Abstract
Broad phenotypic variations were induced in derivatives of an asymmetric somatic hybridization of bread wheat (Triticum aestivum) and tall wheatgrass (Thinopyrum ponticum Podp); however, how these variations occurred was unknown. We explored the nature of these variations by cytogenetic assays and DNA profiling techniques to characterize six genetically stable somatic introgression lines. Karyotyping results show the six lines similar to their wheat parent, but GISH analysis identified the presence of a number of short introgressed tall wheatgrass chromatin segments. DNA profiling revealed many genetic and epigenetic differences, including sequences deletions, altered regulation of gene expression, changed patterns of cytosine methylation, and the reactivation of retrotransposons. Phenotypic variations appear to result from altered repetitive sequences combined with the epigenetic regulation of gene expression and/or retrotransposon transposition. The extent of genetic and epigenetic variation due to the maintenance of parent wheat cells in tissue culture was assessed and shown to be considerably lower than had been induced in the introgression lines. Asymmetric somatic hybridization provides appropriate material to explore the nature of the genetic and epigenetic variations induced by genomic shock.
Keywords: bread wheat; asymmetric somatic hybridization; introgression line; genomic shock, genetic and epigenetic alteration
WITH the world’s population continuing to increase, achieving a sustainable mode of food production represents an ever-growing challenge. Plant breeding has narrowed the genetic base of many crop species, but as yet has had little impact on the genetic diversity present in their wild relatives. In principle, this diversity can be introgressed into crops via sexual hybridization and subsequent backcrossing. However, in practice, wild-crop manipulation has been severely restricted by difficulties in creating the initial sexual hybrid and by sterility issues in the early backcross generations (Xia 2009). Asymmetric somatic hybridization is a viable alternative to introgression, especially where wide crosses are not feasible. It has been successfully exploited in bread wheat to transfer chromosomal segments from a number of related species (Xia et al. 2003; Xiang et al. 2003, 2004; Cheng et al. 2004; Zhou and Xia 2005; Xia 2009). More importantly, asymmetric somatic hybridization offers smaller alien chromatin introgression, thereby overcoming a significant problem in wheat sexual hybrids where the Ph1 gene prevents homeologous recombination (Griffiths et al. 2006).
Newly synthesized allopolyploids have provided an opportunity to explore the nature of the genetic and epigenetic changes triggered by polyploidization (Song et al. 1995; Comai et al. 2000; Ozkan et al. 2001; Shaked et al. 2001; Madlung et al. 2002; Han et al. 2003; Ma et al. 2004; Wang et al. 2004a; Salmon et al. 2005; Tate et al. 2006; Bassene et al. 2010; Xu et al. 2014), although a few allopolyploids were not accompanied with such changes (Liu et al. 2001). Experimental results indicate that the majority of events are highly reproducible (Bento et al. 2010). In particular, sequence deletion is common (Feldman et al. 1997; Liu et al. 1998a,b; Ozkan et al. 2001; Shaked et al. 2001; Kashkush et al. 2002; Ma et al. 2004; Ma and Gustafson 2006). Epigenetic modifications, such as changes in the pattern of cytosine methylation, have also been shown to induce changes in gene expression and activate transposon transcription (Comai et al. 2000; Shaked et al. 2001; Kashkush et al. 2002, 2003). However, these changes induced by “genomic shock” during polyploid synthesis do not represent the changes in somatic introgressions. Allopolyploids represent a combination of nuclear genomes in a fixed cytoplasmic context, while somatic hybrids combine both the nuclear and cytoplasmic genomes within a single cell. The introgression of chromatin segments by asymmetric somatic hybridization likely occurs via nonhomologous end-joining of fragmented genome pieces rather than by homologous recombination, which would show specific genetic and epigenetic changes in these materials. Moreover, the epigenetic state of somatic cells tend to be distinct from gametal cells given that the mutagenesis in gametal cells is more tightly controlled to ensure genetic fidelity (Bird 1997, 2002). Thus, the variations induced by “somatic genomic shock” likely have unique characteristics compared with allopolyploids.
A number of hybrid progenies regenerated from asymmetric somatic hybrids [bread wheat cultivar Jinan 177 (JN177) and tall wheatgrass (Thinopyrum ponticum)] have proven phenotypically stable for a number of generations (Xia et al. 2003; Chen et al. 2004a; Wang et al. 2004b; Liu and Xia 2014). The heterocytoplasmic nature of these hybrid lines were confirmed, with the chloroplast genomic components dominated by wheat, while a few sequences of the chloroplast genome of tall wheatgrass were also detected in these lines (Chen et al. 2004b). DNA comparison of a well-characterized set of glutenin proteins among parents and derivatives shows that all novel glutenin genes in hybrid progenies originated from alien genes of tall wheatgrass and allelic variation of parent wheat genes (Liu et al. 2007, 2009). Such de novo alleles do not arise simply as a result of UV-induced mutagenesis, as high frequency of the glutenin alleles were also found in symmetric somatic hybrids without UV pretreatment (Gao et al. 2010). Moreover, somaclonal variation of parent wheat is too rare to account for the observed high frequency of novel glutenin alleles in the somatic hybrids (Feng et al. 2004). It is more likely that they derive from genomic shock triggered by introgression of alien chromosome fragments during somatic hybridization. Therefore, the suggestion is that somatic hybridization provides a means of introgression distinct from sexual wide crossing and such introgression induces genomic variations. However, the details of the genomic variations and mechanism of transfer are unknown. Grass genomes are composed primarily of repetitive sequences, especially transposable elements (Feschotte et al. 2002). Whether somatic hybridization induces broad variations in repetitive sequences deserves further investigation. Moreover, increasing evidence shows that epigenetic modifications, such as DNA methylation, play important roles in a wide range of biological processes, including transposon inactivation and regulation of gene expression (Bird 2002; Zhang et al. 2006). Whether somatic hybridization induces epigenetic variations that affect gene expression and/or transposon activation needs further examination. Here, we used DNA profiling techniques to characterize genetic and epigenetic alterations from somatic genomic shock in six derivatives of bread wheat/tall wheatgrass somatic hybrids with different phenotypes.
Materials and Methods
Plant materials
Shanrong no. 1 (SR1), Shanrong no. 2 (SR2), Shanrong no. 3 (SR3), Shanrong no. 6 (SR6), Shanrong no. 10 (SR10), and Shanrong no. 12 (SR12) are six representative introgression lines derived from asymmetric somatic hybridization between bread wheat and tall wheatgrass (Xia et al. 2003; Chen et al. 2004a,b). By analogy with terminology applied to generations postsexual crossing, mutagenesis, and transformation, the regenerated plant postfusion is referred to as R1; segregating progeny obtained by self-fertilization in successive generations are R2, R3, R4, etc. (Figure 1). Only one to two seeds could produce from R1 and form a few R2 seedlings, but adequate R3 lines can be used for phenotypic and genomic investigation. We chose the R3 and R10 generations of the six introgression lines SR1, SR2, SR3, SR6, SR10, and SR12 for analysis. Four regenerated plants (R1–R4) were derived from protoplasts isolated from cultured embryonic calli of JN177 (R177) by the method previously described by Feng et al. (2004).
