Abstract
Advances in modern optical microscopy have provided unparalleled access to intracellular structure and function, yet visualizing lipid molecules within a cell remains challenging. Stimulated Raman Scattering (SRS) microscopy is a recently developed imaging modality that addresses this challenge. By selectively imaging the vibration of chemical moieties enriched in lipids, this technique allows for rapid imaging of lipid molecules in vivo without the need for perturbative extrinsic labels. SRS microscopy has been effectively employed in the study of fat metabolism, helping uncover novel regulators of lipid storage. This unit provides a brief introduction to the principle of SRS microscopy, and describes methods for its use in imaging lipids in cells, tissues and whole organisms.
Introduction
Modern molecular biology demands sophisticated imaging techniques capable of rapid detection of specific molecules with high sensitivity. In spite of dramatic advances in fluorescence-based optical microscopy, visualizing certain classes of molecules has remained difficult. Lipids, in particular, are challenging targets to image because they lack intrinsic fluorescence and cannot be fluorescently tagged with ease. The size of the fluorophore often exceeds the size of the lipid molecule, and the tags are often chemically invasive and impractical for in vivo applications.
This unit describes a label-free lipid imaging technique based on the principle of vibrational microscopy. Known as Stimulated Raman Scattering (SRS) microscopy, this method does not require fluorescent tagging, instead relying on the characteristic vibrational frequency of the chemical bonds in lipid molecules to identify various lipid species with high sensitivity and specificity. This unit provides a brief introduction to the principle and its application in lipid imaging. Details of instrumental setup and C. elegans sample preparation prior to imaging are provided in “Basic Protocols 1 and 2”. Preparation of frozen tissue sections and live cells are discussed in “Support Protocols 1 and 2”.
The Raman effect and SRS
When light is incident on a sample, the majority of photons interacting with the molecules scatter elastically (Rayleigh scatter) and maintain their incident energy. However, a tiny fraction of photons interact with the vibrational states of the molecules in the sample and scatter inelastically; this is known as spontaneous Raman scattering or the “Raman effect” (Raman, 1928). The chemical bonds of the molecule may absorb some of the energy of an incident photon (known as the “pump” photon) and get excited to a higher vibrational energy level. The scattered photon (known as the “Stokes” photon) will thus be of lower energy than the incident “pump” photon. Alternatively, a “pump” photon may interact with chemical bonds existing at a higher vibrational state; in this case the scattered photon (known as the “Anti-Stokes” photon) will have higher energy than the “pump” photon. The difference in frequency between incident and scattered photons is known as the Raman shift. Each chemical bond in the molecule has a characteristic and quantifiable Raman shift. The CH2 bond, abundant in lipid molecules, has a Raman shift of 2845 cm−1 (Thomas, 1999).
In conventional spontaneous Raman microscopy, an excitation laser beam at a single frequency is utilized to generate spontaneous Raman scattering. However, the signal generated from the spontaneous vibrational transition is extremely weak (>8 orders of magnitude weaker than fluorescence), making imaging a time-consuming process. Instead, the weak Raman signals can be amplified when two coherent laser beams (set at “pump” and “Stokes” frequencies) are used to excite the sample. If the difference in their frequencies matches the vibrational frequency of a molecule in the sample, the molecular vibration transitions to an excited state due to stimulated excitation. As a result of this coherent excitation, a “pump” photon is absorbed by the sample and a “Stokes” photon is generated. The intensity of the transmitted “Stokes” beam experiences a gain (stimulated Raman gain, SRG) and the intensity of the transmitted “pump” beam experiences a loss (stimulated Raman loss, SRL). However, if the frequency difference between the two laser beams does not match the vibrational frequency of the molecule, neither SRG nor SRL signals will be generated. Therefore, capturing SRG or SRL signals provides the basis for Stimulated Raman Scattering (SRS) microscopy imaging of specific molecules. In principle, both SRG and SRL signals can be utilized for SRS microscopy imaging (Freudiger et al., 2008). In practice, the instrumental set-up designed for SRL signals (described in this protocol) is much more popular than that for SRG signals. The imaging speed of SRS microscopy based on SRL signals is around 2 µsec per pixel, which is much faster than spontaneous Raman microscopy.
Imaging lipids by SRS microscopy
All eukaryotes store calories in the form of neutral lipids, comprised primarily of triacylglycerols (TAGs) and cholesterol esters (CEs). The fatty acid chains comprising a TAG/CE molecule are each 14-24 carbons in length, and may be saturated (made entirely of CH2 groups) or unsaturated (containing one or more C=C groups). The neutral lipids are stored in single-membrane organelles known as lipid droplets, which are highly dynamic and can rapidly expand during caloric excess and shrink with starvation. The high concentration of CH2 bonds within lipid droplets generates a characteristic and strong SRS signal at 2845cm−1 when excited. Neutral lipid molecules can thus be specifically detected and easily quantified using SRS microscopy in a rapid, label-free, non-invasive and high-throughput fashion in intact cells, tissue sections and whole organisms (Wang et al., 2011; Wei et al., 2014; Wei et al., 2013). The two-photon optical process of SRS microscopy is comparable to that of two-photon fluorescence microscopy, allowing for sub-cellular spatial resolution definition even in relatively thick tissue samples up to 1 mm. Unlike canonical lipid imaging methods that employ dye stains or fluorescent analogs, SRS microscopy is capable of distinguishing between various classes of lipid molecules based on their characteristic Raman shifts in a label-free manner.
