Abstract
Obesity has become one of the major public health problems all over the world. Recent novel eras of research are opening for the effective management of obesity though gene and nutrient intake interactions because the causes of obesity are complex and multifactorial. Through GWASs (genome-wide association studies) and genetic variations (SNPs, single nucleotide polymorphisms), as the genetic factors are likely to determine individuals’ obesity predisposition. The understanding of genetic approaches in nutritional sciences is referred as “nutrigenomics”. Nutrigenomics explores the interaction between genetic factors and dietary nutrient intake on various disease phenotypes such as obesity. Therefore, this novel approach might suggest a solution for the effective prevention and treatment of obesity through individual genetic profiles and help improve health conditions.
Keywords: diet, genome-wide association studies, nutrigenomics, single nucleotide polymorphisms, obesity
INTRODUCTION
The increasing prevalence of obesity is a dramatic public health burden (1); simply, individuals’ health problems are associated with a great number of chronic diseases (2). The effects of dietary factors (for example, dietary energy, fat or carbohydrate) on obesity have been reported (3,4). Obesity is the consequence of higher dietary energy intake and lower energy expenditure, which results in an imbalance of energy and an increase in body weight (5). However, obesity is influenced by many other factors such as environmental, behavioral, hormonal, metabolic, and genetic predisposition (6–10). Recent researches (11–14) have suggested that genes, environmental factors such as dietary nutrients intake, and their interactions affect obesity. This is important to better understand obesity by individuals’ genetic predisposition and to create a concept of “personalized nutrition” for the effective prevention and treatment of obesity.
FACTS ON OBESITY
Obesity is one of the major health concerns that pose a considerable burden to public health all over the world (1). It has been strongly associated with an increased risk of cardiovascular diseases, type 2 diabetes, metabolic syndrome, and some types of cancer (2). Globally, the body-mass index (BMI, the weight in kilograms divided by the square of the height in meters) of overweight adults equal to or more than 25 was estimated to be 1.5 billion and obese adults with BMI of 30 or greater were about 500 million in 2008 (15). The fifth Korea National Health and Nutrition Examination Survey (KNHANES V-3) reported in 2012, announced that the overall prevalence of obesity (BMI ≥25 kg/m2) in adults is 32.8% (36.1% in men and 29.7% in women) (16).
The primary cause of obesity has been known as an accumulation of excessive body fat resulting from an imbalance of energy intake over physical activity (5). Many studies have reported that high dietary energy is the major contributor to obesity. The intakes of high energy have been associated with consumption of food groups beyond recommended amounts of protein, carbohydrate and fat (13,14). Also, obesity can result from higher consumptions of dietary fat and empty calorie foods, defined as the sum of energy from solid fats and added sugars (17). On the other hand, some studies have shown that diets or foods with high appetite-controlling characters (such as vegetables, fruits, and whole grains) were inversely related to the prevalence of obesity (18,19) This is likely due to the diluted energy density of the diet, the incomplete absorption, and an increase of satiety through delayed gastric emptying of the ingested food. In addition, diet patterns that disturb the regulation of dietary energy intakes, such as skipping a meal, snacking, and away-from-home meals, were also associated with obesity (18). However, low-energy diets with dietary fat restrictions were shown to be unsustainable in maintaining long-term weight loss in obese individuals (20). The role of fruit and vegetable consumption in body weight, as an indicator of obesity, was not clear (21). Intriguingly, the effect of composition or kind of diet on obesity was differently responded by individuals. For example, some individuals were hyper-responsive to beneficial diets (such as diets or foods with high appetite-controlling characters, low-energy diets with dietary fat restrictions) and were maintaining healthy body weights. Others groups were more prone to gain weight to similar diets (19).
Multiple studies of families, adoptions, and twins suggest that genetic factors accounted for 45 to 75% of the inter-individual variations in BMI, the commonly used measure of adiposity (6). In the Swedish twins study, within-pair correlations for BMI were 0.70 for men and 0.66 for women among identical twins reared apart (7). In Korea, heritability of BMI was 82% for men and 87% for women in adolescent twins (8) and heritability of body fat mass was 45% in a strong familial aggregation (9). With the completion of the Human Genome Project in 2003, the single nuleotide polymorphisms (SNPs), which constitute the most frequent DNA sequence variants in the genome were identified (10).