Figure 1.
Generation of somatic hybridization introgression lines. Circles represent nuclei; lines are chromosomes and chromosome segments.
Mitotic and meiotic chromosome analyses
Root-tip meristems were squashed in 45% glacial acetic acid for GISH karyotyping according to previously described methods by Xiang et al. (2003). Tall wheatgrass genomic DNA was labeled as probes and genomic DNA from the bread wheat cv. Chinese Spring was used for blocking. A probe:block ratio of 1:150 was used. For meiotic chromosome analysis, anthers were fixed, squashed, and stained according to previously described methods by Wang et al. (2004b).
Amplified fragment length polymorphism and methylation-sensitive amplification polymorphism (MSAP) fingerprinting
Amplified fragment length polymorphism (AFLP) and methylation-sensitive amplification polymorphism (MSAP) were performed according to Shaked et al. (2001), with minor modifications. A set of 44 AFLP and 13 MSAP primer combinations were used (Supporting Information, Table S1). Three independent technical replicates were performed for each of the three biological replicates for AFLP and MSAP reaction, and only intensely stained fragments >100 bp were scored.
RT–PCR analysis
RT–PCR was performed with cDNA templates prepared from 2 μg RNA extracted from seedling leaves and roots of cv. JN177 and the R3 of six introgression lines and four R177 plants treated with DNase. The first cDNA strand was synthesized with M-MLV reverse transcriptase (Invitrogen), following the manufacturer’s instructions. The PCR amplicons were separated using SSCP (single-strand conformation polymorphism) electrophoresis through 6% nondenaturing polyacrylamide gels at 4° and visualized by silver staining. The set of 80 primer pairs (Table S2) was developed by Bottley et al. (2006). Three independent technical replicates were performed for each of the three biological replicates for SSCP analysis.
Interretrotransposon amplified polymorphism and retrotransposon microsatellite amplified polymorphism genotyping
Interretrotransposon amplified polymorphism (IRAP) and retrotransposon microsatellite amplified polymorphism (REMAP) PCR were performed in 20-µl reactions, as described by Kalendar et al. (1999). The 10 IRAP and 12 REMAP primer pairs were developed by Bento et al. (2008) (Table S3). The amplicons were electrophoresed through 2% agarose gels and visualized by ethidium bromide staining. Three independent technical replicates were performed for each primer combination.
Cloning and sequencing of gel fragments
Selected gel fragments were excised from gels and reamplified using the primers from which they were generated. The amplicons were separated by electrophoresis through 1% agarose gels, gel purified, and ligated into the pEASY-T1 plasmid (Transgene), which was transformed into Escherichea coli strain DH105 for sequencing based on universal M13 primers. The sequences were BLAST against the NCBI and/or the TREP (the Triticeae Repeat Sequence Database) database.
Southern blot
Fragments with polymorphisms from parent wheat cv. JN177 and the introgression lines in AFLP, SSCP, REMAP, and IRAP analysis were reamplified from the corresponding reconstituted plasmid and labeled with DIG High Prime DNA Labeling and Detection Starter Kit I (Roche) and used as hybridization probes for DNA gel-blot analysis. Genomic DNA from the introgression lines, cv. JN177 and R177 plants, was digested with restriction enzyme EcoRI and blotted onto nylon membranes through vacuum transfer. Prehybridization, hybridization, and color detection were performed according to protocols provided by the DIG High Prime DNA labeling and detection starter kit I (Roche).
Bisulfite genomic sequencing
For bisulfite genomic sequencing, genomic DNA was processed with an EpiTect Bisulfite kit (Qiagen). The amplification primer pairs for each of MSAP-isolated fragments (MIFs) were designed using MethPrimer software (Li and Dahiya 2002). Primer sequences are given in Table S4. The PCR products were ligated with the pEASY-T Vector (TransGen) and at least 30 clones per insert processed for sequencing. The ratio of C methylation at each cytosine site was calculated and transformed into a percentage using CyMATE software (http://www.gmi.oeaw.ac.at/research-groups/cymate/cymate/).
Results
Generation of the introgression lines
Our asymmetric somatic hybridization between bread wheat cv. JN177 and tall wheatgrass produced hundreds of derivatives (Figure 1) classified into five groups according to their phenotypes (Xia et al. 2003; Zhang et al. 2005; Xia 2009). We chose the six introgression lines SR1, SR2, SR3, SR6, SR10, and SR12 to represent the five phenotypic groups (SR2 and SR3 belong to the same group), which has proven to be phenotypically stable over many generations. The first group showed high grain yield represented by SR1, and the grain yield of SR1 was 35% higher compared with JN177 under nonnutrient limiting conditions. These data correlate with the fact that SR1 is registered in China as a high yielding cultivar. The second group showed higher abiotic stress tolerance represented by SR2 and SR3; SR2 showed high levels of drought tolerance, while SR3 was noticeably more salinity tolerant than cv. JN177. In addition, SR3 showed higher grain yield compared with JN177 under nonnutrient limiting conditions; it is designated as a salinity tolerant cultivar in Shandong Province. The third group derivatives showed higher disease resistance represented by SR6; this derivative was highly resistant to powdery mildew and stripe rust. The fourth group derivatives were all dwarfs represented by SR10, and the stems of SR10 were 48.20 ± 1.31 cm, but they possessed strong tillering ability and end-use quality. The last group of derivatives showed large ears and grains. The 1000 grain weight of one member, SR12, was nearly 60 g. All measured agricultural traits of the six introgression lines are listed in Table 1.