Basic Protocol 1: Instrumental setup for stimulated Raman Scattering (SRS) microscopy
Introduction
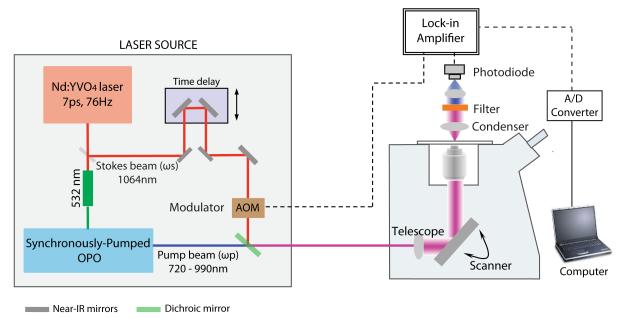
A variety of approaches to imaging by SRS microscopy have been described in the literature for various applications. This protocol describes the basic setup and operation of the SRS system for quantitative lipid imaging. A typical setup for an SRS microscope is shown in Figure 1. Two laser beams, “pump” (ωp) and “Stokes” (ωs) are used to scan the sample under a laser-scanning microscope, and their frequency difference (Δω) can be tuned to match a particular vibrational frequency (Ω). The SRL signal can be specifically picked up, de-modulated and amplified via a lock-in amplifier. The SRL signal intensity is directly proportional to the concentration of the target molecule in the sample, and can be easily used for direct quantification. Comprehensive descriptions of the underlying physical principles are available elsewhere for interested researchers (Min et al., 2011). Detailed information on the various components of an SRS microscope setup is also available for further reference (Freudiger et al., 2008; Saar et al., 2010; Yu et al., 2014).
Figure 1.
Schematic representation of instrumental setup for SRS microscopy. The “Stokes” beam wavelength is held constant at 1064 nm and modulated by an acousto-optic modulator (AOM). The optic parametric oscillator (OPO) is synchronously pumped by frequency-doubled wavelength from the laser source, which serves as the “pump” beam. The transmitted beam is collected by a condenser, filtered and detected by a photodiode. The stimulated Raman Loss (SRL) is demodulated and amplified by a lock-in amplifier. The electric signal from the lock-in amplifier is subsequently converted into a digital signal and used by a computer for image construction.
EQUIPMENT
Laser system and associated components
Laser-scanning microscope
Mirror and filter sets
Detector
Lock-in amplifier
A/D converter
Software for imaging (e.g. Fluoview®) and quantification (ImageJ®)
1) Laser system and associated components
The laser source (Figure 1) is the most crucial and complex component of an SRS microscopy system. Proper setup and calibration are essential for accurate and reproducible imaging. Excitation of SRS microscopy requires two laser wavelengths, one of which must be tunable. Typical laser properties include:
Pulse width: 1-10 picoseconds (ps)
Repetition rate: 40-100 MHz
Power: 30-100 mW at the sample plane, for each laser beam
A commonly used pump laser system for SRS microscopy is the picoTRAIN (High-Q, 532nm, 7 ps, 76 MHz).
Other components coupled to the laser system include:
- Optical Parametric oscillator (OPO) (Levante Emerald, APE Germany): required to tune the “pump” laser beam into different frequencies. The OPO is synchronously pumped by frequency-doubled wavelength from the laser. The idler-beam of the OPO is blocked by an interferometric filter (Chroma Technology, 890/220m).
- Dichroic beam-combiner (Chroma Technology, 1064 DCRB): to achieve spatial overlap with the “Stokes” beam.
- Delay-stage: to adjust the time delay of one laser beam and achieve precise temporal overlap of “pump” and “Stokes” pulses.
- Autocorrelator (APE GmbH, PulseCheck): to measure pulse width and ensure temporal overlap.
- Acousto-optic modulator (AOM) or electro-optic modulator (EOM): necessary for high-frequency modulation of the “Stokes” beam. SRL signals are typically weak and often masked by low-frequency background laser noise. The “Stokes” beam is therefore modulated at high frequency (>2 MHz) to allow the weak high frequency SRL signal to be distinguished from low frequency laser noise. Of the two options, AOM is less expensive but cannot be used for very high-frequency modulation (usually limited to <10 MHz) and is less efficient at frequencies greater than 5 MHz. EOM can achieve rapid modulation (up to 20 MHz for video rate SRS microscopy) and full modulation depth, but the modulation frequency is not adjustable.
NOTE: picoEMERALD (A.P.E. GmbH) (Figure 1) is an all-in-one “turn-key” laser system designed specifically as a light source for SRS and other coherent Raman scattering microscopy systems. It consists of the laser, frequency-doubling unit, AOM, dichroic mirror and delay-stage. For biological imaging, the system can be set to provide the “Stokes” pulse at 1,064 nm with 7 ps pulse width and 80 MHz repetition rate. The frequency-doubled beam at 532 nm is used to synchronously seed a picosecond OPO, to produce a mode-locked pulse train with 5-6 ps pulse width. The wavelength of the OPO is tunable from 720 to 990 nm, which serves as the “pump” beam. The intensity of the “Stokes” beam is modulated by a built-in AOM at 10 MHz, with a modulation depth of more than 70%. The “pump” beam is spatially overlapped with the “Stokes” beam with a dichroic mirror inside picoEMERALD. The temporal overlap between “pump” and “Stokes” beams is ensured with a built-in delay stage. picoEMERALD thus provides fully automated spatiotemporally overlapping “pump” and “Stokes” beams. Overlap can be further optimized using the SRS signal of pure dodecane (See ‘Steps: Calibration’).