SNPs AS A MAIN FORM OF GENETIC VARIATIONS
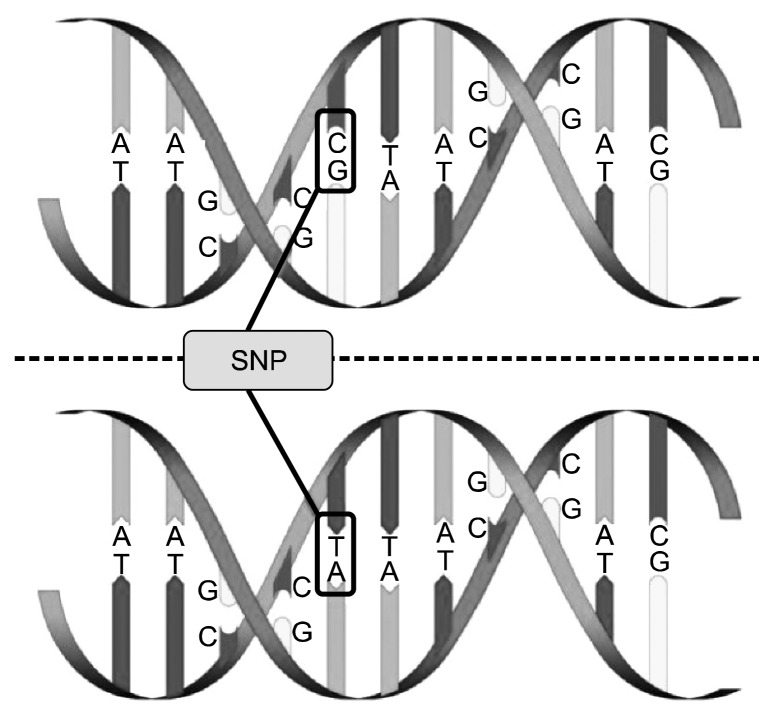
As a main form of human genetic variations, SNPs are single nucleotides—A (adenine), T (thymine), C (cytosine), or G (guanine)—differences in DNA sequences both within and among populations (22). For example, two sequenced DNA fragments from different individuals, CCTAC to CTTAC (forward; GGATG to GAATG as reverse), contain a difference in a single nucleotide (Fig. 1). These C and T are called alleles. Almost all common SNPs have only two alleles (biallelic); one for each of the 22 autosomal chromosomes inherited independently from his or her parents. SNPs, as distinguished from the term “mutation (<1% in the human population)”, exist at a frequency of 1% or higher in the human population (23).
Fig. 1.
Description of single nucleotide polymorphisms (SNPs). As a main form of human genetic variations, single nucleotide polymorphisms (SNPs) are single nucleotides–A (adenine), T (thymine), C (cytosine), or G (guanine)–differences in DNA sequences both within and among populations. For example, two sequenced DNA fragments from different individuals contain a difference at a single nucleotide location (C-T polymorphism).
The SNPs occur throughout the genome, on average every 300 base pairs (24). If two humans are selected at random, the difference in SNPs between them will be around 10 million nucleotide differences because all humans have almost the same sequence of 3 billion nucleotides distributed between their 23 pairs of chromosomes (3×109 base pair÷300 base pair≒10×106 SNPs).
A genotype is an observed set of particular alleles at specified loci (25). In the former example (Fig. 1), there are 3 potential genotypes: C/C, C/T, and T/T; the genotypes of individuals with the C/C or T/T alleles and that of individuals with C/T are assigned as being homozygous and heterozygous, respectively. The expression of genotype results in the individual’s observable traits, the phenotype. Also, the differences of genotype have certain functional consequences, such as altering the activity/function of a protein, and it is followed by the cause of common diseases (22). In other words, the differences in SNPs between individuals may explain why some individuals are more susceptible to common diseases.
GENOME-WIDE ASSOCIATION STUDIES (GWASs) IN OBESITY
The GWASs have become a common method for examining genetic variations, such as finding heritable risk factors associated with a particular complex disease (26,27). The GWASs are based on the “common disease, common variant” hypothesis, which suggests that common allelic variants present in more than 1% to 5% of the population are the major contributors to genetic susceptibility towards diseases (28,29). The entire human genome was scanned by a high-throughput approach, which helped GWASs to determine associations between chromosomal loci and given diseases genome in an unbiased manner (30). The GWASs have been performed for several complex diseases including type I and type II diabetes, inflammatory bowel disease, prostate cancer, breast cancer, asthma, and coronary artery disease (27). Novel loci contributing to BMI and obesity have also been found (14,31,32). The results from such studies have demonstrated that genetic variations could provide valuable insights for several diseases as well as obesity.
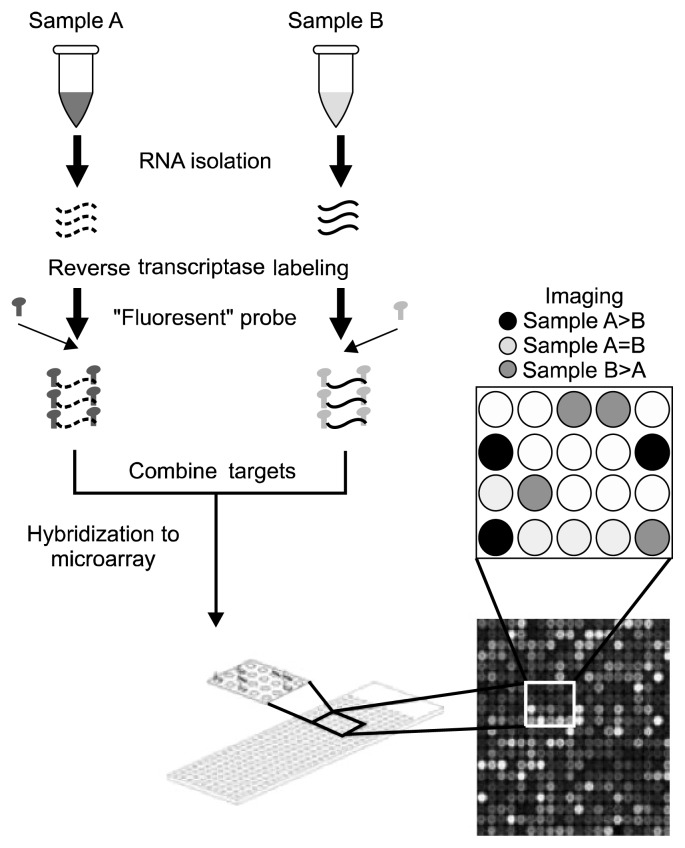
The GWASs genotyped by DNA microarrays are commonly called DNA SNP chips (8,33). The DNA microarrays, which were initially developed in the 1990s, are short DNA probes attached to small glass slides. When fluorescent-labeled DNA fragments of a sample are added to the microarray, they are hybridized to the chip and can then be detected by scanning software (Fig. 2).
