Abstract
In Neurospora, genes not paired during meiosis are targeted by meiotic silencing by unpaired DNA (MSUD). Here, our bimolecular fluorescence complementation (BiFC) study suggests that RNA-directed RNA polymerase, Dicer, Argonaute, and others form a silencing complex in the perinuclear region, with intimate interactions among the majority of them. We have also shown that SAD-2 is likely the anchor for this assembly.
Keywords: RNA interference, Neurospora crassa, meiotic silencing by unpaired DNA (MSUD), bimolecular fluorescence complementation (BiFC), fluorescent protein tagging
THE filamentous fungus Neurospora crassa consists of a hyphal network where nuclei and other cellular components share a common cytoplasm. This trait, while beneficial for distributing resources, may promote the spread of detrimental elements such as transposons and viruses. Perhaps for this reason, Neurospora possesses several surveillance mechanisms that operate during different phases of its life cycle (Aramayo and Selker 2013; Billmyre et al. 2013; Nicolás and Ruiz-Vázquez 2013). For example, quelling silences tandem transgenes during vegetative growth (Romano and Macino 1992). Repeat-induced point mutation (RIP), a premeiotic system functioning before nuclear fusion, introduces C → T mutations to duplicated DNA sequences (Cambareri et al. 1989). Finally, meiotic silencing by unpaired DNA (MSUD) targets genes not paired with a homologous partner during meiosis (Shiu et al. 2001). MSUD begins inside the nucleus when an unpaired gene is detected (Samarajeewa et al. 2014) and acts as a template for the production of an aberrant RNA (aRNA; abnormal RNA that would enter the silencing pathway). A working model for MSUD holds that the aRNA is exported to the perinuclear region, where it is met by a host of RNAi-related proteins, including SAD-1, an RNA-directed RNA polymerase (RdRP) that converts aRNAs into double-strand RNAs (dsRNAs) (Shiu and Metzenberg 2002); SAD-3, a helicase thought to increase SAD-1’s processivity on RNA templates (Hammond et al. 2011a); DCL-1, an RNAse III Dicer that cuts dsRNAs into small interfering RNAs (siRNAs) (Alexander et al. 2008); QIP, an exonuclease that converts siRNAs into single strands (Xiao et al. 2010); and SMS-2, an Argonaute protein that uses siRNAs to guide selective destruction of homologous mRNAs (Lee et al. 2003). SAD-2, unlike the others, is not found in other RNA silencing systems but is essential for the perinuclear localization of SAD-1 (Shiu et al. 2006).
Understanding how proteins interact is a key aspect to revealing how complex cellular processes, like MSUD, function at the molecular level. Numerous methods exist to study protein–protein interactions, e.g., the yeast two-hybrid system (Y2H), co-immunoprecipitation (Co-IP), fluorescence resonance energy transfer (FRET), and bimolecular fluorescence complementation (BiFC) (Syafrizayanti et al. 2014). BiFC is capable of revealing the location of direct protein interactions in vivo (Hu et al. 2002). It is based on the reconstitution of a fluorescent protein when its nonfluorescing halves are brought within close proximity to one another by the association of two interacting proteins. Previously, we adopted the BiFC technology for use in Neurospora (Bardiya et al. 2008; Hammond et al. 2011a,b). In this study, we set out to define the precise interactions among six MSUD proteins formerly shown to localize at the nuclear periphery, a presumptive center for silencing activity. In addition, we explored the role of SAD-2 in the complex formation of these MSUD factors.