2) Laser-scanning microscope
The SRS system is based on a two-photon laser scanning microscopy system. The optical components of the microscope must be optimized for infrared (IR) lasers (750 −1100 nm). The objectives require achromatic compensation, high numerical aperture (N.A.), and high near-IR throughput. Either a water lens or a water dipping lens will work as excitation objectives. The overall throughput from the laser source to the sample plane is typically around 15%-25%, depending on variations in lens, objectives and/or wavelengths.
3) Mirror and filter sets
Several near-IR mirrors with adjustable holders are required to deliver laser beams from the laser source box to the scanning unit of the microscope. A set of telescopes helps fit the laser beam size to the back aperture of the objective.
4) Detector
A silicon photodiode (FDS1010, Thorlabs) with a large area (typically around 1 cm2) is widely used for detecting SRL signals, owing to its high linear dynamic range, fast response, and high quantum efficiency. Furthermore, a reverse DC voltage at 30-60 V, pre-filtered by a low pass filter (Mini-Circuits, BLP-1.9) is applied to the photodiode for reducing its capacitance and increasing response speed (Freudiger et al., 2008). The transmission light from the sample is collected by a condenser with a higher N.A. than the objective, reflected by a mirror, and then projected to the photodiode through a telescope (for adjusting beam size and avoiding beam movement) and a high OD band-pass filter (to block the “Stokes” beam and selectively transmit the “pump” beam for SRL detection, Chroma Technology, 890/220m) (Freudiger et al., 2008).
5) Lock-in amplifier
The detected SRL signals are demodulated and amplified by a lock-in amplifier before used for image construction. An amplifier with the following properties is required:
High-modulation frequency (to demodulate SRL signals at high frequency)
Fast output time-constant (to allow fast image construction)
High dynamic reserve (>70dB) (to allow fast image construction)
Long-term baseline stability (to ensure imaging consistency)
Input signals from the photodiode detector are pre-filtered with a band-pass electric filter (specifically selected for the frequency of “Stokes” beam modulation) to remove low-frequency noise (due to laser scanning) and high-frequency noise (from laser rate repetition), and then fed into the lock-in amplifier. The signal from the modulator is input into the lock-in amplifier as a reference for modulation frequency (Freudiger et al., 2008).
6) A/D converter
An A/D converter is used to convert electric analog signals from the lock-in amplifier to digital signals, which will be subsequently input into a computer. Laser scanning microscopes are typically pre-installed with an A/D converter.
Imaging parameters
As shown above in a typical SRS microscopy system, the “Stokes” beam has a fixed wavelength and the “pump” beam can be tuned to a different wavelength to match the vibrational frequency of the chemical bonds/groups of the molecules to be imaged. The following equation can be used to calculate the wavelength of the “pump” beam for molecules of interest:
| (1) |
where Δω is the Raman shift expressed in wavenumber, λp is the wavelength of the “pump” beam, and λs is the wavelength of the “Stokes” beam. Wavenumber is typically expressed in units of inverse centimeter (cm−1), whereas wavelength is commonly expressed in units of nanometer (nm). Rearranging the equation to obtain λp:
| (2) |
In a common SRS microscopy system for lipid imaging, the wavelength of the “Stokes” beam is typically held constant at 1064 nm. The CH2 bond (enriched for neutral lipids in lipid droplets) has a characteristic Raman shift of 2845 cm−1.
| (3) |
Solving for λp with these numbers, the calculated “pump” beam wavelength for imaging CH2 bonds in lipids is 816.76 nm. For optimization of the imaging system, image a pure reference chemical (such as triolein) and scan a range of wavelengths (calculated wavelength ± 2 nm, in 0.3 nm steps) in order to identify the wavelength that provides the strongest signal.
NOTE: The wavelength calculation outlined above in (Equation 3) is specific to the CH2 bonds that are commonly used to measure total neutral lipids. To differentiate different lipid classes (or image other chemical groups in different molecules), the wavelength of the “pump” beam must be calculated accordingly to the characteristic Raman shifts of those molecules (Thomas, 1999). Depending on the nature of the sample, possible interference must also be considered from other chemicals.
Verifying SRS signals
When first using an SRS microscope, it is necessary to confirm the observed signal is indeed an SRS signal. To verify the identity of the signal, use the following checklist.
- The signal must correspond linearly to both “pump” and “Stokes” laser powers.
- The signal must disappear completely when the frequency difference between the laser beams does not match the vibrational frequency of the molecule of interest.
- The signal must disappear completely when tuning off one laser beam.
- The excitation spectrum obtained when scanning across a range of frequencies within the band of interest should be consistent with the Raman spectrum reported in the literature.
STEPS
Calibration
1) Turn the power on for the laser source. Allow approximately 20 minutes for the laser to power up and equilibrate before image acquisition. In the meantime, turn on the associated components of the laser-scanning microscope and open the software for image acquisition.
