Abstract
Signal transducer and activator of transcription 3 (STAT3) regulates diverse cellular processes including cell growth, differentiation, and apoptosis, and is frequently activated during tumorigenesis. Recently, putative glioblastoma stem cells (GBM-SC) have been isolated and characterized. These cells can self-renew indefinitely in culture, are highly tumorigenic, and retain the ability to differentiate in culture. We have found that treatment of GBM-SC with two chemically distinct small molecule inhibitors of STAT3 DNA-binding inhibits cell proliferation and the formation of new neurospheres from single cells. Genetic knockdown of STAT3 using an shSTAT3-containing lentivirus also inhibits GBM-SC proliferation and neurosphere formation, confirming that these effects are specific to STAT3. While STAT3 inhibition can induce apoptosis in serum-derived GBM cell lines, this effect was not observed in GBM-SC grown in stem cell media. Markers of neural stem cell multipotency also decrease upon STAT3 inhibition, suggesting that STAT3 is required for maintenance of the stem-like characteristics of these cells. Strikingly, even a transient inhibition of STAT3 leads to irreversible growth arrest and inhibition of neurosphere formation. These data suggest that STAT3 regulates the growth and self-renewal of GBM-SC and is thus a potential target for cancer stem cell-directed therapy of glioblastoma multiforme.
Introduction
Glioblastoma multiforme (GBM), the most common adult brain tumor, is a highly malignant and aggressive disease. GBM tumors are invasive and highly vascularized and patients diagnosed with GBM have a mean survival time of only 12-14 months [1]. Glioblastoma can arise de novo or from lower grade astrocytomas. GBMs are composed of multiple cell types, including cells expressing astrocytic, neuronal, or both astrocytic and neuronal lineage markers, suggesting they may originate from a multipotent stem cell.
Recent work has led to the identification, in several cancer types, of a putative tumor “stem cell” with distinct properties from the bulk tumor and from traditional serum-derived lines. Tumor stem cells display an undifferentiated phenotype and an enhanced ability to initiate tumor formation relative to other cells from the bulk tumor in mouse xenograft models. Tumor stem cells have been isolated from human glioblastoma. These cells share many properties with normal neural stem cells [2-5]. Glioblastoma-derived stem cells (GBM-SC) can self-renew, proliferate, and differentiate to form multiple cell types, including cells expressing neuronal and glial markers. Unlike normal neural stem cells, GBM-SC are highly tumorigenic in mice and display aberrant proliferative capacity and gene expression patterns [4]. Tumors initiated by GBM-SC recapitulate the phenotype of the original tumor from which they are isolated, and microarray analysis has shown that GBM-SC have a gene expression signature that more closely resembles the tumor of origin than do serum-derived cell lines from the same tumor [6].
STAT3, a member of the STAT (signal transducers and activators of transcription) family of transcription factors, is important in glioblastoma, tumorigenesis, central nervous system development, and embryonic stem cell biology. STAT3 is activated by a wide variety of cytokines or growth factors. Upon tyrosine phosphorylation by receptor-associated tyrosine kinases, STAT3 translocates to the nucleus and regulates transcription of target genes [7]. STAT3 target genes regulate many cellular processes, including proliferation and apoptosis [8-10]. Constitutive activation of STAT3 has been observed in many human cancers, including breast, head and neck, prostate, melanoma, and thyroid cancer [11]. Knockout of STAT3 in the mouse epithelium completely abrogates the induction of skin tumors by the carcinogen DMBA [12]. Mice overexpressing constitutively activated STAT3 in alveolar epithelial cells develop spontaneous lung tumors [13].
STAT3 is also activated in a high percentage of glioblastomas [14]. We have previously used RNAi knockdown of STAT3 in serum-derived glioblastoma cell lines to demonstrate that STAT3 knockdown induces apoptosis in GBM cell [9]. We have also shown that STAT3 knockdown inhibits the expression of telomerase, Bcl-xl, and survivin in serum grown glioblastoma cell lines [9,15]. Thus, STAT3 plays an anti-apoptotic role in glioblastoma cell lines.
In addition to its role in tumorigenesis, STAT3 is also an important regulator of stem cells and the developing nervous system. Mouse embryonic stem cells are dependent on LIF, a potent activator of STAT3, and dominant negative STAT3 leads to ES cell differentiation and loss of pluripotency [16-18]. In both embryonic and neural stem cells, STAT3 is important in maintaining self-renewal. Deletion of STAT3 in murine embryonic neural stem cells inhibits neurosphere formation and self-renewal{[19,20]. In contrast, other data indicates that STAT3 plays a critical role in the differentiation of neural stem cells intro astrocytes [21,22]. Thus, STAT3 may have multiple roles in neural stem cell function.
The function of STAT3 in glioblastoma stem cells, however, has not been determined. Here we show that inhibition of STAT3 in GBM stem cells irreversibly abrogates neurosphere formation and inhibits proliferation. In addition, we find that inhibition of STAT3 causes the downregulation of genes associated with the neural stem cell phenotype, providing evidence that STAT3 regulates multipotency in these cells. These results suggest that STAT3 could be an effective therapeutic target for the treatment of glioblastoma.
Results
Establishment of GBM-derived stem cell lines
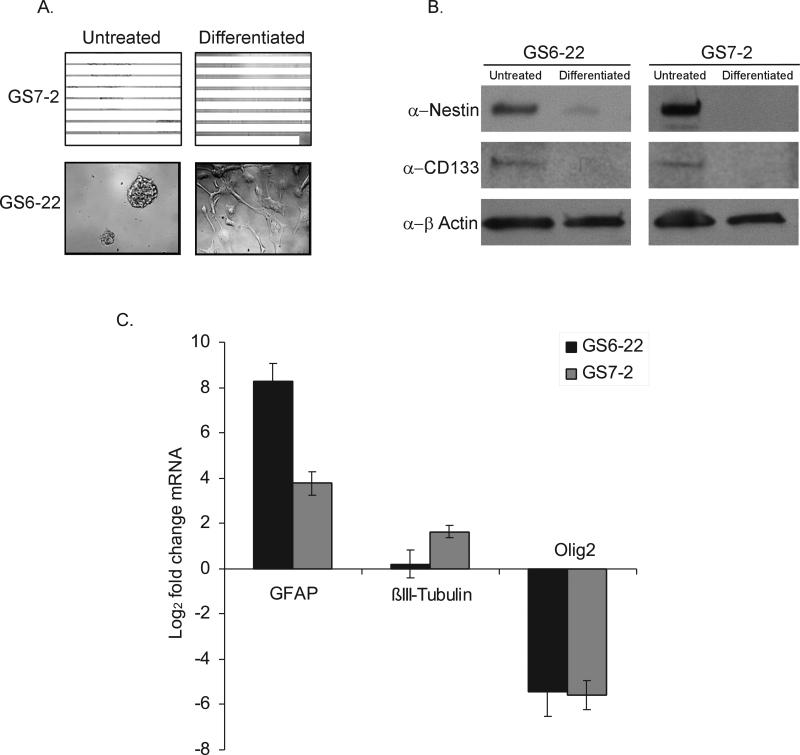
In order to determine what role STAT3 plays in glioblastoma stem cells, we first established glioblastoma stem cell lines from human glioblastoma multiforme tumor specimens. Primary tumor samples were dissociated and plated as single cells in serum-free media containing FGF-2 and EGF, culture conditions that favor the growth of glioblastoma-derived stem cells [4]. While we have established several GBM-SC lines, this work focuses on two lines which we have designated GS7-2 and GS6-22. Both stem cell lines are derived from right temporal glioblastomas; GS7-2 is derived from the tumor of an 83 year old male while the GS6-22 cells were isolated from an 82 year old female. These cell lines have the expected characteristics of glioblastoma stem cells [4,6]. Both cell lines grow as non-adherent neurospheres in culture and can be continuously passaged (Figure 1A). Both GBM-SC lines express the neural stem cell markers CD133 and nestin (Figure 1B). 98% of GS6-22 cells and 94% of GS7-2 cells express CD133, as determined by immunostaining (data not shown). When plated on poly-L-ornithine/laminin in the presence of low (2%) serum and without EGF and FGF-2, conditions established for the differentiation of neural and glioblastoma stem cells, both GBM-SC lines grow as adherent monolayers and begin to exhibit morphology consistent with that of astrocytes and neurons (Figure 1A). Importantly, under differentiation conditions, the expression of the neural stem cell markers CD133, nestin, and olig2 in both the GS6-22 and GS7-2 cells decreased (Figure 1B-C). Concurrent with the decrease in stem cell markers, we observed an induction of the astrocytic marker GFAP and the neural marker βIII-tubulin (Figure 1C). This demonstrates that the tumor-derived cell lines retain the ability to differentiate, a defining characteristic of both normal and tumor-derived neural stem cells. We have confirmed that GS7-2 gives rise to tumors whenas few as 1000 cells are implanted in the brains of nude mice. Additional characterization of these lines reveals that neither stem cell line harbors the EGFRvIII mutation present in a subset of glioblastomas (Supplementary Figure 1). The loss of PTEN expression, which is often found in glioblastoma patient samples, is observed in the GS6-22 line, but not in the GS7-2 stem cell line (Supplementary Figure 2)[23,24].
