Abstract
Success with genome editing by the RNA-programmed nuclease Cas9 has been limited by the inability to predict effective guide RNAs and DNA target sites. Not all guide RNAs have been successful, and even those that were, varied widely in their efficacy. Here we describe and validate a strategy for Caenorhabditis elegans that reliably achieved a high frequency of genome editing for all targets tested in vivo. The key innovation was to design guide RNAs with a GG motif at the 3′ end of their target-specific sequences. All guides designed using this simple principle induced a high frequency of targeted mutagenesis via nonhomologous end joining (NHEJ) and a high frequency of precise DNA integration from exogenous DNA templates via homology-directed repair (HDR). Related guide RNAs having the GG motif shifted by only three nucleotides showed severely reduced or no genome editing. We also combined the 3′ GG guide improvement with a co-CRISPR/co-conversion approach. For this co-conversion scheme, animals were only screened for genome editing at designated targets if they exhibited a dominant phenotype caused by Cas9-dependent editing of an unrelated target. Combining the two strategies further enhanced the ease of mutant recovery, thereby providing a powerful means to obtain desired genetic changes in an otherwise unaltered genome.
Keywords: CRISPR, Cas9, genome editing, co-conversion, C. elegans
THE use of site-specific nucleases with programmable target specificity has transformed the art of genome editing and thereby revolutionized the dissection and manipulation of genome function (reviewed in Mali et al. 2013; Carroll 2014; Doudna and Charpentier 2014; Hsu et al. 2014). Most widely used is the CRISPR-associated nuclease Cas9, whose RNA-programmed DNA cleaving activities create DNA double-strand breaks (DSBs). These DSBs can be repaired imprecisely by nonhomologous end joining (NHEJ) to generate random insertions and deletions or repaired precisely by homology-directed repair (HDR) templated from exogenous DNA to generate custom-designed insertions, deletions, or substitutions (Gasiunas et al. 2012; Jinek et al. 2012; Cong et al. 2013; Jinek et al. 2013; Mali et al. 2013). Modified variants of Cas9 that lack DNA cleaving activity have also been utilized to regulate transcription of designated gene targets and to cytologically mark and track genomic loci in living cells (Bikard et al. 2013; Chen et al. 2013; Larson et al. 2013; Maeder et al. 2013; Perez-Pinera et al. 2013; Qi et al. 2013; Gilbert et al. 2014; Tanenbaum et al. 2014).
The Cas9 protein is targeted to a specific genomic locus by a guide RNA that encodes a 20-nt region of homology to the DNA target (Figure 1A) (Mojica et al. 2009; Garneau et al. 2010; Jinek et al. 2012). The most commonly used guide RNAs are chimeric fusions between the CRISPR RNA (crRNA), which encodes the 20-nt target-specific sequence, and the tracer RNA (trRNA), which enables the formation of active Cas9–guide RNA complexes (Figure 1A) (Jinek et al. 2012). Few constraints are known for Cas9 DNA targets. The most critical requirement is for an NGG trinucleotide motif to reside adjacent to the protospacer, the region of the DNA target with identity to the target-specific region of the guide (Figure 1A). This precisely positioned NGG, called the protospacer adjacent motif (PAM), is essential for Cas9 DNA cleaving activity (Garneau et al. 2010; Sapranauskas et al. 2011; Jinek et al. 2012; Anders et al. 2014; Sternberg et al. 2014). The minimal requirement for target design and the straightforward construction of guide RNAs have made Cas9 a very attractive alternative to other site-specific nucleases, which have more stringent requirements for target selection and require more effort to design and construct.
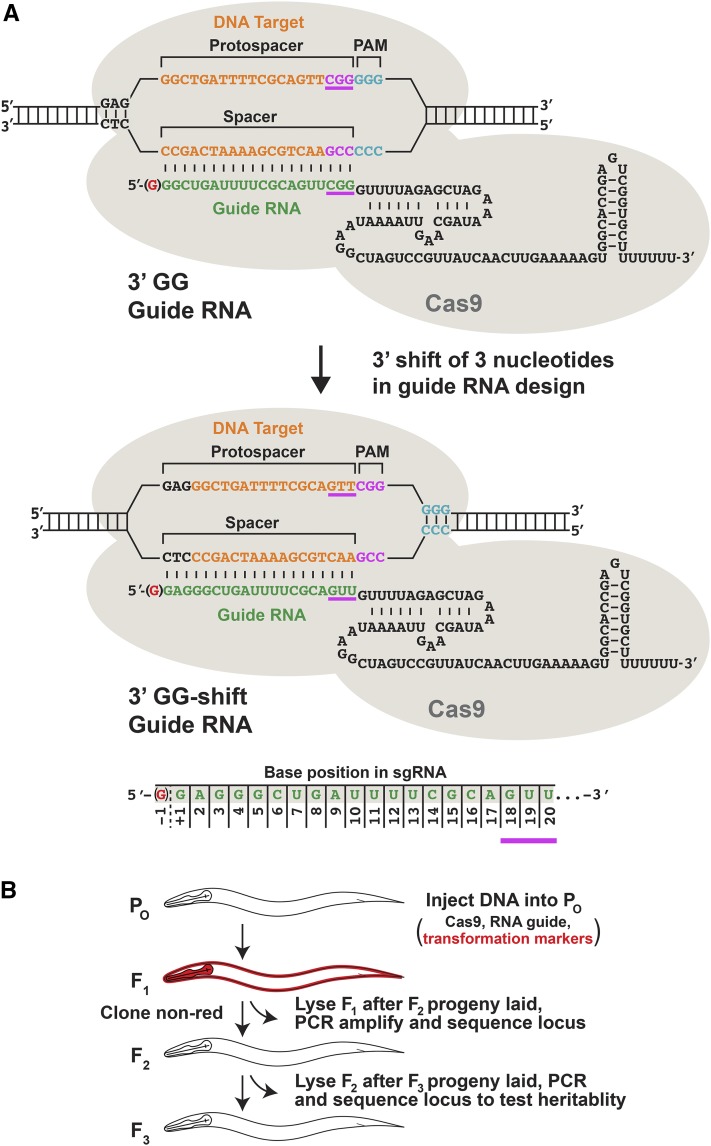
Figure 1.
Guide RNA designs to assess the impact of an NGG motif at the 3′ end of protospacers in Cas9-directed mutagenesis. (A) Shown are diagrams of two related guide RNAs bound in a complex with Cas9 to their DNA target sites. The 3′ GG-guide RNAs have the NGG motif at the 3′ end of the protospacers. The 3′ GG-shift guides shift the NGG motif in the 3′ direction so that the shifted NGG acts as the PAM rather than a feature of the guide RNA. Below the diagrams is a schematic showing the numbering system for spacer-complementary base positions in the guide RNAs. A non-base-paired “G” was also added to the 5′ end of each guide RNA to improve transcription from the polymerase III promoter (−1 position of the guide). (B) Efficient experimental design to isolate mutants. P0 animals were microinjected with plasmids encoding Cas9, a guide RNA, and two mCherry transgene markers. Array-bearing, mCherry-positive F1s were picked to individual plates and allowed to lay F2 progeny. The F1s were lysed, their genomic target site amplified by PCR, and DNA sequence of the PCR products determined to screen for mutations. If an F1 animal tested positive for a mutation, F2 animals were cloned and assessed by PCR and DNA sequence analysis to identify homozygous mutants and to confirm the heritability of the mutation.
Despite the successful application of Cas9 technology to Caenorhabditis elegans, predicting DNA targets and guide RNAs that will support efficient genome editing has been problematic. Moreover, even when guides promote genome editing, the frequency of editing has been unpredictably variable from target to target. The initial report describing the successful use of Cas9 in C. elegans demonstrated a range of editing frequencies from 0.5 to 80%, with only two targets exceeding 4% (Friedland et al. 2013). Subsequent publications also reported variably low mutagenesis rates that required molecular screening of hundreds of animals or required the desired mutation to cause an easily detectable mutant phenotype or to be introduced in tandem with a co-selection marker (Chiu et al. 2013; Cho et al. 2013; Dickinson et al. 2013; Katic and Grosshans 2013; Lo et al. 2013; Tzur et al. 2013; Waaijers et al. 2013; Chen et al. 2014; Liu et al. 2014; Paix et al. 2014; Shen et al. 2014; Zhao et al. 2014). More recent studies greatly improved the odds of detecting a targeted mutation through the simultaneous co-conversion of a mutation in an unrelated target that causes a visible phenotype (Arribere et al. 2014; Kim et al. 2014; Ward 2014). Nevertheless, the overall frequency of editing at most targets remained relatively low.