Fig. 2.
DNA microarrays for GWASs genotyping. DNA is extracted from both samples A and B are differentially tagged with “fluorescent” probe. They are combined and hybridized to the DNA microarray spotted with the candidate genes. Finally, the hybridized array is scanned, and the scanned image is interpreted.
The first GWAS for obesity and related traits identified a variant 10 kb upstream of INSIG2 (insulin-induced gene-2) in the Framingham Heart Study (34). However, the results from subsequent studies have been inconsistent (35–37). Afterward, FTO (fat mass and obesity associated gene), an obesity susceptibility locus identified, has remained the initial gene to be regarded as a well established and thoroughly replicated risk factor for common obesity (38,39). The FTO is a locus with the largest effect on BMI, so its locus is recognized as a powerful genetic susceptibility locus for obesity. In a meta-analysis data of 7 GWASs for obesity and related traits in 16,876 individuals of which 11,012 were from 4 European population-based cohorts, and 5,864 from 3 disease-specific case-series, a variant mapped 188 kb downstream of MC4R (melanocortin-4 receptor) was identified to play a role in the monogenic forms of obesity (40). Besides the FTO and near-MC4R loci, the three other GWASs in 2009 from the GIANT (genomic investigation of anthropometric traits) consortium (41), deCODE genetics (42), and GWAS in European populations (43) identified 13 new loci, near NEGR1 (neuronal growth regulator 1), near TMEM18 (transmembrane protein 18), in SH2B1 (SH2B adaptor protein 1), near KCTD15 (potassium channel tetramerization domain containing 15), near GNPDA2 (glucosamine-6-phosphate deaminase 2), in MTCH2 (mitochondrial carrier homologue 2), in SEC16B (SEC16 homologue B), between ETV5 (ets variant gene 5) and DGKG (diacylglycerol kinase), in BDNF (brain-derived neurotrophic factor), between BCDIN3D (BCDIN3 domain containing) and FAIM2 (Fas apoptotic inhibitory molecule 2), in NPC1 (Niemann-Pick disease, type C1), near MAF (v-maf musculoaponeurotic fibrosarcoma oncogene homologue), and near PTER (phosphotriesterase related).
DIET AND GENETIC VARIATION ON OBESITY
In spite of a number of recent studies in identifying genetic variants on obesity using the GWASs, it is well established that solely those variants do not result in obesity without the exposure of an obesogenic environment. Also, an interaction between genetic and environmental factors on obesity might exceed the effect of specific genetic variants (11). In other words, obesity should be considered as a complex multifactoral disease affected by genetic factors as well as environmental influences, such as diet and physical activity (12–14). The genetic factors and genetic variations (SNPs) are likely to determine an individual’s susceptibility to obesity through the full series of potential mechanisms governing pathways and regulatory systems at different levels, including the intake and expenditure of energy and the controlling and partitioning of nutrients between fat and lean mass tissue.
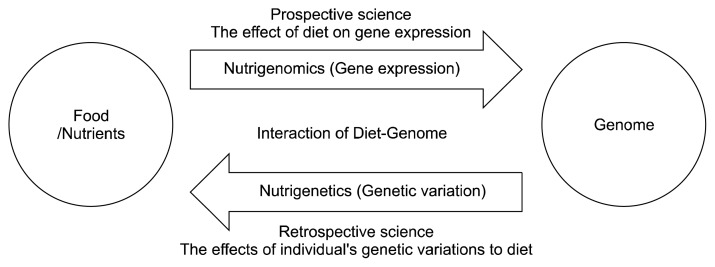
The understanding of genomic approaches in nutritional sciences has created a new field, called “nutrigenomics” (44–47). Nutrigenomics consists of nutrigenomics and nutrigenetics, which explores the interaction between nutrients and genes. However, it is distinctly discriminated through mechanisms of how interactions between nutrients and genes influence the risk for developing a health condition or disease (Fig. 3) (44,45). Nutrigenomics describes the effect of nutrients (food-based diets, dietary restriction, or nutritional supplements) on gene expression; it will increase interest in the genome-wide influences of nutrients on transcriptome, proteome, and metabolome in cells, tissues, or organisms, as well as confirm the genes that influence the risk of diet-related diseases (46). It may also be helpful in the understanding of how nutrients influence metabolic pathways and how these regulations are inhibited in the early phase of a diet-related disease (44).
Fig. 3.
The understanding of genomic approaches in nutritional science has created a new field, called “nutrigenomics”. Nutrigenomics consists of nutrigenomics and nutrigenetics: nutrigenomics is the effect of nutrients or nutritional supplements on gene expression, and nutrigenetics is the effect of interaction between genetic variations and nutrients on diseases.