Physical Associations Among MSUD Factors
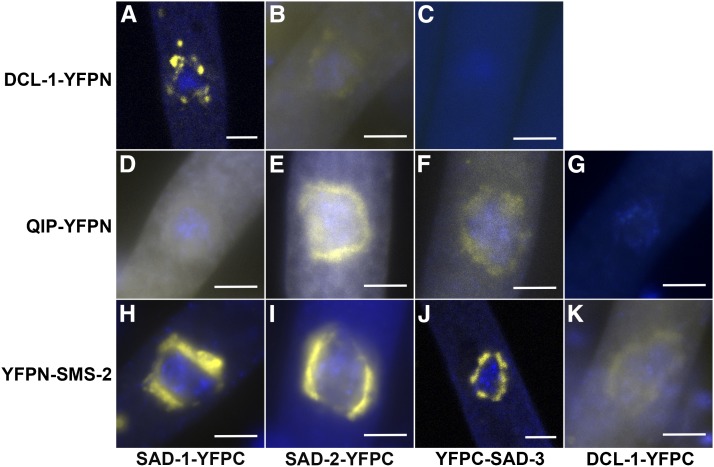
In this work, N-terminal (YFPN) or C-terminal (YFPC) fragments of the yellow fluorescent protein (YFP) were linked to six individual MSUD proteins, and each possible interaction was examined. Our results, as shown in Figure 1 and Table 1, indicate close associations among various perinuclear MSUD proteins. One noticeable exception is DCL-1, which appears to show affinity for SAD-1, SAD-2, and SMS-2 only.
Figure 1.
Assay of interactions among MSUD proteins using BiFC. As demonstrated by the reconstituted YFP signal in prophase asci (spore sacs), the six MSUD proteins are closely associated with each other with the exception of DCL-1, which has intimate interaction with only SAD-1, SAD-2, and SMS-2 (and not SAD-3 or QIP; see C and G). The BiFC temporal expression patterns are similar to those observed in individual MSUD proteins. Construction of YFP tagging vectors was as described by Bardiya et al. (2008) and Hammond et al. (2011b). Preparation of asci from perithecia (fruiting bodies) and visualization of YFP/DAPI (using Olympus BX61 and Zeiss LSM710) were performed as previously reported (Alexander et al. 2008; Xiao et al. 2010; Hammond et al. 2011a). (A) P7-26 × P13-34. (B) P18-29 × P18-52. (C) P21-18 × P21-19. (D) P22-05 × P22-06. (E) P16-37 × P16-38. (F) P21-20 × P21-21. (G) P18-37 × P18-39. (H) P18-19 × P18-21. (I) P16-35 × P16-36. (J) P15-37 × P16-06. (K) P22-03 × P22-04. See Table 2 for full genotypes. Bars, 5 μm.
Table 1. Matrix examination of BiFC interactions among MSUD proteins.
| SAD-1-YFPC | SAD-2-YFPC | YFPC-SAD-3 | DCL-1-YFPC | QIP-YFPC | YFPC-SMS-2 | |
|---|---|---|---|---|---|---|
| SAD-1-YFPN | ||||||
| SAD-2-YFPN | +a | |||||
| YFPN-SAD-3 | +b | +b | ||||
| DCL-1-YFPN | + (A) | + (B) | − (C) | |||
| QIP-YFPN | + (D) | + (E) | + (F) | − (G) | ||
| YFPN-SMS-2 | + (H) | + (I) | + (J) | + (K) | +c |
A positive interaction between two proteins (i.e., one that leads to the reconstitution of the yellow fluorophore) is denoted by +. A negative result (−) may indicate that either the two proteins do not have intimate interaction or the BiFC assay is not sensitive enough to detect their weak/transient interaction. A–K in parentheses refer to Figure 1, A–K.
Table 2. Neurospora strains used in this study.