2) Place a small volume of pure dodecane (CH3(CH2)10CH3, ~10 µl) in the center of a glass slide, covered with a coverslip and place it at the center of view.
3) Turn the bright field light source on and direct light to the eyepiece.
4) Ensure that the condenser is positioned such that the image is in sharp focus and the aperture is located at the center of the view.
5) Start scanning. Adjust the laser power and the focus to obtain a uniformly bright (but not saturated) image. When the alignment of the system is optimized, the brightest spot of the dodecane image should be round and located at the center of the view.
6) Once calibration is complete, begin imaging samples. Recalibrate every time when the system is restarted.
NOTE: Important parameters such as laser power, throughput, modulation depth, and SRS signal strength should be measured and recorded for fine-tuning and troubleshooting.
NOTE: It is critical that the same laser power and imaging parameters are used across all samples throughout the experiment. A change in any of these properties can significantly alter signal strength and complicate subsequent interpretation of results.
Sample Imaging
7) Mount the slide with a coverslip facing the objective lens. If oil immersion lens will be used at the magnification at which the image is obtained, add a droplet of immersion oil to the coverslip and to the lens at this step.
8) Turn the bright field light source on and direct light to the eyepiece of the microscope.
9) At the lowest magnification, gradually adjust the position of the stage to bring the sample to the center of view. Bring the sample in focus using the coarse or fine focus adjustment.
10) Switch to the desired magnification. Re-focus the sample.
11) Turn off the bright field light source.
12) Switch on the laser scanning unit and the diaphragm. Confirm that the “pump” and “Stokes” beams pass through the laser-scanning unit to the sample, then past the condenser and filter, to the photodiode detector.
13) Begin scanning the sample. Set a fast scanning rate (e.g. 512 x 512 pixels) at first, and adjust the fine focus to find the area of interest.
14) Switch to a slower scanning rate and higher resolution (e.g. 1024 x 1024 pixels) without altering the focus. A scanning rate typically used for SRS imaging is 2 µs/pixel.
15) Obtain an SRS signal image, and save it as a high-resolution image (such as a .tiff file).
16) If there are multiple samples on the same slide, switch back to the faster scanning rate and slowly adjust the position of the stage to bring the next sample in the center of view. Repeat steps 8 – 10 to finish imaging all the samples on the same slide.
17) Once the desired samples on the slide have been imaged, close the diaphragm below the stage to block the laser beams (do not turn off the laser source). Carefully remove the slide.
18) Place the next sample slide, and repeat steps 2 – 12.
19) Once all samples have been imaged, turn off the laser source and associated equipment, the microscope and the computer.
Analysis of lipid images with ImageJ®
20) Open the image files to be quantified in ImageJ [go to “File → Open” to select files individually, or select all files (WINDOWS: “Control” + mouse click; OS X: “Command” + mouse click), drag the files and drop into the ImageJ window. If several files are opened simultaneously, cascade or stack the image windows (select “Window → Cascade” or “Window → Tile”).
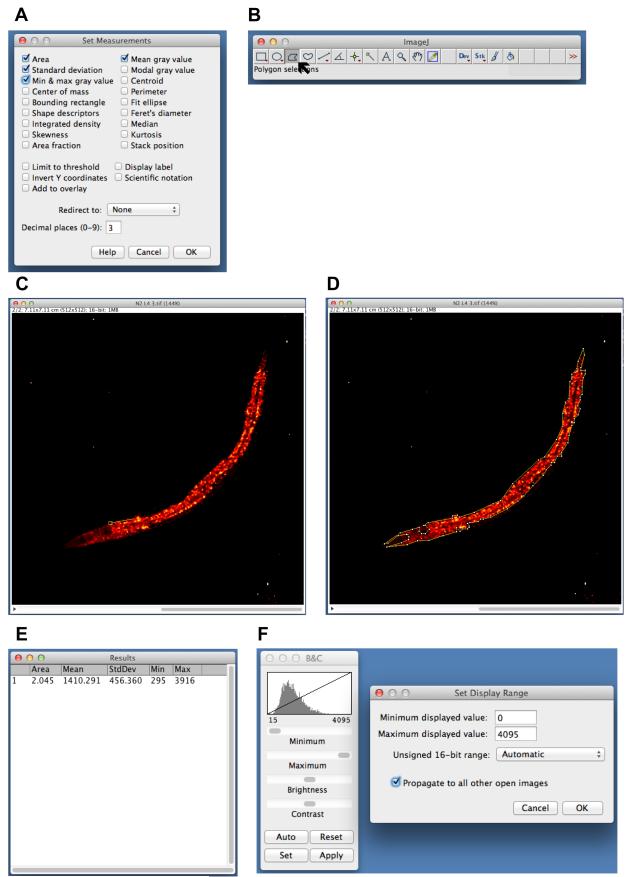
21) Choose the properties to be analyzed, under “Analyze → Set Measurements”. Typical parameters to be measured include “Area”, “Standard deviation”, “Min and max gray value” and “Mean gray value” (Figure 2A), although several other measurements can be made with ImageJ. Check the desired parameters and click “OK” to close the window.
Figure 2.
Quantification of neutral lipid stores using ImageJ®. Refer text for details.
22) In the ImageJ window, click on the polygon selection tool (Figure 2B). On one opened image window, carefully outline the region to be quantified as follows:
- Click once on an outer edge of the desired region. A selection anchor is placed at that point.