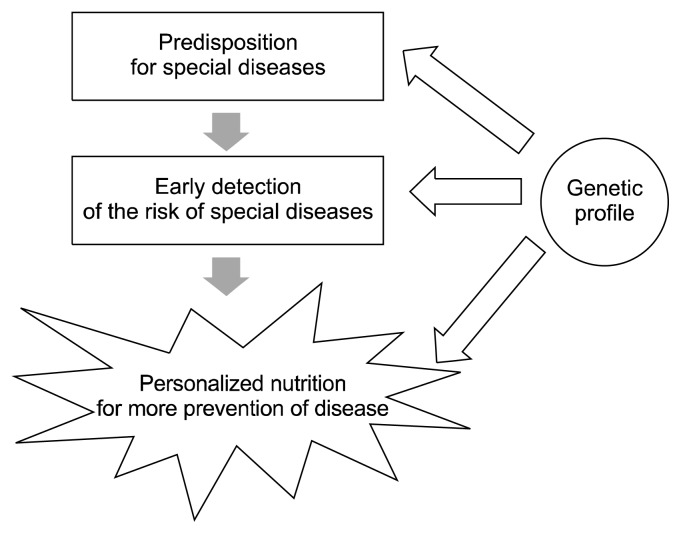
In contrast, nutrigenetics is focused on how the interaction between genetic variations and nutrients influence metabolism, health conditions, and the risk for diet-related diseases (44,47). In other words, the influence of nutrients on diseases is determined by an individual’s genetic variations, which can affect digestion, absorption, metabolism, partitioning, and cellular responsiveness to nutrients (47). Nutrigenetics is useful in providing genetic profiles for the early detection of disease risks and may provide clues for personalized diet recommendations for effective prevention strategies or therapies for individual with genetic predispositions to diseases (Fig. 4) (47). This approach, along with individuals’ motivation to adapt lifestyle changes, may provide health benefits to individuals.
Fig. 4.
Personalized nutrition in maintenance of health condition and prevention of diseases.
STUDIES OF INTERACTIONS BETWEEN DIET AND SNPs ON OBESITY
Several studies examined the interactions between dietary nutrient intake and genetic variations (SNPs) involved in obesity related variables. Sonestedt et al. (48) reported interactions between energy-adjusted fat (P= 0.04) or carbohydrate intake (P=0.001) and a variant of the FTO gene (rs9939609) on BMI among 4,839 subjects in a cross-sectional study; among the participants with an intake of high fat or low carbohydrate, the AA carrier showed a higher BMI than the TT carrier of FTO rs9939609. In the GOLDN (Genetics of Lipid Lowering Drugs and Diet Network) study, Warodomwichit et al. (49) found that subjects who were carriers of the ADIPOQ (adiponectin, C1Q and collagen domain containing)-11391A allele (AA+GA) had significantly lower body weight (P=0.029), BMI (P=0.019), waist circumference (P=0.003), and hip circumference (P=0.004) compared to noncarriers. In addition, in subjects with high monounsaturated fatty acids intake, carrier of the -11391A allele resulted in lower BMI (P=0.002) and decreased risk of obesity (odd ratios=0.52, 95% CI=0.28 ~0.96, P=0.031). Also, it was reported that gene-dietary fat interaction might modulate the risk of obesity. The APO B (apolipoprotein B) SNP rs512535 (A/G) minor allele carrier (G) has been associated with obesity-related phenotypes (BMI and waist circumference). Interestingly, in habitual high fat consumers (>35% of energy), the GG homozygotes of APO B rs512535 have shown higher BMI than the A allele carriers, however those differences were not shown in low fat consumers (<35% of energy) (50).
Recently, several studies have been conducted on the various roles of obesity-related genotype in Korea. We previously observed that the ESR1 (estrogen receptor 1) rs1884051 polymorphism (C>T) was associated with obesity-related variables (body weight, BMI, waist-hip ratio, fat body mass, and body fat percentage), together with their modulations by dietary intake in 3,039 Korean men aged 40~59 years from the KoGES database (51). Moreover, among the subjects with low total energy intake, the minor allele of ESR1 SNP resulted in a lower BMI (P=0.003) compared to the subjects carrying the major allele. Interestingly, among subjects with a high plant protein intake, carriers of the minor allele of ESR1 SNP had a lower BMI (P=0.044) compared to subjects carrying the major allele. Cha et al. (52) reported that the increased BMI was associated with the FTO haplotypes and MC4R variants in two populations using 1,370 Korean subjects before and after Sasang constitutional medicine (SCM) typing, and found the BMI in 538 individuals lowering with a lifestyle intervention for one month. After lifestyle changes such as a low-calorie diet, daily exercise, an electrolipolysis treatment, and the administration of Chegamuiyiin-tang containing 17 herbs, carriers of the haplotype by the minor allele of rs1075440 had a lower waist-to-hip ratio (0.76%) compared to the non-carriers. The rs9939973 and rs9939609 of FTO were recently shown to modulate obesity in 711 Korean children and 8,842 adults. A significant association was identified between rs9939609 and dietary fat intake in children (P=0.008) but not in adults (53). Among the 6 SNPs of WNT10 B gene, known as a potential regulator of adipogenesis in obesity models, -607G>C (rs833840) was significantly associated with body fat mass by bio-impedance analysis and abdominal fat (total and subcutaneous fat areas) by abdominal computed tomography in 1,029 Korean women. However, no significant associations between -607G>C genotype and body weight changes or composition were confirmed among 576 subjects with very low calorie diet for one month (54).
CONCLUSION
The cause of obesity is the complex impact of genetic and environmental factors. It is associated with increased risks of developing many chronic disease including cardiovascular disease, type 2 diabetes, arthritis, hypertension, and certain cancers such as esophagus, breast, endometrium, colorectal, gallbladder and possibly other types of cancers. As dietary nutrient intake is an important environmental factor, many studies have shown that dietary intake plays a key role in the development of the obesity. Although many studies have observed the effects of quantity and quality of dietary nutrients on obesity, intervention studies are inconsistent.