Table 1. Phenotypic traits associated with the six introgression lines and JN177.
| Sample | Growth habit of seedling | Plant height (cm) | Spike length (cm) | No. of spikelet/spike | No. of grain/spike | Effective tillers | 1000 grain weight (g) |
|---|---|---|---|---|---|---|---|
| JN177 | Arrest | 66.90 ± 3.07 | 9.30 ± 0.63 | 19.7 ± 0.95 | 42.8 | 4.7 ± 0.95 | 37.88 |
| SR1 | Creep | 75.60 ± 2.27 | 12.95 ± 0.69 | 22.9 ± 0.74 | 77.1 | 8.4 ± 3.72 | 46.96 |
| SR2 | Creep | 74.30 ± 3.74 | 11.40 ± 0.94 | 21.8 ± 1.14 | 71.0 | 6.4 ± 1.71 | 43.17 |
| SR3 | Creep | 75.70 ± 3.05 | 14.00 ± 0.62 | 24.3 ± 1.33 | 66.7 | 5.5 ± 1.18 | 41.99 |
| SR6 | Arrest | 75.20 ± 2.90 | 8.00 ± 0.33 | 18.5 ± 0.97 | 50.8 | 6.8 ± 1.62 | 33.03 |
| SR10 | Creep | 48.20 ± 1.31 | 9.30 ± 0.48 | 16.6 ± 0.97 | 41.5 | 13.5 ± 3.54 | 38.65 |
| SR12 | Creep | 74.30 ± 3.27 | 12.90 ± 0.39 | 19.8 ± 0.79 | 54.8 | 4.9 ± 1.29 | 57.77 |
Chromosomal constitution of the six introgression lines
We investigated the introgression of Th. ponticum chromosomal segments into wheat chromosomes in the six lines. Most of the root tip mitotic cells in the R3 generation contained 42 chromosomes with a stable somatic chromosome number from generation R3 to R10 (data not shown). Karyotypes were nearly identical from cv. JN177, with some minor variations in arm length ratio and chromosome length (data not shown). According to genomic in situ hybridization (GISH) analysis, we found several small tall wheatgrass chromosomal segments present in each line (Figure 2). We also found >90% ring bivalent and >95% total bivalent frequencies in the six introgression lines at meiosis (Table S5). This suggests that introgressed tall wheatgrass chromosomal segments had little effect on chromosome pairing behavior, likely due to the small segment size. Given that the karyotypes and DNA fingerprinting profiles were stable for each introgression line between the R3 and R10 generations (data not shown), all of the following analysis was restricted to the DNA of R3 plants.
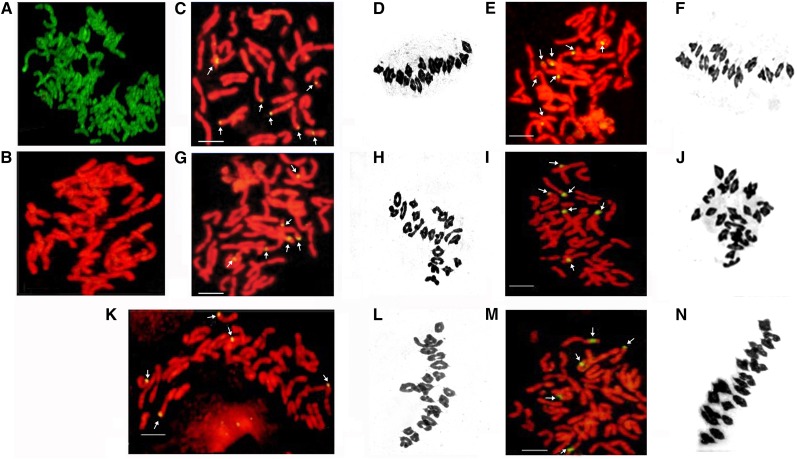
Figure 2.
Cytogenetic analysis of the six introgression lines at R3. GISH karyotypes of (A) tall wheatgrass and (B) bread wheat cv. JN177, with tall wheatgrass genomic DNA as probe. SR1: (C) GISH karyotype, (D) chromosome configuration at meiotic metaphase I. SR2: (E) GISH karyotype, (F) chromosome configuration at meiotic metaphase I. SR3: (G) GISH karyotype, (H) chromosome configuration at meiotic metaphase I. SR6: (I) GISH karyotype, (J) chromosome configuration at meiotic metaphase I. SR10: (K) GISH karyotype, (L) chromosome configuration at meiotic metaphase I. SR12: (M) GISH karyotype, (N) chromosome configuration at meiotic metaphase I.
Frequency of sequence variation as estimated From AFLP profiles
In this study, only a few tall wheatgrass segments were introgressed into the wheat genome. Our expectation was that the AFLP profiles of the introgression lines would be similar to that of cv. JN177, with the addition of some tall wheatgrass-derived fragments. We focused on fragments present in JN177 but lacking in one, or more, of the introgression lines, and the presence of fragments in the introgression line(s) but not in JN177. Only fragments >100 bp were scored.
AFLP profiling revealed 904 cv. JN177 and 777 tall wheatgrass fragments; 388 were shared by both templates (Figure 3, A and B). Given that AFLP assay markers are dominant, the level (percentage) of cv. JN177 fragment loss could be related only to the 516 (904–388) JN177 fragments that did not comigrate with a tall wheatgrass fragment. The number of fragments lost from the profile of the six introgression lines ranged from 22 to 28, with an overall frequency of 4.7% (Table 2). In addition, the six introgression lines displayed 12 to 18 new fragments not present in the cv. JN177 profile, giving an overall frequency of 2.9% (Table 2). AFLP assay results for cv. JN177 and four R177 lines revealed an average of only 3.5 deleted and 2.0 new bands compared with JN177 (Table 2).
Figure 3.
AFLP profile and Southern blot analysis of deleted fragments. (A) AFLP profile based on primer combination EAGC + MCTT, (B) primer combination EAAT + MCTT. Fragments absent from the introgression lines or regenerated plant from JN177 calli (R) are indicated by arrowheads and novel fragments indicated by arrows. Southern blots verified the deletion of genomic DNA in SR6 with probes derived from primer combination EATC + MCTT (C), but not with probes derived from primer combination EAGG + MCTT (D), EAGC + MCTT (E), and EAAA + MCTT (F). 1, SR1; 2, SR2; 3, SR3; 6, SR6; 10, SR10; 12, SR12; 177, parent wheat cv. JN177. R, regenerated JN177 plant R1; Th, tall wheatgrass Th. ponticum; L, the 100-bp DNA ladder marker.
Table 2. AFLP profiles of cv. JN177, the six introgression lines, and regenerated JN177 plants (R177).
| Introgression lines | SR1 | SR2 | SR3 | SR6 | SR10 | SR12 | Average | Frequency (%) |
|---|---|---|---|---|---|---|---|---|
| Deletions in introgression lines | 23 | 22 | 26 | 25 | 28 | 22 | 24.3 | 4.7 |
| New bands in introgression lines | 16 | 15 | 18 | 16 | 12 | 13 | 15.0 | 2.9 |
| Regenerated plants | R1 | R2 | R3 | R4 | ||||
| Deletions in R177 | 5 | 3 | 3 | 3 | 3.5 | 0.7 | ||
| New bands in R177 | 1 | 1 | 2 | 4 | 2.0 | 0.4 |
The AFLP-observed fragment changes may have resulted from several mechanisms including sequence change at restriction sites, methylation alteration, or sequence elimination induced by somatic hybridization. To distinguish between these possibilities, we isolated four bands deleted in some of the introgression lines from acrylamide AFLP gels and used them as probes for Southern blot analysis. The results confirm deletion of one band, with a single-copy fragment (Figure 3C). The remaining three fragments, which had low-copy DNA, showed no obvious differences between JN177 and the introgression lines (Figure 3, D–F). Thus, variations found in AFLP analysis are likely due mainly to sequence changes at restriction sites or from methylation alteration, rather than sequence elimination.
