Abstract
All eukaryotes use three DNA-dependent RNA polymerases (RNAPs) to create cellular RNAs from DNA templates. Plants have additional RNAPs related to Pol II, but their evolutionary role(s) remain largely unknown. Zea mays (maize) RNA polymerase D1 (RPD1), the largest subunit of RNA polymerase IV (Pol IV), is required for normal plant development, paramutation, transcriptional repression of certain transposable elements (TEs), and transcriptional regulation of specific alleles. Here, we define the nascent transcriptomes of rpd1 mutant and wild-type (WT) seedlings using global run-on sequencing (GRO-seq) to identify the broader targets of RPD1-based regulation. Comparisons of WT and rpd1 mutant GRO-seq profiles indicate that Pol IV globally affects transcription at both transcriptional start sites and immediately downstream of polyadenylation addition sites. We found no evidence of divergent transcription from gene promoters as seen in mammalian GRO-seq profiles. Statistical comparisons identify genes and TEs whose transcription is affected by RPD1. Most examples of significant increases in genic antisense transcription appear to be initiated by 3ʹ-proximal long terminal repeat retrotransposons. These results indicate that maize Pol IV specifies Pol II-based transcriptional regulation for specific regions of the maize genome including genes having developmental significance.
Keywords: RNA polymerase IV, transcription, gene regulation, transposons, paramutation
EUKARYOTES use at least three DNA-dependent RNA polymerases (RNAPs) to transcribe their genomes into functional RNAs. RNAP Pol II generates messenger RNAs (mRNAs) and noncoding RNAs involved in various RNA-mediated regulatory pathways (reviewed by Sabin et al. 2013). Flowering plant genomes encode additional RNAP subunits comprising Pol IV and Pol V, which are central to a small interfering RNA (siRNA)-based silencing pathway primarily targeting repetitive sequences such as transposable elements (TEs) (Matzke and Mosher 2014; Matzke et al. 2015). These additional RNAPs derive from duplications of specific Pol II subunits followed by subfunctionalization during plant evolution (Tucker et al. 2011), yet the holoenzyme complexes still share some Pol II subunits (Ream et al. 2009; Haag et al. 2014).
Zea mays (maize) has distinct largest subunits for Pol IV and V and, unlike Arabidopsis thaliana, three second-largest subunits (Erhard et al. 2009; Sidorenko et al. 2009; Stonaker et al. 2009) that in distinct combinations form two Pol IV and three Pol V isoforms (Haag et al. 2014). Genetic analyses of rna polymerase d/e 2a (rpd/e2a) encoding one of the second-largest subunits (Sidorenko et al. 2009; Stonaker et al. 2009) together with recent proteomic data showing association of a putative RNA-dependent RNA polymerase (RDR2) with only RPD/E2a-containing isoforms (Haag et al. 2014) indicate that maize Pol IV isoforms have diverse functional roles in managing genome homeostasis.
Loss of Pol IV function has different consequences in Arabidopsis, Brassica rapa (a close relative of Arabidopsis), and maize, species in which Pol IV mutants have been identified (Herr et al. 2005; Onodera et al. 2005; Erhard et al. 2009; Huang et al. 2013), although mutants in all three species are defective for siRNA production (Zhang et al. 2007; Mosher et al. 2008; Erhard et al. 2009; Huang et al. 2013). Arabidopsis NUCLEAR RNA POLYMERASE D1A (NRPD1A) mutants are late flowering (Pontier et al. 2005) and B. rapa nrpd1a mutants have no obvious phenotypes (Huang et al. 2013), while maize rna polymerase d1 (rpd1) mutants have multiple developmental defects and trans-generational degradation in plant quality compared to nonmutant siblings (Parkinson et al. 2007; Erhard et al. 2009). The disparate impacts of rpd1/NRPD1A mutations in maize vs. Brassicaceae representatives are potentially related to different genomic TE contents as TE sequences are greatly expanded in maize compared to both Arabidopsis (Hale et al. 2009) and B. rapa (Wang et al. 2011). However, recently reported cytosine methylome profiles (Li et al. 2014) indicate maize TEs are as equally well methylated in the absence of Pol IV as their Arabidopsis counterparts (Stroud et al. 2013), predicting that Pol IV-dependent cytosine methylation is not required to maintain TE silencing.
The developmental defects observed in rpd1 mutants are both distinct and nonheritable (Parkinson et al. 2007) and therefore unlikely to be related to TE-derived mutations. These defects also appear unrelated to siRNA-induced silencing because other maize mutants affecting siRNA biogenesis, including rpd/e2a, are developmentally normal (Hale et al. 2007; Stonaker et al. 2009; Barbour et al. 2012). We hypothesize that maize has co-opted RPD1/Pol IV to transcriptionally control specific alleles of genes for which TEs and TE-like repeats act as regulatory elements. Supporting this concept, specific purple plant1 (pl1) alleles having an upstream doppia TE fragment are regulated by RPD1 (Erhard et al. 2013). As the maize genome is composed of >85% TE-like sequences (Schnable et al. 2009), many of which occur within 5 kb of genes (Baucom et al. 2009; Gent et al. 2013), a large number of alleles using TE-like sequences as regulatory elements is possible. Phylogenomic comparisons between A. thaliana and Arabidopsis lyrata also support the idea that gene-proximate TEs represent a source of regulatory diversity (Hollister et al. 2011).
Maize RPD1 was initially identified as a genetic factor required to maintain transcriptional repression of specific alleles subject to paramutation (Hollick et al. 2005)—a process by which meiotically heritable changes in gene regulation are influenced by trans-homolog interactions (Brink 1956, 1958; Hollick 2012). Presumably because detailed pedigree analyses are required to recognize instances of paramutation, only a few clear examples involving endogenous alleles have been described (Brink 1956; Coe 1961; Hagemann and Berg 1978; Hollick et al. 1995; Sidorenko and Peterson 2001; Pilu et al. 2009). Similar behaviors involving transgenes have been noted in both plants and animals (Chandler and Stam 2004; Rassoulzadegan et al. 2006; Khaitová et al. 2011; Ashe et al. 2012; de Vanssay et al. 2012; Shirayama et al. 2012) although it remains unknown whether these examples are due to mechanistically related processes. One strategy for identifying a broader set of alleles subject to paramutation would be to start with a list of candidate genes, the transcriptional regulation of which is affected by RPD1 function.
The evolutionary function(s) of Pol IV remains enigmatic. Because Pol IV is required for siRNA-directed cytosine methylation (reviewed by Matzke and Mosher 2014 and Matzke et al. 2015), it is expected that the regulation of many alleles might be affected by RPD1/NRPD1 action although few such alleles have been identified in maize (Hollick et al. 2005; Parkinson et al. 2007; Erhard et al. 2013) and Arabidopsis (Matzke et al. 2007; Ariel et al. 2014). To date there have been no reports of genome-wide effects of RPD1/NRPD1 on gene regulation in any species although several studies have noted correlations between siRNA profiles and nearby genic mRNA abundance (Hollister et al. 2011; Eichten et al. 2012; Greaves et al. 2012). Template competitions between Pol IV and Pol II have been proposed (Hale et al. 2009) to account for RPD1-based transcriptional repression seen at the Pl1-Rhoades allele of pl1 (Hollick et al. 2005) and for increases in polyadenylated transcripts of some long terminal repeat (LTR) retrotransposons that specifically accompany loss of RPD1 but not loss of two other siRNA biogenesis factors (Hale et al. 2009). These results indicate that RPD1-containing Pol IV complexes directly interfere with Pol II transcription of RPD1-targeted genomic regions.
Here we use global run-on sequencing (GRO-seq) (Core et al. 2008) to identify genome-wide targets of Pol IV-based transcriptional regulation. This technique profiles RNAs from RNAPs incorporating a brominated UTP ribonucleotide during a short nuclear run-on reaction (Core et al. 2008). Maize Pols IV and V can extend transcripts in vitro, but ribonucleotide incorporation is attenuated compared to Pol II (Haag et al. 2014). Because Arabidopsis Pol IV products are rapidly processed to siRNAs (Li et al. 2015), transcription rates of maize Pol IV are relatively slow in vitro (Haag et al. 2014), and no appreciable maize Pol IV RNAs are detected in vivo in short run-on experiments (Erhard et al. 2009), most non-ribosomal RNA (rRNA), non-transfer RNA (tRNA) transcription detected by GRO-seq is expected to represent Pol II function. Our results show that loss of Pol IV affects transcription profiles at the 5′ and 3′ gene ends and at a discrete set of unique TEs and genes, the dysregulation of which may contribute to rpd1 mutant developmental defects.
Materials and Methods
Genetic stocks
The rpd1-1 null mutation (originally designated rmr6-1) (Erhard et al. 2009) was introgressed into the B73 inbred background to ∼97% by repeated backcrosses of F2 rpd1-1 / rpd1-1 mutant pollen to a recurrent B73 female parent. Families segregating 1:2:1 for rpd1-1/rpd1-1 mutants, heterozygotes, and homozygous Rpd1-B73 individuals were used for nuclei isolations and RNA isolations for reverse transcriptase polymerase chain reaction (RT-PCR) and quantitative real-time PCR (qRT-PCR) analysis.
GRO-seq library preparation
Ten homozygous wild-type (WT; Rpd1-B73/Rpd1-B73) and 10 homozygous rpd1-1 mutant (rpd1 mutant) siblings were identified with a dCAPs marker for the rpd1-1 lesion (Erhard et al. 2013) and used for nuclei isolations. Nuclei were isolated from whole shoots (roots removed) of 8-day-old seedlings. Seedling tissues and dry ice were pulverized in a blade coffee grinder and transferred to a ceramic mortar with 15 ml of ice-cold isolation buffer (40% glycerol, 250 mM sucrose, 20 mM Tris, pH 7.8, 5 mM MgCl2, 5 mM KCl, 0.25% Triton X-100, 5 mM β-mercaptoethanol). Pulverized tissue in isolation buffer was ground further with a ceramic pestle and filtered through cheesecloth into a 50-ml conical tube. Grindates were filtered again through 40-μm nylon cell strainers (BD Biosciences, San Jose, CA) into 35-ml centrifuge tubes. Nuclei were centrifuged at 6000 × g for 15 min at 4°, and pellets were washed with 15 ml isolation buffer. Washes were repeated two more times, and pellets were resuspended in 100 μl resuspension buffer (50 mM Tris, pH 8.5, 5 mM MgCl2, 20% glycerol, 5 mM β-mercaptoethanol). Transcription run-ons were performed as described (Hollick and Gordon 1993) with the following changes: 0.5 mM 5-bromouridine 5′-triphosphate (Sigma) was substituted for UTP and 2 μM cold cytidine triphosphate (CTP) was added in addition to 10 μl of α-32P-CTP (3000 Ci/mmol, 10 mCi/mL; Perkin-Elmer). RNA isolation was as described (Hollick and Gordon 1993) with the following changes: DNase I and Proteinase K digestions were performed for 1 hr each at 37° and 42°, respectively; one acid phenol/chloroform extraction was performed to isolate RNA. High-throughput sequencing libraries were prepared from in vitro-labeled RNA as described (Core et al. 2008) using agarose bead-conjugated α-bromodeoxyuridine antibody (Santa Cruz Biotechnology).
GRO-seq library processing
Fifty nucleotide (nt) single-end raw reads (54,318,135 for WT library and 54,873,783 for rpd1 mutant library) were generated on the Illumina HiSeqII platform (Vincent J. Coates Genome Sequencing Laboratory, University of California at Berkeley). Based on FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) Phred score analysis, 10 nt were trimmed from the relatively lower quality 3′ end of all reads from both libraries using the FASTQ/A Trimmer script (http://hannonlab.cshl.edu/fastx_toolkit/). The Cutadapt program (Martin 2011) was used to trim adapter sequences and any additional low-quality bases (option -q 10) from the 3′ end of all reads; reads shorter than 20 nt after adapter trimming (2,253,657 and 2,488,839, respectively) were excluded (option -m 20). The Fastx Quailty Filter (http://hannonlab.cshl.edu/fastx_toolkit/) removed reads with <97% of their bases (option -p 97) above the Phred quality of 10 (option -q 10); 654,703 WT reads and 667,412 rpd1 mutant reads were removed. Finally, 7546 and 5443 sequencing artifacts were removed from the WT and rpd1 mutant libraries, respectively, using the Fastx Artifacts Filter (http://hannonlab.cshl.edu/fastx_toolkit/). The resulting libraries consisted of 51,402,229 and 51,712,089 high-quality WT and rpd1 mutant reads, respectively, with an average length of 32 nt, and these were used for subsequent mapping, computational, and statistical analyses.