Which factors make a difference on obesity with the same dietary intake in individuals? The answer is the difference in genetic variation. In other words, the features of nutrigenomics are explained by the relationship between specific dietary nutrient intake and gene variations on obesity. Now, nutrigenomics is extensively used for researching obesity as well as diet-related disorders. Therefore, nutrigenomics may shift towards the effective management of obesity, through “personalized” nutritional advice and consultation by individuals’ genetic profiles.
Even though many genetic variations to affect the development of obesity are identified in all over world, the unique differences in the genetic variations on obesity by races or populations exist. So, further investigations are still necessary in various genetic variations on obesity, association between various dietary nutrients intake and genetic variation on obesity for effective management tools of personalized nutrition on obesity for various race or populations. And those investigations may provide a solution to a public health problem, as well as personalized obesity prevention.
ACKNOWLEDGEMENTS
This article is based on a part of the first author’s PhD thesis from Ewha Womans University.
Footnotes
AUTHOR DISCLOSURE STATEMENT
The authors declare no conflict of interest.
REFERENCES
- 1.Formiguera X, Cantón A. Obesity: epidemiology and clinical aspects. Best Pract Res Clin Gastroenterol. 2004;18:1125–1146. doi: 10.1016/S1521-6918(04)00091-5. [DOI] [PubMed] [Google Scholar]
- 2.Akabas S, Lederman SA, Moore BJ. Textbook of obesity: biological, psychological and cultural influences. 1st ed. Wiley-Blackwell publishing; Chichester, UK: 2012. pp. 5–41. [Google Scholar]
- 3.Bujnowski D, Xun P, Daviglus ML, Van Horn L, He K, Stamler J. Longitudinal association between animal and vegetable protein intake and obesity among men in the United States: the Chicago Western Electric Study. J Am Diet Assoc. 2011;111:1150–1155.e1. doi: 10.1016/j.jada.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foster GD, Wyatt HR, Hill JO, McGuckin BG, Brill C, Mohammed BS, Szapary PO, Rader DJ, Edman JS, Klein S. A randomized trial of a low-carbohydrate diet for obesity. N Engl J Med. 2003;348:2082–2090. doi: 10.1056/NEJMoa022207. [DOI] [PubMed] [Google Scholar]
- 5.Walker CG, Zariwala MG, Holness MJ, Sugden MC. Diet, obesity and diabetes: a current update. Clin Sci (Lond) 2007;112:93–111. doi: 10.1042/CS20060150. [DOI] [PubMed] [Google Scholar]
- 6.Sharma AM, Padwal R. Obesity is a sign–over-eating is a symptom: an aetiological framework for the assessment and management of obesity. Obes Rev. 2010;11:362–370. doi: 10.1111/j.1467-789X.2009.00689.x. [DOI] [PubMed] [Google Scholar]
- 7.Stunkard AJ, Foch TT, Hrubec Z. A twin study of human obesity. JAMA. 1986;256:51–54. doi: 10.1001/jama.1986.03380010055024. [DOI] [PubMed] [Google Scholar]
- 8.Hur YM. Sex difference in heritability of BMI in South Korean adolescent twins. Obesity (Silver Spring) 2007;15:2908–2911. doi: 10.1038/oby.2007.346. [DOI] [PubMed] [Google Scholar]
- 9.Cho EY, Park HY, Jee SH, Jang YS, Bae SJ, Lee JH, Jang YS. Korean familial resemblance in fat mass and fat free mass: cardiovascular genome study. Korean J Lipidol. 2001;11:537–547. [Google Scholar]
- 10.The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10:241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 12.Loktionov A. Common gene polymorphisms and nutrition: emerging links with pathogenesis of multifactorial chronic diseases. J Nutr Biochem. 2003;14:426–451. doi: 10.1016/S0955-2863(03)00032-9. [DOI] [PubMed] [Google Scholar]
- 13.Amato R, Pinelli M, D’Andrea D, Miele G, Nicodemi M, Raiconi G, Cocozza S. A novel approach to simulate gene-environment interactions in complex diseases. BMC Bioinformatics. 2010;11:8. doi: 10.1186/1471-2105-11-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinney A, Vogel CI, Hebebrand J. From monogenic to polygenic obesity: recent advances. Eur Child Adolesc Psychiatry. 2010;19:297–310. doi: 10.1007/s00787-010-0096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finucane MM, Stevens GA, Cowan MJ, Danaei G, Lin JK, Paciorek CJ, Singh GM, Gutierrez HR, Lu Y, Bahalim AN, Farzadfar F, Riley LM, Ezzati M Global Burden of Metabolic Risk Factors of Chronic Diseases Collaborating Group (Body Mass Index) National, regional, and global trends in body-mass index since 1980: systematic analysis of health examination surveys and epidemiological studies with 960 country-years and 9.1 million participants. Lancet. 2011;377:557–567. doi: 10.1016/S0140-6736(10)62037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ministry of Health and Welfare. The Korea National Health and Nutrition Examination Survey (KNHANES V-3) Chungbuk, Korea: 2012. [Google Scholar]