- Move the pointer along to an adjacent point on the border, and click again to place an anchor. A straight line is drawn between the first and second anchors (Figure 2C).
- Repeat the process, drawing a series of small lines that outline the region to be quantified.
- The polygon is completed once the first anchor is clicked again (Figure 2D).
23) To measure the parameters selected in Step 2, go to “Analyze → Measure” or use the keyboard shortcut (WINDOWS: “Control + M”, OS X: “Command + M”). A new “Results” window opens to display the measured values (Figure 2E).
24) Close the image window.
25) Repeat Steps 3 to 5 for each open image.
26) In the results window, select all the measurements (click on the “Results” window, then “Edit → Select all”). Copy all measurements to the clipboard (“Edit → Copy”)
27) Paste the results into a new Microsoft Excel file. Proceed with quantification and statistical analysis as desired.
NOTE: To ensure that all images have the same distribution of pixel intensities, set the window of displayed brightness and contrast to maximum. Go to “Image → Adjust → Brightness and Contrast”. In the window that opens, click on “Set” and choose the following (Figure 2F):
Minimum displayed value: 0
Maximum displayed value: 4095
Check the box to select “Propagate to all other open images”
Click “OK” to close the window.
This is important for displaying comparative images consistently across samples, for instance when presenting images of wild type/control and mutant/treated samples in publication. Note that altering the displayed values will not affect the results of measurement.
BASIC PROTOCOL 2: Preparation of C. elegans samples for SRS imaging
INTRODUCTION
Caenorhabditis elegans is an excellent model organism for multiple imaging techniques. Since it is whole-body transparent, various tissues can be imaged in the intact animal without the need for dissection. They are also useful for high-throughput screens owing to the availability of genetic tools, small size and easy handling.
In C. elegans, neutral lipids are largely stored in the intestine, which consists of 20 epithelial cells located bisymmetrically around the intestinal lumen. Hypodermal cells and oocytes also store lipids. Many regulatory factors involved in lipid storage and distribution are conserved between C. elegans and higher organisms. We found that C. elegans lipid droplets consist mainly of triacylglycerols (TAGs), which generate strong SRS signals due to the abundance of CH2 bonds present in fatty acid chains. Applying the SRS system to this genetically tractable model organism has discovered new genetic regulatory factors of lipid metabolism (Wang et al., 2011).
This section describes the preparation of C. elegans samples for SRS imaging.
MATERIALS
C. elegans
Glass slides and coverslips
General purpose laboratory labeling tape
Agarose
Heat block
Glass Pasteur pipettes
Pipette bulb
Dissection microscope
Worm picker (optional: platinum wire fixed at the tip of a Pasteur pipette)
Anaesthetizing agent (sodium azide or levamisole)
M9 buffer
STEPS
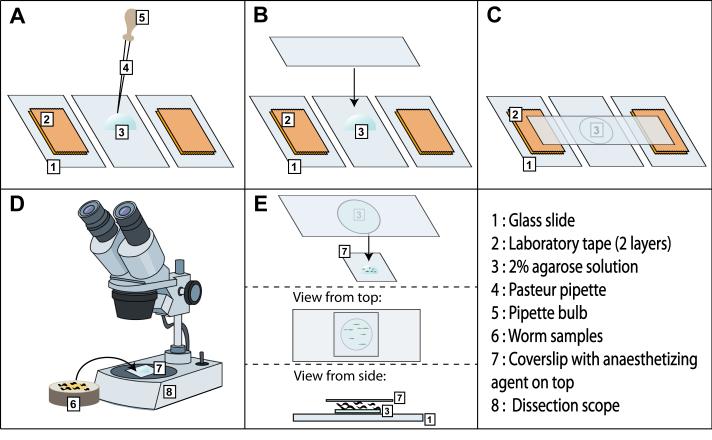
Briefly, prepare the slides with agarose pads and mount the worms onto the pads. The agarose pads help to prevent worms from being crushed between the coverslip and the glass slide, and also help the worms stay moist and avoid desiccation. Follow the steps depicted in Figure 3.
Figure 3.
Preparation of agarose pads and C. elegans samples for SRS imaging. Refer text for details.
Preparing the agarose pads
1) Prepare 2% agarose solution in distilled H2O. Place the melted agarose on a heat block (at approximately 200°C) while preparing the slides.
2) Prepare two support slides by placing two layers of laboratory tape on each. The support slides will ensure that the pad thickness (~0.2 mm) will stay the same across all of the samples. Place these support slides on each side of a blank slide (Figure 3A).
3) Place one drop of warm 2% agarose onto the blank slide using glass Pasteur pipette (Figure 3A).
4) Quickly place a second slide on top of the agarose drop, at a 90° angle to the three slides (blank slide and two supporting slides) (Figure 3B), and gently press the edges of this slide such that the agarose drop spreads in a circle (Figure 3C). Set these slides aside; do not separate the two glass slides holding the agarose pad until the samples are ready.
NOTE: Make fresh agarose pads for each imaging session.