Somatic hybridization induced changes in methylation pattern
The isoschizomers MspI and HpaII share the same recognition sequence, but differ in their sensitivity to C methylation. HpaII activity is suppressed when the internal cytosine at the CCGG recognition site is methylated. Therefore, C methylation differences can be detected by MSAP analysis, comparing profiles generated by EcoRI–MspI and EcoRI–HpaII digestion (Shaked et al. 2001). The standards applied to scoring MSAP profiles and AFLP analyses are similar. The 13 MSAP primer combinations amplified 347 fragments, of which 210 were present in cv. JN177 but not in the tall wheatgrass parent (Figure 4). Of the 210, the number of hypermethylated loci in different introgression lines ranged from 16 to 27, giving an overall frequency of 10.5% (Table 3). Among the loci methylated in cv. JN177, 35 were hypomethylated in SR1, 33 in SR3, 29 in SR6, 22 in SR10, and 23 in both SR2 and SR12, giving an overall frequency of 13.1% (Table 3). The C methylation in cv. JN177 and four R177 lines were also detected by MSAP analysis; the results indicated that only 5.5 loci hypermethylated in the regenerated plant and 8.0 loci hypomethylated (Table 3). The sequencing of five of the fragments hypomethylated in the introgression lines showed that three were high-copy-number retrotransposons (TREP accession nos. TREP255, TREP1418, TREP3251), while the other two showed no significant similarity to any known sequence. None of the five sequenced fragments that were hypermethylated in the introgression lines showed any significant similarity to DNA sequences of publicly available databases.
Figure 4.
MSAP profiling based on primer combinations EATC + HMTCAA. M, DNA restricted by EcoRI + MspI; H, EcoRI + HpaII. Arrows indicate hypermethylated fragments and arrowheads indicate hypomethylated fragments. 1, SR1; 2, SR2; 3, SR3; 6, SR6; 10, SR10; 12, SR12; 177, parent wheat cv. JN177. R, regenerated JN177 plant R1; Th, tall wheatgrass Th. ponticum; L, the 100-bp DNA ladder marker.
Table 3. Variation in C methylation in the introgression lines and regenerated JN177 plants (R177).
| Introgression lines | SR1 | SR2 | SR3 | SR6 | SR10 | SR12 | Average | Frequency (%) |
|---|---|---|---|---|---|---|---|---|
| Hypermethylation in introgression lines | 27 | 23 | 24 | 23 | 16 | 19 | 22.0 | 10.5 |
| Hypomethylation in introgression lines | 35 | 23 | 33 | 29 | 22 | 23 | 27.5 | 13.1 |
| Regenerated plants | R1 | R2 | R3 | R4 | ||||
| Hypermethylation in R177 | 5 | 9 | 5 | 3 | 5.5 | 2.6 | ||
| Hypomethylation in R177 | 5 | 7 | 9 | 11 | 8.0 | 3.8 |
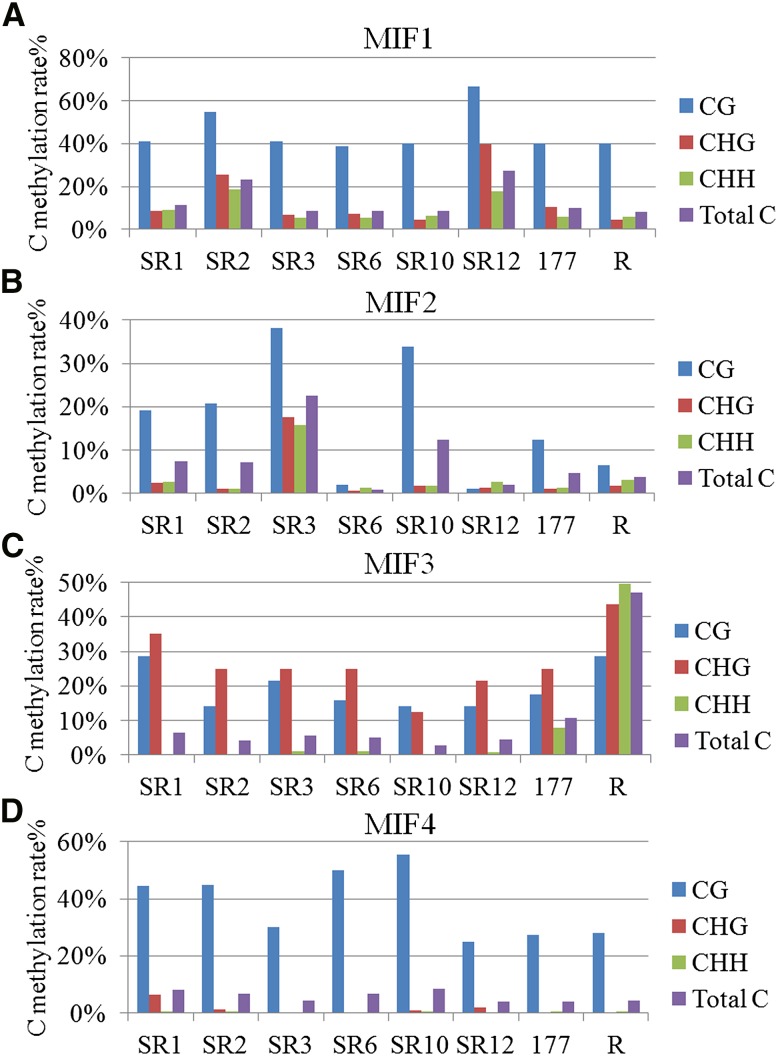
To further investigate the C-methylation changes induced by somatic hybridization in the introgression lines, C-methylation status in several variant MSAP bands was identified by bisulfite sequencing. We found the methylation rates at CG, CHG, and CHH sites of MSAP-isolated fragment 1 (MIF1) were all elevated in SR2 and SR12 (Figure 5A). The level of C methylation at CG sites of MIF2 was elevated in SR1, SR2, SR3, and SR10 but reduced in SR6 and SR12, although the CHG and CHH sites were also hypermethylated in SR3 (Figure 5B). Moreover, the level of C methylation at CG and CHG sites of MIF3 was elevated in SR1, and those at CG, CHG, and CHH sites were all elevated in R177 lines (Figure 5C). The level of C methylation at CG sites of MIF4 was elevated in SR1, SR2, SR6, and SR10 (Figure 5D).
Figure 5.