- 17.Reedy J, Krebs-Smith SM. Dietary sources of energy, solid fats, and added sugars among children and adolescents in the United States. J Am Diet Assoc. 2010;110:1477–1484. doi: 10.1016/j.jada.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giskes K, van Lenthe F, Avendano-Pabon M, Brug J. A systematic review of environmental factors and obesogenic dietary intakes among adults: are we getting closer to understanding obesogenic environments? Obes Rev. 2011;12:e95–e106. doi: 10.1111/j.1467-789X.2010.00769.x. [DOI] [PubMed] [Google Scholar]
- 19.Deram S, Villares SM. Genetic variants influencing effectiveness of weight loss strategies. Arq Bras Endocrinol Metabol. 2009;53:129–138. doi: 10.1590/S0004-27302009000200003. [DOI] [PubMed] [Google Scholar]
- 20.Summerbell CD, Cameron C, Glasziou PP. WITHDRAWN: Advice on low-fat diets for obesity. Cochrane Database Syst Rev. 2008;16:CD003640. doi: 10.1002/14651858.CD003640.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alinia S1, Hels O, Tetens I. The potential association between fruit intake and body weight–a review. Obes Rev. 2009;10:639–647. doi: 10.1111/j.1467-789X.2009.00582.x. [DOI] [PubMed] [Google Scholar]
- 22.Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, Nemesh J, Ziaugra L, Friedland L, Rolfe A, Warrington J, Lipshutz R, Daley GQ, Lander ES. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- 23.den Dunnen JT, Antonarakis SE. Nomenclature for the description of human sequence variations. Hum Genet. 2001;109:121–124. doi: 10.1007/s004390100505. [DOI] [PubMed] [Google Scholar]
- 24.Kruglyak L, Nickerson DA. Variation is the spice of life. Nat Genet. 2001;27:234–236. doi: 10.1038/85776. [DOI] [PubMed] [Google Scholar]
- 25.Vakili S, Caudill MA. Personalized nutrition: nutritional genomics as a potential tool for targeted medical nutrition therapy. Nutr Rev. 2007;65:301–315. doi: 10.1111/j.1753-4887.2007.tb00308.x. [DOI] [PubMed] [Google Scholar]
- 26.Lewis SN, Nsoesie E, Weeks C, Qiao D, Zhang L. Prediction of disease and phenotype associations from genome-wide association studies. PLoS One. 2011;6:e27175. doi: 10.1371/journal.pone.0027175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 28.Delahanty RJ, Beeghly-Fadiel A, Xiang YB, Long J, Cai Q, Wen W, Xu WH, Cai H, He J, Gao YT, Zheng W, Shu XO. Association of obesity-related genetic variants with endometrial cancer risk: a report from the Shanghai Endometrial Cancer Genetics Study. Am J Epidemiol. 2011;174:1115–1126. doi: 10.1093/aje/kwr233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gandhi S, Wood NW. Genome-wide association studies: the key to unlocking neurodegeneration? Nat Neurosci. 2010;13:789–794. doi: 10.1038/nn.2584. [DOI] [PubMed] [Google Scholar]
- 30.Chial H, Craig J. Genome-wide association studies (GWAS) and obesity. Nature Education. 2008;1:80. [Google Scholar]
- 31.Gresham D, Dunham MJ, Botstein D. Comparing whole genomes using DNA microarrays. Nat Rev Genet. 2008;9:291–302. doi: 10.1038/nrg2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pearson TA, Manolio TA. How to interpret a genome-wide association study. JAMA. 2008;299:1335–1344. doi: 10.1001/jama.299.11.1335. [DOI] [PubMed] [Google Scholar]
- 33.Collins FS, Guyer MS, Chakravarti A. Variations on a theme: cataloging human DNA sequence variation. Science. 1997;278:1580–1581. doi: 10.1126/science.278.5343.1580. [DOI] [PubMed] [Google Scholar]
- 34.Herbert A, Gerry NP, McQueen MB, Heid IM, Pfeufer A, Illig T, Wichmann HE, Meitinger T, Hunter D, Hu FB, Colditz G, Hinney A, Hebebrand J, Koberwitz K, Zhu X, Cooper R, Ardlie K, Lyon H, Hirschhorn JN, Laird NM, Lenburg ME, Lange C, Christman MF. A common genetic variant is associated with adult and childhood obesity. Science. 2006;312:279–283. doi: 10.1126/science.1124779. [DOI] [PubMed] [Google Scholar]