Computational analyses of GRO-seq libraries
Alignments to genomic features:
For filtering and downstream comparisons, high-quality GRO-seq reads were mapped using Bowtie alignment software (version 0.12.7; Langmead et al. 2009) to annotated maize sequence features (see Supporting Information, Table S1, for file types and origins): rRNAs and tRNAs (kindly provided by Blake Meyers, University of Delaware), maize B73 AGPv2 filtered gene set (FGS), Maize Transposable Element Consortium (MTEC) consensus sequences, and maize pseudochromosomes 1–10 representing the sequenced B73 reference genome (AGP version 2, build 5b). Bowtie indices were built from the above annotated maize feature sequences using the bowtie-build command with default parameter settings. Reads that failed to match to the rRNA/tRNA indices with up to two mismatches (option -v 2) comprise the filtered non-rRNA/non-tRNA alignments (41,169,885 and 42,498,964 for WT and rpd1 mutant libraries, respectively), which were aligned to the maize pseudo-chromosomes 1–10 allowing two mismatches (option -v 2) and to match only once in the genome (option -m 1). These alignments define uniquely mapping reads (12,239,069 and 11,739,444 for WT and rpd1 mutant libraries, respectively), which were used for the metagene and differential expression analyses described below.
Distribution of reads over genomic features:
To measure the relative contribution of introns and exons in WT and rpd1 mutant GRO-seq libraries, we first isolated the subset of 26,987 (68%) maize FGS models with no predicted alternatively spliced transcript isoforms, which we define as single-transcript genes. As a control, we determined the contribution of introns and exons to all single-transcript genes using intron and exon chromosomal coordinates contained in the FGS position annotation file (Table S1). We compared the control distribution to percentages of uniquely mapping GRO-seq reads overlapping introns or exons within single transcript genes, which were identified using the intersectBed tool, part of the BEDTools suite (Quinlan and Hall 2010), with the following parameters: -f 1 -u -wa. Only reads contained entirely within a gene, exon, or intron were reported (no untranslated regions or exon/intron boundaries are reported by this method). Percentages of introns and exons were calculated as a proportion of unique reads contained within single-transcript genes.
For direct Bowtie alignments of filtered non-rRNA/non-tRNA reads to TE and FGS sequence indices, we allowed two mismatches (option -v 2) and reads to map more than once to the respective set of sequences, but counted each multi-mapping read only once (default option -k 1 to report only one random alignment per read for TEs and option --best to report the best FGS alignment). Analyses of differentially transcribed TE superfamilies compared numbers of reads aligned directly to MTEC consensus TE sequences (with the same alignment parameters) normalized by total mappable reads of each respective library. For a control alignment to TE sequences, we generated a random sampling of genomic sequences using the Sherman program (http://www.bioinformatics.babraham.ac.uk/projects/sherman/). Specifically, a set of 51,402,229 (the number of high-quality reads in the WT GRO-seq library; option -n 51,402,229) random 32-mers (the average length of high-quality GRO-seq reads for both the WT and rpd1 mutant libraries, weighted for abundances: option -l 32) was generated from the maize genome, inhibiting in silico bisulfite conversion with option -cr 0. The resulting 32-mers were aligned directly to TE and FGS sequences using identical Bowtie parameters as GRO-seq read alignments.
To determine overlaps of uniquely and repetitively mapping reads to genomic features (genes, TEs, and intergenic regions), we created alignment tables from the raw Bowtie alignment files (SAM formatted). For each read, alignment characteristics (unmappable, maps uniquely, or maps repetitively) were extracted from the unique alignment of non-rRNA/non-tRNA reads to the B73 genome. These were compared to mapping sense/mapping antisense/not mapping values for the same reads to indices built from the FGS sequences, MTEC TE sequences, or custom sequences extending 5 kb upstream (FGS −5 kb) or downstream (FGS +5 kb) of the original FGS sequences. In all cases, two mismatches were allowed for each reference; for the FGS-only alignment, the best alignment was reported (option --best); for the rest, a random alignment was reported (default option -k 1). The resulting dataset was used to collapse reads into different groups; for example, uniquely mapping TE reads within 5 kb of gene starts would map only once to the B73 genome and map to the MTEC and FGS −5 kb indices, but not to the FGS or FGS +5 kb indices. Only the original B73 genome alignment determined uniquely vs. repetitively mapping. Therefore, when a uniquely mapping read maps within 5 kb of an annotated transcription start site (TSS) and within 5 kb of a 3′ gene end, the read is likely between two genes that are <10 kb apart.
Metagene and heatmap profiles of gene boundaries:
Uniquely mapping reads overlapping TSSs and 3′ gene ends were tallied and binned using a metagene analysis pipeline of custom Python scripts (https://github.com/HollickLab/metagene_analysis). Using metagene_count.py, the 5′ read ends (option --count_method start) were tallied against the positions of TSSs (option --feature_count start) and 3′ ends (option --feature_count end) of FGS models (Table S1, FGS positions) >1 kb in length (31,794 of 39,656 total models). The tallies extended ±5 or ±1 kb by changing the padding option (--padding) to 5000 or 1000, respectively; in both cases counting was strand-specific relative to the feature orientation by default. For the 5-kb tallies, only the first 1 kb of genic windows at each gene were kept by excluding windows with starting positions > +1000 from the TSS or < −1000 from the 3′ end in R. Tallies from metagene_count.py were strand-specifically binned (option --separate_groups) with metagene_bin.py in either 10-nt nonoverlapping windows (--window_size 10 --step_size 10) for detailed metagene plots or 50-nt nonoverlapping windows (--window_size 50 --step_size 50) for the heatmap plots. The resulting count tables were imported to R (version 2.15.2; http://www.r-project.org/) for normalization, statistical testing, and plotting.
To view the normalized coverage (reads per million uniquely mapped), a heatmap-like plot was made using image, a base R command, to plot coverage (z-axis) on a color scale at each position along the gene model (x-axis) for groups of 60 genes (y-axis). Gene models were ordered by their maximum contribution to a 10-nt window’s total abundance (Maximum Sum Contribution) for all heatmap and metagene plots. At each window, a gene’s sum contribution represents its influence on the total coverage via the calculation: gene’s coverage at the window/total coverage of all genes at the window. The Maximum Sum Contribution was also used to exclude the upper and lower 5% of genes from the metagene plots described below. Heatmap plots were binned into 50-nt nonoverlapping windows and neighboring gene models after sorting by Maximum Sum Contribution were averaged in groups of 60 genes.
To summarize the average behavior of GRO-seq coverage over the FGS models, we created metagene plots summarizing the coverage of each gene using either the total (sum) or the mean coverage at each 10-nt window. Welch’s two-sample t-tests and 95% confidence intervals were calculated for each 10-nt window across all inner 90% quantile (by Maximum Sum Contribution) gene models. To identify the windows with statistically significant coverage differences ±RPD1, the individual Welch’s t-test results were corrected for multiple sampling using the Holm–Bonferroni method (α = 0.05) across all 800 windows comprising the regions around the TSSs and 3′ ends on both sense and antisense strands. Final plots used base R commands to plot mean or sum abundance as lines (or bars) and 95% confidence intervals as polygons and to highlight statistically significant windows with horizontal line segments.
Correlations between WT and rpd1 mutant libraries:
To determine the correspondence between WT and rpd1 mutant libraries, the number of uniquely mapping reads per kilobase per million uniquely mapped reads were tallied across various regions. Near-genic analysis of the 31,794 genes analyzed by the metagene profiles divided the region around each gene model into constant 1 kb upstream of TSS, 1 kb downstream of TSS, 1 kb upstream of gene end, and 1 kb downstream of gene end regions. The internal portion (>1 kb away from each gene boundary) of those genes >2 kb in length (23,050 of 31,794) was used to represent the interior gene region. All counts used the 5′-most base of each GRO-seq read. For each region, zero values were artificially set to 1/10 of the lowest nonzero value in either data set; this allowed both the inclusion of zero vs. non-zero data and had minimal (linear regression) to no (Spearman’s rank correlation) effect on the summary statistics. Resulting normalized coverages were log10-transformed for plotting, fitting a linear regression “Data fit” line, and calculating the Spearman’s rank correlation coefficient, all of which were performed in R (version 3.0.2).
siRNA analyses:
National Center for Biotechnology Information (NCBI)-sourced profiles of maize 16- to 35-nt RNAs were pooled and overlapped with the gene models used in the GRO-seq metagene analysis. Because no profiles represented 8-day-old B73 seedling shoots, we pooled WT B73 siRNAs from seedling (day 3) root tips (SRR218319: Gent et al. 2012), seedling (day 11) shoot apices (SRR488770 and SRR488774: Barber et al. 2012), ovule-enriched unfertilized cob (at silk emergence) (SRR408793: Gent et al. 2013), and developing ear (SRR1583943 and SRR1583944: Gent et al. 2014). Pooled siRNAs from B73 rdr2 mutant developing ears (SRR1583941 and SRR1583942: Gent et al. 2014) represent 24-nt RNA deficiency that should mimic Pol IV loss as RDR2 is required with Pol IV for 24-nt RNA biogenesis (Matzke and Mosher 2014; Matzke et al. 2015).
All raw sequences were downloaded from the NCBI Sequence Read Archive and processed to a high-quality set that all had trimmable 3′ adapters and neither low-quality (Phred scores <30) nor ambiguous bases. The resulting high-quality reads (11,163,623 for rdr2 mutant and 74,787,717 for WT) were filtered against rRNA/tRNA sequences and aligned to the maize B73 v2 genome with 551,194 (rdr2 mutant) and 11,466,496 (WT) reads mapping uniquely. Uniquely mapping 24-nt reads (154,509 or 28% for rdr2 mutant and 8,611,691 or 75% for WT) were subjected to metagene analysis as described above, using all size classes for library normalization in reads per million uniquely mapped per region length. For consistency, the order of genes in the heatmaps followed the same order (by Maximum Sum Contribution to total GRO-seq coverage) as the GRO-seq heatmaps.
Definition of alternative transcription start sites:
As an alternative TSS definition, we used the 5′-end sequence of full-length complementary DNAs (flcDNAs) from predominately 7-day-old seedling tissues (ZM_BFc set: http://www.ncbi.nlm.nih.gov/nucest/?term=ZM_BFc; see Soderlund et al. 2009). Positions for the flcDNA 5′ ends were defined by perfectly (0 mismatches) and uniquely (only 1 B73 alignment) aligning the first 50 nt of the cDNA sequence to the maize B73 reference genome (version 2, build 5b) with Bowtie (version 0.12.7). Identical TSS positions were collapsed. Metagene counting, binning, and plotting of total normalized GRO-seq read abundance were performed ±1 kb from all of the flcDNA-defined TSSs in 10-nt nonoverlapping windows.
Identification of differentially transcribed genes and TEs:
For differential transcription analysis of genes (gene body only) and TEs with defined genomic coordinates (positionally defined TEs), the DESeq package (Anders and Huber 2010) was used with uniquely aligning reads and the maize FGS GFF3 and MTEC TE GFF3 (Table S1) annotation files. The counts table for DESeq used tallies of raw reads overlapping features in sense and antisense orientations (-s and -S options, respectively) generated by intersectBed (additional options -c -wa; Quinlan and Hall 2010). The sense and antisense counts tables were processed with the following DESeq parameters: fit = “local”; method = “blind”; and sharingMode = “fit-only”. The Benjamini–Hochberg correction (Benjamini and Hochberg 1995) for multiple sampling adjusted the P-values adjusted (padj) for False Discovery Rate (FDR) control, and a 10% FDR threshold was applied to the padj values (Anders and Huber 2010). The list of features passing the DESeq thresholds were further curated and trimmed as described below. Sequence polymorphisms between the introgressed haplotype containing the rpd1-1 mutation and the homologous B73 chromosome 1 region likely contribute to differences in the abundances of reads aligning to this region between the WT and rpd1 mutant libraries. While the size of the rpd1-1–containing haplotype is unknown, we estimated it to be at most 20 Mb. Therefore, we excluded 12 TEs and 26 genes located within 20 Mb on either side of the rpd1 locus from the respective lists of features classified as having decreased transcription in rpd1 mutants. We manually curated genes with increased or decreased sense transcription based on visual inspection for GRO-seq read coverage consistent with the transcription unit defined by the gene annotation; coverage localized to only a portion of the gene model were tagged as unlikely to be related to that gene’s expression (nonbold entries in Table S3). Genes with increased antisense transcription were visually inspected, and TEs <2 kb beyond the genic 3′ end were tallied. We determined whether positionally defined, differentially transcribed TEs were located within annotated genic regions or <5 kb from their 5′ or 3′ ends using the closestBed tool (Quinlan and Hall 2010) with the -d parameter, which in this case reports the distance of the closest gene to each TE analyzed.