Figure 1. Glioblastoma stem cells derived from primary tumor samples form neurospheres in culture, express neural stem cell markers, and retain the ability to differentiate.
A. Images of GS7-2 and GS6-22 neurospheres grown in serum-free media with EGF and FGF-2 (Untreated) or differentiated by plating on poly-L-ornithine and laminin in the presence of 2% serum without growth factors (Differentiated). B. Immunoblot of lysates of GS7-2 and GS-22 neurospheres in serum-free media or after 7 days under differentiation conditions with antibodies to CD133, nestin, and β-actin. C. RT-qPCR analysis of GFAP, βIII-tubulin, and olig2 mRNA in GS7-2 and GS6-22 neurospheres exposed to differentiation conditions for 7 days. Fold change values were calculated relative to untreated control and normalized by comparison to β-actin and plotted as log2. Bars represent mean of three separate experiments; bars SD.
STAT3 is present and activated in GBM-SC
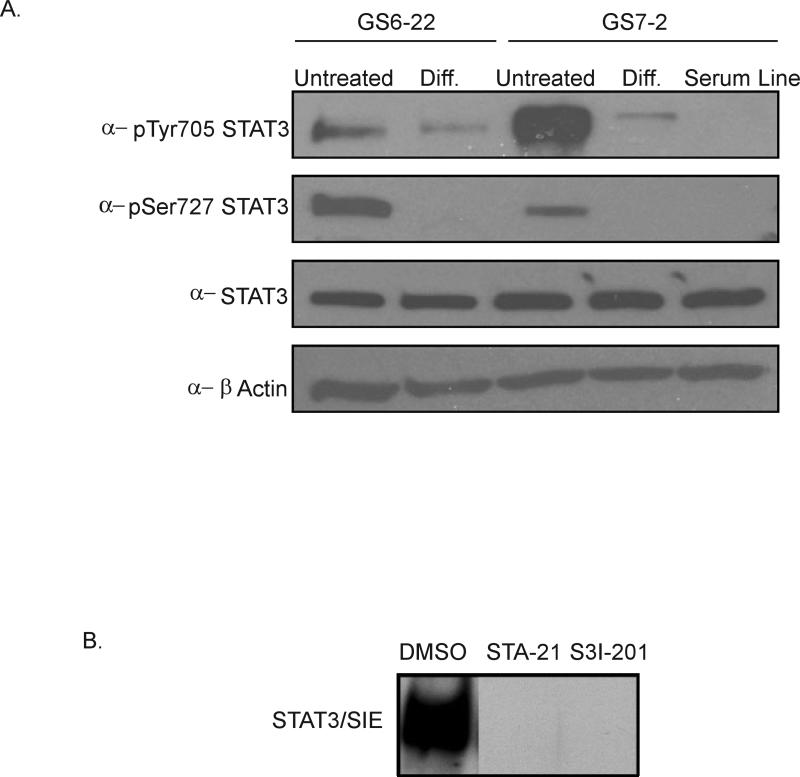
Next, we characterized the expression and phosphorylation status of STAT3 in GBM-SC. STAT3 is expressed in these cells as shown by immunoblotting (Figure 2A). Immunoblotting with phospho-specific antibodies demonstrated that STAT3 is phosphorylated on both tyrosine-705 and serine-727 in the GS6-22 and GS7-2 GBM-SC lines. Since phosphorylation of tyrosine-705 is required for DNA-binding, and serine-727 phosphorylation has been shown to stimulate maximal transcriptional activity [25,26], we conclude from our immunoblots that STAT3 is present and activated in GBM-SC under neural stem cell culture conditions. We found that STAT3 is also activated in three additional glioblastoma stem cell lines that we have isolated as shown by immunostaining with anti-phospho-Y705 staining (Supplementary Figure 3). Thus, STAT3 activation is a common feature of GBM-SC.
Figure 2. STAT3 is expressed and activated in GBM stem cells.
A. Immunoblot of STAT3, pSer727 STAT3, and pTyr705 STAT3 in GBM-SC untreated and differentiated for 7 days. The serum line derived from the GS7-2 parent tumor was also probed. B. Bandshift assay of GS7-2 neurosphere lysates treated with inhibitors of STAT3 DNA binding (STA-21 30μM, S3I-201 100 μM), incubated with radiolabeled high affinity SIE probe, and separated by PAGE.
Interestingly, differentiation of the GS6-22 and GS7-2 lines for 7 days leads to a decrease in phospho-tyrosine levels, while phospho-serine STAT3 becomes undetectable (Figure 2A). A cell line derived from the GS7-2 tumor sample in the presence of serum has comparable levels of total STAT3, but undetectable phospho-tyrosine or phospho-serine STAT3. Thus, both the differentiated GBM-SC and a matched serum-derived cell line have reduced STAT3 phosphorylation.
In order to assess the function of STAT3 in GBM-SC, we first analyzed two structurally distinct small molecule inhibitors of STAT3 DNA-binding. These inhibitors, STA-21 and S3I-201, both target the SH2 domain of STAT3, thereby preventing STAT3 dimers from binding DNA and activating transcription of their target genes [27,28]. To confirm their STAT3 inhibitory activity in GBM-SC as measured by DNA binding, each drug was used in a bandshift assay with a radiolabeled SIE probe that binds STAT3 with high affinity [29]. As shown in Figure 2B, both STA-21 and S3I-201 abolished the ability of STAT3 to bind the SIE probe in GS7-2 cells. STA-21 and S3I-201 also inhibit STAT3 DNA-binding in GS6-22 cells (Supplementary Figure 4). This confirms that STA-21 and S3I-201 can inhibit STAT3 DNA binding in our GBM-SC lysates, which is consistent with published reports of their efficacy against STAT3 in other cell lines in vitro and in vivo [27,28]. Neither STA-21 nor S3I-201 affects ERK or AKT activation, as shown by immunoblotting of the U251 glioblastoma serum line (Supplementary Figure 5).