- 35.Lyon HN, Emilsson V, Hinney A, Heid IM, Lasky-Su J, Zhu X, Thorleifsson G, Gunnarsdottir S, Walters GB, Thorsteinsdottir U, Kong A, Gulcher J, Nguyen TT, Scherag A, Pfeufer A, Meitinger T, Brönner G, Rief W, Soto-Quiros ME, Avila L, Klanderman B, Raby BA, Silverman EK, Weiss ST, Laird N, Ding X, Groop L, Tuomi T, Isomaa B, Bengtsson K, Butler JL, Cooper RS, Fox CS, O’Donnell CJ, Vollmert C, Celedón JC, Wichmann HE, Hebebrand J, Stefansson K, Lange C, Hirschhorn JN. The association of a SNP upstream of INSIG2 with body mass index is reproduced in several but not all cohorts. PLos Genet. 2007;3:e61. doi: 10.1371/journal.pgen.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dina C, Meyre D, Samson C, Tichet J, Marre M, Jouret B, Charles MA, Balkau B, Froguel P. Comment on “a common genetic variant is associated with adult and childhood obesity”. Science. 2007;315:187. doi: 10.1126/science.1129402. [DOI] [PubMed] [Google Scholar]
- 37.Andreasen CH, Mogensen MS, Borch-Johnsen K, Sandbaek A, Lauritzen T, Sørensen TI, Hansen L, Almind K, Jørgensen T, Pedersen O, Hansen T. Non-replication of genome-wide based associations between common variants in INSIG2 and PFKP and obesity in studies of18,014 Danes. PLoS ONE. 2008;3:e2872. doi: 10.1371/journal.pone.0002872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hinney A, Nguyen TT, Scherag A, Friedel S, Brönner G, Müller TD, Grallert H, Illig T, Wichmann HE, Rief W, Schäfer H, Hebebrand J. Genome wide association (GWA) study for early onset extreme obesity supports the role of fat mass and obesity associated gene (FTO) variants. PLoS ONE. 2007;2:e1361. doi: 10.1371/journal.pone.0001361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hunt SC, Stone S, Xin Y, Scherer CA, Magness CL, Iadonato SP, Hopkins PN, Adams TD. Association of the FTO gene with BMI. Obesity (Silver Spring) 2008;16:902–904. doi: 10.1038/oby.2007.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, Prokopenko I, Inouye M, Freathy RM, Attwood AP, Beckmann JS, Berndt SI, Jacobs KB, Chanock SJ, Hayes RB, Bergmann S, Bennett AJ, Bingham SA, Bochud M, Brown M, Cauchi S, Connell JM, Cooper C, Smith GD, Day I, Dina C, De S, Dermitzakis ET, Doney AS, Elliott KS, Elliott P, Evans DM, Sadaf Farooqi I, Froguel P, Ghori J, Groves CJ, Gwilliam R, Hadley D, Hall AS, Hattersley AT, Hebebrand J, Heid IM, Lamina C, Gieger C, Illig T, Meitinger T, Wichmann HE, Herrera B, Hinney A, Hunt SE, Jarvelin MR, Johnson T, Jolley JD, Karpe F, Keniry A, Khaw KT, Luben RN, Mangino M, Marchini J, McArdle WL, McGinnis R, Meyre D, Munroe PB, Morris AD, Ness AR, Neville MJ, Nica AC, Ong KK, O’Rahilly S, Owen KR, Palmer CN, Papadakis K, Potter S, Pouta A, Qi L, Randall JC, Rayner NW, Ring SM, Sandhu MS, Scherag A, Sims MA, Song K, Soranzo N, Speliotes EK, Syddall HE, Teichmann SA, Timpson NJ, Tobias JH, Uda M, Vogel CI, Wallace C, Waterworth DM, Weedon MN, Willer CJ, Wraight, Yuan X, Zeggini E, Hirschhorn JN, Strachan DP, Ouwehand WH, Caulfield MJ, Samani NJ, Frayling TM, Vollenweider P, Waeber G, Mooser V, Deloukas P, McCarthy MI, Wareham NJ, Barroso I, Jacobs KB, Chanock SJ, Hayes RB, Lamina C, Gieger C, Illig T, Meitinger T, Wichmann HE, Kraft P, Hankinson SE, Hunter DJ, Hu FB, Lyon HN, Voight BF, Ridderstrale M, Groop L, Scheet P, Sanna S, Abecasis GR, Albai G, Nagaraja R, Schlessinger D, Jackson AU, Tuomilehto J, Collins FS, Boehnke M, Mohlke KL Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial, KORA, Nurses’ Health Study, Diabetes Genetics Initiative, SardiNIA Study, Wellcome Trust Case Control Consortium, FUSION. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, Berndt SI, Elliott AL, Jackson AU, Lamina C, Lettre G, Lim N, Lyon HN, McCarroll SA, Papadakis K, Qi L, Randall JC, Roccasecca RM, Sanna S, Scheet P, Weedon MN, Wheeler E, Zhao JH, Jacobs LC, Prokopenko I, Soranzo N, Tanaka T, Timpson NJ, Almgren P, Bennett A, Bergman RN, Bingham SA, Bonnycastle LL, Brown M, Burtt NP, Chines P, Coin L, Collins FS, Connell JM, Cooper C, Smith GD, Dennison EM, Deodhar P, Elliott P, Erdos MR, Estrada K, Evans DM, Gianniny L, Gieger C, Gillson CJ, Guiducci C, Hackett R, Hadley D, Hall AS, Havulinna AS, Hebebrand J, Hofman A, Isomaa B, Jacobs KB, Johnson T, Jousilahti P, Jovanovic Z, Khaw KT, Kraft P, Kuokkanen M, Kuusisto J, Laitinen J, Lakatta EG, Luan J, Luben RN, Mangino M, McArdle WL, Meitinger T, Mulas A, Munroe PB, Narisu N, Ness AR, Northstone K, O’Rahilly S, Purmann C, Rees MG, Ridderstråle M, Ring SM, Rivadeneira F, Ruokonen A, Sandhu MS, Saramies J, Scott LJ, Scuteri A, Silander K, Sims MA, Song K, Stephens J, Stevens S, Stringham HM, Tung YC, Valle TT, Van Duijn CM, Vimaleswaran KS, Vollenweider P, Waeber G, Wallace C, Watanabe RM, Waterworth DM, Watkins N, Witteman JC, Zeggini E, Zhai G, Zillikens MC, Altshuler D, Caulfield MJ, Chanock SJ, Farooqi IS, Ferrucci L, Guralnik JM, Hattersley AT, Hu FB, Jarvelin MR, Laakso M, Mooser V, Ong KK, Ouwehand WH, Salomaa V, Samani NJ, Spector TD, Tuomi T, Tuomilehto J, Uda M, Uitterlinden AG, Wareham NJ, Deloukas P, Frayling TM, Groop LC, Hayes RB, Hunter DJ, Mohlke KL, Peltonen L, Schlessinger D, Strachan DP, Wichmann HE, McCarthy MI, Boehnke M, Barroso I, Abecasis GR, Hirschhorn JN Wellcome Trust Case Control Consortium, Genetic Investigation of ANthropometric Traits Consortium. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, Styrkarsdottir U, Gretarsdottir S, Thorlacius S, Jonsdottir I, Jonsdottir T, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Jonsson F, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T, Lauritzen T, Aben KK, Verbeek AL, Roeleveld N, Kampman E, Yanek LR, Becker LC, Tryggvadottir L, Rafnar T, Becker DM, Gulcher J, Kiemeney LA, Pedersen O, Kong A, Thorsteinsdottir U, Stefansson K. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 43.Meyre D, Delplanque J, Chèvre JC, Lecoeur C, Lobbens S, Gallina S, Durand E, Vatin V, Degraeve F, Proença C, Gaget S, Körner A, Kovacs P, Kiess W, Tichet J, Marre M, Hartikainen AL, Horber F, Potoczna N, Hercberg S, Levy-Marchal C, Pattou F, Heude B, Tauber M, McCarthy MI, Blakemore AI, Montpetit A, Polychronakos C, Weill J, Coin LJ, Asher J, Elliott P, Järvelin MR, Visvikis-Siest S, Balkau B, Sladek R, Balding D, Walley A, Dina C, Froguel P. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41:157–159. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 44.Raqib R, Cravioto A. Nutrition, immunology, and genetics: future perspectives. Nutr Rev. 2009;67:S227–S236. doi: 10.1111/j.1753-4887.2009.00244.x. [DOI] [PubMed] [Google Scholar]
- 45.Fenech M. Genome health nutrigenomics and nutrigenetics-diagnosis and nutritional treatment of genome damage on an individual basis. Food Chem Toxicol. 2008;46:1365–1370. doi: 10.1016/j.fct.2007.06.035. [DOI] [PubMed] [Google Scholar]
- 46.Panagiotou G, Nielsen J. Nutritional systems biology: definitions and approaches. Annu Rev Nutr. 2009;29:329–339. doi: 10.1146/annurev-nutr-080508-141138. [DOI] [PubMed] [Google Scholar]
- 47.Rimbach G, Minihane AM. Nutrigenetics and personalised nutrition: how far have we progressed and are we likely to get there? Proc Nutr Soc. 2009;68:162–172. doi: 10.1017/S0029665109001116. [DOI] [PubMed] [Google Scholar]
- 48.Sonestedt E, Roos C, Gullberg B, Ericson U, Wirfält E, Orho-Melander M. Fat and carbohydrate intake modify the association between genetic variation in the FTO genotype and obesity. Am J Clin Nutr. 2009;90:1418–1425. doi: 10.3945/ajcn.2009.27958. [DOI] [PubMed] [Google Scholar]
- 49.Warodomwichit D, Shen J, Arnett DK, Tsai MY, Kabagambe EK, Peacock JM, Hixson JE, Straka RJ, Province MA, An P, Lai CQ, Parnell LD, Borecki IB, Ordovas JM. ADIPOQ polymorphisms, monounsaturated fatty acids, and obesity risk: the GOLDN study. Obesity (Silver Spring) 2009;17:510–517. doi: 10.1038/oby.2008.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phillips CM, Goumidi L, Bertrais S, Field MR, McManus R, Hercberg S, Lairon D, Planells R, Roche HM. Gene-nutrient interactions and gender may modulate the association between ApoA1 and ApoB gene polymorphisms and metabolic syndrome risk. Atherosclerosis. 2011;214:408–414. doi: 10.1016/j.atherosclerosis.2010.10.029. [DOI] [PubMed] [Google Scholar]
- 51.Doo MA, Kim YH. Association between ESR1 rs1884051 polymorphism and dietary total energy and plant protein intake on obesity in Korean men. Nutr Res Pract. 2011;5:527–532. doi: 10.4162/nrp.2011.5.6.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cha SW, Koo IH, Park BL, Jeong SK, Choi SM, Kim KS, Shin HD, Kim JY. Genetic effects of FTO and MC4R polymorphisms on body mass in constitutional types. Evid Based Complement Alternat Med. 2011;2011:106390. doi: 10.1093/ecam/nep162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee HJ, Kim IK, Kang JH, Ahn YJ, Han BG, Lee JY, Song JH. Effects of common FTO gene variants associated with BMI on dietary intake and physical activity in Koreans. Clin Chim Acta. 2010;411:1716–1722. doi: 10.1016/j.cca.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 54.Kim IC, Cha MH, Kim DM, Lee HY, Moon JS, Choi SM, Kim KS, Yoon YS. A functional promoter polymorphism –607G>C of WNT10B is associated with abdominal fat in Korean female subjects. J Nutr Biochem. 2011;22:252–258. doi: 10.1016/j.jnutbio.2010.02.002. [DOI] [PubMed] [Google Scholar]