Expression analysis
RT-PCR expression analysis:
Three homozygous Rpd1-B73 (WT) and three homozygous rpd1-1 mutant 8-day-old seedlings from accessions described in Genetic stocks, were identified by genotyping as described above and used for RNA isolations. Seedling tissues (whole shoots, as described above) were pulverized separately in ceramic mortars in 1 ml of Trizol reagent (Invitrogen), and RNAs were isolated following the manufacturer’s protocol. cDNA synthesis using oligo(dT) primers (New England Biolabs) and Superscript II (Invitrogen) followed manufacturer’s protocols using 1 μg RNA as templates for reverse transcription reactions. cDNAs were amplified using gene-specific primers (Table S2) designed from sequences corresponding to the predicted coding regions of ocl2 (AC235534.1_FG007), GRMZM2G043242 (ATPase-domain containing protein), and GRMZM2G161658 (Epoxide hydrolase2), as well as primers matching a control gene alanine aminotransferase (aat) as described (Woodhouse et al. 2006). RT-PCR products were sized on a 2% agarose gel and stained with ethidium bromide for visualization and quantified with ImageJ software (http://rsbweb.nih.gov/ij/) to normalize to aat products. For each gene-specific primer pair, PCR amplicons were gel-extracted (Qiaquick Gel Extraction kit) and Sanger-sequenced (University of California at Berkeley Sequencing Facility) to verify that correctly spliced products were being amplified (sequences available upon request).
qRT-PCR expression analysis:
Seedling tissues (whole shoots, as described above) were harvested from 8-day-old sibling homozygous Rpd1-B73 (WT) and rpd1-1 mutant plants, flash-frozen in liquid nitrogen, and stored at −80° for subsequent RNA extraction. A total of nine seedlings for each genotype were prepared in pools of three seedlings each. These pools of three were pulverized with dry ice in a coffee grinder and subsequently ground in a mortar and pestle with 5 ml of ice-cold isolation buffer (40% glycerol, 250 mM sucrose, 20 mM Tris, pH 7.8, 5 mM MgCl2, 5 mM KCl, 0.25% Triton X-100, 5 mM β-mercaptoethanol). RNAs were then extracted from 1 ml of the grindate with 1 ml of TRIzol reagent (Invitrogen) following the manufacturer’s protocol. An aliquot (2 µg) of total RNA was DNaseI (Roche) treated for 20 min at 37° and heat-inactivated (75° for 10 min) in 5 mM EDTA. The Tetro cDNA synthesis kit (Bioline, Taunton, MA) was used for the first-strand cDNA synthesis, followed by RNA degradation with an RNase A/TI/H cocktail. qRT-PCR reactions each used cDNA from 20 ng of starting total RNA in a 1× SYBR Sensimix (Bioline) reaction with 0.25 µM of each primer (Table S2). Each biological sample had three technical replicates for both the aat and ocl2 primer sets. The qRT-PCR conditions used 40 cycles with 30 sec at 60° annealing and 45-sec extension steps, the melt curve ramped from 60° to 95° in 20 min. The technical (three per template) and biological (three pools of three seedlings each) replicates were combined to calculate the average ocl2 abundance relative to aat as 2(aat Ct - ocl2 Ct).
Results
Global run-on sequencing profiles nascent transcription in maize seedlings
Additional Pol II-derived RNAPs in multicellular plants (Luo and Hall 2007) may affect transcription dynamics across the genome, particularly as all of these RNAPs share accessory subunits with Pol II (Ream et al. 2009; Tucker et al. 2011; Haag et al. 2014). Pol II transcription of both LTR retrotransposons (Hale et al. 2009) and certain pl1 alleles (Erhard et al. 2013) is increased in rpd1 mutants. However, dysregulation of these specific loci cannot explain all the developmental phenotypes observed in rpd1 mutants (Parkinson et al. 2007; Erhard et al. 2009). We used GRO-seq to view nascent transcription profiles ± RPD1 to identify both particular (haplotype specific) and general (locus independent) effects of Pol IV loss on maize genome transcription.
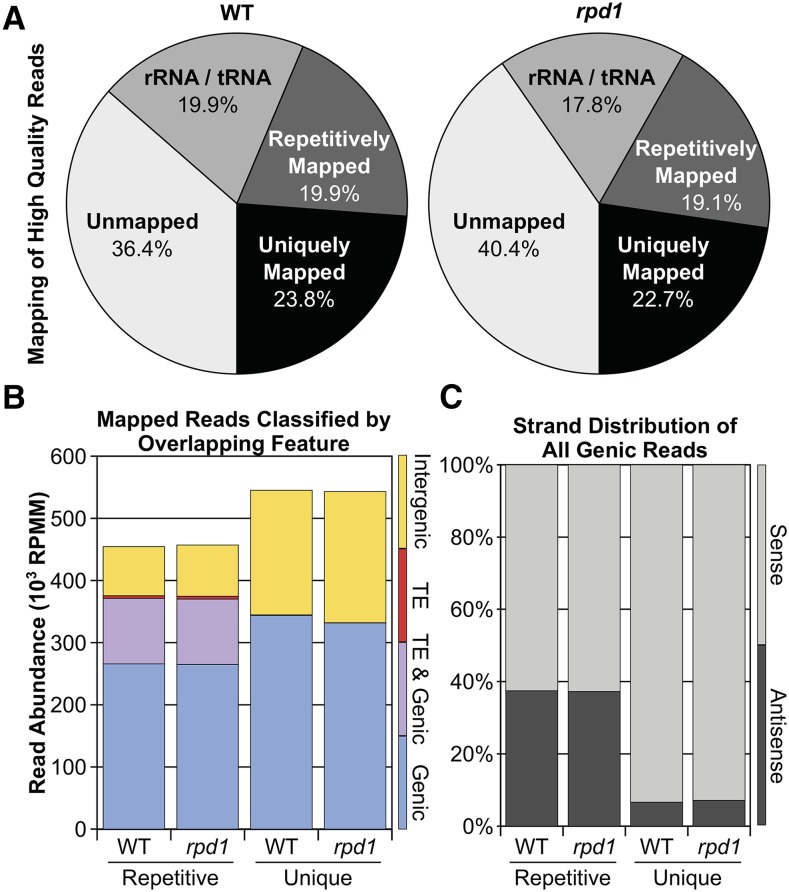
GRO-seq libraries were prepared using sibling rpd1 mutant and nonmutant (WT) seedlings with each library representing nuclei from 10 separate individuals (see Materials and Methods). Sequencing reads from these libraries were mapped to the B73 reference genome (Schnable et al. 2009) (Figure 1A). Transcripts from all five classes of maize RNAPs (Pols I–V) could be represented in the WT GRO-seq library, whereas only those requiring Pol IV function (including Pol IV transcripts) would be absent in the rpd1 mutant library. To focus predominantly on Pol II, IV, and V transcription, we removed most Pol I and III products by excluding rRNA- and tRNA-aligning reads from subsequent analysis (see Materials and Methods). Genomic non-rRNA/non-tRNA reads separated into repetitively aligning and uniquely aligning groups show similar distributions ± RPD1 (Figure 1A). To evaluate whether the libraries are enriched for nascent transcripts as opposed to spliced mRNAs, we compared the exonic/intronic distributions of genic reads (Figure S1). Both repetitively and uniquely mapping reads include intronic sequences (∼90 and ∼40%, respectively), confirming the enriched representation of nascent, unspliced transcripts.
Figure 1.
GRO-seq reads are similarly distributed in WT and rpd1 mutant libraries. (A) Percentages of WT and rpd1 mutant GRO-seq reads that are unmappable, map to rRNA/tRNA sequences, map repetitively (>1 alignment), or map uniquely to the B73 reference genome. (B) Distribution of repetitively and uniquely mapped reads [reads per million mapped (RPMM)] from A to annotated genes (blue), TEs (red), both (purple), or neither (intergenic; yellow). (C) Strandedness of the best alignment to gene models of all potentially genic reads (those that align with genes only or with both genes and TEs; blue and purple regions from B, respectively).
Because genes and TEs are predicted to be differentially affected by Pol IV loss, we categorized each read as having possibly originated from a TE, a gene (including annotated UTRs and introns), both, or neither (intergenic). Most uniquely mapping reads originate from genic or intergenic loci having little overlap with TEs (Figure 1B, Unique). Repetitively mapping reads align to TE-like sequences (Figure 1B, Repetitive), although these TE-like reads usually align to genes as well (Figure 1B, purple boxes). Approximately 96% of repetitive reads aligning to TE sequences could originate from genic transcripts. To ensure that this enrichment is not biased by our categorical analysis, we repeated the analysis on a set of in silico sequences randomly generated from the maize genome (see Materials and Methods; Figure S2) and found 97.3% of the TE-aligning, in silico-generated 32mers also aligned to genes. These results indicate that the majority of TE sequences represented in nascent transcription profiles are likely found within introns and untranslated regions of gene-derived Pol II RNAs.
Exclusively genic reads in both libraries (61 and 59.7% of all mappable WT and rpd1 mutant reads, respectively) are highly enriched compared to the prevalence of genic sequences in the maize genome (8%) (Figure S3), indicating that these GRO-seq profiles represent largely genic transcription. Because GRO-seq reads provide strand-specific information, we could also identify a sense-oriented strand bias among all genic reads, particularly the uniquely mapping reads that originate from the mapped locus (Figure 1C). Even reads representing TEs embedded in host genes appear biased toward the sense strand: 95% of uniquely aligning TE-like reads (5937 and 5641 reads in WT and rpd1 mutant libraries) are sense-oriented relative to their host genes. In all comparisons, the read distributions remain similar between WT and rpd1 mutant libraries, consistent with prior run-on transcription results showing that Pol IV contributes <5% of the transcribed nuclear RNA pool (Erhard et al. 2009) and the recent finding that Pol IV transcripts are rapidly processed to siRNAs (Li et al. 2015). These results indicate that loss of RPD1 does not generally shift transcription away from genes and toward TEs, suggesting that Pol IV has a more precise mechanism for regulating transcription of its targets.
Maize seedling transcription is focused on genic regions
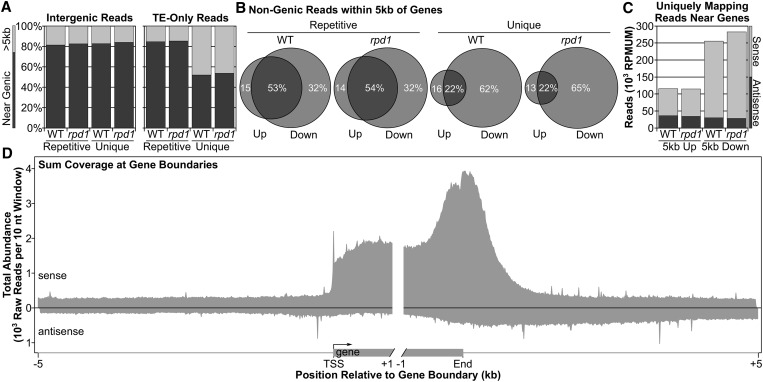
Mappable GRO-seq reads not aligning to either gene or TE features represented the “intergenic” category. These reads could have several origins including unannotated genes and TEs or noncoding DNA sources for siRNAs. However, we find that most nongenic non-TE reads represent pretermination transcription downstream of currently annotated polyadenylation addition sites (PAS) (Figure 2). Nascent transcription profiles in Homo sapiens (Core et al. 2008), Drosophila melanogaster (Core et al. 2012), and Caenorhabditis elegans (Kruesi et al. 2013) also identify transcription beyond the PAS and, in some cases, antisense transcription upstream of the TSS. To test whether nascent transcription is enriched near maize genes, we queried nongenic read alignments at regions 5 kb upstream of annotated TSSs and 5 kb downstream of annotated 3′ ends. Both intergenic and TE-only reads consist largely (>80%) of sequences that can align to within 5 kb of a gene model, representing potential genic reads (Figure 2A). The one exception is uniquely mapping TE-only reads, where only half of the reads align within 5 kb of genes. These intergenic and TE-only reads >5 kb from the nearest gene comprise ∼5% of the maize seedling non-rRNA/non-tRNA transcriptome (see Materials and Methods). At our current depth of sequence coverage we cannot confidently determine if transcription of those loci far from genes is influenced by RPD1 loss. However, our datasets are enriched over genes (∼65% of uniquely mapping reads in both WT and rpd1 mutant datasets) with an average sense-strand coverage of 173 (rpd1 mutant) to 187 (WT) raw reads per gene; we therefore continued our focus predominantly on near-genic regions, which are enriched in our dataset.
Figure 2.
Nongenic GRO-seq reads are enriched near genes. (A) Percentage of nongenic/non-TE (intergenic) or TE-only reads that align near genes (within 5 kb; dark gray) or >5 kb (light gray) from genes. (B) Nongenic reads within 5 kb of genes found exclusively upstream (Up, left circle), downstream (Down, right circle), or at both ends (intersection) of a nearby gene. (C) Distribution of uniquely mapping near-genic reads [reads per million uniquely mapped (RPMUM)] by strand orientation relative to the nearby gene model. (D) Metagene profile of uniquely mapping WT GRO-seq reads summed over 10-nt windows ±5 kb from FGS models.