Previously both STA-21 and S3I-201 have been shown to inhibit STAT3 preferentially as compared to STAT1 and STAT5. [27,28]. From immunoblots of GS6-22 and GS7-2 GBM-SC, it is clear that while these cell lines express STAT5 and STAT1, STAT3 is the major activated STAT. Phosphotyrosine STAT1 and STAT5 are undetectable under neural stem cell culture conditions (Supplementary Figure 6). Thus, STAT3 is the principal activated STAT protein in these cells.
Because STA-21 and S3I-201 directly target the SH2 domain of STAT3 and not the upstream activating kinases, their efficacy is not necessarily correlated with a decrease in STAT3 tyrosine phosphorylation in contrast with DNA binding activity. However, in some cases, STAT3 tyrosine phosphorylation may decrease with these drugs if targeting of STAT3 to receptors by the SH2 domain is disrupted. In fact, a time course of both GS6-22 and GS7-2 GBM-SC treated with each inhibitor reveals that STAT3 tyrosine phosphorylation is more strongly inhibited in the GS6-22 line than in the GS7-2 line. (Supplementary Figure 7). This is probably due to distinct modes of STAT3 activation between the two cell lines [30].
Inhibition of STAT3 prevents neurosphere formation and decreases proliferation in GBM-SC
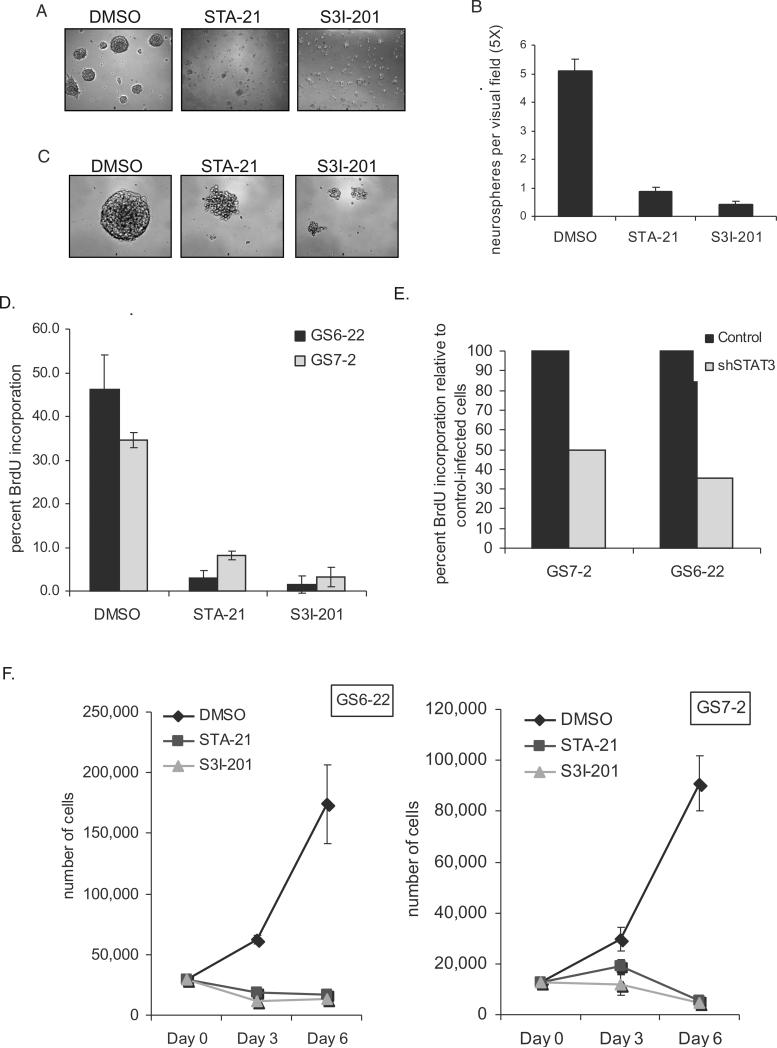
We next investigated the effect of STAT3 inhibition on GBM-SC. Cultures of both GS7-2 and GS6-22 cells were dissociated and plated as single cells in the presence of either STA-21, S3I-201, or DMSO control. After several days, a dramatic difference was observed between the drug and control-treated cultures. While neurospheres formed in the control wells, very few spheres formed from cells treated with either of the STAT3 inhibitors (Figure 3A and Supplementary Figure 8). While an average of 5 neurospheres could be seen in each field of view in DMSO-treated cultures, fewer than 1 neurosphere per field could be seen in wells treated with either STA-21 or S31-201 (Figure 3B). Cells were viable, however, as determined by trypan blue exclusion (data not shown). Thus, inhibition of STAT3 abrogates neurosphere formation in GBM-SC. Interestingly, treatment of intact neurospheres with either STAT3 inhibitor causes the dissociation of pre-existing neurospheres (Figure 3C).
Figure 3. STAT3 inhibitors prevent neurosphere formation and proliferation of GBM stem cells.
A. GS7-2 neurospheres mechanically dissociated into single cells, seeded into 96 well plates, and treated with the indicated STAT3 inhibitors or DMSO control. Representative images after 4 days in culture are shown (50X magnification). B. Quantitation of experiment described in A. The number of neurospheres in a single visual field (50X) per well was determined for 21 wells per treatment. Values represent mean; bars SD. C. STAT3 inhibitors cause dissociation of existing neurospheres. Intact GS6-22 spheres were treated with STAT3 inhibitors or DMSO control for 4 days. D. Inhibition of STAT3 prevents BrdU incorporation in GBM stem cells. GS6-22 and GS7-2 neurospheres were treated with STAT3 inhibitors for 24 hours and then pulsed for 16 hours with 10 μM BrdU. Cells were fixed and stained with FITC-conjugated anti-BrdU using the FITC-anti-BrdU Kit (BD Biosciences) and analyzed via flow cytometry. Bars represent SD of the mean of three separate experiments. E. Knockdown of STAT3 decreases BrdU incorporation in glioblastoma stem cells. GS6-22 and GS7-2 cells were infected with lentiviruses containing either shRNA to STAT3 or a non-targetting control shRNA. 3 days post-infection and puromycin selection, cells were pulsed with 10 μM BrdU for 3 days, fixed, stained, and analyzed as described above. Data is represented as the percent BrdU incorporation compared to control-infected cells. F. STAT3 inhibition prevents GBM-SC growth. GS6-22 and GS7-2 cells were dissociated into single cells and plated in equal numbers. 24 hours after plating, cells were counted and treated with DMSO, STA-21 and S3I-201. At day 3 and day 6 post-drug treatment cells were counted in triplicate, and plotted as SD of the mean.
To determine whether the inhibition of neurosphere formation could be explained by decreased proliferation, GS6-22 and GS7-2 cells were cultured in the presence of STA-21, S3I-201, or DMSO for 24 hours and then pulsed with BrdU for 16 hours. Cells were analyzed by flow cytometry following staining with an anti-BrdU antibody to assess the percentage of cells that incorporated BrdU during the 16 hour pulse. Treatment with either STAT3 inhibitor significantly decreased BrdU incorporation as compared to the DMSO control cells (Figure 3D). In the GS6-22 cell line an average of 46% of DMSO-treated cells incorporated BrdU compared to only 3% of STA-21-treated cells and 2% of S3I-201-treated cells. A similar decrease in BrdU incorporation was observed in the GS7-2 line. The GS7-2 cells continue to proliferate under differentiation conditions and upon STAT3 inhibition with STA-21 or S3I-201, a decrease in BrdU incorporation from 32% for DMSO-treated cells to 13 and 10% for inhibitor-treated cells was observed (Supplementary Figure 9A). The GS6-22 cells, in contrast, do not continue to proliferate under differentiation conditions. Interestingly, BrdU incorporation of the serum-derived cell line derived from the GS7-2 tumor is not affected by STAT3 inhibitor treatment (Supplementary Figure 10A). This is consistent with the observation that the serum line does not have detectable activated STAT3 as determined by immunoblotting for phospho-STAT3 (Figure 2A). This further confirms that these STAT3 inhibitors are not non-specifically inhibiting cellular proliferation.