Mounting the worms
5) Pick the worms to be imaged under the dissection microscope and add them to a droplet of M9 buffer with anaesthetizing agent such as sodium azide or levamisole, placed on a coverslip (Figure 3D). Generally, 4-5 µL of 75 µM sodium azide/ 1 mM levamisole is sufficient for 10-20 worms. Adjust the droplet size as needed. Once all the worms are added to the drop of anaesthetizing agent, move them around within the droplet using a picker so that they do not overlap with each other.
NOTE: Levamisole is recommended if the worms need to be recovered after imaging.
6) Once the worms are anaesthetized, separate the glass slides (from Step 4) without perturbing the agarose pad. Take the glass slide containing the agarose pad and cover the worms with this slide with the pad facing down towards the worms (Figure 3E). Alternatively, place a droplet of sodium azide/levamisole on the agarose pad, pick worms onto the droplet, and cover the droplet with the coverslip.
7) To easily locate the worms when imaging, mark the position of the worms on the coverslip with a permanent marker. Proceed with imaging.
NOTE: For total lipid content quantification, there is no need to fix or stain the worms. If fixing is the preferred method, the use of plastic, paraffin or oil (all rich in CH2 bonds and bearing strong Raman signals) must be avoided. Formaldehyde solutions may be used for fixing, but ethanol or other organic solvents that may dissolve lipids from samples should be avoided for sample preparation.
SUPPORT PROTOCOL 1: Sample preparation for live cell imaging by SRS microscopy
INTRODUCTION
SRS microscopy is a non-invasive, label-free imaging modality that is amenable to live cell imaging with minimal preparatory steps. Cells are grown on glass bottom dishes and imaged directly, with no fixation or staining required. Care must be taken to maintain cellular integrity before and during imaging.
MATERIALS
Cells of interest
Appropriate growth medium
Uncoated glass bottom dishes (P35G-1.0-14-C, MatTek)
60x water objective
Water condenser
STEPS
1) Grow cells on uncoated glass bottom dishes with the appropriate growth medium and culture conditions.
2) Culture cells to the required degree of confluency. A very high degree of confluency may affect the ability to visualize single cells.
NOTE: Components of the growth medium may interfere with the SRS signal. To check for this possibility, image a small volume of fresh growth medium at the difference frequency to be used during cell imaging. If an SRS signal is obtained from fresh growth medium, test other alternative/minimal media. If no alternative is available, replace growth medium with 1x PBS and image the cells as quickly as possible.
3) Place the dish on the stage.
4) If using an upright microscope: position the stage such that the water objective is in contact with the medium, as close to the cells as possible. If using an inverted microscope: add 1-2 drops of water to the objective lens and ensure contact with the bottom of the glass dish.
5) Install the water condenser. If using an upright microscope: add 1-2 drops of water to the condenser lens and ensure contact with the bottom of the glass dish. If using an inverted microscope: position the stage such that the condenser is in contact with the medium, as close to the cells as possible.
6) Proceed with imaging.
NOTE: Imaging should be performed within 30 minutes to prevent sample deterioration/degradation.
SUPPORT PROTOCOL 2: Preparation of frozen tissue sections for imaging by SRS microscopy
INTRODUCTION
Both fresh and frozen tissue slices can be imaged by SRS microscopy. For example, fresh mice brain slices are commonly imaged using SRS microscopy (Ji et al., 2013). Preparation of fresh brain slices is straightforward. Once the brain is harvested, place it in a brain mold that matches the age of the mice. Slice sections manually at regular size intervals using a razor blade. Depending on the mold, these intervals range from 0.5 mm to 3 mm. After slicing, mounting sections onto glass slides and cover them with coverslips. Image as soon as possible.
Frozen sections can be unfixed or formaldehyde fixed. However, usage of organic solvents other than formaldehyde should be avoided because they can dissolve lipids, disrupt the lipid storage compartments, and generate imaging artifacts. The use of plastic, paraffin or oil (all rich in CH2 bonds and bearing strong Raman signals) must also be avoided. If the tissue is not going to be imaged freshly, the dissected tissue should be frozen or fixed promptly before the cell destruction starts. Slow freezing can distort the tissue due to ice crystal formation and expansion. The goal should be to freeze rapidly such that water does not have time to form crystals, and will thus not expand when solidified. This technique is often referred to as “snap-freezing” or “flash-freezing” (Scouten and Cunningham, 2012). After freezing, the tissue can be stored at −80°C until sectioning using cryostat. The thickness of frozen sections can range from 5 µm to 300 µm.
MATERIALS
Tissue sample
Liquid nitrogen
Liquid nitrogen canister
Metal beaker
Isopentane (2-methyl-butane)
Dry ice
Petri dish
Tissue freezing medium (Triangle Biomedical Sciences)
Gum tragacanth (Sigma-Aldrich)
Cryomolds
Long metal forceps
Cryostat (Leica CM3000)
Treated glass slides (e.g. Superfrost® Plus Sides from VMR)
Coverslips
PBS
STEPS
1) At least 10 minutes before freezing the sample, prepare cold isopentane. Fill two-thirds of a metal beaker with isopentane and place the metal beaker in a liquid nitrogen canister. At the same time, prepare a bucket with dry ice.
2) Place the dissected tissue in a Petri dish and cover it with 3:1 tissue freezing medium and gum tragacanth mixture. This step will acclimate the tissue to freezing medium.
3) Transfer the tissue to a cryomold and cover it with 3:1 tissue freezing and gum tragacanth mixture. Orient the tissue in the preferred position for sectioning. The bottom of the cryomold will be the sectioning surface. Avoid formation of bubbles, especially near the tissue.