Here we describe and thoroughly validate a method for guide RNA design that significantly and reliably enhances the frequency of genome editing by Cas9 in C. elegans. All guides designed for all targets supported robust genome editing, both imprecise NHEJ events and precise, templated HDR events. The median frequency of editing at all targets was 51% without any coselectable markers, a 10-fold increase above previous studies that also reported numerous guide RNAs in the nonfunctional class (Friedland et al. 2013; Waaijers et al. 2013; Kim et al. 2014). Combining our already effective guide RNA design with the co-conversion/co-CRISPR strategy of others (Arribere et al. 2014; Kim et al. 2014; Ward 2014) enhanced the ease of mutant recovery and boosted our median for both precise and imprecise genome editing to 86%. Our strategy for guide RNA design should be widely applicable to diverse organisms and cell lines.
Materials and Methods
Strains
Nematode strains were maintained at 20° or 25°, as described previously (Brenner 1974). N2 Bristol was used as wild type.
Plasmid construction
Plasmids encoding the Streptococcus pyogenes Cas9 protein (Peft-3::Cas9-SV40_NLS::tbb-2 3′ UTR; Addgene plasmid designation: 46168) and the klp-12 guide RNA (pU6::klp-12 sgRNA; Addgene plasmid designation: 46170) were gifts from J. Calarco. The co-injection marker plasmids pCFJ90 (Pmyo-2::mCherry::unc-54 3′ UTR; Addgene plasmid designation: 19327) and pCFJ104 (Pmyo-3::mCherry::unc-54 3′ UTR; Addgene plasmid designation: 19328) were gifts from C. Frøkjær-Jensen and E. Jorgensen. The rol-6(su1006) HDR oligo AF-JA-53, the rol-6 guide RNA plasmid pJA42, and the empty guide RNA plasmid pRB1017 were gifts from J. A. Arribere and A. Z. Fire.
For the pU6::sgRNA vector, which utilizes the K09B11.12 small nuclear RNA (snRNA) promoter, the guide RNA plasmid was made by using Gibson assembly (Gibson 2011) to replace the 20-bp protospacer of klp-12 with the 20-bp protospacers of new targets and an additional 5′ G (Friedland et al. 2013). For the guide RNA vector pRB1017, which utilizes the R07E5.16 snRNA promoter, complementary oligos encoding the desired guide RNA sequences were annealed and ligated into BsaI-digested pRB1017, as described in Arribere et al. (2014).
Plasmid templates for HDR were constructed in the pGEM7z backbone using Gibson assembly. The repair templates included the new sequences to be inserted and 500 bp of homologous sequence on either side of the predicted DSB. The PAM was mutated in the repair plasmid to prevent the plasmid and the repaired genomic locus from being cleaved by Cas9 cleavage. Mutations that disrupted the PAM were designed to maintain protein sequence fidelity.
DNA sequences of the single-stranded oligo repair templates are as follows:
rol-6(gof): 5′ TGTGGGTTGATATGGTTAAACTTGGAGCAGGAACCGCTTCCAACCGTGTGCGCtGcCAACAATATGGAGGATATGGAGCCACTGGTGTTCAGCCACCAGCACCAAC 3′
cpsf-2::avitag: 5′ TTGAATATTTTCAGAACAGCGGGAAAACCTAAAAGACACATCAAATGACCggacttaatgatatttttgaagctcagaagattgaatggcatgagggtggaaccTCCATTATCAAATTGAAAGTGTTTTCCGGTGCAAAAGACGAAGGACCTCT 3′
sex-1(ΔH12): 5′ AACAGCAACTCAATATTCTTCGAGAAAATTTGTCGTTTTTAAACCTGCCTtaaatagatgaCCTCTCGTTGTGGAAATGTTCCAACTCTCAACACTTCCGTTGCCTGTTAA 3′
For rol-6, the two lowercase letters are the changes needed for the rol-6(gof) phenotype. For cpsf-2::avitag and sex-1(ΔH12), the uppercase letters are the homology arms, and the lowercase letters are the new sequences to be inserted.
DNA microinjection
All plasmids for microinjection were purified using Qiagen’s Midi Plasmid Purification kit. Plasmid cocktails were prepared and injected into the gonad arms of wild-type adult hermaphrodite worms as previously described (Mello and Fire 1995). For injections that mirrored the Friedland et al. (2013) protocol using “high” concentrations of plasmids, the following DNA cocktails were injected: 250 ng/μl Peft-3::cas9-SV40_NLS::tbb-2, 225 ng/μl pU6::sgRNA, and 25 ng/μl pCFJ104. All other injections using the pU6 guide RNA vector to generate mutations via imprecise DSB repair were performed with the following plasmid concentrations: 50 ng/μl Peft-3::cas9-SV40_NLS::tbb-2, 250 ng/μl pU6::sgRNA, 2.5 ng/μl pCFJ90, and 5 ng/μl pCFJ104.
The guide RNA plasmid concentration is important, since an attempt at using the lower concentration of 25 ng/μl for guide pU6-Y62E10A.17 rather than 250 ng/μl reduced the proportion of red transformants carrying Y62E10A.17 mutations from 57 to 20%. For experiments using the pU6 vector to generate mutations through homology-directed repair, the following plasmid concentrations were used: 50 ng/μl Peft-3::cas9-SV40_NLS::tbb-2, 50 ng/μl pGEM7z-(HDR construct), 200 ng/μl pU6::sgRNA, 2.5 ng/μl pCFJ90, and 5 ng/μl pCFJ104.
For the co-CRISPR/co-conversion experiments, the plasmid DNA concentrations were as described previously: 25 ng/μl pRB1017-derived guide RNAs, 50 ng/μl pDD162 (Cas9), and 500 nM for each single-stranded oligo (Arribere et al. 2014). Oligos were ordered from IDT at the 4-nmol Ultramer scale.
Isolation of mutants using mCherry co-injection markers
For NHEJ experiments, P0 worms were allowed to recover after microinjection at 20° for 3 hr on NG-agar plates with OP50 bacteria and then transferred to 25° for 2 days. After 2 days, mCherry-positive worms or embryos (F1s) were cloned to plates and returned to 25° for 2 days to enable them to lay progeny. After 2 days, each F1 animal was picked to a single well of a 96-well plate for lysis, single-worm PCR to generate an amplicon spanning the Cas9/guide RNA target site, and DNA sequence analysis. When a heterozygous (or homozygous) mutant F1 was identified, its F2 and F3 progeny were also examined by DNA sequence analysis to assess the heritability of the mutation and its segregation pattern.
For HDR experiments, lysates from mCherry-positive F1s were screened by PCR to identify clones that possessed the epitope-tag sequence in appropriate genomic locations. Oligonucleotides used for PCR were complementary to the epitope-tag sequence and to nearby genomic sequence located outside the bounds of the HDR template. Further PCR and DNA sequence analysis was performed on F2 and F3 progeny from positive F1s to ensure they carried the epitope-tagged allele and had lost the transgenic arrays (indicated by the lack of fluorescence and the absence of a PCR product specific to the Bla gene encoded by all plasmids in the array). The PCR oligos flanked the target site and lacked complementarity to sequence in the plasmid repair template, ensuring the PCR amplicon was made from a genomic locus. The amplicon was sequenced to identify homozygous mutants and to determine the frequency of allele segregation. Oligonucleotide sequences used for screening are available upon request.
Isolation of mutants using the co-CRISPR/co-conversion approach with rol-6(gof)
P0 worms were injected with the following cocktail of plasmids, as described in Arribere et al. (2014) and recovered as above: 50 ng/μl Peft-3::cas9-SV40_NLS::tbb-2, 25 ng/μl pRB1017::sex-1 guide, 500 nM sex-1 single-stranded DNA oligo repair template (three-frame stop), and 25 ng/μl pJA42 (rol-6 guide: Addgene plasmid designation 59930), 500 nM rol-6(su1006) single-strand repair template (AF-JA-53). After 3 days at 25°, plates were screened for Rol animals. Rol F1 animals were picked to individual NG-agar plates and allowed to produce F2 progeny. F1s were then lysed and their sex-1 locus amplified by PCR and sequenced to identify sex-1 insertions. F2 progeny from positive F1s were analyzed to identify homozygous mutants.