To confirm that STAT3 is required for the growth of the GBM stem cells, the effects of knockdown of STAT3 by RNAi were investigated. Cells infected with a lentivirus containing an shRNA to STAT3 showed a decrease in BrdU incorporation compared to cells expressing a control shRNA. In the GS6-22 cells, infection with the STAT3 knockdown virus decreased BrdU incorporation by 65% compared to cells infected with a non-targeting shRNA control virus (Figure 3E). In the GS7-2 cells, BrdU incorporation was decreased by 50% in the cells infected with the STAT3 knockdown virus. Consistent with STAT3 small molecule inhibition, knockdown of STAT3 by RNAi decreases the ability of dissociated GS6-22 cells to form neurospheres (Supplementary Figure 8B-C). While this phenotype is not as pronounced as the inhibition caused by the STAT3 small molecule inhibitors, this is likely due to incomplete knockdown of STAT3 protein expression by this shRNA (Supplementary Figure 11). It is clear, however, that treatment with either of two distinct small molecule inhibitors as well as genetic knockdown of STAT3 inhibits GBM-SC proliferation.
To confirm that the STAT3 inhibitors do inhibit cellular proliferation, GS7-2 and GS6-22 cells were treated with each inhibitor, and cell number was counted 3 and 6 days later. STA-21 or S3I-201 treatment dramatically inhibited proliferation over the 6 day period (Figure 3F). In the GS6-22 cells, STA-21 and S3I-201-treated cell numbers remained constant over the 6 day period. This suggests that STAT3 inhibition blocks cell growth rather than induces cell death. In the GS7-2 cells a slight decrease in cell number was observed in STAT3 inhibitor-treated cells. While this decrease was statistically significant, no decrease in live cells between control and STAT3 inhibitor-treated cells was observed by trypan blue exclusion of cells before counting (data not shown). Overall, however, it is clear that both small molecule inhibitors and shRNA knockdown of STAT3 lead to a dramatic decrease in GBM-SC proliferation.
Inhibition of STAT3 causes apoptosis in GBM serum-derived cell lines, but not in GBM-SC
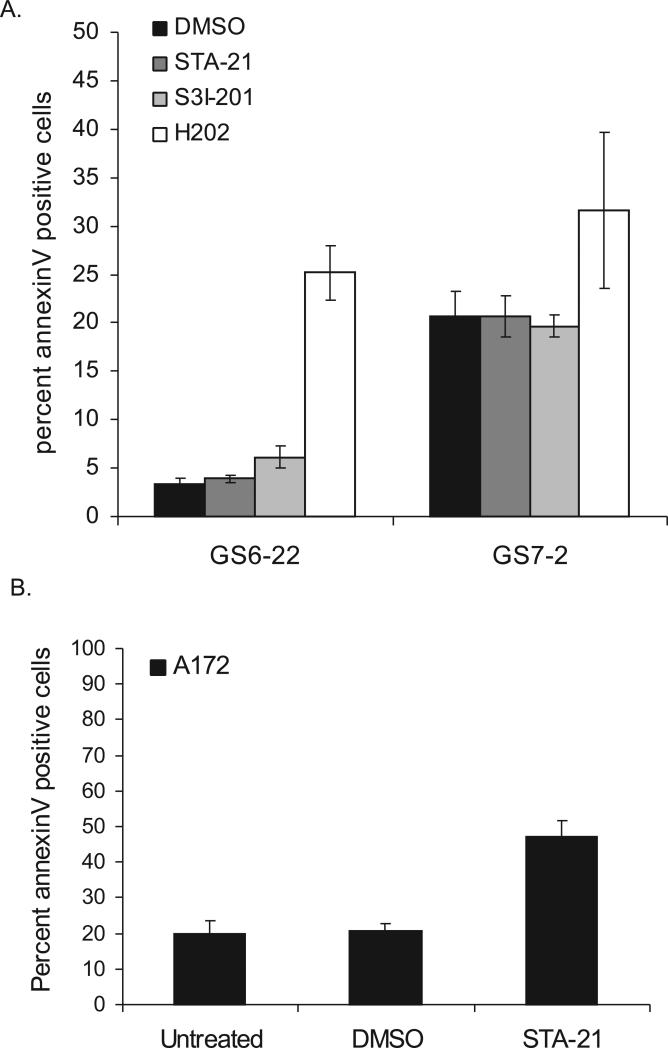
Because we have previously shown that STAT3 regulates survival of traditional serum-derived GBM cell lines, we asked whether STAT3 inhibitor treatment would have a similar effect on GBM-SC [9]. As judged by annexinV staining (Figure 4A), GS7-2 and GS6-22 GBM-SC treated with either STA-21 or S3I-201 showed no increase in apoptosis over untreated or DMSO-treated cells over a three day period. Treatment with hydrogen peroxide, however, did cause an increase in the percentage of annexinV positive cells, demonstrating that the GBM-SC are capable of undergoing apoptosis. Thus, the lack of apoptotic induction with STAT3 inhibitor treatment does not reflect a complete resistance of the stem cells to apoptosis. The percentage of dead cells staining positive for propidium iodide also did not change with STAT3 inhibitor treatment (Supplementary Figure 12), further indicating that cell viability is also not affected by drug treatment. The GBM-SC lines, then, do not undergo increased apoptosis upon STAT3 inhibitor treatment. It is possible that the modest decrease in cell number in the GS7-2 line in response to STAT3 inhibitors is due to increased autophagic cell death instead of apoptosis.
Figure 4. STAT3 inhibitor treatment causes increased apoptosis in serum-derived GBM cell lines, but not in GBM stem cells.
A. GS7-2 and GS6-22 cells were treated with STAT3 inhibitor or DMSO control for 3 days, stained with annexinV-FITC, and analyzed via flow cytometry. H2O2 treatment (4.4 mM) was performed for 4 hours as a positive control. Cells were stained using the AnnexinV-FITC Apoptosis Detection Kit (BD Biosciences) according to the manufacturer's instructions. Values represent mean of three samples; bars SE. B. The A172 serum-derived glioma cell line was treated for 3 days with STA-21 or DMSO and analyzed as described above. Values represent mean of three experiments; bars SE.
Though STAT3 inhibitor treatment failed to induce apoptosis in glioblastoma-derived stem cells, it did cause an increase in apoptosis in the A172 serum-derived glioblastoma cell line. Cells treated with STA-21 for three days had a two-fold increase in annexinV positive cells compared to DMSO and untreated controls (Figure 4B). The A172 cell line harbors constitutively activated STAT3 and has been shown to undergo apoptosis when STAT3 is knocked down using RNAi [9]. Interestingly, the GS6-22 and GS7-2 stem cell line become sensitive to apoptosis induced by STAT3 inhibition when differentiated for 7 days (Supplementary Figure (9B). The STAT3 inhibitors also induce extensive cell death when placed into differentiation media in the absence of growth factors.
In contrast, the serum line derived from the GS7-2 tumor lacks activated STAT3 (Figure 2) and fails to undergo apoptosis in response to STAT3 inhibitor treatment (Supplementary Figure 10B). Thus, as expected, STAT3 inhibition leads to apoptosis and cell death in serum-derived GBM lines containing activated STAT3, but not in serum-derived lines lacking tyrosine-phosphorylated STAT3. In glioblastoma stem cells, STAT3 inhibitor treatment induces apoptosis only when removed from stem cell media. This suggests that the role of STAT3 in GBM-SC is distinct from its role in serum-derived cell glioma lines and in glioblastoma stem cell cultures that have been induced to differentiate.