4) Using long metal forceps freeze the tissue by partially submerging the cryomold into isopentane. Do not fully submerge. After the freezing medium mixture is frozen, place the cryomold on dry ice until all of the tissue samples are frozen.
5) When freezing of all of the samples are completes, wrap the cryomolds with aluminum foil individually and store in a plastic bag at −80°C until sectioning.
6) When ready to section, use a cryostat to create 10 µm- thick sections and mount on glass slides.
7) Add a drop of PBS onto the tissue section and cover with a coverslip. The slide can then be directly imaged. SRS imaging should avoid using mounting media containing glycerol, which would emanate a strong SRS signal.
NOTE: Fixing the tissue sections directly is also possible and is recommended for long time imaging and preserving samples. To fix sections that are mounted onto slides, place frozen slides to 3.7% formaldehyde (in PBS) for 30 min, and then cover with a coverslip.
COMMENTARY
Background Information
Obesity has reached epidemic proportions worldwide and is a significant risk factor for expensive health problems such as type II diabetes, cardiovascular disease and certain types of cancer (Lawrence and Kopelman, 2004; Ogden et al., 2014). To defend against the obesity epidemic, significant research efforts are underway to better understand the regulatory mechanisms of lipid metabolism and storage. Lipids lack inherent fluorescence and cannot be tagged with fluorophores due to their small size. For quantitative detection of lipid molecules in samples, the label-free imaging technique of SRS microscopy is a reliable alternative to the traditional methods.
Traditional approaches to lipid storage analysis have involved biochemical assays or fixation and staining protocols. Comparative analysis of label and label-free methods is available in (Yen et al., 2010; Yu et al., 2014). Biochemical quantification assays such as TLC or GC/LC-MS can provide detailed analysis of the composition of various lipid molecules. However, they require very large sample amounts and sample preparation usually takes several hours. Another major limitation of these assays is that no spatial information on the distribution of lipid droplets is obtained. Lipid dyes such as Oil Red O and Sudan Black provide useful cellular information of lipid droplets. The assays are of low cost and the protocols relatively straightforward. However, these staining protocols require fixation, which can sometimes generate artificial morphological changes of lipid droplets and result in inconsistency between samples.
The usage of coherent Raman scattering (CRS) microscopy in biomedical research has increased rapidly in recent years. The two most commonly used CRS microscopy modalities are Coherent Anti-Stokes Raman Scattering (CARS) microscopy and SRS microscopy. They have slightly different working principles, but both are label-free imaging techniques that only require a small sample size that can be prepared for imaging in minutes. CARS signals generate non-resonant background signals even in the absence of vibrational resonance that limits the detection sensitivity and specificity (Evans and Xie, 2008). In addition, they have non-linear relationship with the molecule concentration, which complicates quantification processes. Unlike CARS microscopy, SRS microscopy does not generate non-resonant background signal and offers direct linear quantification of analyte concentration. Currently, the major limitation of this technique is the cost of the equipment. We anticipate that new technological advances and commercial availability will lower the cost and expand use of SRS microscopy in biomedical research.
Critical Parameters and Troubleshooting
Building the SRS microscopy system and finding the SRS signal
The SRS system is based on a two-photon laser scanning confocal system. For compatibility with SRS microscopy, ensure that the optical components of the microscope are optimized for IR lasers. The most critical added component of the system is the laser source. It is essential that the pulse width, power and repetition rate of the laser meet the criteria described in the “Basic Protocol.” Similarly, the lock-in amplifier must meet all the properties described in order to extract the signal efficiently and construct the image. Also, ensure that the “Stokes” beam is modulated at high frequency (> 2 MHz) to minimize background.
Sample preparation for SRS imaging
Preparing samples for SRS imaging is not very different than for other imaging urposes. However, certain factors need special attention. In general, fresh samples should be imaged as soon as possible after mounting them onto the slide to prevent sample deterioration and lipid droplet coalescence. If the sample will be fixed, then the use of ethanol or other organic solvents should be avoided since they may dissolve lipids from samples. Formaldehyde is safe to use with SRS microscopy. For cell imaging, the choice of medium is important; the medium itself may interfere with the SRS signal. The medium should be imaged independently for any SRS signal at the same difference frequency used for sample imaging.
Anticipated Results
SRS microscopy can be utilized to selectively visualize lipid stores in tissue sections, in live cells and in whole organisms such as C. elegans. Cells/animals with genetic mutations, or those exposed to different environmental conditions (such as starvation, temperature changes or diet), can be imaged, quantified and compared to their corresponding controls. The spatial resolution of SRS microscopy is comparable to that of two-photon fluorescence microscopy (~ 300 nm in the x–y plane and ~ 1000 nm in the z-axis, depending on objectives and wavelengths used), allowing for lipid molecules to be visualized at the subcellular level. In live cells and C. elegans, SRS microscopy can reveal individual lipid droplets, including nascent droplets that may be particularly small. Furthermore, the use of near-infrared lasers allows for high penetration depths of up to 1 mm (Helmchen and Denk, 2005). This is particularly useful for imaging the spatial distribution of lipid molecules in thick tissue samples such as mouse skin, liver and brain.