In support of the idea that near-genic reads in the maize GRO-seq profiles represent Pol II pretermination extensions of genic transcription units, uniquely mapping near-genic reads align predominantly in the downstream 5 kb (up to 84 and 87% of WT and rpd1 mutant near-genic reads, respectively; Figure 2B). The overlap of uniquely mapping reads between the upstream 5 kb and downstream 5 kb (22% in both genotypes) likely represents reads aligning between genes separated by <10 kb. Repetitively mapping near-genic reads have a more even distribution between upstream and downstream 5 kb, which could represent an artifact of alignments to nonorigin loci. With uniquely mapping reads, where alignments likely correspond to the originating locus, near-genic reads are predominantly sense-stranded (Figure 2C) in accord with the hypothesis that they represent continuation of genic transcription.
We next used pile-ups of uniquely mapped WT GRO-seq reads across genic loci to profile the typical maize genic transcription unit at higher resolution. Combining all sense and antisense profiles (Figure S4) into a metagene composite (Figure 2D) confirms the sense-strand enrichment downstream of currently annotated gene models. Additionally, the composite profile indicates that most pretermination transcription occurring 3′ of PASs concludes within 1–1.5 kb of currently annotated gene ends. This result agrees with estimates from metazoan profiles (Core et al. 2008, 2012; Kruesi et al. 2013). Beyond presumptive genic transcription termination points and upstream of TSSs there is remarkably little evidence of transcription. Although our results do not distinguish RNAs produced from different RNAPs, the nature of the read enrichment at genes (starting near the TSS and extending beyond the 3′ PAS) and the sense-strand bias supports the prediction that most of these reads derive from gene-associated nascent Pol II RNAs.
Promoter-proximal transcription in maize is distinct from that seen in metazoans
The composite metagene profile (Figure 2D) highlighted unexpected features of typical maize transcription initiation. The beginning of composite genic transcription is marked with a prominent narrow peak of GRO-seq reads nearly coincident with the TSS (Figure 2D). Sense-oriented peaks located ∼50 bp downstream of TSSs are identified by GRO-seq profiles in humans (Core et al. 2008) and Drosophila (Core et al. 2012) and under stress conditions in C. elegans (Kruesi et al. 2013; Maxwell et al. 2014). These peaks are cited as evidence of promoter-proximal Pol II pausing, a transcriptional regulatory mechanism first described at the hsp70 gene in Drosophila (Rougvie and Lis 1988). It is unclear whether the more upstream maize TSS-proximal peak (Figure 2D) represents Pol II pausing. Over 1/4 of maize gene models used in the original metagene profile (Figure 2D) begin with the triplet ATG sequence (27%), compared to only 2% when triplet sequences are sampled 5 kb upstream of the gene models (an approximation of ATG enrichment genome-wide) (Figure S5). This finding indicates that translation, rather than transcription, initiation sites define many current annotations of maize gene start positions. To test if an alternative gene start definition would shift the TSS-proximal peak, we first defined a set of genomic TSS annotations using maize seedling and young leaf full-length cDNA sequences (see Materials and Methods; Soderlund et al. 2009). A similar metagene analysis using this validated set of TSSs places the sense-oriented GRO-seq peak upstream of the TSS (Figure S6). This result indicates that the peak positions relative to TSSs are dependent on annotation methods but likely do not represent canonical Pol II pausing as described in metazoans.
Nascent transcription profiling (Core et al. 2008) and short RNA cDNA libraries (Seila et al. 2008) identified evidence of divergent antisense transcription peaking at ∼ −250 bp at mammalian promoters. Similar to Drosophila GRO-seq profiles (Core et al. 2012), our metagene profile has no evidence of such a broad antisense peak (Figure 2D). The only peak at ∼ −250 bp is due to antisense-biased GRO-seq coverage of a single gene (Figure S7), which likely represents a transcription unit unrelated to the annotated gene model. Together, these two characteristics of near-promoter transcription—a TSS-proximal peak and lack of divergent transcription—distinguish the maize seedling transcriptional environment at promoters from that of currently profiled metazoans.
Pol IV affects nascent transcription at gene boundaries
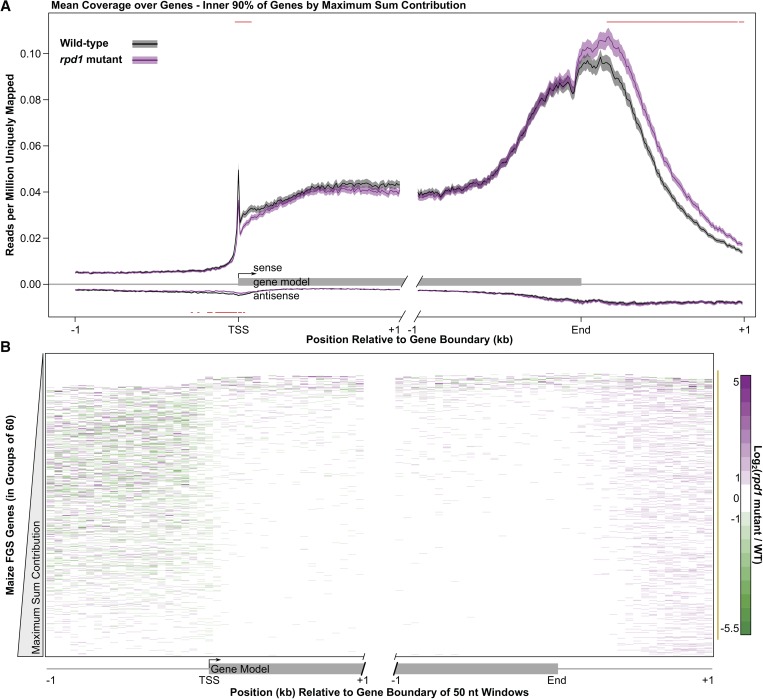
Additional Pol II-derived plant RNAPs (Pols IV and V) represent a key distinction between the transcriptional landscape of multicellular plants and other eukaryotes. To evaluate Pol IV effects on transcriptional activity at functionally important sites surrounding genes, such as promoters and transcription termination sites, we generated and compared metagene profiles displaying the mean GRO-seq read abundance across a composite of annotated gene edges and their flanking genomic regions (Figure 3A). To exclude outliers, we sorted gene models based on their maximum read count contribution to the metagene plot and included only the inner 90% (∼28.6 thousand) of gene models in the metagene profiles (see Materials and Methods).
Figure 3.
Pol IV loss alters global transcription profiles at gene boundaries. (A) WT and rpd1 mutant mean GRO-seq read coverage (black and purple lines, respectively) of 90% of the maize genes within 1 kb of gene start (TSS) or 3′ end (End). Gray and purple shading represent 95% confidence intervals; red horizontal bars highlight 10-nt nonoverlapping windows that significantly differ between libraries (Welch’s t-test by window, corrected to α = 0.05 with the Holm–Bonferroni method for multiple sampling). (B) Variation in coverage between WT and rpd1 mutant libraries for all FGS genes. Fold change was calculated from the average coverage (reads per million uniquely mapped) of 60 neighboring genes when sorted by their maximum sum contribution. Fifty-nucleotide windows with zero coverage in either library are plotted in white. The gold bar highlights the inner 90% of genes used in A.
Profile comparisons reveal changes in transcription at gene boundaries ± RPD1, while transcription of genic and upstream regions remain unaffected (Figure 3A and Figure S6). Near TSSs, the rpd1 mutant library has significantly lower read coverage in both strand orientations (Figure 3A, Welch’s t-tests in 10-nt windows, corrected for multiple sampling by the Holm–Bonferroni method at α = 0.05). Heatmap representations of read abundance differences across individual gene boundaries indicate that the trends observed in the metagene summary apply to most genes (Figure 3B). Elsewhere, the WT and rpd1 mutant GRO-seq profiles are remarkably similar, particularly for regions having high coverage (sense strand over gene bodies and 1 kb downstream of gene ends) (Figure S8). The sense-strand gene body coverage between libraries has Spearman’s rank correlation coefficients (ρ) of 0.971, 0.963, and 0.977 (first 1 kb, middle, and last 1 kb, respectively), which approximate the ρ = 0.967 observed between biological replicates in the original GRO-seq analysis (Core et al. 2008). Together, the metagene summary, fold-change heatmap, and Spearman’s correlations highlight that RPD1 has no general impact over the coding region of genes. More striking, the pretermination region beyond the PAS has significantly increased read coverage in the absence of RPD1. Downstream of the PAS there is evidently increased transcription for most genes relative to upstream of the PAS, and this 3′ transcription is even more pronounced in rpd1 mutants (Figure 3B). Upstream of currently annotated gene models, the fold differences in read abundances are variable as expected for regions of relatively slow or underrepresented transcription; fold changes in read abundances representing antisense-oriented transcription (Figure S9) show similarly variable patterns. Although window sizes used for generating fold-change heatmap data are larger (50 vs. 10 nt) than in the metagene profiles, most of these larger windows directly above TSSs still show negative fold changes, indicating increased transcription in WT vs. rpd1 mutant samples is a common feature of many genes (Figure 3B).
The RPD1-dependent changes in the GRO-seq profiles observed near gene boundaries implicate Pol IV activity near genes. Because Pol IV is required to generate 24-nt RNAs, presumably through downstream processing of Pol IV transcripts by RNA interference-like machinery (Li et al. 2015; Matzke and Mosher 2014; Matzke et al. 2015), we looked for 24-nt read (24mer) evidence supporting Pol IV action nearby recognized Pol II transcription units. Previous genome-wide profiling of maize 24mers identified enrichment 1.5 kb upstream of genes (Gent et al. 2014). To profile 24mers within 1 kb of gene boundaries, we pooled ∼75 million B73 16- to 35-nt RNA reads from published datasets for metagene analysis. To increase effective sequencing depth, we pooled four distinct datasets: 3-day-old seedling root (Gent et al. 2012), unfertilized cobs (Gent et al. 2013, 2014), and 11-day-old seedling shoot apices (Barber et al. 2012), all representing the B73 inbred background, and subjected them to similar coverage analysis near genes (see Materials and Methods). Most 24mers represent repetitive features so their originating loci cannot be determined. We therefore limited analysis to uniquely mapping 24mers (8.6 million). These 24mers are enriched both upstream and downstream of genes (Figure S10A and B), and because they are uniquely mapping, they are presumably generated from Pol IV transcription occurring in regions immediately flanking genes. Biogenesis of 24-nt RNAs also requires RDR2 to create a double-stranded RNA from Pol IV transcripts (Li et al. 2015; Matzke et al. 2015). In Arabidopsis, RDR2 functions only in physical association with Pol IV (Haag et al. 2012). While the maize RDR2 ortholog (Alleman et al. 2006) has not been tested for a similar requirement, it physically associates with Pol IV (Haag et al. 2014) and is required for 24-nt RNA biogenesis (Nobuta et al. 2008). We therefore used an existing siRNA dataset from an rdr2 mutant (Gent et al. 2014) as a proxy for identifying RPD1-dependent 24-nt RNAs. Although this comparative analysis is more limited (11.1 million total reads, ∼551,000 unique 16-35mers, and ∼154,000 unique 24mers), the 24mer coverage across all genes showed no enrichment flanking gene boundaries (Figure S10A, C, and D), indicating that the near-genic 24-nt RNAs are RDR2-dependent. These 24mer analyses support the presence of Pol IV immediately upstream and downstream of Pol II genes. Together, this analysis of maize rpd1 mutants at gene boundaries identifies previously unknown interactions by which Pol IV affects Pol II transcription at discrete positions near genic regions. In addition, these results identify both conserved and novel features of transcription in higher plants vs. metazoans.
Transcription of specific genes is affected by RPD1
RPD1 is required for restriction of silkless1 gene expression from apical inflorescences, thus ensuring proper male flower development (Parkinson et al. 2007), although how RPD1 regulates developmentally important genes is unknown. We employed a computational method (DESeq; Anders and Huber 2010) that assigns statistical significance to regions annotated as genes over- or underrepresented in uniquely mapping GRO-seq reads from either WT or rpd1 mutant libraries. This approach robustly controls for false positives (type I errors) across a dynamic range of coverage levels and, by pooling information from similarly represented loci, can estimate the required mean and variance values for each locus even when the total sequencing depth and/or number of biological replicates is small (Anders and Huber 2010). Using this method, we could treat the WT and rpd1 mutant libraries as effective biological replicates (see Materials and Methods), assuming that at most loci there was no differential transcription, an assumption supported by the trends observed in the global analysis (Figure 1), the metagene profiling (Figure 3), and the Spearman’s rank correlation coefficients (Figure S8). As a result, we have higher confidence in our identified differentially transcribed loci than if we treated each library individually, although the method likely underestimates the total number of differentially transcribed loci.