Inhibition of STAT3 depletes markers of multipotency in GBM-SC
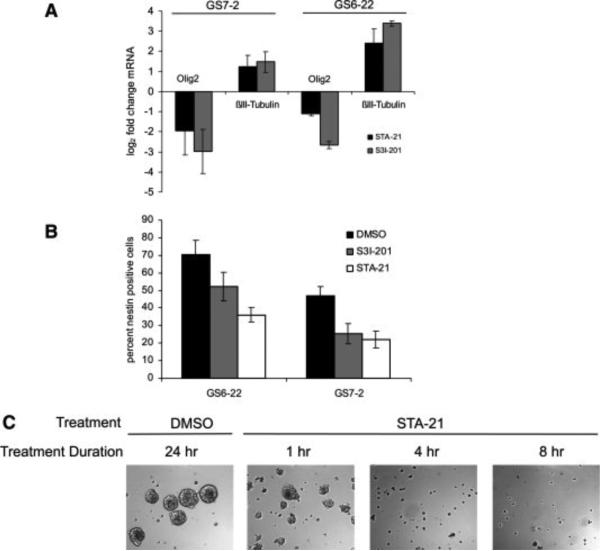
GBM-SC treated with STAT3 inhibitors fail to proliferate, but do not undergo apoptosis. In order to determine whether this effect signifies a loss of self-renewal potential, we examined the expression of known stem cell markers in GBM-SC treated with either STAT3 inhibitor. After 7 days of treatment with STA-21 or S3I-201, the expression of the neural stem cell marker Olig2 in GS6-22 and GS7-2 cells was decreased at the mRNA level (Figure 5A). Staining for the neural stem cell marker nestin also decreased in GBM-SC treated with STAT3 inhibitors, consistent with a loss of multipotency (Figure 5B). Expression of the neuronal marker βIII-tubulin increased slightly in either cell line treated with the STAT3 inhibitors (Figure 5A), suggesting that STAT3 inhibition over a period of 7 days leads to a partial induction of differentiation in addition to the observed loss of multipotency. However, the STAT3-inhibitor treated cells do not attach to the tissue culture plate and morphologically continue to resemble undifferentiated cells. Other differentiation markers such as GFAP are not upregulated (data not shown), suggesting that STAT3 inhibition alone is not sufficient to induce complete differentiation of the glioblastoma stem cells.
Figure 5. STAT3 inhibition depletes markers of stem-ness in GBM stem cells.
A. Olig2 and βIII-tubulin transcript levels in GBM-SC treated with STAT3 inhibitors or DMSO control for 7 days. mRNA was quantified by RT-qPCR. Data is shown as fold change relative to DMSO-treated cells and normalized for β-actin. Values represent mean of three experiments and bars SD. B. Nestin expression in GS7-2 and GS6-22 GBM-SC treated for 7 days with STAT3 inhibitors or DMSO control. Expression was quantified by immunostaining of fixed cells with mouse α-nestin antibody followed by flow cytometry. C. Treatment with STAT3 inhibitor STA-21 irreversibly inhibits neurosphere formation. GS6-22 cells were treated with inhibitor for the indicated periods of time, washed thoroughly, and replated in fresh media. Representative images were taken one week after drug removal.
Since loss of stem cell markers suggests that inhibition of STAT3 initiates differentiation of GBM-SC, we sought to determine whether the inhibition observed was reversible. Cells were treated transiently for various periods of time with either STA-21 (Figure 5C) or S3I-201 (Supplementary Figure 13). A single treatment of GS6-22 cells with STA-21 for as little as 4 hours prevented neurosphere formation from single cells for several weeks despite re-plating cells in fresh media lacking STA-21. Transient treatment with S3I-201 also irreversibly prevented neurosphere formation (Supplementary Figure 13). Cells remained viable as judged by trypan blue exclusion and by luciferase transgene expression (data not shown) but remained single cells. STAT3 activity was not permanently blocked by inhibitor treatment, as is demonstrated by the recovery of its nuclear localization as quickly as 1 hour following inhibitor-removal (Supplementary Figure 14). The inability to produce neurospheres for as long as 2 weeks post drug removal, along with the downregulation of stem cell markers, suggests that STAT3 inhibition was able to induce a sustained loss of self-renewal with respect to growth in culture. Even a transient treatment with STAT3 inhibitor was able to induce this loss, despite re-activation of STAT3 after drug removal. Both the loss of neural stem cell markers and the permanent nature of the growth arrest induced by transient STAT3 inhibition suggest that activated STAT3 is critical for maintaining GBM-SC in a proliferative and self-renewing state.
Discussion
STAT3 is regulated by a wide variety of growth factors and cytokines [7]. It has been shown to be activated in many different human cancers including glioblastoma [14,31,32]. Dominant negative STAT3 can inhibit transformation by v-src and activated STAT3 can transform 3T3 cells [33,34]. In mice, directed expression of activated STAT3 to the lung induces lung cancer and knockout of STAT3 in the skin epithelium renders mice resistant to skin tumor induction by phorbol esters [12,13]. STAT3 regulates a remarkable number of key cancer genes including proliferation genes c-myc, p21, cyclin D; the anti-apototic genes bcl-xl and survivin; the angiogenic gene VEGF; the EMT transition genes twist and LIV1; and the immortalization gene telomerase [9,15,35-38].
Recently, there has been increasing evidence that a minority of cells in a tumor with stem cell like properties called cancer stem cells are responsible for most of the tumorigenic potential. The evidence for this is particularly compelling for glioblastoma where most of the tumor initiating ability resides in the CD133+ fraction of cells [2,6]. These cells have many properties of neural stem cells and can differentiate in culture, but unlike normal neural stem cells can be maintained in culture indefinitely and form tumors in xenografts with high efficiency. Interestingly, STAT3 is known to play an important role in stem cells. It is required for the maintenance of pluripotency in murine ES cells [16-18,39]. There is also evidence that it is necessary for self-renewal of murine neural stem cells [19,20].
Here we have examined the function of STAT3 in glioblastoma stem cells. We have found that GBM-SC express STAT3 and that it is phosphorylated on the activating tyrosine and serine residues. Inhibition of STAT3 in these cells with either small molecular inhibitors or RNAi results in inhibition of growth and neurosphere formation. Unlike what we have found for serum derived glioma cell lines, the GBM-SC do not undergo apoptosis upon STAT3 inhibition in stem cell media. This is likely a reflection of the finding that tissue stem cells are relatively resistant to apoptosis. Consistent with this, the GBM-SC lines become sensitive to apoptosis induced by STAT3 inhibition when they are differentiated. It is possible that STAT3 inhibition sensitizes glioblastoma stem cells to apoptotic stimuli as it does for some non-transformed cell lines, but this remains to be determined [40,41].
We have also observed that in addition to inhibiting the formation of neurospheres from single GBM stem cells, inhibition of STAT3 leads to dissociation of pre-formed neurospheres. While it is not clear how self-adhesion of GBM-SC or NSC is mediated, STAT3 is known to regulate various intercellular adhesion molecules including I-CAMs [42].
In addition to inhibiting proliferation of GBM-SC, we have found that inhibition of STAT3 leads to loss of expression of the neural stem cell markers nestin and olig2. However, unlike cells grown in differentiation media, the STAT3 inhibitor-treated cells fail to adhere to the culture dish and show no clear signs of morphological differentiation, though there may be a modest induction of some differentiation markers such as βIII-tubulin. The astroctyic marker GFAP is not upregulated. This suggests that the GBMSC lose their ability to self renew in culture, but do not fully execute the differentiation program following STAT3 inhibition. An upregulation of the neuronal marker βIII-tubulin but not the astrocytic marker GFAP suggests that the partial differentiation we observe is biased towards the neuronal lineage, which is consistent with reports that STAT3 is necessary for neural stem cell differentiation to astrocytes [21,22].