C-methylation status in the MSAP-isolated fragments (MIFs) as identified by bisulfite sequencing. (A) The level of C methylation at CG, CHG, and CHH sites of MIF1 was all elevated in SR2 and SR12. (B) The level of C methylation at CG sites of MIF2 was elevated in SR3 and SR10 but reduced in SR6 and SR12 compared to JN177; those at CHG and CHH sites were also elevated in SR3. (C) The level of C methylation at CG and CHG sites of MIF3 was elevated in SR1 and those at CG, CHG, and CHH sites were all elevated in regenerated JN177 plant R1. (D) The level of C methylation at CG sites of MIF4 was elevated in SR1, SR2, SR6, and SR10 compared to JN177. 177, parent wheat cv. JN177. R, regenerated JN177 plant R1.
Gene expression alterations as estimated by SSCP
To survey the homeologous gene silencing or activation induced by somatic hybridization, we used SSCP to compare the amplification profiles of the six introgression lines with those of seedling shoot and root RNA extracted from cv. JN177 and R177. Expression variation was noted for 11 of the 80 targeted sequences. Eight of these involved the loss of a specific homeologous product in at least one introgression line (Figure 6, A–D). In two cases, a specific mRNA was present in some introgression lines but not in cv. JN177 (Figure 6C). In three cases, the introgression line profiles included a variant of one of the cv. JN177 fragments (Figure 6D). To test whether genetic variations were responsible for the changes of the expression, genomic DNA was also included as a template. In four of the eight genes involved in homeologous gene silencing, the corresponding fragment was also absent from the genomic DNA profile (Figure 6, A and D). Southern blot analysis based on genomic DNA showed that one of the homeologues of BE490384, BE494911, and BE606965 had been deleted in the introgression lines (Figure 6, E–G). But, this was not the case for BE399113 (Figure 6H). No variation was found between cv. JN177 and the four R177 lines in all 80 targeted sequences.
Figure 6.
RT–PCR and genomic DNA characterization of differentially expressed genes. (A) One homoeoallele of BE494911 is silenced in SR3 and SR10 while another homoeoallele silenced in SR6 and root of SR12; the silencing of BE494911 in SR3, SR6, and SR10 is caused by a deletion in the coding region. (B) BE202265 silenced in shoot and root of SR1, SR2, SR3 and shoot of SR12. (C) One homoeoallele of BE606719 is activated in root of SR2, while another homoeoallele silenced in SR10 root. (D) For the gene, BE606965, one homeologous copy differs from that of cv. JN177 in SR1, SR2, SR6, and SR10, while another homoeoallele is deleted in the coding region of SR2, SR6, and SR10. Southern blots demonstrate the deletion of genomic DNA in (E) BE490384, (F) BE494911, and (G) BE606965, but not in (H) BE399113. Arrows indicate the genomic fragments deleted in the introgression lines. 1, SR1; 2, SR2; 3, SR3; 6, SR6; 10, SR10; 12, SR12; 177, parent wheat cv. JN177; R, regenerated JN177 plant R1; Th, tall wheatgrass Th. ponticum.
Genomic rearrangements in retrotransposon-adjacent regions
IRAP and REMAP assays were developed to detect variation in sequence-flanking retrotransposons (Kalendar et al. 1999). We used these two techniques to identify genome rearrangements from retrotransposition or inter- and intra-element recombination. Here, 219 wheat specific loci were detected. Five were lost in SR1, SR2, and SR3; 8 were lost in SR6 and SR12; 11 were lost in SR10, with an overall mean frequency of lost fragments across the six introgression lines at 3.2% (Figure 7, A–D and Table 4). An average of 1.5 fragments was lost in R177 plants when compared with cv. JN177 (Table 4). Seven novel fragments were found in SR1, SR2, and SR12 that were not amplified in both cv. JN177 and tall wheatgrass. Additionally, 6, 5, and 12 novel fragments also appeared in SR3, SR6, and SR10, respectively. These values give an overall new-band frequency of 3.3% (Figure 7, A-D, Table 4). Only 1.0 novel fragment appeared in R177 plants when compared with cv. JN177 (Table 4).
Figure 7.
REMAP and IRAP analysis, based on primer combinations (A) (GA)9C + sukkula, (B) (GA)9C + sabrina, (C) nikita + sukkula, (D) nikita + sabrina. Southern blots demonstrate the retrotransposon-related deletions in the introgression lines with probes derived from (CA)9G + sabrina (E) and nikita + sukkula (F). Arrowheads indicate retrotransposon-related deletions in the introgression lines induced by asymmetric somatic hybridization, while arrows indicate retrotransposon-related insertions in the introgression lines induced by asymmetric somatic hybridization. 1, SR1; 2, SR2; 3, SR3; 6, SR6; 10, SR10; 12, SR12; 177, parent wheat cv. JN177; R, regenerated JN177 plant R1; Th, tall wheatgrass Th. ponticum.
Table 4. Genomic rearrangements in retrotransposon related regions in the introgression lines and regenerated JN177 plants (R177).
| Introgression lines | SR1 | SR2 | SR3 | SR6 | SR10 | SR12 | Average | Frequency (%) |
|---|---|---|---|---|---|---|---|---|
| Bands lost in introgression lines | 5 | 5 | 5 | 8 | 11 | 8 | 7.0 | 3.2 |
| New bands in introgression lines | 7 | 7 | 6 | 5 | 12 | 7 | 7.3 | 3.3 |
| Regenerated plants | R1 | R2 | R3 | R4 | ||||
| Bands lost in R177 | 2 | 2 | 1 | 1 | 1.5 | 0.7 | ||
| New bands in R177 | 1 | 0 | 1 | 2 | 1.0 | 0.5 |
Southern blot analysis based on genomic DNA using polymorphic fragments isolated from IRAP and REMAP gels as probes shows that the lost fragments were deleted in some of the introgression lines (Figure 7, E and F). We sequenced a sample of these fragments to identify the genes associated with these genomic rearrangements. A fragment present in cv. JN177, but absent in some of the introgression lines, resembled a Ty3-gypsy subclass retrotransposon (ABA97230.1). A second polymorphic fragment (present in some of the introgression lines, but not in cv. JN177) appeared to be highly homologous to a wheat pore-forming toxin-like protein Hfr-2 (AAW48295.1) involved in plant defense (Puthoff et al. 2005).