Applying this stringent statistical method identified a total of 209 annotated genes whose seedling-stage transcription across the entire gene body (annotated UTRs, introns, and exons) is significantly increased or decreased by loss of RPD1. We excluded a cluster of 26 gene models having significantly reduced GRO-seq representation in the rpd1 mutant profiles found within 20 Mb of the rpd1-1 introgressed haplotype as these were likely identified because of an inability of some polymorphic rpd1 mutant reads to align to B73 sequences in this interval. The remaining 183 genes represent potential direct or indirect targets of RPD1/Pol IV regulation (Figure 4A and Table S3). To determine whether or not genic TEs were related to RPD1-affected transcription, we re-annotated the 183 genes and compared their TE content with 200 randomly selected genes (Table S3 and Table S4). This analysis indicates that RPD1-regulated genes have an average TE content: 64% of the differentially transcribed genes vs. 66% of the randomly selected genes.
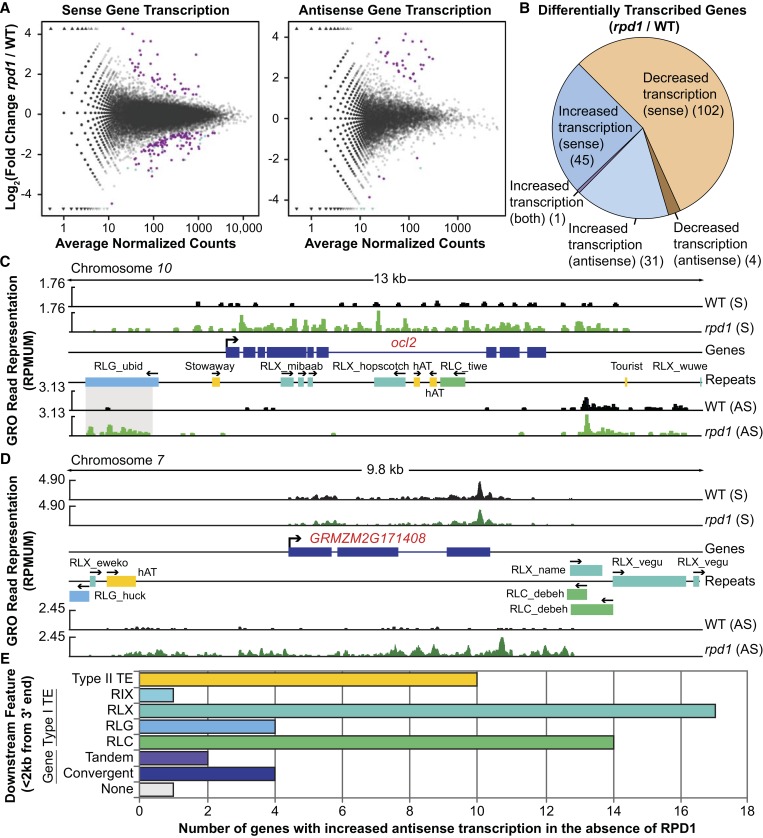
Figure 4.
Specific alleles are susceptible to Pol IV-induced changes in gene expression. (A) Across FGS gene bodies, the log2 fold change (rpd1 mutant/WT) of uniquely mapping read coverage vs. total coverage (average of WT and rpd1 mutant reads). Triangles represent genes with infinite fold change due to zero coverage from WT (top) or rpd1 mutant (bottom) uniquely mapping reads. Of the 39,656 FGS gene bodies analyzed, those with zero coverage in both WT and rpd1 mutant datasets (7783 and 9667 for sense and antisense strand transcription, respectively) were excluded from the plots. Purple dots represent genes with significantly (by the DESeq statistical method of Anders and Huber 2010; see Materials and Methods) increased or decreased GRO-seq read representation in rpd1 mutants. Teal dots represent genes within 20 Mb of the rpd1 locus whose decreased transcription in rpd1 mutants may reflect alignment artifacts (see Materials and Methods) and are excluded from subsequent analysis. (B) Distribution of categories (by direction of the change and strand) among transcriptionally altered genes in rpd1 mutants. (C) Genome browser view of WT (black peaks) and rpd1 mutant (green peaks) GRO-seq reads [normalized to reads per million uniquely mapped (RPMUM)] in sense (S) and antisense (AS) orientation over the ocl2-coding region and ∼3 kb of flanking genomic sequences on chromosome 10. Gray-shaded area highlights the ubid TE fragment 5′ of ocl2 having increased transcription in rpd1 mutants. (D) Gene browser view of GRMZM2G171408 showing increased antisense transcription in rpd1 mutants. Sense (S) and antisense (AS) transcription occur in distinct units of GRO-seq coverage in both WT (black peaks) and rpd1 mutant (green peaks) libraries. (E) Distribution of downstream features within 2 kb by type. Type I TEs are subdivided into LINE-like elements (RIX) and Copia (RLC), Gypsy (RLG), and Unknown (RLX) classes of LTR TEs. Color coding in E applies to TEs in browser views. Arrows indicate orientation of gene and TE features.
Visual inspection of the differentially transcribed genes having sense-strand changes (148 in total) using genome browser displays identified two classes: those with consistent GRO-seq read coverage across the entire gene model and those having more biased or localized coverage. In total, 32 of 46 (70%) having increased transcription (Table 1) and 96 of 102 (94%) with decreased transcription (boldface entries in Table S3) matched current gene annotations and are likely related to RPD1-dependent effects on genic transcription. The remaining 20 genes identified as differentially expressed by DESeq analysis have changes in GRO read coverage localized to subgenic regions, sometimes overlapping TEs or the beginning of the gene with persistent coverage extending from the 3′ end of an upstream gene.
Table 1. Curated genes with increased sense-oriented transcription in rpd1 mutants.
| Gene | Annotation | Fold change (rpd1/WT) | P-valuea |
|---|---|---|---|
| GRMZM2G303010 | NBS-LRR disease resistance protein | 9.113 | 7.07E-05 |
| AC235534.1_FG007 | ocl2 (HD-ZIP IV) | 7.932 | 2.65E-09 |
| GRMZM2G161658 | Epoxide hydrolase 2-like | 7.893 | 3.01E-22 |
| GRMZM2G132763 | Putative LRR receptor-like protein kinase | 6.681 | 2.27E-02 |
| GRMZM2G062716 | Defense-related protein (type 1 glutamine amidotransferase domain) | 5.867 | 2.49E-02 |
| GRMZM2G047105 | Hypothetical, unknown protein | 5.867 | 2.49E-02 |
| GRMZM2G088413 | Hypothetical, unknown protein | 5.098 | 1.47E-02 |
| GRMZM5G830269 | Hypothetical, unknown protein | 4.639 | 4.71E-02 |
| GRMZM2G333140 | Hypothetical, unknown protein | 4.490 | 6.73E-02 |
| GRMZM2G045155 | B12D protein | 4.243 | 2.55E-02 |
| GRMZM2G147724 | Phosphotidic acid phosphatase | 4.023 | 1.51E-04 |
| GRMZM2G043242 | Putative ATP-binding, ATPase-like domain-containing protein | 3.963 | 9.42E-08 |
| GRMZM2G147399 | Early nodulin 93 | 3.897 | 2.49E-02 |
| GRMZM2G028677 | Putative cytochrome P450 superfamily protein | 3.824 | 7.34E-02 |
| GRMZM2G009080 | Hypothetical, unknown protein | 3.542 | 7.05E-03 |
| GRMZM2G131421 | Early nodulin 93 | 3.421 | 8.64E-04 |
| GRMZM2G174449 | Hypothetical, unknown protein | 3.329 | 1.26E-02 |
| AC197705.4_FG001 | Pyruvate decarboxylase isozyme 1 | 3.206 | 1.26E-02 |
| GRMZM2G045560 | WRKY DNA-binding domain-containing protein | 3.102 | 1.89E-02 |
| GRMZM2G300965 | Respiratory burst oxidase-like protein B | 2.897 | 3.37E-05 |
| GRMZM2G053503 | Ethylene-responsive factor-like protein (ERF1) | 2.548 | 2.63E-02 |
| GRMZM2G087063 | Hypothetical, unknown protein, DUF 2930 | 2.444 | 2.54E-02 |
| GRMZM2G051683 | Anthocyanidin 5,3-O-glucosyltransferase | 2.415 | 1.48E-03 |
| GRMZM2G145213 | 14-3-3-like protein | 2.406 | 1.27E-03 |
| GRMZM2G024996 | Pseudogene, transposon relic, upregulated | 2.385 | 1.21E-03 |
| GRMZM5G814164 | Peroxisome biogenesis protein 3-2-like | 2.360 | 2.80E-03 |
| GRMZM2G168747 | Nrat1 aluminum transporter 1 | 2.182 | 2.49E-02 |
| GRMZM2G031827 | Splicing factor U2af 38-kDa subunit | 2.174 | 2.65E-02 |
| GRMZM2G392791 | Epoxide hydrolase 2-like | 2.107 | 4.14E-02 |
| GRMZM2G083538 | Amino-acid-binding protein (ACR5) | 2.058 | 2.54E-02 |
| GRMZM2G021369 | Putative AP2/EREBP transcription factor | 1.986 | 2.95E-02 |
| GRMZM2G013448 | 1-Aminocyclopropane-1-carboxylate oxidase | 1.981 | 3.24E-02 |
P-values were adjusted by the Benjamini–Hochberg method for multiple testing (Benjamini and Hochberg 1995) as part of the DESeq analysis (Anders and Huber 2010).
Three examples of differentially transcribed genes were subsequently examined and validated at mRNA levels. Results of oligo(dT)-primed RT-PCR analyses using similar biological materials confirmed increased sense-oriented mRNA levels of all three genes tested from the set identified by computational analysis of WT and rpd1 mutant GRO-seq reads (Figure S11). As an example, the predicted fold changes ± RPD1 of outer cell layer 2 (ocl2) nascent transcripts and mRNAs are ∼7.9- and 3-fold increases, respectively. Additional qRT-PCR results estimate a 10-fold increase in ocl2 mRNAs in the absence of RPD1 (Figure S12). These results validate the GRO-seq technique and DESeq analyses in discovering genes whose Pol II transcription is affected by RPD1.
We expected to detect primarily increased sense-specific transcription by comparing WT and rpd1 mutant GRO-seq profiles because the Pl1-Rhoades allele is transcriptionally repressed by RPD1 (Hollick et al. 2005). However, the largest fraction (∼0.56) of genes affected by loss of RPD1 has lower sense-oriented transcription in rpd1 mutants (Figure 4B and Table S3). One possible explanation for this result is that genomic features transcriptionally repressed in an rpd1 mutant background represent indirect effects. Potentially related to this idea, the presumed maize ortholog of the Arabidopsis REPRESSOR OF SILENCING 1 (ROS1) gene, which encodes a DNA glycosylase that facilitates demethylation of cytosine residues (Morales-Ruiz et al. 2006), is transcriptionally repressed in rpd1 mutants (2.9-fold decrease) although this differential representation does not pass our statistical cutoff after adjustment for multiple testing by our DESeq test (raw P-value: 0.002, adjusted P-value: 0.2). Maize ros1 RNA levels are also reduced in meristems of rdr2 mutants (Jia et al. 2009), although the mechanism by which any genes are repressed in the absence of RPD1 or Pol IV-dependent siRNAs remains unknown.
Among differentially transcribed gene models in rpd1 mutants (Table S3), we identified several candidates whose dysregulation could result in developmental abnormalities and potentially contribute to rpd1 mutant phenotypes (Parkinson et al. 2007). The candidate showing the second greatest increased transcription is ocl2 (Figure 4C), a member of the plant-specific homeodomain leucine zipper IV (HD-ZIP IV) family of transcription factors predicted to have leaf epidermis-related functions in maize (Javelle et al. 2011). Mature rpd1-2 mutant plants often exhibit problems maintaining proper leaf polarity (adaxialized leaf sectors) (Parkinson et al. 2007). Interestingly, ocl2 is not normally expressed in epidermal or mesophyll cells (Javelle et al. 2011), which comprise the majority of cells used for GRO-seq library generation, indicating that this gene is transcribed outside its normal expression domain in rpd1 mutants.
Genome browser visualization of GRO-seq reads uniquely mapping to the genomic region containing ocl2 (Figure 4C) highlights coincident transcription profiles of a proximate TE and this RPD1-regulated gene. A fragment of an LTR retrotransposon of the Gypsy class (RLG) assigned to the ubid family located ∼1.3 kb upstream of the ocl2 gene is also transcribed in rpd1 mutants, but in antisense orientation with respect to the ocl2-coding region (Figure 4C). These results identify the ubid fragment upstream of ocl2 as a putative controlling element for this allele, with the absence of RPD1 corresponding with transcriptional increases of both the ubid fragment and the adjacent gene.