Consistent with this, we have found that even a transient inhibition of STAT3 leads to an irreversible loss of the ability of the GBM-SC to form neurospheres. Along with growth inhibition and loss of stem cell markers, this suggests that STAT3 inhibition permanently induces a loss of self-renewal potential with respect to the ability to proliferate in culture as undifferentiated cells. This may or may not correlate with the ability of these cells to initiate tumors in mice. A possible mechanism for the permanent loss of this stem cell characteristic is that STAT3 inhibition leads to epigenetic changes that are not readily reversed. This would explain the rapid and irreversible nature of this phenotype, and would be consistent with findings in ES cells and murine NSC that inhibition of STAT3 leads loss of self-renewal and induction of differentiation [16-20,39]. Similarly, in olfactory bulb cells knockdown of STAT3 induces terminal neuronal differentiation [43]. In contrast, we have found that inhibition of STAT3 in the glioblastoma stem cells does not induce a fully differentiated phenotype likely due to aberrant signaling caused by tumor specific mutations. Indeed, inhibition of STAT3 in these cells under differentiation conditions leads to cell death. This suggests that STAT3 might be a particularly good target for therapeutic inhibition of cancer stem cells. Short term treatment could possibly be translated into long term effects on growth, self-renewal, and resistance to apoptosis of the cancer stem cell population within the tumor. However, it will be important to determine whether STAT3 inhibition would deplete the normal, but quiescent NSC population as well.
We have found that STAT3 in the GBM stem cells is phosphorylated on both tyrosine and serine residues. The interaction of the STAT3 pY705 with the STAT3 SH2 domain is required for high affinity DNA binding of the STAT3 dimer [7]. Phosphorylation of serine 727 is generally thought to be necessary for maximal transcriptional activity [26]. Previously, the serine phosphorylation of STAT3 has been implicated in the self-renewal of neural stem cells [44]. Indeed, recent evidence has indicated that there are important functions of the unphosphorylated form of STAT3 as well [45]. Our experiments do not distinguish clearly between any of these possibilities, though the loss of serine phosphorylation upon GBM-SC differentiation is consistent with a relationship between self-renewal potential and STAT3 serine 727 phosphorylation. The two drugs we have used, STA-21 and S3I-201, both bind to the STAT3 SH2 domain presumably preventing the STAT3 phosphotyrosine residue from interacting with it and thus abrogating the DNA binding activity. This suggests a requirement for the STAT3 tyrosine phosphorylation, but does not directly address whether serine phosphorylation is necessary for GBM-SC proliferation.
Previous studies have implicated STAT3 in the differentiation of neural precursor cells into astrocytes [21,22,46]. Our data here do not conflict with this data, but suggest an additional function for STAT3 in maintaining glioblastoma stem cell self-renewal. Our findings are consistent with previous work in mice that indicate that STAT3 maintains NSC self renewal [19,20]. From our data, this function of STAT3 appears to be maintained in cancer stem cells. It has been suggested that STAT3 promotes self-renewal of neural stem cells by regulating the Notch ligand DLL1 [20]. However, we have not observed changes in DLL1 expression in GBM-SC after STAT3 inhibition suggesting another mechanism for maintaining self-renewal of these cells (data not shown). Future experiments will examine the mechanism by which STAT3 regulates self-renewal in GBM-SC cells in culture.
Previous work has suggested that epigenetic changes that occur during astrocytic differentiation lead to changes in STAT3 function during neural stem cell differentiation. For instance, there is a critical STAT3 binding site in the GFAP promoter that is methylated in precursor cells that becomes demethylated and therefore STAT3 accessible during differentiation [47]. Lee et at. (2008) have similarly found that induction of astrocytes in GBM-SC depends partly on demethylation and expression of the BMPR1B receptor and that this differentiation depends on STAT3 activation [46]. Thus, it may be that STAT3 regulates different genes in astrocytes than it does in GBM-SC or NSC due to epigenetic differences between the cell types. This is consistent with our observations that STAT3 inhibition has a distinct phenotype in GBM-SC and in differentiated cells. While STAT3 inhibition blocks proliferation and self-renewal of GBM-SC, upon differentiation these cells are sensitized to apoptosis upon STAT3 inhibition. Thus, our findings are in agreement with studies in normal neural stem cells and astrocytes, which have both been shown to depend on STAT3 activation for distinct cellular processes.
For most human tumors, STAT3 is believed to oncogenic [11,32,48]. Most studies have also found STAT3 to be pro-oncogenic in serum-derived glioma cell lines as well [9,14,43,49-52]. However, two recent papers suggest that in some cases STAT3 may be a tumor suppressor gene in glioblastomas that have lost PTEN expression[53,54]. In contrast, our GS6-22 stem cell line has lost PTEN expression, but remains sensitive to STAT3 inhibition. One possible explanation of these seemingly conflicting observations may be that some glioma cell lines that are no longer in the STAT3 dependent stem cell state, but are blocked from terminal astrocytic differentiation by the loss of STAT3 and thereby stuck in a proliferative progenitor cell state. This would be consistent with the phenotype of the aberrant GBM-SC line of Lee et al (2008) that proliferates instead of differentiates in response to BMP signaling [46]. The relative importance or frequency of these two distinct functions of STAT3 (stem cell self-renewal versus astrocytic differentiation) in primary human glioblastoma remains to be determined, but is an important therapeutic question. Nonetheless, our findings that STAT3 is required for tumor stem cell self-renewal in glioblastoma suggest that it might be an effective target for tumor stem cell directed therapy.
Materials and Methods
Establishment and Culture of Glioblastoma Stem Cell lines
Glioblastoma tumor samples were obtained from recent surgical resections in accordance with the Tufts Medical Center IRB. Tumor samples were dissociated to form a single cell suspension, and plated in serum-free DMEM/F12 supplemented with B27 (Invitrogen, Carlsbad, CA), EGF (20 ng/mL, Peprotech, Rocky Hill, NJ), and FGF-2 (20 ng/mL, Peprotech). Neurosphere cultures were passaged approximately every week after initial neurosphere formation was observed. Sphere dissociation was performed according to an acid base dissociation protocol [55]. When indicated, differentiation of neurosphere cultures was obtained following protocols established for the differentiation of neural progenitor cells [4]. In short, cells were plated overnight on poly-L-ornithine- and laminin-coated plates in DMEM/F12 supplemented with B27 but lacking EGF and FGF-2. After 2 days of growth factor withdrawal, DMEM/F12 was replaced with DMEM supplemented with 2% FBS. Cells were lysed for western blotting or RT-qPCR analysis after 5 days in 2% FBS/DMEM. Inhibitor experiments were performed using 100 μM S3I-201 (NCI) or 30 μM STA-21 (BioMol International, NCI) unless otherwise indicated.