Discussion
Merging divergent genomes into a single nucleus can trigger a “highly-programmed sequence of events within the cell that serves to cushion the effect of [genomic] shock” (McClintock 1984). However, the mechanisms of genomic shock are not well understood. The introgression of alien chromosome fragments can also lead to severe effects in the accepting genome, which trigger genetic and genomic changes. Exploring genetic and genomic changes induced by somatic genomic shock in the somatic introgression lines will be helpful for understanding the nature of genomic shock. Here, we demonstrated that the somatic hybridization process induced widespread genetic and epigenetic changes including sequence absence, modified regulation of gene expression, alteration of cytosine methylation patterns, and the activation of quiescent retrotransposons. Similar genetic and/or epigenetic alterations occurred following the formation of de novo polyploids involving Arabidopsis spp. (Comai et al. 2000; Madlung et al. 2002; Wang et al. 2004a), Brassica spp. (Song et al. 1995), Spartina spp. (Salmon et al. 2005), Tragopogon spp. (Tate et al. 2006), and Triticeae spp. (Ozkan et al. 2001; Shaked et al. 2001; Han et al. 2003; Ma et al. 2004). However, the somatic genomic shock induced by somatic hybridization might be stronger than those in allopolyploids given the former resulted in more genetic and epigenetic instabilities. In somatic hybrids, the requirement for a period of in vitro culture adds an additional potential source of variation. However, the contrast made between the DNA extracted from cv. JN177 and R177 plants implies that somaclonal variation was responsible for only a small proportion of the overall variation induced in the introgression lines. Thus, genome introgression was likely responsible for most genetic and epigenetic variations. Although we cannot exclude the effect of organellar genome introgression, considering the limited amount of alien organellar DNA in the hybrids (Chen et al. 2004b), the introgression of nuclear DNA was likely responsible for most genetic and epigenetic variations.
Sequence absence
One of the most common responses to polyploidization is the rapid elimination of DNA sequences (Feldman et al. 1997; Liu et al. 1998a,b; Ozkan et al. 2001; Shaked et al. 2001; Kashkush et al. 2002; Ma et al. 2004; Ma and Gustafson 2006). The frequency of elimination is particularly marked in synthetic tetraploid wheat and triticale (Shaked et al. 2001; Ma et al. 2004; Ma and Gustafson 2006). In our study, sequence loss was also observed following somatic hybridization (Figure 3), but at a rather lower rate. Only 4.7% of the cv. JN177 AFLP fragments were not recovered in the somatic introgression lines, while 0.7% were concluded to have been induced by somaclonal variation (Table 2). Moreover, Southern blot analysis of the lost AFLP fragments indicated that only a few were eliminated from the genome of the introgression lines. Most of the band absence may be due to sequence changes of restriction sites or from methylation alterations. Sequence elimination during allopolyploidization provides the physical basis for the diploid-like meiotic behavior of newly formed allopolyploids (Ozkan et al. 2001). Unlike allopolyploidization, where the whole parental genomes are combined, the size of the individual donor chromosome fragments introgressed into cv. JN177 via asymmetric somatic hybridization is too small to affect meiotic chromosome pairing (Figure 2). Thus, meiotic pressure is likely to be lower compared with a polyploid. This may partly explain why sequence elimination is not a predominant effect in somatic hybridization, unlike in polyploidization.
Epigenetic alterations
Patterns of gene expression are affected by allopolyploidization, particularly in wheat, where 1–5% of the donor genes have altered expression in a synthetic allotetraploid, largely via epigenetic changes (Kashkush et al. 2002). Changes in gene expression are mediated at either the transcriptional and/or the post-transcriptional levels. Both transcriptional and post-transcriptional gene silencing are associated with alterations to DNA methylation (Paszkowski and Whitham 2001). In newly synthesized allohexaploid wheat, ∼13% of the loci experience an alteration in cytosine methylation (Shaked et al. 2001). A commonality between the effect of somatic hybridization and allopolyploidization appears to be that both result in changes to cytosine methylation, which results in the perturbation of gene expression (Comai et al. 2000; Shaked et al. 2001; Kashkush et al. 2002; Han et al. 2003). However, the epigenetic variations induced by somatic hybridization are stronger than those by allopolyploidization. Based on the analysis of our sample of 80 cDNAs, altered expression was induced by the somatic hybridization procedure in 10% of the introgression line transcripts (Figure 6). The notable frequency of cytosine methylation changes observed in the asymmetric somatic hybrids (23.6%) is consistent with large-scale epigenetic regulation (Table 3). Thus, we concluded that somatic hybridization induced a broad spectrum of C-methylation changes that perturbed gene expression to a larger extent than allopolyploidization.
Activation and repression of transposon activity
Genomic shock activates quiescent transposons and causes genome restructuring (McClintock 1984). In fact, transcriptional activation of transposable elements has been shown in some newly synthesized allopolyploids (Kashkush et al. 2002, 2003). IRAP and REMAP both represent multilocus PCR-based methods designed to detect retrotransposon related genome rearrangements and have been used to uncover sequence rearrangements associated with retrotransposon-rich regions in triticale (Bento et al. 2008, 2010). Here, 6.5% of the IRAP and REMAP fragments were polymorphic between cv. JN177 and the introgression lines (Figure 7 and Table 4). The introgression events were associated with novel sequence polymorphisms at the junctions between the donor and the recipient DNA. However, given the overall number of wheatgrass segments present in the introgression lines, frequency of polymorphisms revealed by IRAP and REMAP was too high to merely reflect introgression (Figure 2). Instead, the polymorphisms detected by IRAP and REMAP likely arose from de novo transposition events. These have been detected in the vicinity of genes encoding certain endosperm storage proteins by Liu et al. (2009).
Evidence suggests that C methylation has evolved as a means of repressing transposon activity. Clearly, highly repetitive DNA—much of which is contributed by retrotransposons in the large genome grass species—in plant genomes is heavily methylated (Bennetzen et al. 1994). The activity of retrotransposons needs to be tightly controlled to ensure viability and survival of the host, and a major control step involves the inhibition of transcription. Reactivation of transposons has been observed either as a response to stress or to genomic shock (Bennetzen 2000; Liu and Wendel 2000; Kashkush et al. 2002, 2003; Shan et al. 2005, 2009; Bento et al. 2008, 2010). Our MSAP analysis of the introgression lines indicates that alteration in C methylation involves repetitive sequences. In particular, the hypomethylation of retrotransposons revealed is suggestive of retrotransposon activation in the introgression lines.
Although considerable transposition occurred during the somatic hybridization event, the introgression lines were highly stable from R3 to R10 generation. The implied repression of retrotransposon activity closely resembles the behavior of rice introgression lines (Liu and Wendel 2000), although the underlying mechanism is unknown. Wolffe and Matzke (1999) have proposed the involvement of homology-dependent gene silencing (HDGS), an epigenetic phenomenon whereby one gene is suppressed by the action of a homologous gene (Zaratiegui et al. 2007). When transposon activity is activated by hypomethylation, HDGS is triggered and similar sequences become targets for suppression. How important the rapid repression of retrotransposon activity is for the stabilization of the introgression lines following somatic hybridization clearly needs further investigation.