In initial attempts at co-conversion experiments, we found that the success of converting the wild-type rol-6(+) allele to the rol-6(gof) allele depended critically on the preparation of the rol-6(su1006) single-stranded oligo repair template and on the vector used to express the guide RNA in vivo. No Rol progeny were recovered from 130 P0s injected with 25 ng/μl of the 3′ GG pU6::rol-6 guide RNA plasmid (K09B11.12 snRNA promoter) and a rol-6(gof) single-stranded oligo (500 nM, IDT Ultramer). Subsequently, no Rol progeny were recovered from each set of 30 P0s injected with the same 3′ GG pU6::rol-6 guide RNA plasmid and one of three different preparations of the rol-6(gof) oligo (IDT Ultramer). However, two of the four rol-6(gof) oligos produced abundant Rol progeny when each was co-injected into a set of 30 P0s with the identical 3′ GG rol-6 guide above (25 ng/μl), but expressed from the pRB1017 guide vector, which uses the R07E5.16 snRNA promoter. Even the combination of a successful rol-6(gof) oligo with a higher concentration of the 3′ GG pU6::rol-6 guide RNA (250 ng/μl) gave unsatisfactory results. Of 40 injected P0s, only 1 gave six Rol progeny, a result that represents a 10-fold reduction in Rol progeny from a typical experiment with the pRB1017 guide RNA vector. As a consequence, all subsequent co-conversion experiments were performed with the 3′ GG rol-6 guide expressed from pRB1017. The variability in success with different preparations of the same oligo is reminiscent of findings from Ward (2014), that oligo effectiveness was improved by PAGE gel purification to eliminate the incompletely synthesized oligos that contaminate the preparation.
We also noticed an incompatibility when using the successful rol-6(gof) oligo and successful 3′ GG rol-6 single guide RNA (sgRNA) expressed from the pRB1017 vector with previously successful sgRNAs expressed from the pU6::sgRNA vector. As examples, when we co-injected the 3′ GG lir-2 guide expressed from the pU6 guide vector (either 250 ng/µl or 25 ng/µl) with the successful rol-6 guide expressed from pRB1017 and rol-6(gof) oligo, no Rol progeny were produced from 36 or 57 P0s, respectively. Similarly, just co-injecting the pU6 vector plasmid (25 ng/µl) that lacked a 20-bp protospacer failed to yield Rol progeny from 29 injected P0s. Thus, for all subsequent co-conversion experiments, we used guide RNAs for rol-6 and the second target that were expressed solely from the pRB1017 guide vector.
Comparison of cotransformation strategy with 3′ GG guide and co-conversion strategy
For the genome editing strategy in which we co-injected mCherry transformation markers and plasmids encoding Cas9 and guide RNAs, with or without a single-stranded DNA oligo repair template, ∼50% of all injected P0s produced red fluorescent F1s carrying the injected DNAs. Of those red F1s, an average of 50% had mutations in the target of interest. Thus, ∼25% of all injected P0s produced F1 progeny with the desired mutations. The most difficult aspect of this strategy was that the mCherry-expressing animals exhibited variable fluorescence, making them time consuming to identify.
For the genome editing strategy combining our 3′ GG guide RNA design and the co-conversion approach using the rol-6(gof) oligo (Arribere et al. 2014), only 10% of P0s produced Rol progeny, but most of those P0s had jackpot broods of several Rol animals. In these experiments, ∼80% of the Rol animals had either a targeted mutation or insertion. Thus, ∼8% of injected P0s produced F1 progeny with a mutation or insertion in the designated target. Even though this combined strategy required more P0s to be injected than the mCherry co-transformation strategy, the Rol animals were easier to detect than the red fluorescent animals.
Results
A guide RNA design that dramatically improves Cas9-directed genome editing in vivo
Our initial application of the CRISPR/Cas9 protocol by Friedland et al. (2013) successfully reproduced the published mutagenesis rate for the target gene klp-2, but our subsequent attempts to edit other genes failed. More specifically, microinjection of their more effective concentration of DNAs encoding Cas9 (250 ng/μl) and guide RNAs (225 ng/μl) yielded rare somatic mutations but no heritable mutations for cpsf-2, lir-2, and sex-1, as assessed by CEL-1 digestion assays and DNA sequencing (Figure 2A, 3′ non-GG guides at high [Cas9]). Moreover, the injected worms were often sterile or produced only a few transgenic F1 progeny, making the recovery of candidates for mutant analysis very inefficient (Figure 2A).
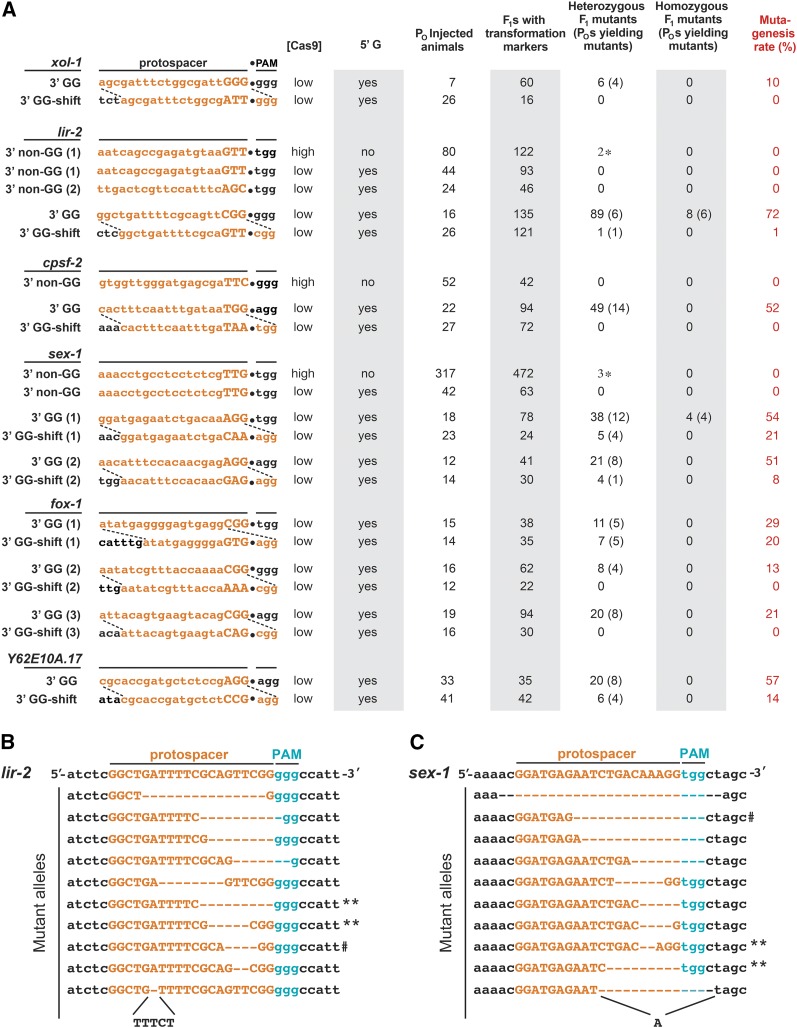
Figure 2.