Immunoblotting and Immunocytochemistry
Cells were harvested by centrifugation and lysed in radioimmunoprecipitation assay buffer [0.15 mol/L NaCl, 1% NP40, 0.01 mol/L desoxycholate, 0.1% SDS, 0.05 mol/L Tris-HCl (pH 8.0)]. Lysates were electrophoresed on SDS-PAGE gels and transferred to nitrocellulose membranes. Membranes were probed with each antibody according to the manufacturer's instructions and immunoreactive proteins were visualized using Western Lightning™ chemiluminescence reagent (Perkin Elmer, Waltham, MA ). Anti-β-Actin (1:2000) was obtained from Sigma (St. Louis, MO); anti-STAT3 (1:1000), anti pSer-STAT3 (1:200), anti-pTyr STAT3 (1:500), anti p-AKT (1:1000) and anti-PTEN (1:1000) were obtained from Cell Signaling Technologies (Beverly, MA). Anti-nestin (1:2000) and anti-CD133 (1:300) were obtained from Abcam (Cambridge, MA). Anti p-ERK (1:1000) was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). For immunocytochemistry, cells were allowed to attach to laminin-coated coverslips, fixed with 4% parafomaldehyde for 20 minutes, washed, permeabilized and blocked in 1% BSA/ 0.2% Tween-20/ 10% goat serum/ PBS, and stained with anti-pTyrSTAT3 (1:50; Cell Signaling Technologies). After washing, cells were stained with a secondary goat anti-rabbit antibody (1:50, Jackson) and counterstained with DAPI.
RT-qPCR
RNA was isolated using the RNeasy mini-kit (Qiagen, Valencia, CA) according to the manufacturer's instructions and treated with DNAse I (Invitrogen) for 30 minutes in order to digest genomic DNA. RNA was reverse-transcribed with Supercript III (Invitrogen). Quantitative PCR was performed using the Quantitect SYBR Green PCR Kit (Quiagen) and the MX4000 real time cycler (Stratagene, La Jolla, CA). Data presented is the average of three individual experiments; within each experiment technical triplicates were performed. Fold changes were calculated relative to control by the 2-ΔΔCt method [56]. All primer sets amplify a single product of expected size; the following sequences were used: Olig2 forward: GGACAAGCTAGGAGGCAGTG; Olig2 reverse: ATGGCGATGTTGAGGTCGTG; GFAP forward: GGCAAAAGCACCAAAGACGG; GFAP reverse: GGCGGCGTTCCATTTACAAT; βIII-tubulin forward: GCCTCTTCTCACAAGTACGTG, βIII-tubulin reverse: CCCCAC T C T G A C C A A A G A T G A A ; β-Ac t i n f o r w a r d : CCTGGGCATGGAGTCCTGTGG; β-Actin reverse: CTGTGTTGGCGTACAGGTCTT.
Bandshift Assay
GS7-2 or GS6-22 cells were treated for 24 hours with DMSO control or STAT3 inhibitor and lysed in a high salt lysis buffer [500 mM NaCl, 50 mM Tris, 1% Triton X-100, 10% glycerol, 1mM EDTA]. Lysates were treated with STAT3 inhibitors (STA-21 30 μM, S3I-201 100μM) for 30 min at 37°C. 12 μg total protein was incubated with 32P-endlabeled hSIE STAT3 binding probe (approximately 40,000 cpm per reaction), 5μg/μL poly didc, 0.1M DTT, 10mg/mL BSA, and 50% glycerol and incubated for 30 min at 30°C. Lysates were loaded on a 5% PAGE gel and electrophoresed; gel was fixed and exposed to film overnight.
Nestin Staining
GS7-2 and GS6-22 cells were treated with STA-21, S3I-201, or DMSO control for 7 days. On day 7 the cells were trypsinized, washed, and fixed in paraformaldehyde in suspension. Cells were incubated with Stem Cell Technologies mouse anti-nestin antibody (1:50) for 30 minutes, washed, and incubated with Jackson FITC-rabbit anti-mouse secondary antibody (1:50) for 20 minutes in the dark. Cells were washed 4 times BD Biosciences Perm/Wash buffer, resuspended in PBS, and analyzed via flow cytometry.
Lentiviral shRNA Infection
Cells were infected with either PLKO.1 non-targeting control lentivirus (Sigma) or shSTAT3 PLKO.1 lentivirus (clone number TRCN0000020843) (Sigma) at an approximate MOI of 35 in the presence of 8 μg/mL polybrene. 24 hours post-infection the virus containing media was removed and replaced with fresh stem cell media for 24 hours. Cells were then treated with 2.5 μg/mL puromycin. For BrdU assays, cells were selected in puromycin for 2 days, plated in stem cell media without puromycin for 24 hours, then pulsed with 10 μM BrdU for 3 days. Cells were stained using the FITC BrdU flow kit (BD Biosciences) and analyzed on a flow cytometer.
Supplementary Material
Acknowledgements
This work was supported in part by a grant from the Brain Tumor Society to BHC. MMS was supported by NIH training grant T32 DK07542. We would like to thank the Tufts Neuroscience core for the use of the Stratagene qPCR machine, P30 NS047243. We thank Anton Cochran, Forrest Jones, and James Krasker for their help with the proliferation experiments and Hans-Peter Biemann for critical reading of the manuscript.
Footnotes
Author Contributions:
Maureen M. Sherry- Conception and design, collection of data, data analysis and interpretation, manuscript writing, final approval of manuscript.
Andrew Reeves- Collection of data, data analysis and interpretation, final approval of manuscript.
Julian K. Wu- Provision of study material and patients, final approval of manuscript.
Brent H. Cochran- Conception and design, collection of data, data analysis and interpretation, manuscript writing, final approval of manuscript.
References
- 1.Filippini G, Falcone C, Boiardi A, et al. Prognostic factors for survival in 676 consecutive patients with newly diagnosed primary glioblastoma. NEURO ONCOL. 2008;10:79–87. doi: 10.1215/15228517-2007-038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. CANCER RES. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 3.Yuan X, Curtin J, Xiong Y, et al. Isolation of cancer stem cells from adult glioblastoma multiforme. ONCOGENE. 2004;23:9392–9400. doi: 10.1038/sj.onc.1208311. [DOI] [PubMed] [Google Scholar]
- 4.Galli R, Binda E, Orfanelli U, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. CANCER RES. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 5.Ignatova TN, Kukekov VG, Laywell ED, et al. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. GLIA. 2002;39:193–206. doi: 10.1002/glia.10094. [DOI] [PubMed] [Google Scholar]
- 6.Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. CANCER CELL. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 7.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. NAT REV MOL CELL BIOL. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 8.Leslie K, Lang C, Devgan G, et al. Cyclin D1 is transcriptionally regulated by and required for transformation by activated signal transducer and activator of transcription 3. CANCER RES. 2006;66:2544–2552. doi: 10.1158/0008-5472.CAN-05-2203. [DOI] [PubMed] [Google Scholar]
- 9.Konnikova L, Kotecki M, Kruger MM, et al. Knockdown of STAT3 expression by RNAi induces apoptosis in astrocytoma cells. BMC CANCER. 2003;3:23. doi: 10.1186/1471-2407-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simeone-Penney MC, Severgnini M, Rozo L, et al. PDGF-induced human airway smooth muscle cell proliferation requires STAT3 and the small GTPase Rac1. AM J PHYSIOL LUNG CELL MOL PHYSIOL. 2008;294:L698–704. doi: 10.1152/ajplung.00529.2007. [DOI] [PubMed] [Google Scholar]
- 11.Bromberg J Stat proteins and oncogenesis. J CLIN INVEST. 2002;109:1139–1142. doi: 10.1172/JCI15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan KS, Sano S, Kiguchi K, et al. Disruption of Stat3 reveals a critical role in both the initiation and the promotion stages of epithelial carcinogenesis. J CLIN INVEST. 2004;114:720–728. doi: 10.1172/JCI21032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Du H, Qin Y, et al. Activation of the signal transducers and activators of transcription 3 pathway in alveolar epithelial cells induces inflammation and adenocarcinomas in mouse lung. CANCER RES. 2007;67:8494. doi: 10.1158/0008-5472.CAN-07-0647. [DOI] [PubMed] [Google Scholar]