Genetic alterations driven by repetitive sequences
Repetitive sequences are a major source of genome instability as they are susceptible to expansion and contraction (Heidenfelder and Topal 2003). As the most abundant repetitive sequences in wheat genome, the retrotransposon-related sequences showed high frequency of alteration (6.5%) in the introgression lines (Table 4). Similarly, high-frequency sequence variation through indels of repetitive motifs (14/37, 37.8%) was also observed in the glutenin gene family (Feng et al. 2004; Liu et al. 2007, 2009; Gao et al. 2010). This supports the notion that repetitive sequences are a driving force behind the de novo genetic variation generated in the introgression lines. Double-strand breaks (DSBs) appear to be common in regions enriched for repetitive sequences, and these “fragile sites” represent hotspots for recombination (Wicker et al. 2010). The suggestion is therefore that repetitive sequences drive the formation of DSBs or other genetic alterations that occur as a result of genomic shock and subsequent introgression.
Conclusions
We generated a set of introgression lines displaying a variety of genetic and epigenetic changes relative to their parents using somatic hybridization between wheat and tall wheatgrass. The somatic hybridization process mimics many of the genetic alterations induced by polyploidization or sexual wide hybridization, in a remarkably short time frame and with stronger extent. Therefore, we suggest the somatic hybridization approach not only provides an effective and time-efficient means of achieving introgression into a crop species from its wild relatives, but also provides a means to explore the nature of the genetic and epigenetic events induced by somatic genomic shock. An understanding of the underlying mechanisms may shed light on the variation released in sexual wild hybrids, which has been broadly described as being due to genomic shock.
Supplementary Material
Acknowledgments
We thank Dr. Austin Cape for careful reading and feedback. This work was supported by the funds of the Natural Science Foundation of China (no. 30871320; 31000568), Major Program of the Natural Science Foundation of China (no. 31030053), Shandong Province Program (no. Q2006D02).
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.174094/-/DC1.
Communicating editor: J. A. Birchler
Literature Cited
- Bassene J. B., Froelicher Y., Dubois C., Ferrer R. M., Navarro L., et al. , 2010. Non-additive gene regulation in a citrus allotetraploid somatic hybrid between C. reticulata Blanco and C. limon (L.) Burm. Heredity 105: 299–308. [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L., 2000. Transposable element contributions to plant gene and genome evolution. Plant Mol. Biol. 42: 251–269. [PubMed] [Google Scholar]
- Bennetzen J. L., Schrick K., Springer P. S., Brown W. E., SanMiguel P., 1994. Active maize genes are unmodified and flanked by diverse classes of modified, highly repetitive DNA. Genome 37: 565–576. [DOI] [PubMed] [Google Scholar]
- Bento M., Pereira H. S., Rocheta M., Gustafson P., Viegas W., et al. , 2008. Polyploidization as a retraction force in plant genome evolution: sequence rearrangements in triticale. PLoS ONE 3: e1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento M., Gustafson P., Viegas W., Silva M., 2010. Genome merger: from sequence rearrangements in triticale to their elimination in wheat–rye addition lines. Theor. Appl. Genet. 121: 489–497. [DOI] [PubMed] [Google Scholar]
- Bird A. P., 1997. Does DNA methylation control transposition of selfish elements in the germline. Trends Genet. 13: 469–470. [DOI] [PubMed] [Google Scholar]
- Bird A. P., 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16: 6–21. [DOI] [PubMed] [Google Scholar]
- Bottley A., Xia G. M., Koebner R. M. D., 2006. Homoeologous gene silencing in hexaploid wheat. Plant J. 47: 897–906. [DOI] [PubMed] [Google Scholar]
- Chen S. Y., Xia G. M., Quan T. Y., Xiang F. N., Jin Y., et al. , 2004a Introgression of salt-tolerance from somatic hybrids between common wheat and Thinopyrum ponticum. Plant Sci. 167: 773–779. [Google Scholar]
- Chen S. Y., Liu S. W., Xu C. H., Chen Y. Z., Xia G. M., 2004b Heredity of chloroplast and nuclear genomes of asymmetric somatic hybrid lines between wheat and couch grass. Acta Bot. Sin. 46: 110–115. [Google Scholar]
- Cheng A. X., Xia G. M., Zhi D. Y., Chen H. M., 2004. Intermediate fertile Triticum aestivum (+) Agropyron elongatum somatic hybrids are generated by low doses of UV irradiation. Cell Res. 14: 86–91. [DOI] [PubMed] [Google Scholar]
- Comai L., Tyagi A. P., Winter K., Holmes-Davis R., Reynolds S. H., et al. , 2000. Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell 12: 1551–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M., Liu B., Segal G., Abbo S., Levy A. A., et al. , 1997. Rapid elimination of low-copy DNA sequences in polyploid wheat: A possible mechanism for differentiation of homoeologous chromosomes. Genetics 147: 1381–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D. S., Xia G. M., Zhao S. Y., Chen F. G., 2004. Two quality-associated HMW glutenin subunits in a somatic hybrid line between Triticum aestivum and Agropyron elongatum. Theor. Appl. Genet. 110: 136–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C., Jiang N., Wessler S. R., 2002. Plant transposable elements: where genetics meets genomics. Nat. Rev. Genet. 3: 329–341. [DOI] [PubMed] [Google Scholar]
- Gao X., Liu S. W., Sun Q., Xia G. M., 2010. High frequency of HMW-GS sequence variation through somatic hybridization between Agropyron elongatum and common wheat. Planta 231: 245–250. [DOI] [PubMed] [Google Scholar]
- Griffiths S., Sharp R., Foote T. N., Bertin I., Wanous M., et al. , 2006. Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439: 749–752. [DOI] [PubMed] [Google Scholar]
- Han F. P., Fedak G., Ouellet T., Liu B., 2003. Rapid genomic changes in interspecific and intergeneric hybrids and allopolyploids of Triticeae. Genome 46: 716–723. [DOI] [PubMed] [Google Scholar]
- Heidenfelder B. L., Topal M. D., 2003. Effects of sequence on repeat expansion during DNA replication. Nucleic Acids Res. 31: 7159–7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush K., Feldman M., Levy A. A., 2002. Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 160: 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush K., Feldman M., Levy A. A., 2003. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 33: 102–106. [DOI] [PubMed] [Google Scholar]
- Kalendar R., Grob T., Regina M., Suoniemi A., Schulman A., 1999. IRAP and REMAP: two new retrotransposon-based DNA fingerprinting techniques. Theor. Appl. Genet. 98: 704–711. [Google Scholar]