A guide RNA design that yields reproducibly high frequencies of Cas9-directed mutagenesis. (A) Comparison of Cas9-mediated mutagenesis frequencies using the two guide RNA designs in Figure 1. Mutations from these experiments were generated through the NHEJ pathway. Shown are the matched sets of protospacers corresponding to the 3′ GG and 3′ GG-shift guides for 9 different targets. Also shown are the 3′ non-GG guides that failed in initial attempts at Cas9 mutagenesis. Protospacers (hence guide RNAs) with a GG in the 19th and 20th positions were highly efficient at generating mutations. The 18th–20th bases of the protospacer are capitalized. Dashed lines show the shift in guide design that redirects the 3′ GG-shift guides in the 3′ direction by 3 bp of the successful 3′ GG guides. Shifted guides produce no or few mutants, indicating that efficient DNA cleavage and mutagenesis require the GG dinucleotide to reside in the protospacer, hence guide RNA, not simply adjacent to the bona fide PAM. Mutagenesis rates were calculated by the formula: (total number of mutants/transgenic F1s) × 100. An asterisk (*) marks the mutations that were identified in F1 progeny but proved not to be heritable in later generations. These mutations were likely to be somatic. Cas9 plasmid concentrations ([Cas9]), indicated as “high” or “low” correspond to 250 ng/μl and 50 ng/μl, respectively, in the DNA cocktails injected. The figure provides the total number of injected P0s, the number of red transformed F1s carrying Cas9 and guide RNA plasmids, the number of F1s that were heterozygous or homozygous for the target mutations, and the number of P0s that produced those mutants in parentheses. (B and C) High rates of mutagenesis with the 3′ GG guides produced a wide variety of heritable mutant alleles in a single experiment. Shown for lir-2 (B) and sex-1 (C) are DNA sequences for nine F2 progeny produced from F1 hermaphrodites that tested positive for the target mutations. All nine F1 clones yielded heritable mutations. A double asterisk (**) indicates the two mutant alleles that were isolated from the same heterozygous F1 mutant. Wild-type F2 animals were also isolated from these heterozygous F1 animals, providing evidence that the F1 wild-type chromosome continued to be a target for Cas9 cleavage. The hashtag symbol (#) denotes an allele that was homozygous in the F1 and heritable in F2 progeny. Dashed lines indicate deleted bases.
To improve the efficiency of Cas9-directed mutagenesis in vivo, we performed experiments stimulated by the observation of Sternberg et al. (2014) that the effectiveness of competitor DNA at disrupting Cas9 cleavage of target DNA in vitro was positively correlated with the density of NGG motifs on the competitor. They found that the residence time of Cas9–RNA complexes on competitor DNA increased with an increase in NGG motifs, even when the competitor lacked sequences complementary to the guide RNA. This finding led us to test whether the frequency of genome editing in vivo could be improved by using DNA targets with additional NGG motifs adjacent to the PAM.
To increase the local concentration of NGG sequences near the DNA target site, we tried two different positions for the extra NGG (Figure 1A). The first placed the NGG motif at the 3′ end of the protospacer, immediately adjacent to the PAM. In this configuration, the extra NGG is part of the guide RNA as well as the DNA target. Guide RNAs for these targets, called “3′ GG guides,” had the NGG motifs at nucleotide positions 18–20. The second placed the NGG 3′ to the protospacer, making it part of the DNA target site but not the guide RNA. This placement was achieved by using targets related to the first targets but shifted in the 3′ direction by three nucleotides. As a result, the NGG at the 3′ end of the protospacer in the first set of targets became the bona fide PAM in the second set of targets. RNAs for these new targets lacked an NGG at positions 18–20 and were called “3′ GG-shift guides.”
The use of such closely related targets enabled us to maintain the density of NGG motifs near the target site and minimize the differences between the guide RNAs, while allowing us to correlate mutagenesis rates with the location of NGG sequences. If the feature that enhanced DNA cleavage in vivo were the density of NGG motifs adjacent to the PAMs, then both the 3′ GG and 3′ GG-shift guide RNAs would facilitate high mutagenesis rates. However, if the important parameter was the presence of NGG motifs at the 3′ end of the DNA protospacers, and hence within the target-specific sequences of the guide RNAs, the 3′ GG guides would promote high rates of mutagenesis, but the 3′ GG-shift guides would not.
We also changed two other parameters. We reduced the concentration of the Cas9 plasmid DNA (50 ng/μl) to help improve the viability and fertility of injected animals (Figure 2A). We added a nontemplate G to the 5′ end of the guide, just prior to the target-specific 20 nucleotides, to ensure efficient transcription from the U6 RNA polymerase III promoter (Figure 1A).
We first examined the extreme case of the gene xol-1, for which the 3′ GG guide RNAs placed the 3′ NGG of the protospacer directly adjacent to 15 Gs, hence 16 potential PAMs, making the region a possible thermodynamic sink for Cas9 binding. Our protocol was to co-inject worms with plasmids encoding Cas9, the guide RNA, and two transformation markers (Pmyo-2::mCherry and Pmyo-3::mCherry). Red transformed F1s were then assayed for mutations in the target gene by DNA sequence analysis (Figure 1B). With this 3′ GG guide design, 10% of the transgenic F1 progeny had a xol-1 mutation (Figure 2A). All the mutations were heritable. However, when the target and guide were shifted by three nucleotides, making ATT the terminal three nucleotides of the protospacer, the mutagenesis rate dropped to zero. The decrease in mutagenesis efficiency caused by the shift in guide RNA design suggested that the enhanced mutagenesis rate observed with the 3′ GG guide was due to the composition of the protospacer and guide RNA rather than a general increase in NGG motif density near the DNA target.
To test further the hypothesis that the presence of a GG dinucleotide at the 3′ end of the protospacer, and hence in the guide RNA, enhances Cas9-mediated DNA cleavage in vivo, we reexamined the three target genes previously refractory to mutagenesis by using 3′ GG and 3′ GG-shift guide RNAs. All experiments using 3′ GG guides yielded high rates of mutagenesis (Figure 2A). In contrast, the mutagenesis rates plummeted when 3′ GG-shift guides were used (Figure 2A).
For lir-2, 72% of all transgenic F1 progeny (97 of 135) from P0 animals injected with the 3′ GG guide carried mutations in lir-2. Analysis of subsequent nontransgenic progeny showed that the lir-2 mutations were heritable. Two of the heterozygous F1 lir-2 mutant progeny produced F2 progeny that carried two different lir-2 mutant alleles (Figure 2B, see double asterisk), showing that the wild-type chromosome of lir-2/+ mutants continued to be a target for Cas9 cleavage and imprecise DSB repair. The mutagenesis frequency using the 3′ GG guides was sufficiently high that 8% (8 of 97) of the F1 mutants were homozygous for the same mutant allele of lir-2. In contrast, the 3′ GG-shift guide was much less effective at yielding mutations for lir-2: the mutagenesis rate dropped to 1% (Figure 2A).
For cpsf-2, 52% (49 of 94) of the transgenic F1 progeny carrying the 3′ GG guide had a heterozygous mutation (Figure 2A). CPSF-2 is an essential RNA cleavage and polyadenylation factor. Our attempt to isolate a null allele using Cas9 showed the gene to be haploinsufficient: a heterozygous null mutation causes complete embryonic lethality, a finding that would have been difficult to obtain by more classical genetic screens. Mutant alleles were only identified from the dead F1 red embryos, thereby preventing us from assessing the heritability of the mutations. This result is consistent with the extensive lethality and sterility observed in animals treated with cpsf-2 RNAi (data not shown) and with mutational studies that failed to recover deficiencies for this region of chromosome V but did recover deficiencies of neighboring intervals (Rosenbluth et al. 1985). The lethality is not the result of an off-target mutation, as assessed by our ability (shown later) to use the same guide RNA to insert a benign tag into the gene. In contrast, the 3′ GG-shift guide was ineffective at yielding mutations for cpsf-2: the mutagenesis rate dropped to 0% (Figure 2A).
For sex-1, two different 3′ GG guides were used and both were effective in generating heritable mutations (Figure 2A). For the first, 54% (42 of 78) of transgenic F1 progeny carried sex-1 mutations. Ten percent of the mutants (4 of 42) were homozygous for the identical mutant allele in the F1 generation. In contrast, the corresponding 3′ GG-shift guide reduced the mutagenesis rate to 21% (Figure 2A). For the second 3′ GG guide, 51% (21 of 41) transgenic F1 progeny had sex-1 mutations. The corresponding 3′ GG-shift guide reduced the mutagenesis rate to 8% (Figure 2A). The frequencies found with the 3′ GG-shift guides demonstrate that the 3′ GG guides substantially improve the frequencies of genome editing even at closely related targets that already exhibit modest levels of mutagenesis.
Thus, for all three gene targets that were refractory to mutagenesis in initial attempts using non-3′ GG guides, the 3′ GG guides generated mutations at high frequencies. Furthermore, the frequencies were severely reduced by shifting those guides by only three nucleotides to create overlapping guides that lacked the 3′ GG. All the mutations had very similar molecular defects: 85% had small deletions (1–27 bp), 9% had insertions, and 6% had insertions and deletions (Figure 2, B and C and data not shown). The dramatic decrease in mutagenesis efficiency that correlated with the shift in guide design suggests that the higher mutagenesis rates using the 3′ GG guides was due to the GG in the protospacer and/or in the guide RNA, rather than an increase in NGG density around the DNA target site. Hence, it seems no coincidence that the most successful guide in the study by Friedland et al. (2013) was that for klp-12, which also had a 3′ terminal GG dinucleotide and yielded a mutagenesis rate of 80%.