| Strain | Genotype |
|---|---|
| F2-23 | rid; fl A |
| F4-30 | sad-3-gfp::hph rid; fl; mus-52Δ::bar a |
| F5-06 | fl; gfp-sms-2::hph a |
| F6-30 | rid his-3; fl; sad-2Δ::hph; gfp-sms-2::hph A |
| F6-31 | sad-3-gfp::hph rid his-3; fl; sad-2Δ::hph A |
| P7-26 | sad-1Δ::hph rid his-3+::sad-1-yfpc a |
| P13-14 | sad-1-gfp::hph; mus-51Δ::bar a |
| P13-15 | sad-1-gfp::hph A |
| P13-34 | rid his-3+::dcl-1-yfpn; mus-52Δ::bar; dcl-1Δ::hph A |
| P14-59 | sad-3-gfp::hph rid his-3 A |
| P15-14 | rid his-3; mus-52Δ::bar; gfp-sms-2::hph A |
| P15-37 | rid; yfpn-sms-2::hph A |
| P15-62 | rid; mus-52Δ::bar qip-gfp::hph a |
| P15-63 | rid his-3; mus-52Δ::bar qip-gfp::hph A |
| P16-06 | yfpc-sad-3::hph rid; sms-2-yfpn::hph a |
| P16-35 | rid his-3+::sad-2-yfpc; inv sad-2RIP; yfpn-sms-2::hph A |
| P16-36 | rid his-3+::sad-2-yfpc; inv sad-2RIP; yfpn-sms-2::hph a |
| P16-37 | rid his-3+::sad-2-yfpc; qip-yfpn::hph; inv sad-2RIP a |
| P16-38 | rid his-3+::sad-2-yfpc; qip-yfpn::hph; inv sad-2RIP A |
| P18-19 | rid his-3+::sad-1-yfpc; yfpn-sms-2::hph A |
| P18-21 | sad-1Δ::hph rid his-3+::sad-1-yfpc; yfpn-sms-2::hph a |
| P18-29 | rid his-3+::sad-2-yfpc; mus-51Δ::bar dcl-1-yfpn::hph; inv sad-2RIP a |
| P18-37 | rid his-3; qip-yfpn::hph; dcl-1-yfpc::hph A |
| P18-39 | rid; qip-yfpn::hph a |
| P18-52 | rid his-3+::sad-2-yfpc; mus-51Δ::bar dcl-1-yfpn::hph; inv sad-2RIP A |
| P21-18 | yfpc-sad-3::hph rid; mus-52Δ::bar a |
| P21-19 | yfpc-sad-3::hph rid his-3+::dcl-1-yfpn; mus-52Δ::bar A |
| P21-20 | yfpc-sad-3::hph rid his-3; mus-52Δ::bar qip-yfpn::hph a |
| P21-21 | yfpc-sad-3::hph rid his-3; qip-yfpn::hph A |
| P21-26 | rid his-3; mus-52Δ::bar qip-gfp::hph; sad-2Δ::hph a |
| P21-27 | rid his-3; mus-52Δ::bar qip-gfp::hph; sad-2Δ::hph A |
| P21-28 | rid his-3; sad-2Δ::hph; gfp-sms-2::hph a |
| P21-30 | rid his-3; mus-51Δ::bar gfp-dcl-1::hph; sad-2Δ::hph A |
| P21-31 | rid his-3; mus-51Δ::bar gfp-dcl-1::hph; sad-2Δ::hph a |
| P21-32 | sad-3-gfp::hph rid his-3; sad-2Δ::hph a |
| P21-33 | sad-3Δ::hph rid A |
| P21-34 | sad-3Δ::hph; sad-2-gfp::hph a |
| P21-35 | sad-1-gfp::hph sad-3Δ::hph A |
| P21-36 | sad-1-gfp::hph sad-3Δ::hph a |
| P21-37 | sad-1-gfp::hph rid his-3; sad-2Δ::hph a |
| P21-38 | sad-1-gfp::hph; sad-2Δ::hph A |
| P21-39 | rid his-3; mus-51Δ::bar gfp-dcl-1::hph A |
| P21-40 | rid; mus-51Δ::bar gfp-dcl-1::hph a |
| P21-41 | rid; sad-2-gfp::hph a |
| P22-03 | rid; dcl-1-yfpc::hph; yfpn-sms-2::hph a |
| P22-04 | rid his-3; dcl-1-yfpc::hph; yfpn-sms-2::hph A |
| P22-05 | sad-1Δ::hph rid his-3+::sad-1-yfpc; qip-yfpn::hph a |
| P22-06 | rid his-3+::sad-1-yfpc; qip-yfpn::hph A |
Genetic markers and knockout mutants are originated from the Fungal Genetics Stock Center (FGSC; Mccluskey et al. 2010) and the Neurospora Functional Genomics Group (Colot et al. 2006), and their descriptions can be found in the e-Compendium (http://www.bioinformatics.leeds.ac.uk/~gen6ar/newgenelist/genes/gene_list.htm). Strains used in this study were constructed essentially as described (Bardiya et al. 2008; Hammond et al. 2011b). Fungal manipulations were performed as described in the Neurospora protocol guide (http://www.fgsc.net/Neurospora/NeurosporaProtocolGuide.htm).
