle progression and cell proliferation in mitotic cells. Journal of cell science. 2004;117:6043–6051. doi: 10.1242/jcs.01523. [DOI] [PubMed] [Google Scholar]
- Zheng L, Dominski Z, Yang XC, Elms P, Raska CS, Borchers CH, Marzluff WF. Phosphorylation of stem-loop binding protein (SLBP) on two threonines triggers degradation of SLBP, the sole cell cycle-regulated factor required for regulation of histone mRNA processing, at the end of S phase. Molecular and cellular biology. 2003;23:1590–1601. doi: 10.1128/MCB.23.5.1590-1601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]