Different classes of lipid molecules are structurally diverse, and canonical lipid imaging methods that use dye staining or fluorescent analogs cannot distinguish these different classes. In contrast, SRS microscopy can differentiate specific lipids based on differences in their fatty acid chains (Fu et al., 2014).
Time Considerations
Generally, at least 20 worms should be analyzed for each mutant or condition. With practice, it is possible to image worms from 3-4 different mutants/conditions or a total 60-80 worms per hour. For the purpose of a high-throughput genetic screen, the imaging parameters can be suitably adjusted to increase this number; for instance, scanning speed can be increased at the cost of lower image resolution for first-pass screening.
Troubleshooting
| PROBLEM | CAUSE | SOLUTION | |
|---|---|---|---|
| Problems caused during imaging |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Problems caused during sample preparation |
|
|
|
|
|
|
|
|
|
|
Acknowledgements
We would like to thank Dr. Y. Yu for critical reading and helpful discussions. M.C.W. is supported by grants from the National Institutes of Health (R01AG046582, R00AG034988) and the Ellison Medical Foundation. P.V.R. is supported by a pre-doctoral fellowship award from the American Heart Association. A.S.M. is a Howard Hughes Medical Institute International Student Research fellow.
Literature Cited
- Brooks KK, Liang B, Watts JL. The influence of bacterial diet on fat storage in C. elegans. PloS one. 2009;4:e7545. doi: 10.1371/journal.pone.0007545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans CL, Xie XS. Coherent anti-stokes Raman scattering microscopy: chemical imaging for biology and medicine. Annual review of analytical chemistry. 2008;1:883–909. doi: 10.1146/annurev.anchem.1.031207.112754. [DOI] [PubMed] [Google Scholar]
- Freudiger CW, Min W, Saar BG, Lu S, Holtom GR, He C, Tsai JC, Kang JX, Xie XS. Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science. 2008;322:1857–1861. doi: 10.1126/science.1165758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Yu Y, Folick A, Currie E, Farese RV, Jr., Tsai TH, Xie XS, Wang MC. In vivo metabolic fingerprinting of neutral lipids with hyperspectral stimulated Raman scattering microscopy. Journal of the American Chemical Society. 2014;136:8820–8828. doi: 10.1021/ja504199s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmchen F, Denk W. Deep tissue two-photon microscopy. Nature methods. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- Ji M, Orringer DA, Freudiger CW, Ramkissoon S, Liu X, Lau D, Golby AJ, Norton I, Hayashi M, Agar NY, et al. Rapid, label-free detection of brain tumors with stimulated Raman scattering microscopy. Science translational medicine. 2013;5:201ra119. doi: 10.1126/scitranslmed.3005954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence VJ, Kopelman PG. Medical consequences of obesity. Clinics in dermatology. 2004;22:296–302. doi: 10.1016/j.clindermatol.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Min W, Freudiger CW, Lu S, Xie XS. Coherent nonlinear optical imaging: beyond fluorescence microscopy. Annual review of physical chemistry. 2011;62:507–530. doi: 10.1146/annurev.physchem.012809.103512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke EJ, Soukas AA, Carr CE, Ruvkun G. C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell metabolism. 2009;10:430–435. doi: 10.1016/j.cmet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden CL, Carroll MD, Kit BK, Flegal KM. Prevalence of childhood and adult obesity in the United States, 2011-2012. JAMA : the journal of the American Medical Association. 2014;311:806–814. doi: 10.1001/jama.2014.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman CV. A new radiation. Indian Journal of Physics. 1928;2:387–398. [Google Scholar]
- Saar BG, Freudiger CW, Reichman J, Stanley CM, Holtom GR, Xie XS. Video-rate molecular imaging in vivo with stimulated Raman scattering. Science. 2010;330:1368–1370. doi: 10.1126/science.1197236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scouten CW, Cunningham M. Freezing Biological Samples (Leica Biosystems) 2012 [Google Scholar]
- Thomas GJ., Jr. Raman spectroscopy of protein and nucleic acid assemblies. Annual review of biophysics and biomolecular structure. 1999;28:1–27. doi: 10.1146/annurev.biophys.28.1.1. [DOI] [PubMed] [Google Scholar]
- Wang MC, Min W, Freudiger CW, Ruvkun G, Xie XS. RNAi screening for fat regulatory genes with SRS microscopy. Nature methods. 2011;8:135–138. doi: 10.1038/nmeth.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Hu F, Shen Y, Chen Z, Yu Y, Lin CC, Wang MC, Min W. Live-cell imaging of alkyne-tagged small biomolecules by stimulated Raman scattering. Nature methods. 2014;11:410–412. doi: 10.1038/nmeth.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Yu Y, Shen Y, Wang MC, Min W. Vibrational imaging of newly synthesized proteins in live cells by stimulated Raman scattering microscopy. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:11226–11231. doi: 10.1073/pnas.1303768110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K, Le TT, Bansal A, Narasimhan SD, Cheng JX, Tissenbaum HA. A comparative study of fat storage quantitation in nematode Caenorhabditis elegans using label and label-free methods. PloS one. 2010;5 doi: 10.1371/journal.pone.0012810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Ramachandran PV, Wang MC. Shedding new light on lipid functions with CARS and SRS microscopy. Biochimica et biophysica acta. 2014 doi: 10.1016/j.bbalip.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]