The six proteins examined include those that are common in many organisms (SAD-1, SAD-3, DCL-1, and SMS-2) and those that are seemingly found only in fungi (QIP and SAD-2). These proteins are localized in the perinuclear region regardless of the presence/absence of artificially unpaired genes (Xiao et al. 2010; Hammond et al. 2011a,b). QIP was first isolated as a QDE-2-interacting protein (Maiti et al. 2007). Since QDE-2 is the Argonaute responsible for vegetative silencing, it is not surprising that QIP also interacts with the meiotic phase Argonaute (SMS-2). The fact that DCL-1 and QIP are important for both vegetative and sexual silencing (Alexander et al. 2008; Xiao et al. 2010) suggests that there is a cross-talk between the two genome surveillance systems.
In yeast’s RNAi-mediated heterochromatin formation, the model for silencing complex assembly involves an RdRP-helicase-Argonaute association (Motamedi et al. 2004). These three proteins (SAD-1-SAD-3-SMS-2) also have interactions in Neurospora, suggesting that the complex formations in MSUD and RNAi-induced heterochromatin assembly may be evolutionarily related. The interaction between SAD-1 RdRP and DCL-1 Dicer is also not surprising, as the latter functions to process dsRNAs produced by the former. This finding is consistent with those observed in other organisms, in which the two common RNAi proteins conglomerate (Duchaine et al. 2006; Lee and Collins 2007). The affinity between these two proteins may be of special importance, as their physical association actually stimulates Dicer activity on dsRNA substrates in Tetrahymena (Lee and Collins 2007). Finally, the observed interaction between Dicer and Argonaute is not unexpected (Dueck and Meister 2014), as the small RNA has to be transferred from the site of origin (DCL-1 in this case) to the effector protein (SMS-2).
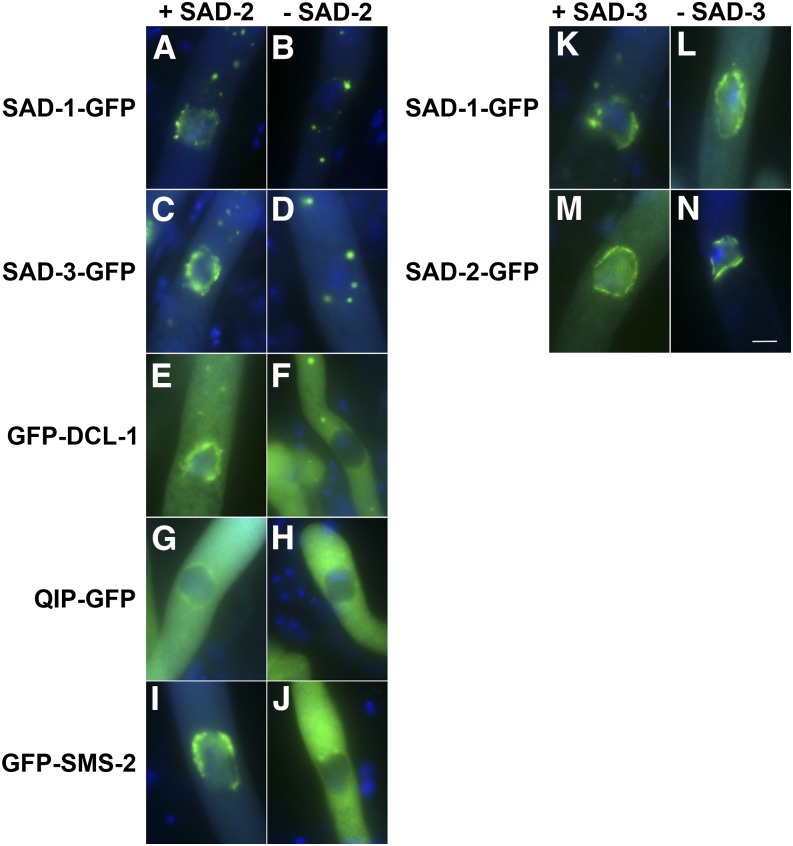
SAD-2 Is Required for the Localization of Perinuclear Silencing Proteins
SAD-2 appears to be restricted to the fungal kingdom, and it has not been shown to mediate RNAi other than in Neurospora. SAD-2 is absolutely required for the silencing of all unpaired markers tested, suggesting its importance in the MSUD pathway (Shiu et al. 2006). It was first identified as a silencing protein required for the proper localization of SAD-1. Without SAD-2, SAD-1 RdRP mislocalizes in the cytoplasm. In this study, we asked whether SAD-2 is also crucial for the localization of SAD-3, DCL-1, QIP, and SMS-2. As shown in Figure 2, A–J, their affinity for the perinuclear region is also hampered by the absence of SAD-2.