We also identified 36 genes transcriptionally altered in the antisense orientation in rpd1 mutants (Figure 4B). Pol IV is implicated in the production of an antisense precursor transcript and corresponding 24-nt siRNAs, homologous to the 3′ end of the Arabidopsis gene FLOWERING LOCUS C (FLC) (Swiezewski et al. 2007), which encodes an epigenetically regulated MADS Box factor important for vernalization and the regulation of flowering time (Dennis and Peacock 2007). However, only four genes (Table S3) show decreased antisense transcription in the rpd1 mutant, indicating that Pol IV-dependent antisense transcription of genes is unlikely a primary mechanism of its action on a genome-wide scale. Many more genes (32 total) were recognized having increased transcription in antisense orientation (Figure 4B), yet only one of these (GRMZM2G045560; a gene model encoding a WRKY DNA-binding domain-containing protein) had a significant (DESeq method of Anders and Huber 2010) increase in sense transcription as well. Most of these examples appear to represent transcription of noncoding RNAs initiated 3′ of the annotated genes (Figure 4D). Additionally, visual inspections indicate that most (22 of 32, 69%) of these transcription units begin at downstream TEs (Figure 4D). Counting MTEC TE annotations within 2 kb of the 3′ ends of these 32 genes identifies a large number of LTR retrotransposons immediately downstream (Figure 4E). These results support previous findings showing that Pol IV loss allows increased transcription of certain LTR retrotransposons (Hale et al. 2009), and they highlight how such promiscuous Pol II transcription could affect gene regulation (Kashkush and Khasdan 2007). Genes encoding a histidine kinase receptor for cytokinin, an important phytohormone, and a homolog of an Arabidopsis HD-ZIP factor ATHB-4 also have elevated antisense transcription profiles in the absence of RPD1 (Table S3), indicating the potential for a biologically significant role for this novel mechanism of RPD1 gene regulation in maize.
Transcription of TE families and specific TEs is affected by RPD1
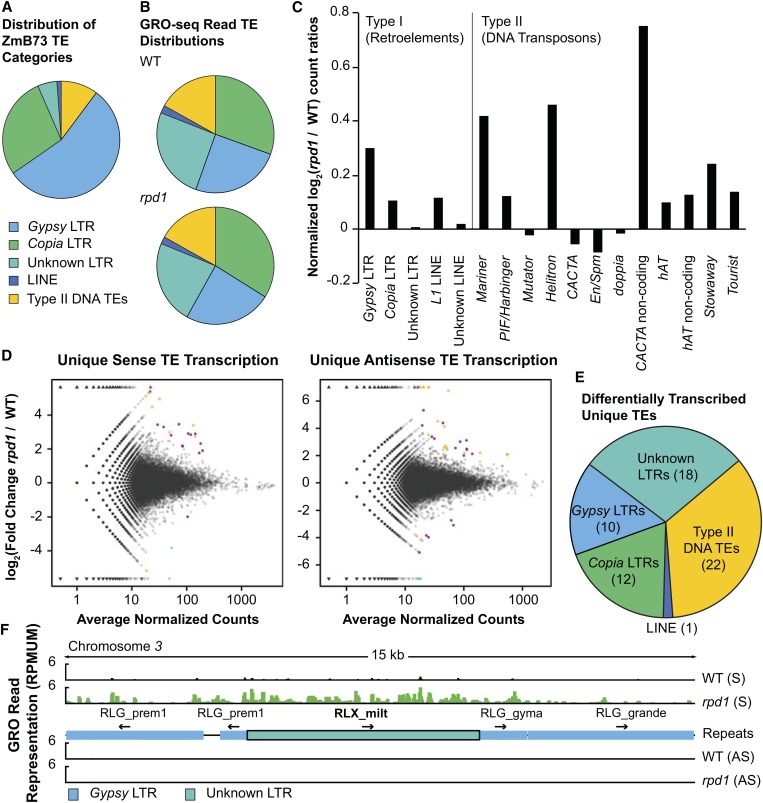
While overall TE transcription appears modest (Figure 1B and Figure 2A), we compared GRO-seq read representations among specific TE families and at individual TE loci to determine if certain types are preferentially transcribed. Direct alignments of total WT and rpd1 mutant GRO-seq reads (unique and non-unique) to consensus sequences of annotated maize TE classes and major superfamilies allowed us to compare transcription of these features to their relative abundance in the maize genome (Figure 5A) (Schnable et al. 2009). Although the RLG class of LTR TEs is the most abundant TE superfamily in the maize genome (Figure 5A) (Schnable et al. 2009), it is underrepresented in both GRO-seq profiles as compared to the LTR Unknown class (RLX) and to type II DNA TEs (Figure 5B).
Figure 5.
Pol IV loss affects both entire TE families and individual elements. (A) Distribution of TE categories within the B73 genome (Schnable et al. 2009). (B) Distribution of total (unique and repetitive) WT and rpd1 mutant GRO-seq reads within the different TE categories shown in A. (C) Log2 ratios (rpd1 mutant/WT) of GRO-seq reads, normalized to total mappable reads, mapping to annotated TE superfamilies. (D) Log2 fold change (rpd1 mutant/WT) of uniquely mapping reads in sense and antisense orientation to genomic regions annotated as TEs vs. total coverage (averages of WT and rpd1 mutant reads) to those regions. Triangles represent TEs with infinite fold change due to zero coverage from WT (top) or rpd1 mutant (bottom) uniquely mapping reads. Of the 1,612,638 TE annotations analyzed, those with zero coverage in both WT and rpd1 mutant datasets (1,392,382 and 1,399,008 for sense and antisense strand transcription, respectively) were excluded from the plots. Purple dots represent TEs with significantly (by the DESeq statistical method of Anders and Huber 2010; see Materials and Methods) increased or decreased GRO-seq read representation in rpd1 mutants; orange stars or triangles represent those differentially transcribed TEs farther than 5 kb from the nearest FGS gene. Teal dots represent TEs within 20 Mb of the rpd1 locus whose decreased transcription in rpd1 mutants may reflect alignment artifacts (see Materials and Methods) and are excluded from subsequent analysis. (E) Distribution of transcriptionally altered unique TEs among TE categories shown in A. (F) Genome browser view of WT (black peaks) and rpd1 mutant (green peaks) GRO-seq reads [normalized to reads per million uniquely mapped (RPMUM)] in sense (S) and antisense (AS) orientation over a 15-kb interval on chromosome 3 containing an RLX_milt type I element with increased transcription in rpd1 mutants. Only the element outlined in black has significantly altered GRO-seq read representation in rpd1 mutants based on the statistical threshold used (Anders and Huber 2010; see Materials and Methods).
To determine if distinct TE groups were differentially represented in nascent transcriptomes, we compared normalized abundances of WT and rpd1 mutant reads mapping to annotated TE superfamilies (Figure 5C). This comparison identified several TE superfamilies with elevated transcription in the rpd1 mutant background (Figure 5C), including Gypsy, Mariner, Helitron, and CACTA noncoding elements. The majority of TE-derived GRO-seq reads cannot be mapped uniquely to specific genomic coordinates, limiting the detection of individual rpd1-affected TE loci to those TEs harboring significant sequence polymorphisms with respect to their family members. We thus employed the same method (DESeq; Anders and Huber 2010) used with genes to identify genomic regions annotated as TEs (Baucom et al. 2009; Schnable et al. 2009) having statistically significant differences in uniquely mapping GRO-seq read coverage. This method identifies 63 individual TEs (Figure 5D and Table S5) representing several different superfamilies (Figure 5E) whose transcription is either increased (Figure 5F) or decreased in rpd1 mutants in either sense or antisense directions. Only 28 of these unique TEs are not within or nearby (±5 kb) genic regions, and some identify larger TE regions affected by RPD1 (Figure 5F and Table S5). These results agree with previous analyses (Hale et al. 2009) indicating that RPD1 prohibits mRNA accumulation of certain LTR retrotransposons by interfering with normal Pol II transcription and RNA processing.
Discussion
Our GRO-seq profiling of WT and rpd1 mutant maize seedlings represents the first genome-wide nascent transcription analysis in plants and of a Pol IV mutant. The GRO-seq technique facilitates future studies of RNAP dynamics relevant to basic mechanisms of gene control in higher plants. Our results identify Pol IV effects on transcription at most gene boundaries, indicating that distinctions between higher plant and metazoan transcription may be partly related to the plant-specific expansion of RNAP diversity. We also identified specific TE and genic alleles that show significant changes in nascent transcription ± RPD1, a dataset that should prove useful for better understanding the role(s) of Pol IV function in TE silencing, paramutation, and maize development.
The GRO-seq method captures snapshots of active transcription, which can identify entire transcription units from initiation to termination, helping to identify alternative TSSs and cryptic transcripts. Through comparisons of GRO-seq and RNA-seq profiles, it should be possible to identify the extent to which regulation of gene expression occurs at the level of post-transcriptional RNA stability. As GRO-seq reveals aspects of transcriptional regulation absent from the mature mRNA, nascent transcriptome profiles in metazoans, plants, and fungi will continue to define and distinguish RNAP functions across eukaryotes.
General Pol IV effects on genic transcription
Similar to metazoans, maize transcription extends beyond the PAS with termination occurring within ∼1–1.5 kb downstream. However, because of its additional RNAPs, plants may have alternative mechanisms to terminate Pol II transcription. At maize 3′ gene ends, we speculate that Pol IV plays a role in attenuating aberrant readthrough transcription by Pol II into neighboring genes or TEs. This model is supported by the enrichment of RDR2-dependent 24-nt RNAs immediately downstream of PASs. Another possibility is that the kinetics of cotranscriptional mRNA splicing and/or polyadenylation are affected by Pol IV, perhaps related to the sharing of specific holoenzyme subunits (Haag et al. 2014). Together, the GRO-seq profiles and rpd1 mutant analysis indicate that Pol II termination in maize is unique relative to metazoans.
Maize Pol IV also affects transcription at 5′ gene boundaries. Our results show that rpd1 mutants have decreased transcription at most gene TSSs, identifying a previously unknown role for Pol IV at Pol II initiation sites. Enrichment of 24-nt RNAs immediately (this article) and further upstream (on average 1.5 kb in maize; Gent et al. 2014) of genes supports the presence of Pol IV at genic promoters. Because Pol IV is predicted to engage transcription bubble-like DNA templates (Haag et al. 2012) and appears to initiate at AT-rich and nucleosome-depleted regions (Li et al. 2015), perhaps Pol IV holoenzymes synthesize RNA, either abortively or productively, at loci undergoing Pol II transcription initiation. Such behaviors could account for the relatively higher abundance of both sense and antisense 5′ GRO-seq reads found in WT although this idea seems inconsistent with the observed patterns of 24mer vs. GRO-seq enrichment upstream of genes (Figure 3A and Figure S10). These discordant distributions indicate that the decrease in GRO-seq coverage near the TSS is an indirect effect of Pol IV loss affecting transcription from another RNAP. It remains formally possible that Pol V contributes to this TSS-proximate transcription as Arabidopsis Pol V associates with TE-proximal promoters and more transiently with other promoters (Zhong et al. 2012). Alternatively, the decrease in promoter proximal GRO-seq read coverage could be due to titration of Pol II to other initiation sites in the absence of Pol IV (Hale et al. 2009), such as to the LTR TEs downstream of genes showing increased antisense transcription in rpd1 mutants (Figure 4D).
The maize TSS-proximal peak of GRO-seq reads appears distinct from the promoter-proximal peak associated with canonical Pol II pausing in metazoans. Whether Pol II pausing, as described in humans and Drosophila (Core et al. 2008; Core et al. 2012), regulates transcription elongation of some maize genes remains unknown, although we observe no strong evidence for Pol II pausing in our datasets. Our experimental design focused on capturing a broad view of nascent transcription ± RPD1, and as such, we chose seedling tissue in which >90% of the genes produce detectable mRNAs via ultradeep sequencing (Martin et al. 2014). Seedlings grown under laboratory conditions may have no need for Pol II elongation regulation; C. elegans tends to show evidence of Pol II pausing only under stress (Kruesi et al. 2013; Maxwell et al. 2014). Additionally, we omitted Sarkosyl from the run-on reactions not knowing how this detergent might affect Pol IV function. This omission may also prevent detection of promoter-proximal Pol II pausing peaks as Sarkosyl can dissociate inhibitory factors holding Pol II at a canonical paused gene, hsp70, in Drosophila (Rougvie and Lis 1988). However, it should be noted that GRO-seq profiles in Drosophila indicate that the pausing peak, although greatly diminished, can still be detected in the absence of Sarkosyl (Core et al. 2012). While the nature of the maize TSS-proximal peak remains unclear, there is still a significant difference in transcription behavior at these regions ± RPD1.