The dramatic change in editing success with only a three-nucleotide shift in the protospacer makes it unlikely that the chromatin status of the endogenous targets accounts for the success of the 3′ GG guides. More likely, the interactions among the DNA target, guide RNA, and Cas9 protein were enhanced. For none of these 3′ GG guides is it yet known whether the frequency of potential off-target editing events is greater than that for other less-efficient guides.
The lower concentration of Cas9 DNA reduced the lethality and increased the brood sizes, but neither the concentration change nor the addition of a nontemplate G to the guide RNAs was sufficient to make the previously unsuccessful guides useful. All attempts at genome editing using the three original 3′ non-GG guides with an added nontemplate G failed at the lower Cas9 DNA concentrations (Figure 2A).
To test further the efficacy of a GG dinucleotide at the 3′ terminus of the protospacer in achieving reliable and efficient Cas9-directed genome editing in vivo, we tested multiple 3′ GG guides for two other genes: Y62E10A.17 and fox-1. As before, the 3′ GG guides yielded high rates of mutagenesis: 57% for Y62E10A.17 and 29, 20, and 13% for three different targets within fox-1 (Figure 2A).
For the three fox-1 targets, we also tested 3′ GG derivatives of an alternative guide RNA design (Supporting Information, Figure S1) that was successful for Cas9 targeting in mammalian cell lines (Chen et al. 2013). This alternative guide had a longer stem and loop structure and an A–U flip that improved its efficacy in mammalian cell Cas9 experiments. We found the mutagenesis rate for each alternative 3′ GG guide RNA to be very similar to that of the original 3′ GG guide (Figure S1, A and B), so we continued to use the original guide design.
The frequency of mutagenesis declined substantially for the Y62E10A.17 and fox-1 targets when 3′ GG-shift guides were used. For Y62E10A.17, the mutagenesis frequency dropped from 57 to 14%. For two fox-1 targets, the 3′ GG-shift guides caused the mutagenesis rate to drop from either 20% or 13% down to 0%. The third fox-1 guide, which was shifted by six bases instead of three to prevent the new guide from acquiring an alternate 3′ GG in the protospacer, was less successful than its 3′ GG counterpart (29%), but still yielded a mutagenesis rate of 20%.
Remarkably, all guide RNAs (9/9) selected solely by the criterion that they include a 3′ GG in the target-specific sequences were highly successful in generating mutants (median frequency of 51%, range of 10–72%) (Table S1). The frequency was so high that mutations could be identified by DNA sequence analysis of a small number of transgenic F1 animals. The mutants did not need to have a visible phenotype. In all cases, shifting the 3′ GG out of the protospacer universally reduced the frequency of mutagenesis, but to a variable and unpredictable level (median frequency 1%, range of 0–21%) reflective of the more common range of frequencies found in most C. elegans studies using non-3′ GG guides and related screening approaches (Table S1) (Friedland et al. 2013; Waaijers et al. 2013; Kim et al. 2014).
To understand the variability in success of the 3′ GG-shift guides, we compared our guide sequences and editing frequencies to those of other organisms. Analysis of guide RNAs found to be effective in zebrafish (Gagnon et al. 2014), Drosophila (Ren et al. 2014), and mammalian cell lines (Doench et al. 2014; Wang et al. 2014; Wu et al. 2014) suggests that the partial success of some 3′ GG-shift guides in C. elegans might be due either to a G at position 20 of the target-specific sequences of the guide or to any combination of two purines at positions 19 and 20. Although those features are positively correlated with the effectiveness of guides in other species and might contribute to the partial success of guides in C. elegans, they cannot account for all the success of either the 3′ GG guides or 3′ GG-shift guides. Several of our 3′ GG-shift guides and 3′ non-GG guides had a 3′ G or some combination of two 3′ purines other than GG, but supported no or very low mutagenesis, while their 3′ GG counterparts achieved robust mutagenesis.
Analysis of guide RNAs in mammalian cell lines also showed that the presence of Us in guide RNA positions 18–20 was detrimental for Cas9 binding and cleavage (Doench et al. 2014; Wang et al. 2014; Wu et al. 2014). However, Us cannot be the sole reason for the failure of our 3′ GG-shift guides and 3′ non-GG guides, because some successful 3′ GG guides had a U at position 18 or 19. Lastly, the simple presence of 3′ purines and absence of Us does not correlate directly with the efficacy of our guides, since several unsuccessful guides had both features.
Our studies revealed important principles for achieving efficient Cas9-directed genome editing at desired targets. We found that a 3′ terminal GG dinucleotide in the protospacer, and hence positions 19 and 20 of the guide RNA, is the most reliable predictor for whether or not a guide will promote genome editing at a designated target. Our efforts at Cas9-directed editing succeeded with all 3′ GG guides. In contrast, success was infrequent and unpredictable with guides lacking a 3′ GG in our study and in published studies (Table S1) (Chiu et al. 2013; Cho et al. 2013; Dickinson et al. 2013; Friedland et al. 2013; Katic and Grosshans 2013; Lo et al. 2013; Tzur et al. 2013; Liu et al. 2014; Paix et al. 2014; Shen et al. 2014; Waaijers and Boxem 2014; Zhao et al. 2014). We also found that a 3′ GG dinucleotide reliably increased the frequency of genome editing at all targets, even related targets for which the 3′ GG-shift guides supported a low to modest frequency of editing.
The median frequency of NHEJ-mediated mutagenesis for successfully edited C. elegans targets was 4.3% (range of 0.2–100%) in published studies using DNA sequence to screen targets in transgenic F1 animals expressing Cas9 (Table S1) (Friedland et al. 2013; Waaijers et al. 2013; Kim et al. 2014). In contrast, the median frequency of editing for targets in our study was 51% (range of 10–72%) (Table S1), a 10-fold increase (P ≤ 0.02. Mann–Whitney U-test). Consistent with this observation, we found that the highest rates of mutagenesis in published C. elegans experiments also correlate with guide RNAs fortuitously having a 3′ GG in target-specific sequences (Table S1) (Friedland et al. 2013; Kim et al. 2014).
Given the range in mutant frequencies achieved by successful 3′ GG guides, the 3′ GG dinucleotide cannot be the only factor in determining a guide’s efficacy in directing Cas9-mediated mutagenesis, but it is the strongest and most reliable predictor of guide RNA success and favorable target site selection. Indeed, the presence of the 3′ GG motif in the protospacer is a better predictor of editing outcome than the algorithms derived from mammalian cells (Table S2) (Doench et al. 2014). The overlap of the 3′ GG dinucleotide in our guide design with the 3′ G and/or two 3′ purines of any arrangement in successful mammalian-cell guides (Doench et al. 2014; Wang et al. 2014; Wu et al. 2014) and with the high GC content for the six 3′ nucleotides of successful Drosophila guides (Ren et al. 2014) makes it likely that the two key principles to emerge from our studies will apply to diverse species and cell lines.