Figure 2.
SAD-2 is required for the localization of other perinuclear MSUD proteins. (A–J) In the absence of SAD-2, other MSUD proteins lose their affinity for the nuclear periphery. (K–N) As with SAD-1 (Shiu et al. 2006), SAD-3 does not appear to affect the localization of others. GFP constructs were made according to Hammond et al. (2011b). (A) P13-14 × P13-15. (B) P21-37 × P21-38. (C) F4-30 × P14-59. (D) F6-31 × P21-32. (E) P21-39 × P21-40. (F) P21-30 × P21-31. (G) P15-62 × P15-63. (H) P21-26 × P21-27. (I) F5-06 × P15-14. (J) F6-30 × P21-28. (K) P13-14 × P13-15. (L) P21-35 × P21-36. (M) F2-23 × P21-41. (N) P21-33 × P21-34. Bar, 5 μm. [Note that a perinuclear MSUD protein first emerges as aggregates around the nuclei at the binuclear stage (Shiu et al. 2006). These aggregates coalesce after karyogamy and eventually become two opposite crescents during leptotene. The crescents spread into a ring-like structure during zygotene and pachytene. The ring becomes more patchy and irregular during the diffuse stage, and the perinuclear localization can no longer be seen after diplotene. A perinuclear MSUD protein may also appear as cytoplasmic foci between karyogamy and metaphase I. The exact nature of these foci is still unknown, although they have been speculated as some kind of endomembrane body.]
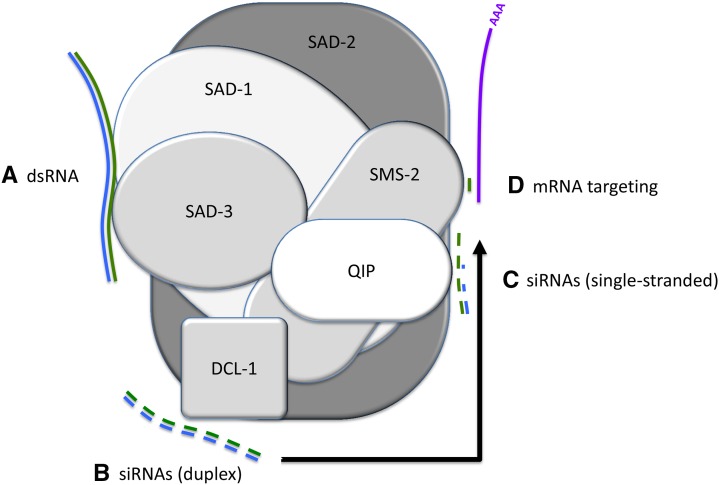
Although SAD-2 is required for the localization of other perinuclear MSUD proteins, the reverse is not true. In the absence of SAD-1 (Shiu et al. 2006) or SAD-3 (Figure 2, K–N), SAD-2 localizes normally (we were not able to perform the same assay for DCL-1, QIP, or SMS-2, as their presence is required for ascus formation). Since (1) SAD-2 has close interaction with all other perinuclear MSUD factors and (2) it is required for their localization (but not vice versa), it is possible that it functions as a scaffold protein that unites silencing factors at the nuclear periphery. A working model for the MSUD activity is shown in Figure 3.
Figure 3.
A model for the reactions performed by the meiotic silencing complex (MSC). (A) Exported aberrant RNA (blue) is turned into double strands by SAD-1 RdRP and SAD-3 helicase. (B) DCL-1 Dicer processes the dsRNA into siRNAs, (C) which (with the help of QIP exonuclease) become single-stranded (green) and (D) are utilized by SMS-2 Argonaute to target homologous mRNAs (purple). SAD-2, a presumptive scaffold protein, holds these silencing factors in the perinuclear region.