A third characteristic identified in metazoan GRO-seq profiles is divergent transcription, which may be a by-product of previous Pol II initiations at the same promoter and/or a mechanism to maintain the nucleosome-free region (reviewed by Seila et al. 2009). These divergent transcripts may have roles as either regulatory scaffolds or sources of small RNAs (Core et al. 2008; Seila et al. 2008). Divergent transcription is prevalent at mammalian genes, but less so at C. elegans and Drosophila promoters, perhaps related to the directional specificity of favored promoter sequences (Core et al. 2012; Kruesi et al. 2013). We found no evidence of divergent transcription at maize promoters, potentially placing them in a similar category as Drosophila, which has a median 32-fold bias for sense-oriented transcription at promoters (Kruesi et al. 2013). This finding is curious because evidence in Arabidopsis indicates that Pol V can be recruited to Pol II promoters (Zhong et al. 2012), and profiles of uniquely mapping 24-nt RNAs (Figure S10) place Pol IV near genes. It may be that plant RNAPs provide divergent transcription to help maintain nucleosome-free regions and that our GRO-seq assay conditions do not detect such nascent transcripts.
If Pol IV and/or Pol V are present immediately upstream of genes, then why is there little evidence of nascent transcription upstream of genic units? The Pol IV and V catalytic cores differ from that of Pol II, leading to relatively slow elongation rates and insensitivity to the Pol II inhibitor α-amanitin in both Arabidopsis (Haag et al. 2012) and maize (Haag et al. 2014). These differences may also affect their sensitivity to the limiting CTP present in the GRO-seq run-on reaction that affects Pol II NTP incorporation rates (Core et al. 2008) or their ability to incorporate the brominated UTP analog. Because only uniquely mapping reads were analyzed at gene boundaries, repetitive Pol IV- and/or Pol V-derived reads would be excluded. Additionally, any nascent Pol IV RNAs cotranscriptionally processed into siRNAs (Haag et al. 2012; Li et al. 2015) would not have been incorporated into the GRO-seq libraries. However, we are still able to identify effects of RPD1 loss on Pol II transcription (Table 1; Figure 4C; Figure S11; and Figure S12). Within limits of the GRO-seq assay, our results indicate that Pol II behaviors are generally affected by Pol IV, and this defines fundamental differences in genic transcription between metazoans and higher plants.
Pol IV affects gene regulation
McClintock referred to TEs as controlling elements because of their potential to affect gene regulation (McClintock 1951). TEs are transcriptionally repressed by Pol IV action either through direct competitions with Pol II (Hale et al. 2009) or through chromatin modifications dictated by Pol IV small RNAs (Matzke and Mosher 2014; Matzke et al. 2015); thus it is not surprising that increasing evidence points to Pol IV as a general source of epigenetic variation affecting gene regulation (Parkinson et al. 2007; Hollister et al. 2011; Eichten et al. 2012; Gent et al. 2012; Greaves et al. 2012; Erhard et al. 2013). Here we found Pol IV responsible for transcriptional control of both TEs and genes, consistent with a role of TEs as regulatory elements for specific alleles.
In accord with prior results (Erhard et al. 2009; Hale et al. 2009), loss of RPD1 results primarily in increased TE transcription, and both type I and type II TEs are among those affected. We identified only 28 unique TEs whose transcription was affected by RPD1 that were farther than 5 kb of annotated genes (Table S5). While specific repetitive TE classes are differentially affected, our results indicate that the majority of the genome-wide nongenic TEs are not transcribed at the seedling stage of development even in the absence of RPD1. Our analyses, however, likely underestimate the number of transcribed TEs because our sequencing depth, particularly in nongenic regions, is insufficient to detect low-abundance transcripts (Martin et al. 2014). Additionally, TE-like reads aligning to the B73 genome representing unannotated TEs, TEs highly divergent from the Maize TE Consortium canonical set, or chimeras from multiple insertion events may have been misclassified as intergenic reads. Our results contrast with RNA-seq data from rdr2 mutant meristems (Jia et al. 2009) showing significant increases in TE RNAs in the absence of this siRNA biogenesis factor. Assuming that TE RNA levels accurately reflect transcription rates, this difference in experimental results indicates that the mechanisms of TE repression among meristematic and differentiated cell types are distinct. Consistent with the limited cytosine methylation changes seen in the absence of maize RPD1 (Parkinson et al. 2007; Erhard et al. 2013; Li et al. 2014), Pol IV plays a potentially redundant role in repressing most TE transcription in whole seedlings although a fraction appear to be directly controlled by Pol IV action(s). The genomic and/or molecular features that distinguish these two general classes remain to be identified.
In addition to TEs, ∼0.5% of all B73 alleles are transcriptionally responsive to RPD1, although loss of RPD1 can result in either increased or decreased transcription and in some cases in antisense orientation. It seems plausible that inappropriate antisense gene transcription could interfere with normal cotranscriptional RNA-processing steps or that sense-antisense RNA pairs could create double-stranded substrates for endonucleases, leading to post-transcriptional degradation. Pol II/Pol IV competitions for gene and TE promoters as previously proposed (Hale et al. 2009) and here exemplified by the B73 ocl2 haplotype profiles (Figure 4C) remain a viable hypothesis to explain RPD1-specific effects. Such competitions could also account for the increased antisense transcription of many genes in the absence of RPD1 where the antisense transcription unit is contiguous with a downstream TE (Figure 4, D and E). It will be important to characterize the make-up of nuclear transcription “factories” in plants to see if and how Pol II and Pol IV potentially compete for specific genomic templates. It will also be necessary to compare whole-genome transcription profiles of mutants deficient for downstream components required for RNA-directed DNA methylation to identify genes whose regulation is associated with modulations of cytosine methylation. Independent of specific mechanisms, our genome-wide analyses indicate that Pol IV plays a significant regulatory role for specific alleles in the grasses. Given that there are multiple functional Pol IV isoforms defined by alternative second largest subunits in the grasses (Sidorenko et al. 2009; Stonaker et al. 2009; Haag et al. 2014; Sloan et al. 2014), and that haplotype diversity in maize is largely based on radically different intergenic TE compositions (Wang and Dooner 2006), the potential for regulatory diversity controlled by alternate RNAPs is immense in the maize pangenome.
Transcriptional control affecting paramutation
One feature of the ocl2 haplotype, transcription of an upstream TE affected by Pol IV, is shared among pl1 alleles affected by Pol IV loss. Several pl1 alleles, including Pl1-Rhoades, have an upstream type II CACTA-like TE fragment belonging to the doppia subfamily, which defines kernel-specific expression after conditioning in an rpd1 mutant background (Erhard et al. 2013). This Pl1-Rhoades doppia fragment may be necessary, but is insufficient, for facilitating paramutations (Erhard et al. 2013). A related doppia fragment serves as a kernel-specific promoter and is partly required for paramutation occurring at R-r:standard haplotypes (Kermicle 1996; Walker 1998). At both Pl1-Rhoades and R-r:standard, other doppia-independent features are clearly important for mediating the trans-homolog interactions characteristic of paramutation (Kermicle 1996; Erhard et al. 2013). At the B1-Intense haplotype, paramutation interactions require a distal 5′ transcriptional enhancer composed of direct repeats (Stam et al. 2002) that loop to the b1 promoter region (Louwers et al. 2009), but there is currently no evidence supporting a functional role of specific TE sequences. In the four most studied examples of paramutation occurring in maize, transcription and/or transcriptional enhancers are functionally implicated in the mechanism required for establishing meiotically heritable repression associated with paramutation (Kermicle 1996; Sidorenko and Peterson 2001; Stam et al. 2002; Gross and Hollick 2007). In the absence of RPD1, high levels of gene expression are restored at R-r:standard, Pl1-Rhoades, and B1-Intense haplotypes previously repressed by paramutation (Hollick et al. 2005). At Pl1-Rhoades, loss of RPD1 results in increased pl1 transcription, and this is often associated with meiotically heritable reversions of Pl1-Rhoades to a stable and highly expressed nonparamutant state (Hollick et al. 2005), resulting in strong plant pigmentation. We purposely excluded Pl1-Rhoades from materials used for the GRO-seq libraries reported here to avoid changes in transcription related to light perception that might be affected by seedling pigmentation. However, now having a list of RPD1-regulated alleles, we can use pedigree analyses to test whether these alleles also exhibit paramutation-like properties. Characterizing nascent transcription in different Pol IV and siRNA mutant backgrounds across Pl1-Rhoades and other haplotypes subject to paramutation promises to uncover important features of the underlying mechanism.
Our findings indicate that RPD1 uses mechanistically diverse actions, some of which may be independent of its catalytic action within the Pol IV holoenzyme, to regulate alleles in different genomic contexts. The identification of alleles affected by RPD1 loss now presents the opportunity to identify specific haplotype structures that have co-opted direct Pol IV action for their regulation. Given that maize Pol IV defines both mitotically and meiotically heritable patterns of gene regulation (Parkinson et al. 2007; Erhard et al. 2013), alterations of its function by developmental, environmental, or genealogical sources might lead to both ontogenetic and phylogenetic changes.
Supplementary Material
Acknowledgments
We thank Leighton Core for consultations regarding the GRO-seq protocol; Blake Meyers for kindly providing maize rRNA and tRNA sequences; and Janelle Gabriel, Brian Giacopelli, Tzuu Fen Lee, Reza Hammond, Blake Meyers, and Keith Slotkin for comments and discussion. Illumina sequencing was supported by the Vincent J. Coates Genome Sequencing Laboratory, University of California at Berkeley. This work was supported by the National Science Foundation (MCB-0920623). The views expressed are solely those of the authors and are not endorsed by the sponsors of this work.
Footnotes
Communicating editor: E. U. Selker
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.174714/-/DC1.
Sequencing data have been deposited in the Gene Expression Omnibus database under accession no. GSE54166.