High-efficiency Cas9 editing in vivo using 3′ GG guide RNAs yielded high frequencies of heritable DNA insertion events through precise, homology-directed DNA repair
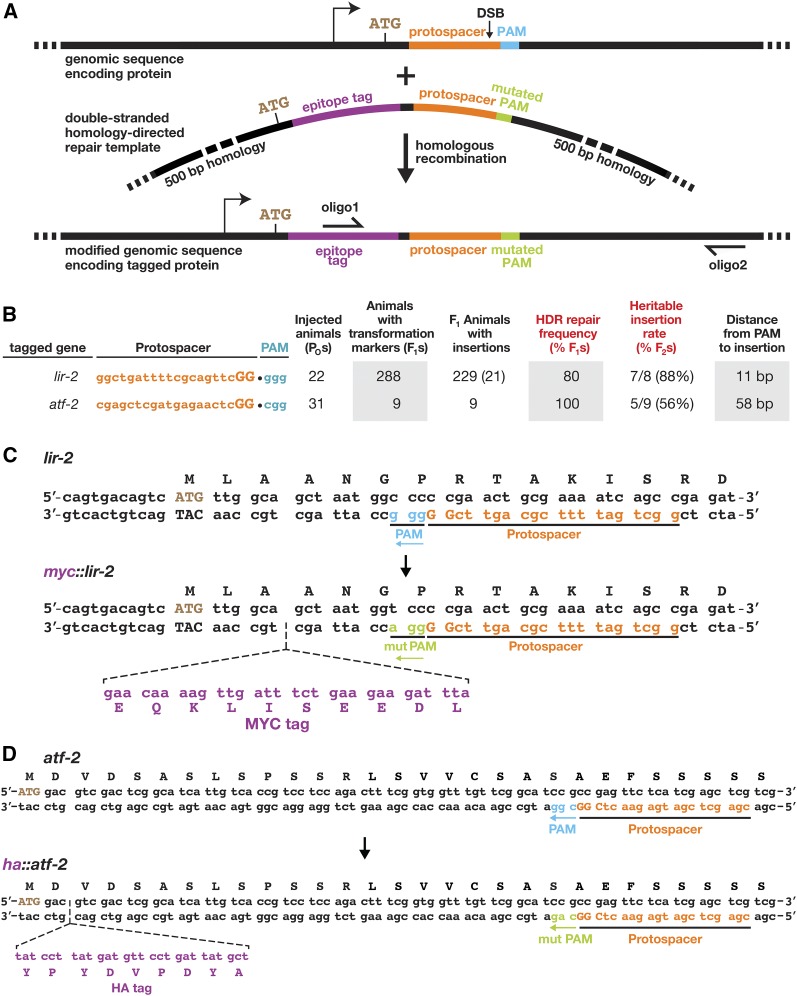
Taking advantage of our highly effective guide RNA design, we assessed the frequency of Cas9-initiated homology-directed repair from an exogenous double-stranded DNA template to insert DNA sequences of choice. We first utilized our most successful guide, lir-2 3′ GG, to initiate a DSB near the 5′ end of lir-2 and trigger the insertion of a Myc epitope tag. For the repair template, we used a plasmid that contained the 30 bp Myc tag flanked on both sides by 500 bp of homology to lir-2 (Figure 3, A–C). The PAM in the HDR repair template was mutated to prevent Cas9 from cleaving either the repaired lir-2::myc endogenous sequence or the extrachromosomal arrays carrying the repair template. Of the transgsenic F1s produced by injected P0 hermaphrodites, 80% tested positive for the Myc tag by PCR analysis (Figure 3B). This high HDR repair rate suggested that nearly all DSB repair events were shunted from NHEJ to HDR by the exogenous repair template. Progeny from a sample of the Myc-positive F1s were followed for several generations, and 88% of positive F1s had heritable insertions, confirming the very high success rate for precise, heritable DNA integrations via HDR using our new guide design.
Figure 3.
Cas9 and 3′ GG-guide RNAs elicit a high frequency of precise, HDR-mediated DNA insertion events. (A) Experimental design for homology-directed repair of Cas9 DSBs using custom-designed, double-stranded DNA repair templates. Shown are diagrams of the DNA target, double-stranded DNA repair template, and desired genomic insertion. The arrow shows the start point of transcription. The double-stranded DNA repair templates had 500 bp of homology on either side of the target site, a silent mutation in the PAM domain to prevent Cas9 cleavage of the repair template, and an in-frame epitope tag. The mutant PAM is labeled “mut PAM” in green. Two oligonucleotides were used to screen for the epitope tag. One oligo annealed to the sequence encoding the tag and the second oligo to a genomic region more than 500 bases away from the tag and not present in the repair template. (B) High frequencies of homology-directed repair at two genomic loci using 3′ GG-guide RNAs. Transgenic F1 animals were examined for DNA insertions via PCR, and tag-positive F1 animals were reexamined by DNA sequence analysis. Heritability of the insertion was tested in the F2 generation by DNA sequence analysis. (C) DNA sequence of the genomic myc-tagged lir-2 locus and (D) ha-tagged atf-2 locus.
To further explore our success with HDR, we also targeted atf-2 for insertion of a 5′ HA epitope tag. While fewer transgenic F1 animals were obtained, all tested positive by PCR for precise insertion of the HA tag (Figure 3, B and D), and 56% of the positive F1 animals proved to have heritable insertions. The other insertions were likely to have been somatic events. In both experiments, the availability of double-stranded DNA repair templates shifted nearly all the DSB repair to HDR rather than NHEJ.
The frequency of insertion by HDR was vastly improved by using 3′ GG guides. In published experiments using a similar screening protocol, the median frequency for insertion by HDR was 2.5% (range of 1.3–17%) (Tzur et al. 2013), compared to our median frequency of 63% (range of 56–70%).
Although the requirement for a 3′ GG in the protospacer and guide RNA places some constraint on the location of the DSB cleavage site within a locus, this constraint should pose little problem for inserting specific DNA changes at desired locations, because double-stranded DNA repair templates can be used to repair in a desired sequence at a desired location, even if somewhat distant (≤500 bp, Q. Bian, E. Anderson, B. Meyer, unpublished data) from the DSB.
Coupling the high-efficiency guide RNA design with the co-CRISPR/co-conversion strategy enhanced the detection and recovery of genome editing events and expedited the mutant screening process
Recent studies improved the detection of Cas9-directed editing events by developing a co-selection strategy termed co-CRISPR/co-conversion (Arribere et al. 2014; Kim et al. 2014; Ward 2014). In this approach, simultaneous introduction of guide RNAs to two different endogenous loci results in double editing events that are not statistically independent. Instead, the occurrence of a cleavage and repair event in one locus enhances the probability of finding a heritable mutagenetic event in a second locus in the same animal. This approach enables one to screen animals that have acquired a visible mutant phenotype due to the editing of one locus for the presence of a phenotypically silent, but molecularly detectable mutation in a separate locus. However, the frequency of co-CRISPR/co-conversion varied widely (Arribere et al. 2014; Kim et al. 2014; Ward 2014).
We combined the 3′ GG guide design with the co-CRISPR/co-conversion strategy to assess whether the two approaches together would enhance the detection of phenotypically silent mutations and thereby improve our Cas9 mutagenesis protocol. For the co-conversion marker, we chose the dominant roller (Rol) phenotype obtained by using a rol-6 3′ GG guide RNA and a single-stranded DNA oligo repair template that inserts the rol-6(su1006) dominant gain-of-function allele (Arribere et al. 2014) (Figure 4, A and B). Our goal for the co-conversion experiment was to insert a translation stop codon in all three reading frames of sex-1, thereby forcing the truncation of SEX-1 at its C terminus, before helix 12 of the ligand-binding domain (Figure 4, A and B).
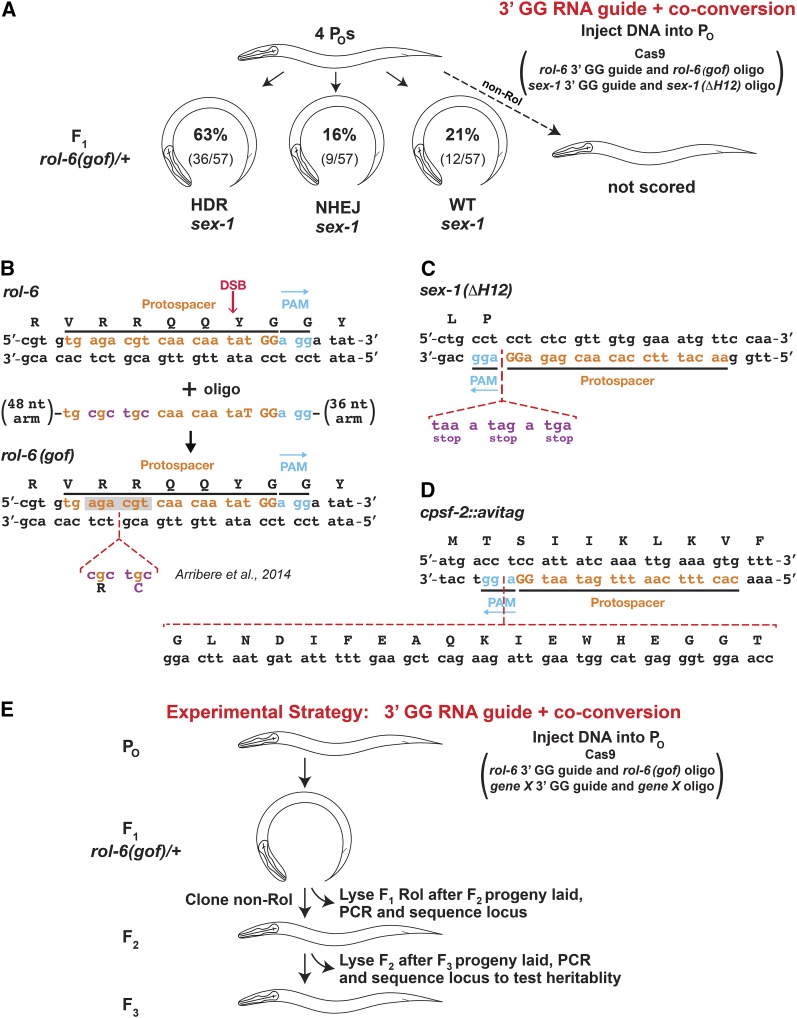
Figure 4.