- Li L. C., Dahiya R., 2002. MethPrimer: designing primers for methylation PCRs. Bioinformatics 18: 1427–1431. [DOI] [PubMed] [Google Scholar]
- Liu B., Wendel J. F., 2000. Retrotransposon activation followed by rapid repression in introgressed rice plants. Genome 43: 874–880. [PubMed] [Google Scholar]
- Liu B., Vega M. J., Feldman M., 1998a Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. II. Changes in low-copy coding DNA sequences. Genome 41: 535–542. [DOI] [PubMed] [Google Scholar]
- Liu B., Vega J. M., Segal G., Abbo S., Rodova M., et al. , 1998b Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. I. Changes in low-copy non-coding DNA sequences. Genome 41: 272–277. [DOI] [PubMed] [Google Scholar]
- Liu B., Brubaker C. L., Mergeai G., Cronn R. C., Wendel J. F., 2001. Polyploid formation in cotton is not accompanied by rapid genomic changes. Genome 44: 321–330. [PubMed] [Google Scholar]
- Liu H., Liu S. W., Xia G. M., 2009. Generation of high frequency of novel alleles of the high molecular weight glutenin in somatic hybridization between bread wheat and tall wheatgrass. Theor. Appl. Genet. 118: 1193–1198. [DOI] [PubMed] [Google Scholar]
- Liu S. W., Xia G. M., 2014. The place of asymmetric somatic hybridization in wheat breeding. Plant Cell Rep. 33: 595–603. [DOI] [PubMed] [Google Scholar]
- Liu S. W., Zhao S. Y., Chen F. G., Xia G. M., 2007. Generation of novel high quality HMW-GS genes in two introgression lines of Triticum aestivum/Agropyron elongatum. BMC Evol. Biol. 7: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X. F., Gustafson J. P., 2006. Timing and rate of genome variation in triticale following allopolyploidization. Genome 49: 950–958. [DOI] [PubMed] [Google Scholar]
- Ma X. F., Fang P., Gustafson J. P., 2004. Polyploidization-induced genome variation in triticale. Genome 47: 839–848. [DOI] [PubMed] [Google Scholar]
- Madlung A., Masuelli R. W., Watson B., Reynolds S. H., Davison J., et al. , 2002. Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allotetraploids. Plant Physiol. 129: 733–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B., 1984. The significance of responses of the genome to challenge. Science 226: 792–801. [DOI] [PubMed] [Google Scholar]
- Ozkan H., Levy A. A., Feldman M., 2001. Alloploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13: 1735–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkowski J., Whitham S. A., 2001. Gene silencing and DNA methylation processes. Curr. Opin. Plant Biol. 4: 123–129. [DOI] [PubMed] [Google Scholar]
- Puthoff D. P., Sardesai N., Subramanyam S., Nemacheck J. A., Williams C. E., 2005. Hfr-2, a wheat cytolytic toxin-like gene, is up-regulated by virulent Hessian fly larval feeding. Mol. Plant Pathol. 6: 411–423. [DOI] [PubMed] [Google Scholar]
- Salmon A., Ainouche M. L., Wendel J. F., 2005. Genetic and epigenetic consequences of recent hybridization and polyploidy in Spartina (Poaceae). Mol. Ecol. 14: 1163–1175. [DOI] [PubMed] [Google Scholar]
- Shaked H., Kashkush K., Ozkan H., Feldman M., Levy A. A., 2001. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13: 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X. H., Liu Z. L., Dong Z. Y., Wang Y. M., Chen Y., et al. , 2005. Mobilization of the active MITE transposons mPing and Pong in rice by introgression from wild rice (Zizania latifolia Griseb.). Mol. Biol. Evol. 22: 976–990. [DOI] [PubMed] [Google Scholar]
- Shan X. H., Ou X. F., Liu Z. L., Dong Y. Z., Lin X. Y., et al. , 2009. Transpositional activation of mPing in an asymmetric nuclear somatic cell hybrid of rice and Zizania latifolia was accompanied by massive element loss. Theor. Appl. Genet. 119: 1325–1333. [DOI] [PubMed] [Google Scholar]
- Song K., Lu P., Tang K., Osborn T. C., 1995. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. USA 92: 7719–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate J. A., Ni Z., Scheen A.-C., Koh J., Gilbert C. A., et al. , 2006. Evolution and expression of homeologous loci in Tragopogon miscellus (Asteraceae), a recent and reciprocally formed allopolyploid. Genetics 173: 1599–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tian L., Madlung A., Lee H.-S., Chen M., et al. , 2004a Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics 167: 1961–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Xiang F. N., Xia G. M., 2004b Transfer of small chromosome fragments of Agropyron elongatum to wheat chromosome via asymmetric somatic hybridization. Sci. China Ser. C. 47: 434–441. [DOI] [PubMed] [Google Scholar]
- Wicker T., Buchmann J. P., Keller B., 2010. Patching gaps in plant genomes results in gene movement and erosion of colinearity. Genome Res. 20: 1229–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A. P., Matzke M. A., 1999. Epigenetics: regulation through repression. Science 286: 481–486. [DOI] [PubMed] [Google Scholar]
- Xia G. M., 2009. Progress of chromosome engineering mediated by asymmetric somatic hybridization. J. Genet. Genomics 36: 547–556. [DOI] [PubMed] [Google Scholar]
- Xia G. M., Xiang F. N., Zhou A. F., Wang H., Chen H. M., 2003. Asymmetric somatic hybridization between wheat (Triticum aestivum L.) and Agropyron elongatum (Host). Nevishi. Theor. Appl. Genet. 107: 299–305. [DOI] [PubMed] [Google Scholar]
- Xiang F. N., Xia G. M., Chen H. M., 2003. Effect of UV dosage on somatic hybridization between common wheat (Triticum aestivum L.) and Avena sativa L. Plant Sci. 164: 697–707. [DOI] [PubMed] [Google Scholar]
- Xiang F. N., Xia G. M., Zhi D. Y., Wang J., Nie H., et al. , 2004. Hybrid plant regeneration in relation to the nuclear and cytoplasmic genomes of wheat and Setaria italica. Genome 47: 680–688. [DOI] [PubMed] [Google Scholar]
- Xu S. X., Cai D. F., Tan F. Q., Fang Y. N., Xie K. D., et al. , 2014. Citrus somatic hybrid: an alternative system to study rapid structural and epigenetic reorganization in allotetraploid genomes. Plant Cell Tissue Organ Cult. 119: 511–522. [Google Scholar]
- Zaratiegui M., Irvine D. V., Martienssen R. A., 2007. Noncoding RNAs and gene silencing. Cell 128: 763–776. [DOI] [PubMed] [Google Scholar]
- Zhang Q. L., Xia G. M., Quan T. Y., Zhang Y. G., Li F. X., et al. , 2005. Trait variation of somatic hybrid progenies from Triticum aestivum/Agropyron elongatum Host (Nevski) (in Chinese with English Abstract). J. Triticeae Crop. 25(6): 11–14. [Google Scholar]
- Zhang X. Y., Yazaki J., Sundaresan A., Cokus S., Chan S. W., et al. , 2006. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126: 1189–1201. [DOI] [PubMed] [Google Scholar]
- Zhou A. F., Xia G. M., 2005. Introgression of the Haynaldia villosa genome into γ-ray-induced asymmetric somatic hybrids of wheat. Plant Cell Rep. 24: 289–296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.