Concluding Remarks
Our results show that the aforementioned MSUD components form a silencing complex in the perinuclear region (the “surveillance checkpoint”) during prophase I of meiosis. In addition, we have established the role of SAD-2 as the likely scaffold protein for this complex. In higher eukaryotes, the perinuclear region in germ cells contains an organelle known as chromatoid body in mammals, nuage in flies, and P-granules in worms (Meikar et al. 2011; Voronina 2013; Kloc et al. 2014). By providing an environment in which RNAs can meet up with their developmental regulators, these perinuclear organelles play an important role in silencing, translation, and/or RNA processing/regulation/organization. Certain silencing proteins (e.g., Argonaute) have been shown to congregate in these perinuclear regions, and their presence is required for specific RNA regulation and meiotic progression (Updike and Strome 2010; Meikar et al. 2011; Kloc et al. 2014). Conglomeration of related proteins may allow the coupling of consecutive reactions, thereby increasing the efficiency of the biochemical process. To our knowledge, this is the most comprehensive examination of RNAi protein interactions in a single organism. Results from this and future studies could be useful in determining if silencing complex formation is conserved among various RNAi processes.
Acknowledgments
We thank James Birchler and Patrice Albert for their equipment sharing and advice. We are indebted to Tony Perdue, Patricia Pukkila, the Fungal Genetics Stock Center, the Neurospora Functional Genomics Group, University of Missouri Core Facilities, colleagues from our community, and members of the Shiu laboratory for their indispensible help. We are pleased to acknowledge use of materials generated by P01 GM068087 “Functional Analysis of a Model Filamentous Fungus”. L.M.D. was supported by a GK-12 Fellowship from the National Science Foundation (DGE1045322). E.C.B. was supported by a Graduate Assistance in Areas of National Need Fellowship from the U.S. Department of Education. T.M.H. was supported by a Life Science Fellowship from the University of Missouri and a Ruth L. Kirschstein National Research Service Award from the National Institute of General Medical Sciences. This work was supported by the National Science Foundation (MCB1157942 to P.K.T.S.).
Footnotes
Communicating editor: N. Hunter
Literature Cited
- Alexander W. G., Raju N. B., Xiao H., Hammond T. M., Perdue T. D., et al. , 2008. DCL-1 colocalizes with other components of the MSUD machinery and is required for silencing. Fungal Genet. Biol. 45: 719–727. [DOI] [PubMed] [Google Scholar]
- Aramayo R., Selker E. U., 2013. Neurospora crassa, a model system for epigenetics research. Cold Spring Harb. Perspect. Biol. 5: a017921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardiya N., Alexander W. G., Perdue T. D., Barry E. G., Metzenberg R. L., et al. , 2008. Characterization of interactions between and among components of the MSUD machinery in Neurospora crassa using bimolecular fluorescence complementation. Genetics 178: 593–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billmyre R. B., Calo S., Feretzaki M., Wang X., Heitman J., 2013. RNAi function, diversity, and loss in the fungal kingdom. Chromosome Res. 21: 561–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambareri E. B., Jensen B. C., Schabtach E., Selker E. U., 1989. Repeat-induced G-C to A-T mutations in Neurospora. Science 244: 1571–1575. [DOI] [PubMed] [Google Scholar]
- Colot H. V., Park G., Turner G. E., Ringelberg C., Crew C. M., et al. , 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103: 10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine T. F., Wohlschlegel J. A., Kennedy S., Bei Y., Conte D., Jr, et al. , 2006. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124: 343–354. [DOI] [PubMed] [Google Scholar]
- Dueck A., Meister G., 2014. Assembly and function of small RNA–Argonaute protein complexes. Biol. Chem. 395: 611–629. [DOI] [PubMed] [Google Scholar]