Efficient production and recovery of custom-designed mutations using 3′ GG-guide RNAs and the co-CRISPR strategy. (A) Results for the co-conversion of the rol-6(gof) insertion and sex-1(ΔH12) insertion from HDR. P0 animals were microinjected with plasmids encoding Cas9, 3′ GG-guide RNA for rol-6 and sex-1 plus two oligonucleotide repair templates, a rol-6(gof) template to induce a Rol phenotype, and a sex-1(ΔH12) template to insert stop codons in all reading frames and thereby truncate SEX-1 protein prior to helix 12 of its ligand binding domain. DNA sequence was determined for all sex-1 alleles of F1 Rol animals to assess the relative proportion of wild-type, NHEJ-repaired, and HDR-repaired alleles. In this experiment, HDR-mediated insertion mutations were enriched by fourfold over NHEJ-mediated mutations. Non-Rol animals were not scored. (B) Steps for HDR-mediated insertion of the rol-6(gof) allele at the rol-6 target site. Shown are the rol-6 target, highlighting the protospacer, PAM, and DSB site; the single-stranded oligo used to insert the gof allele; and the final target sequence after HDR. (C) DNA sequence of the sex-1 target site after HDR-mediated insertion of the sex-1 stop codons. (D) DNA sequence of the cpsf-2 target site after HDR-mediated insertion of DNA sequences encoding the AviTag (15 amino acids) and the three amino acids GGT that serve as a flexible linker between CPSF-2 and the tag. (E) Effective experimental strategy for the production and isolation of mutants with precisely engineered insertion and deletion alleles. P0 animals are microinjected with DNA encoding Cas9, a 3′ GG rol-6 guide RNA, and a rol-6(gof) single-stranded repair template to generate F1s with the easily scorable Rol phenotype, along with DNA encoding a 3′ GG-guide RNA and oligo repair template to a second target to generate co-converted, phenotypically silent mutations in that target. F1 Rol animals were picked onto individual plates, allowed to produce F2 progeny, and then analyzed by PCR or DNA sequence analysis for HDR-mediated insertions and NHEJ-mediated mutations in the second target. F2’s from mutant F1s were allowed to produce F3 progeny and then examined by DNA sequence analysis to confirm the heritability of the mutations and to determine the exact mutant allele sequence.
After many attempts, we had little success in obtaining Cas9-mediated Rol mutants using a rol-6 guide RNA expressed from a guide vector (Friedland et al. 2013) that uses the snRNA gene K09B11.12 promoter. However, for unknown reasons we had strong success in obtaining Rol mutants using the same rol-6 guide RNA expressed from a guide vector (Arribere et al. 2014) that uses the snRNA gene R07E5.16 promoter (see Materials and Methods).
Our subsequent co-CRISPR/co-conversion experiments targeting both rol-6 and sex-1 were highly successful when using 3′ GG guide RNAs expressed from the R07E5.16 promoter. Of 57 F1 Rol animals derived from 4 of the 46 injected P0 animals, 63% had the three-frame stop codon insertion in sex-1, 16% had a sex-1 mutation resulting from DSB repair via NHEJ, and 21% had a wild-type sex-1 allele (Figure 4, A–C). All mutations were heritable. Since our initial NHEJ experiments using the same sex-1 guide (3′ GG guide 2) yielded 51% mutagenesis (Figure 2B), the co-conversion strategy coupled with the 3′ GG guide RNA design produced a marked improvement: 79% of F1 Rol animals were also mutant for sex-1 (HDR and NHEJ events).
We also determined the DNA sequence of the sex-1 locus in half of the non-Rol F1 progeny (103 F1s) of one injected P0 that produced 18 Rol F1’s to determine whether sex-1 had been edited independently of rol-6. We found no sex-1 mutants, suggesting that most of the Cas9 mutagenic events were co-conversion events, as observed by others (Arribere et al. 2014; Kim et al. 2014). Therefore, not only was the combined 3′ GG-guide design and Rol co-conversion scheme more efficient for recovering desired mutations than either approach alone, screening for Rol mutants was much easier than screening for mCherry-expressing worms, which exhibit a variable degree of fluorescence.
We also had good success with the second co-conversion experiment using a 3′ GG guide to insert an AviTag into the cpsf-2 gene (Figure 4D). Of 81 F1 Rol animals derived from 4 of the 54 injected P0s, 30% were heterozygous for the tag. We detected no NHEJ-derived mutations as predicted from the dominant embryonic lethality caused by cpsf-2 loss-of-function mutations in previous experiments. We therefore cannot calculate the true fraction of Rol animals that received an edit in the cpsf-2 locus. The success in tag insertion using the co-conversion strategy represents an improvement over the numerous failures using the same guide but selecting F1s for cpsf-2 screening by the presence of transgenic arrays encoding the mCherry transformation marker, Cas9, and guide RNAs. No insertions were found in 237 red F1s from 121 injected P0s. Thus, this rol-6/cpsf-2 experiment provides further evidence that a 3′ GG guide coupled with the co-conversion strategy promotes strong success and ease in obtaining desired insertions at endogenous loci.
The use of 3′ GG guides elevated the frequency of co-conversion events compared to those in published reports. The median for published co-conversion events involving repair by either NHEJ or HDR was 33% (range of 0–88%) (Arribere et al. 2014; Kim et al. 2014; Ward 2014) compared to our median of 86% (range of 79–93%). For co-conversion involving HDR using a single-stranded oligo, the median for published experiments was 31% (range of 0–61%) (Arribere et al. 2014; Ward 2014) compared to our median of 61% (range of 59–63%).
Throughout our co-conversion experiments, only 10% of the injected P0s produced Rol progeny, and many independent F1 Rol animals were usually produced from a single P0. These jackpot broods had a very high frequency of co-conversion for the second target. Curiously, we never found F1 Rol progeny that carried a homozygous mutation for any target in our co-conversion experiments. In contrast, for experiments using transformation markers to identify potential mutants, 2% of F1 progeny expressing transgenic markers such as mCherry had homozygous mutations in the target genes. In a single, separate Cas9 mutagenesis experiment (Figure S2), we compared directly the combination of the 3′ GG guide and the co-CRSPR/co-conversion scheme with the combination of the 3′ GG guide and the mCherry transformation marker scheme. From 7 of the 61 P0 animals injected with Cas9, the rol-6 and sex-1 guides expressed from the R07E5.16 promoter, the rol-6 and sex-1 oligo repair templates, and the two DNA transformation markers, Pmyo-2::mCherry and Pmyo-3::mCherry, we obtained 32 Rol F1 animals and 40 red F1 animals, with no overlap between the Rol and red animals. Of the Rol animals, 93% percent had a sex-1 mutation: 59% from HDR repair and 34% from NHEJ repair. Of red animals, 21% had a sex-1 mutation: 13% from HDR repair and 8% from NHEJ repair. One red animal was homozygous for the HDR-mediated insertion. From our finding that F1 Rol animals were not red, we infer that the HDR insertion events in co-conversion experiments occurred during meiosis in the P0, and only rare Cas9-mediated DSBs occurred in the F1 because they lacked the Cas9 plasmid or protein. In contrast, for mCherry-marked F1s, the Cas9 plasmid was inherited and thereby provided the chance for Cas9 to cleave its target in the F1 and for the resulting DSBs to be repaired from the homologous mutant chromosome created during meiosis of the P0. This direct comparison allows us to conclude that the most effective strategy to obtain mutations in loci of choice via HDR and NHEJ events is to combine our 3′ GG-guide RNA design with the co-CRISPR/co-conversion strategy (Figure 4D).