Literature Cited
- Alleman M., Sidorenko L., McGinnis K., Seshadri V., Dorweiler J. E., et al. , 2006. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature 442: 295–298. [DOI] [PubMed] [Google Scholar]
- Anders S., Huber W., 2010. Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel F., Jegu T., Latrasse D., Romero-Barrios N., Christ A., et al. , 2014. Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol. Cell 55: 383–396. [DOI] [PubMed] [Google Scholar]
- Ashe A., Sapetschnig A., Weick E.-M., Mitchell J., Bagijn M. P., et al. , 2012. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150: 88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber W. T., Zhang W., Win H., Varala K. K., Dorweiler J. E., et al. , 2012. Repeat associated small RNAs vary among parents and following hybridization in maize. Proc. Natl. Acad. Sci. USA 109: 10444–10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour J. R., Liao I. T., Stonaker J. L., Lim J. P., Lee C. C., et al. , 2012. required to maintain repression2 is a novel protein that facilitates locus-specific paramutation in maize. Plant Cell 24: 1761–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucom R. S., Estill J. C., Chaparro C., Upshaw N., Jogi A., et al. , 2009. Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet. 5: e1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Brink R. A., 1956. A genetic change associated with the R locus in maize which is directed and potentially reversible. Genetics 41: 872–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink R. A., 1958. Paramutation at the R locus in maize. Cold Spring Harb. Symp. Quant. Biol. 23: 379–391. [DOI] [PubMed] [Google Scholar]
- Chandler V. L., Stam M., 2004. Chromatin conversations: mechanisms and implications of paramutation. Nat. Rev. Genet. 5: 532–544. [DOI] [PubMed] [Google Scholar]
- Coe E. H., Jr, 1961. A test for somatic mutation in the origination of conversion-type inheritance at the B locus in maize. Genetics 46: 707–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core L. J., Waterfall J. J., Lis J. T., 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core L. J., Waterfall J. J., Gilchrist D. A., Fargo D. C., Kwak H., et al. , 2012. Defining the status of RNA polymerase at promoters. Cell Reports 2: 1025–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis E. S., Peacock W. J., 2007. Epigenetic regulation of flowering. Curr. Opin. Plant Biol. 10: 520–527. [DOI] [PubMed] [Google Scholar]
- de Vanssay A., Bougé A.-L., Boivin A., Hermant C., Teysset L., et al. , 2012. Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature 490: 112–115. [DOI] [PubMed] [Google Scholar]
- Eichten S. R., Ellis N. A., Makarevitch I., Yeh C.-T., Gent J. I., et al. , 2012. Spreading of heterochromatin is limited to specific families of maize retrotransposons. PLoS Genet. 8: e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhard K. F., Jr, Stonaker J. L., Parkinson S. E., Lim J. P., Hale C. J., et al. , 2009. RNA polymerase IV functions in paramutation in Zea mays. Science 323: 1201–1205. [DOI] [PubMed] [Google Scholar]
- Erhard K. F., Jr, Parkinson S. E., Gross S. M., Barbour J. R., Lim J. P., et al. , 2013. Maize RNA polymerase IV defines trans-generational epigenetic variation. Plant Cell 25: 808–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent J. I., Dong Y., Jiang J., Dawe R. K., 2012. Strong epigenetic similarity between maize centromeric and pericentromeric regions at the level of small RNAs, DNA methylation and H3 chromatin modifications. Nucleic Acids Res. 40: 1550–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent J. I., Ellis N. A., Guo L., Harkess A. E., Yao Y., et al. , 2013. CHH islands: de novo DNA methylation in near-gene chromatin regulation in maize. Genome Res. 23: 628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent J. I., Madzima T. F., Bader R., Kent M. R., Zhang X., et al. , 2014. Accessible DNA and relative depletion of H3K9me2 at maize loci undergoing RNA-directed DNA methylation. Plant Cell 26: 4903–4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves I. K., Groszmann M., Ying H., Taylor J. M., Peacock W. J., et al. , 2012. Trans-chromosomal methylation in Arabidopsis hybrids. Proc. Natl. Acad. Sci. USA 109: 3570–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S. M., Hollick J. B., 2007. Multiple trans-sensing interactions affect meiotically heritable epigenetic states at the maize pl1 locus. Genetics 176: 829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag J. R., Ream T. S., Marasco M., Nicora C. D., Norbeck A. D., et al. , 2012. In vitro transcription activities of Pol IV, Pol V, and RDR2 reveal coupling of Pol IV and RDR2 for dsRNA synthesis in plant RNA silencing. Mol. Cell 48: 811–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag J. R., Brower-Toland B., Krieger E. K., Sidorenko L., Nicora C. D., et al. , 2014. Functional diversification of maize RNA polymerase IV and V subtypes via alternative catalytic subunits. Cell Reports 9: 378–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann R., Berg W., 1978. Paramutation at the sulfurea locus of Lycopersicon esculentum Mill.: VII. Determination of the time of occurrence of paramutation by the quantitative evaluation of the variegation. Theor. Appl. Genet. 53: 113–123. [DOI] [PubMed] [Google Scholar]
- Hale C. J., Stonaker J. L., Gross S. M., Hollick J. B., 2007. A novel Snf2 protein maintains trans-generational regulatory states established by paramutation in maize. PLoS Biol. 5: e275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C. J., Erhard K. F., Jr, Lisch D., Hollick J. B., 2009. Production and processing of siRNA precursor transcripts from the highly repetitive maize genome. PLoS Genet. 5: e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr A. J., Jensen M. B., Dalmay T., Baulcombe D. C., 2005. RNA polymerase IV directs silencing of endogenous DNA. Science 308: 118–120. [DOI] [PubMed] [Google Scholar]
- Hollick J. B., 2012. Paramutation: a trans-homolog interaction affecting heritable gene regulation. Curr. Opin. Plant Biol. 15: 536–543. [DOI] [PubMed] [Google Scholar]
- Hollick J. B., Gordon M. P., 1993. A poplar tree proteinase inhibitor-like gene promoter is responsive to wounding in transgenic tobacco. Plant Mol. Biol. 22: 561–572. [DOI] [PubMed] [Google Scholar]
- Hollick J. B., Patterson G. I., Coe E. H., Jr, Cone K. C., Chandler V. L., 1995. Allelic interactions heritably alter the activity of a metastable maize pl allele. Genetics 141: 709–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick J. B., Kermicle J. L., Parkinson S. E., 2005. Rmr6 maintains meiotic inheritance of paramutant states in Zea mays. Genetics 171: 725–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister J. D., Smith L. M., Guo Y.-L., Ott F., Weigel D., et al. , 2011. Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata. Proc. Natl. Acad. Sci. USA 108: 2322–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Kendall T., Mosher R. A., 2013. Pol IV-dependent siRNA production is reduced in Brassica rapa. Biology 2: 1210–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javelle M., Klein-Cosson C., Vernoud V., Boltz V., Maher C., et al. , 2011. Genome-wide characterization of the HD-ZIP IV transcription factor family in maize: preferential expression in the epidermis. Plant Physiol. 157: 790–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., Lisch D. R., Ohtsu K., Scanlon M. J., Nettleton D., et al. , 2009. Loss of RNA-dependent RNA polymerase 2 (RDR2) function causes widespread and unexpected changes in the expression of transposons, genes, and 24-nt small RNAs. PLoS Genet. 5: e1000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush K., Khasdan V., 2007. Large-scale survey of cytosine methylation of retrotransposons and the impact of readout transcription from long terminal repeats on expression of adjacent rice genes. Genetics 177: 1975–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle J. L., 1996. Epigenetic silencing and activation of a maize r gene, pp. 267–287 in Epigenetic Mechanisms of Gene Regulation, edited by Russo V. E. A., Martienssen R. A., Riggs A. D. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Khaitová L. C., Fojtová M., Křížová K., Lunerová J., J. Fulneček et al, 2011. Paramutation of tobacco transgenes by small RNA-mediated transcriptional gene silencing. Epigenetics 6: 650–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruesi, W. S., L. J. Core, C. T. Waters, J. T. Lis, and B. J. Meyer, 2013 Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. ELife 2: e00808. [DOI] [PMC free article] [PubMed]
- Langmead B., Trapnell C., Pop M., Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q., S. R. Eichten, P. J. Hermanson, V. M. Zaunbrecher, J. Song et al., 2014 Genetic perturbation of the maize methylome. Plant Cell 26: 4602–4616. [DOI] [PMC free article] [PubMed]
- Li S., Vandivier L. E., Tu B., Gao L., Won S. Y., et al. , 2015. Detection of Pol IV/RDR2-dependent transcripts at the genomic scale in Arabidopsis reveals features and regulation of siRNA biogenesis. Genome Res. 25: 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louwers M., Bader R., Haring M., van Driel R., de Laat W., et al. , 2009. Tissue- and expression level-specific chromatin looping at maize b1 epialleles. Plant Cell 21: 832–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Hall B. D., 2007. A multistep process gave rise to RNA polymerase IV of land plants. J. Mol. Evol. 64: 101–112. [DOI] [PubMed] [Google Scholar]
- Martin J. A., Johnson N. V., Gross S. M., Schnable J., Meng X., et al. , 2014. A near complete snapshot of the Zea mays seedling transcriptome revealed from ultra-deep sequencing. Sci. Rep. 4: 4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M., 2011 Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12.
- Matzke M. A., Mosher R. A., 2014. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15: 394–408. [DOI] [PubMed] [Google Scholar]
- Matzke M. A., Kanno T., Huettel B., Daxinger L., Matzke A. J. M., 2007. Targets of RNA-directed DNA methylation. Curr. Opin. Plant Biol. 10: 512–519. [DOI] [PubMed] [Google Scholar]
- Matzke, M. A., T. Kanno, and A. J. M. Matzke, 2015 RNA-directed DNA methylation: the evolution of a complex epigenetic pathway in flowering plants. Ann. Rev. Plant Biol. 66: 9.1–9.25. [DOI] [PubMed]
- Maxwell C. S., Kruesi W. S., Core L. J., Kurhanewicz N., Waters C. T., et al. , 2014. Pol II docking and pausing at growth and stress genes in C. elegans. Cell Reports 6: 455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B., 1951. Chromosome organization and genic expression. Cold Spring Harb. Symp. Quant. Biol. 16: 13–47. [DOI] [PubMed] [Google Scholar]
- Morales-Ruiz T., Ortega-Galisteo A. P., Ponferrada-Marín M. I., Martínez-Macías M. I., Ariza R. R., et al. , 2006. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc. Natl. Acad. Sci. USA 103: 6853–6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher R. A., Schwach F., Studholme D., Baulcombe D. C., 2008. PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc. Natl. Acad. Sci. USA 105: 3145–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobuta K., Lu C., Shrivastava R., Pillay M., De Paoli E., et al. , 2008. Distinct size distribution of endogenous siRNAs in maize: evidence from deep sequencing in the mop1–1 mutant. Proc. Natl. Acad. Sci. USA 105: 14958–14963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y., Haag J. R., Ream T., Costa Nunes P., Pontes O., et al. , 2005. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120: 613–622. [DOI] [PubMed] [Google Scholar]
- Parkinson S. E., Gross S. M., Hollick J. B., 2007. Maize sex determination and abaxial leaf fates are canalized by a factor that maintains repressed epigenetic states. Dev. Biol. 308: 462–473. [DOI] [PubMed] [Google Scholar]
- Pilu R., Panzeri D., Cassani E., Cerino Badone F., Landoni M., et al. , 2009. A paramutation phenomenon is involved in the genetics of maize low phytic acid1–241 (lpa1–241) trait. Heredity 102: 236–245. [DOI] [PubMed] [Google Scholar]
- Pontier D., Yahubyan G., Vega D., Bulski A., Saez-Vasquez J., et al. , 2005. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 19: 2030–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassoulzadegan M., Grandjean V., Gounon P., Vincent S., Gillot I., et al. , 2006. RNA-mediated non-Mendelian inheritance of an epigenetic change in the mouse. Nature 441: 469–474. [DOI] [PubMed] [Google Scholar]
- Ream T. S., Haag J. R., Wierzbicki A. T., Nicora C. D., Norbeck A. D., et al. , 2009. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol. Cell 33: 192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie A. E., Lis J. T., 1988. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54: 795–804. [DOI] [PubMed] [Google Scholar]
- Sabin L. R., Delás M. J., Hannon G. J., 2013. Dogma derailed: the many influences of RNA on the genome. Mol. Cell 49: 783–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P. S., Ware D., Fulton R. S., Stein J. C., Wei F., et al. , 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Seila A. C., Calabrese J. M., Levine S. S., Yeo G. W., Rahl P. B., et al. , 2008. Divergent transcription from active promoters. Science 322: 1849–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila A., Core L. J., Lis J. T., Sharp P. A., 2009. Divergent transcription: a new feature of active promoters. Cell Cycle 8: 2557–2564. [DOI] [PubMed] [Google Scholar]
- Shirayama M., Seth M., Lee H.-C., Gu W., Ishidate T., et al. , 2012. piRNAs initiate an epigenetic memory of non-self RNA in the C. elegans germline. Cell 150: 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorenko L. V., Peterson T., 2001. Transgene-induced silencing identifies sequences involved in the establishment of paramutation of the maize p1 gene. Plant Cell 13: 319–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorenko L., Dorweiler J. E., Cigan A. M., Arteaga-Vazquez M., Vyas M., et al. , 2009. A dominant mutation in mediator of paramutation2, one of three second-largest subunits of a plant-specific RNA polymerase, disrupts multiple siRNA silencing processes. PLoS Genet. 5: e1000725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan A. E., Sidorenko L., McGinnis K. M., 2014. Diverse gene silencing mechanisms with distinct requirements for RNA polymerase subunits in Zea mays. Genetics 198: 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund C., Descour A., Kudrna D., Bomhoff M., Boyd L., et al. , 2009. Sequencing, mapping, and analysis of 27,455 maize full-length cDNAs. PLoS Genet. 5: e1000740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam M., Belele C., Dorweiler J. E., Chandler V. L., 2002. Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev. 16: 1906–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stonaker J. L., Lim J. P., Erhard K. F., Jr, Hollick J. B., 2009. Diversity of Pol IV function is defined by mutations at the maize rmr7 locus. PLoS Genet. 5: e1000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H., Greenberg M. V. C., Feng S., Bernatavichute Y. V., Jacobsen S. E., 2013. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S., Crevillen P., Liu F., Ecker J. R., Jerzmanowski A., et al. , 2007. Small RNA-mediated chromatin silencing directed to the 3′ region of the Arabidopsis gene encoding the developmental regulator, FLC. Proc. Natl. Acad. Sci. USA 104: 3633–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker S. L., Reece J., Ream T. S., Pikaard C. S., 2010. Evolutionary history of plant multisubunit RNA polymerases IV and V: subunit origins via genome-wide and segmental gene duplications, retrotransposition, and lineage-specific subfunctionalization. Cold Spring Harb. Symp. Quant. Biol. 75: 285–297. [DOI] [PubMed] [Google Scholar]
- Walker E. L., 1998. Paramutation of the r1 locus of maize is associated with increased cytosine methylation. Genetics 148: 1973–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Dooner H. K., 2006. Remarkable variation in maize genome structure inferred from haplotype diversity at the bz locus. Proc. Natl. Acad. Sci. USA 103: 17644–17649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang H., Wang J., Sun R., Wu J., et al. , 2011. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43: 1035–1039. [DOI] [PubMed] [Google Scholar]
- Woodhouse M. R., Freeling M., Lisch D., 2006. Initiation, establishment, and maintenance of heritable MuDR transposon silencing in maize are mediated by distinct factors. PLoS Biol. 4: e339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Henderson I. R., Lu C., Green P. J., Jacobsen S. E., 2007. Role of RNA polymerase IV in plant small RNA metabolism. Proc. Natl. Acad. Sci. USA 104: 4536–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Hale C. J., Law J. A., Johnson L. M., Feng S., et al. , 2012. DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat. Struct. Mol. Biol. 19: 870–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.