- Hammond, T. M., H. Xiao, E. C. Boone, T. D. Perdue, P. J. Pukkila et al., 2011a SAD-3, a putative helicase required for MSUD, interacts with other components of the silencing machinery. Genes Genomes Genet. 1: 369–376. [DOI] [PMC free article] [PubMed]
- Hammond T. M., Xiao H., Rehard D. G., Boone E. C., Perdue T. D., et al. , 2011b Fluorescent and bimolecular-fluorescent protein tagging of genes at their native loci in Neurospora crassa using specialized double-joint PCR plasmids. Fungal Genet. Biol. 48: 866–873. [DOI] [PubMed] [Google Scholar]
- Hu C. D., Chinenov Y., Kerppola T. K., 2002. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9: 789–798. [DOI] [PubMed] [Google Scholar]
- Kloc M., Jedrzejowska I., Tworzydlo W., Bilinski S. M., 2014. Balbiani body, nuage and sponge bodies: the germ plasm pathway players. Arthropod Struct. Dev. 43: 341–348. [DOI] [PubMed] [Google Scholar]
- Lee S. R., Collins K., 2007. Physical and functional coupling of RNA-dependent RNA polymerase and Dicer in the biogenesis of endogenous siRNAs. Nat. Struct. Mol. Biol. 14: 604–610. [DOI] [PubMed] [Google Scholar]
- Lee D. W., Pratt R. J., McLaughlin M., Aramayo R., 2003. An argonaute-like protein is required for meiotic silencing. Genetics 164: 821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti M., Lee H. C., Liu Y., 2007. QIP, a putative exonuclease, interacts with the Neurospora Argonaute protein and facilitates conversion of duplex siRNA into single strands. Genes Dev. 21: 590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCluskey K., Wiest A., Plamann M., 2010. The Fungal Genetics Stock Center: a repository for 50 years of fungal genetics research. J. Biosci. 35: 119–126. [DOI] [PubMed] [Google Scholar]
- Meikar O., Da Ros M., Korhonen H., Kotaja N., 2011. Chromatoid body and small RNAs in male germ cells. Reproduction 142: 195–209. [DOI] [PubMed] [Google Scholar]
- Motamedi M. R., Verdel A., Colmenares S. U., Gerber S. A., Gygi S. P., et al. , 2004. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119: 789–802. [DOI] [PubMed] [Google Scholar]
- Nicolás F. E., Ruiz-Vázquez R. M., 2013. Functional diversity of RNAi-associated sRNAs in fungi. Int. J. Mol. Sci. 14: 15348–15360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano N., Macino G., 1992. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 6: 3343–3353. [DOI] [PubMed] [Google Scholar]
- Samarajeewa D. A., Sauls P. A., Sharp K. J., Smith Z. J., Xiao H., et al. , 2014. Efficient detection of unpaired DNA requires a member of the Rad54-like family of homologous recombination proteins. Genetics 198: 895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu P. K. T., Metzenberg R. L., 2002. Meiotic silencing by unpaired DNA: properties, regulation, and suppression. Genetics 161: 1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu P. K. T., Raju N. B., Zickler D., Metzenberg R. L., 2001. Meiotic silencing by unpaired DNA. Cell 107: 905–916. [DOI] [PubMed] [Google Scholar]
- Shiu P. K. T., Zickler D., Raju N. B., Ruprich-Robert G., Metzenberg R. L., 2006. SAD-2 is required for meiotic silencing by unpaired DNA and perinuclear localization of SAD-1 RNA-directed RNA polymerase. Proc. Natl. Acad. Sci. USA 103: 2243–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syafrizayanti C. Betzen, J. D. Hoheisel, D. Kastelic, 2014. Methods for analyzing and quantifying protein-protein interaction. Expert Rev. Proteomics 11: 107–120. [DOI] [PubMed] [Google Scholar]
- Updike D., Strome S., 2010. P granule assembly and function in Caenorhabditis elegans germ cells. J. Androl. 31: 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina E., 2013. The diverse functions of germline P-granules in Caenorhabditis elegans. Mol. Reprod. Dev. 80: 624–631. [DOI] [PubMed] [Google Scholar]
- Xiao H., Alexander W. G., Hammond T. M., Boone E. C., Perdue T. D., et al. , 2010. QIP, an exonuclease that converts duplex siRNA into single strands, is required for meiotic silencing by unpaired DNA. Genetics 186: 119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]