Conclusion
We developed a widely applicable strategy that dramatically increases the efficiency of genome engineering directed by Cas9. This strategy achieved a uniformly high frequency of targeted mutagenesis via NHEJ and precise DNA integration via HDR for all genomic targets tested in vivo. The key innovation was to design guide RNAs with a GG motif at the 3′ end of their target-specific sequences. This new design is an impressively reliable predictor for success and for effective target site selection. All guides (10 of 10) designed using this single principle achieved a high frequency of Cas9-directed mutagenesis. Since this guide design enhances Cas9 binding and DNA cleavage in vivo, it will likely be a universally effective strategy for diverse species and cell lines. Finally, the coupling of the 3′ GG-guide design with the co-CRISPR/co-conversion strategy facilitates mutant detection and enhances the rate of mutant recovery. This combined approach offers a powerful means for obtaining desired genetic changes in an otherwise unmodified genome.
Supplementary Material
Acknowledgments
We thank J. A. Calarco, A. Z. Fire, C. Frøkjær-Jensen, and E. M. Jorgensen for plasmids. We are grateful to S. H. Sternberg, J. H. D. Cate, and J. A. Doudna for helpful discussions. We thank K. Brejc, A. Severson, T. Lee, C. Preston, and T. Cline for critical comments on the manuscript and D. Stalford for assistance with figures. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440). This work was supported in part by NIH grant R01-GM030702. B.J.M. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.175166/-/DC1.
Communicating editor: O. Hobert
Literature Cited
- Anders C., Niewoehner O., Duerst A., Jinek M., 2014. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513: 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere J. A., Bell R. T., Fu B. X., Artiles K. L., Hartman P. S., et al. , 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198: 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D., Jiang W., Samai P., Hochschild A., Zhang F., et al. , 2013. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 41: 7429–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D., 2014. Genome engineering with targetable nucleases. Annu. Rev. Biochem. 83: 409–439. [DOI] [PubMed] [Google Scholar]
- Chen B., Gilbert L. A., Cimini B. A., Schnitzbauer J., Zhang W., et al. , 2013. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155: 1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Xu F., Zhu C., Ji J., Zhou X., et al. , 2014. Dual sgRNA-directed gene knockout using CRISPR/Cas9 technology in Caenorhabditis elegans. Sci. Rep. 4: 7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu H., Schwartz H. T., Antoshechkin I., Sternberg P. W., 2013. Transgene-free genome editing in Caenorhabditis elegans using CRISPR-Cas. Genetics 195: 1167–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. W., Lee J., Carroll D., Kim J. S., Lee J., 2013. Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9-sgRNA ribonucleoproteins. Genetics 195: 1177–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., et al. , 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J., Goldstein B., 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10: 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J. G., Hartenian E., Graham D. B., Tothova Z., Hegde M., et al. , 2014. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 32: 1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J. A., Charpentier E., 2014. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346: 1258096. [DOI] [PubMed] [Google Scholar]
- Friedland A. E., Tzur Y. B., Esvelt K. M., Colaiacovo M. P., Church G. M., et al. , 2013. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10: 741–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon J. A., Valen E., Thyme S. B., Huang P., Ahkmetova L., et al. , 2014. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS ONE 9: e98186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau J. E., Dupuis M. E., Villion M., Romero D. A., Barrangou R., et al. , 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468: 67–71. [DOI] [PubMed] [Google Scholar]
- Gasiunas G., Barrangou R., Horvath P., Siksnys V., 2012. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA 109: E2579–E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G., 2011. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 498: 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L. A., Horlbeck M. A., Adamson B., Villalta J. E., Chen Y., et al. , 2014. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159: 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. D., Lander E. S., Zhang F., 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157: 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., et al. , 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek, M., A. East, A. Cheng, S. Lin, E. Ma et al., 2013 RNA-programmed genome editing in human cells. elife 2: e00471. [DOI] [PMC free article] [PubMed]
- Katic I., Grosshans H., 2013. Targeted heritable mutation and gene conversion by Cas9-CRISPR in Caenorhabditis elegans. Genetics 195: 1173–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Ishidate T., Ghanta K. S., Seth M., Conte D., Jr, et al. , 2014. A co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans. Genetics 197: 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson M. H., Gilbert L. A., Wang X., Lim W. A., Weissman J. S., et al. , 2013. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 8: 2180–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Long L., Xiong K., Yu B., Chang N., et al. , 2014. Heritable/conditional genome editing in C. elegans using a CRISPR-Cas9 feeding system. Cell Res. 24: 886–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo T. W., Pickle C. S., Lin S., Ralston E. J., Gurling M., et al. , 2013. Precise and heritable genome editing in evolutionarily diverse nematodes using TALENs and CRISPR/Cas9 to engineer insertions and deletions. Genetics 195: 331–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder M. L., Linder S. J., Cascio V. M., Fu Y., Ho Q. H., et al. , 2013. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods 10: 977–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Esvelt K. M., Church G. M., 2013. Cas9 as a versatile tool for engineering biology. Nat. Methods 10: 957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C., Fire A., 1995. DNA transformation. Methods Cell Biol. 48: 451–482. [PubMed] [Google Scholar]
- Mojica F. J., Diez-Villasenor C., Garcia-Martinez J., Almendros C., 2009. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155: 733–740. [DOI] [PubMed] [Google Scholar]
- Paix A., Wang Y., Smith H. E., Lee C. Y., Calidas D., et al. , 2014. Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 sites in Caenorhabditis elegans. Genetics 198: 1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera P., Kocak D. D., Vockley C. M., Adler A. F., Kabadi A. M., et al. , 2013. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods 10: 973–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L. S., Larson M. H., Gilbert L. A., Doudna J. A., Weissman J. S., et al. , 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Yang Z., Xu J., Sun J., Mao D., et al. , 2014. Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Reports 9: 1151–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluth R. E., Cuddeford C., Baillie D. L., 1985. Mutagenesis in Caenorhabditis elegans. II. A spectrum of mutational events induced with 1500 r of gamma-radiation. Genetics 109: 493–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapranauskas R., Gasiunas G., Fremaux C., Barrangou R., Horvath P., et al. , 2011. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 39: 9275–9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Zhang X., Chai Y., Zhu Z., Yi P., et al. , 2014. Conditional knockouts generated by engineered CRISPR-Cas9 endonuclease reveal the roles of coronin in C. elegans neural development. Dev. Cell 30: 625–636. [DOI] [PubMed] [Google Scholar]
- Sternberg S. H., Redding S., Jinek M., Greene E. C., Doudna J. A., 2014. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507: 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum M. E., Gilbert L. A., Qi L. S., Weissman J. S., Vale R. D., 2014. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159: 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzur Y. B., Friedland A. E., Nadarajan S., Church G. M., Calarco J. A., et al. , 2013. Heritable custom genomic modifications in Caenorhabditis elegans via a CRISPR-Cas9 system. Genetics 195: 1181–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waaijers S., Boxem M., 2014. Engineering the Caenorhabditis elegans genome with CRISPR/Cas9. Methods 68: 381–388. [DOI] [PubMed] [Google Scholar]
- Waaijers S., Portegijs V., Kerver J., Lemmens B. B., Tijsterman M., et al. , 2013. CRISPR/Cas9-targeted mutagenesis in Caenorhabditis elegans. Genetics 195: 1187–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Wei J. J., Sabatini D. M., Lander E. S., 2014. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343: 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. D., 2014. Rapid and precise engineering of the Caenorhabditis elegans genome with lethal mutation co-conversion and inactivation of NHEJ repair. Genetics 199: 363–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Scott D. A., Kriz A. J., Chiu A. C., Hsu P. D., et al. , 2014. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. 32: 670–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Zhang Z., Ke H., Yue Y., Xue D., 2014. Oligonucleotide-based targeted gene editing in C. elegans via the CRISPR/Cas9 system. Cell Res. 24: 247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.