- 14.Rahaman SO, Harbor PC, Chernova O, et al. Inhibition of constitutively active Stat3 suppresses proliferation and induces apoptosis in glioblastoma multiforme cells. ONCOGENE. 2002;21:8404–8413. doi: 10.1038/sj.onc.1206047. [DOI] [PubMed] [Google Scholar]
- 15.Konnikova L, Simeone MC, Kruger MM, et al. Signal transducer and activator of transcription 3 (STAT3) regulates human telomerase reverse transcriptase (hTERT) expression in human cancer and primary cells. CANCER RES. 2005;65:6516–6520. doi: 10.1158/0008-5472.CAN-05-0924. [DOI] [PubMed] [Google Scholar]
- 16.Niwa H, Burdon T, Chambers I, et al. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. GENES DEV. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boeuf H, Hauss C, Graeve FD, et al. Leukemia inhibitory factor-dependent transcriptional activation in embryonic stem cells. J CELL BIOL. 1997;138:1207–1217. doi: 10.1083/jcb.138.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raz R, Lee CK, Cannizzaro LA, et al. Essential role of STAT3 for embryonic stem cell pluripotency. PROC NATL ACAD SCI U S A. 1999;96:2846–2851. doi: 10.1073/pnas.96.6.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu F, Hata R, Ma YJ, et al. Suppression of Stat3 promotes neurogenesis in cultured neural stem cells. J NEUROSCI RES. 2005;81:163–171. doi: 10.1002/jnr.20561. [DOI] [PubMed] [Google Scholar]
- 20.Yoshimatsu T, Kawaguchi D, Oishi K, et al. Non-cell-autonomous action of STAT3 in maintenance of neural precursor cells in the mouse neocortex. DEVELOPMENT. 2006;133:2553–2563. doi: 10.1242/dev.02419. [DOI] [PubMed] [Google Scholar]
- 21.Bonni A, Sun Y, Nadal-Vicens M, et al. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. SCIENCE. 1997;278:477–483. doi: 10.1126/science.278.5337.477. [DOI] [PubMed] [Google Scholar]
- 22.Rajan P, McKay RD. Multiple routes to astrocytic differentiation in the CNS. J NEUROSCI. 1998;18:3620–3629. doi: 10.1523/JNEUROSCI.18-10-03620.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Yen C, Liaw D, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. SCIENCE. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 24.Huang HS, Nagane M, Klingbeil CK, et al. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J BIOL CHEM. 1997;272:2927–2935. doi: 10.1074/jbc.272.5.2927. [DOI] [PubMed] [Google Scholar]
- 25.Zhong Z, Wen ZDarnell JE., Jr Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. SCIENCE. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 26.Wen Z, Zhong ZDarnell JE., Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. CELL. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 27.Song H, Wang R, Wang S, et al. A low-molecular-weight compound discovered through virtual database screening inhibits Stat3 function in breast cancer cells. PROC NATL ACAD SCI U S A. 2005;102:4700–4705. doi: 10.1073/pnas.0409894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siddiquee K, Zhang S, Guida WC, et al. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. PROC NATL ACAD SCI U S A. 2007;104:7391–7396. doi: 10.1073/pnas.0609757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner BJ, Hayes TE, Hoban CJ, et al. The SIF binding element confers sis/PDGF inducibility onto the c-fos promoter. EMBO J. 1990;9:4477–4484. doi: 10.1002/j.1460-2075.1990.tb07898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stommel JM, Kimmelman AC, Ying H, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. SCIENCE. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 31.Darnell JE. Validating Stat3 in cancer therapy. NAT MED. 2005;11:595–596. doi: 10.1038/nm0605-595. [DOI] [PubMed] [Google Scholar]
- 32.Groner B, Lucks PBorghouts C. The function of Stat3 in tumor cells and their microenvironment. SEMIN CELL DEV BIOL. 2008 doi: 10.1016/j.semcdb.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 33.Turkson J, Bowman T, Garcia R, et al. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. MOL CELL BIOL. 1998;18:2545–2552. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bromberg JF, Wrzeszczynska MH, Devgan G, et al. Stat3 as an oncogene. CELL. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 35.Niu G, Wright KL, Huang M, et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. ONCOGENE. 2002;21:2000–2008. doi: 10.1038/sj.onc.1205260. [DOI] [PubMed] [Google Scholar]
- 36.Barre B, Avril S, Coqueret O. Opposite regulation of myc and p21waf1 transcription by STAT3 proteins. J BIOL CHEM. 2003;278:2990–2996. doi: 10.1074/jbc.M210422200. [DOI] [PubMed] [Google Scholar]
- 37.Yamashita S, Miyagi C, Fukada T, et al. Zinc transporter LIVI controls epithelialmesenchymal transition in zebrafish gastrula organizer. NATURE. 2004;429:298–302. doi: 10.1038/nature02545. [DOI] [PubMed] [Google Scholar]
- 38.Cheng GZ, Zhang WZ, Sun M, et al. Twist is transcriptionally induced by activation of STAT3 and mediates STAT3 oncogenic function. J BIOL CHEM. 2008;283:14665–14673. doi: 10.1074/jbc.M707429200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torres J, Watt FM. Nanog maintains pluripotency of mouse embryonic stem cells by inhibiting NFkappaB and cooperating with Stat3. NAT CELL BIOL. 2008;10:194–201. doi: 10.1038/ncb1680. [DOI] [PubMed] [Google Scholar]
- 40.Burke WM, Jin X, Lin HJ, et al. Inhibition of constitutively active Stat3 suppresses growth of human ovarian and breast cancer cells. ONCOGENE. 2001;20:7925–7934. doi: 10.1038/sj.onc.1204990. [DOI] [PubMed] [Google Scholar]
- 41.Alas S, Bonavida B. Inhibition of constitutive STAT3 activity sensitizes resistant non-Hodgkin's lymphoma and multiple myeloma to chemotherapeutic drug-mediated apoptosis. CLIN CANCER RES. 2003;9:316–326. [PubMed] [Google Scholar]
- 42.Park EJ, Ji KA, Jeon SB, et al. Rac1 contributes to maximal activation of STAT1 and STAT3 in IFN-gamma-stimulated rat astrocytes. J IMMUNOL. 2004;173:5697–5703. doi: 10.4049/jimmunol.173.9.5697. [DOI] [PubMed] [Google Scholar]
- 43.Ren W, Duan Y, Yang Y, et al. Down-regulation of Stat3 induces apoptosis of human glioma cell: a potential method to treat brain cancer. NEUROL RES. 2008;30:297–301. doi: 10.1179/016164107X230784. [DOI] [PubMed] [Google Scholar]
- 44.Androutsellis-Theotokis A, Leker RR, Soldner F, et al. Notch signalling regulates stem cell numbers in vitro and in vivo. NATURE. 2006;442:823–826. doi: 10.1038/nature04940. [DOI] [PubMed] [Google Scholar]
- 45.Yang J, Stark GR. Roles of unphosphorylated STATs in signaling. CELL RES. 2008;18:443–451. doi: 10.1038/cr.2008.41. [DOI] [PubMed] [Google Scholar]
- 46.Lee J, Son MJ, Woolard K, et al. Epigenetic-mediated dysfunction of the bone morphogenetic protein pathway inhibits differentiation of glioblastoma-initiating cells. CANCER CELL. 2008;13:69–80. doi: 10.1016/j.ccr.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takizawa T, Nakashima K, Namihira M, et al. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. DEV CELL. 2001;1:749–758. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- 48.Haura EB, Turkson J, Jove R. Mechanisms of disease: Insights into the emerging role of signal transducers and activators of transcription in cancer. NAT CLIN PRACT ONCOL. 2005;2:315–324. doi: 10.1038/ncponc0195. [DOI] [PubMed] [Google Scholar]
- 49.Iwamaru A, Szymanski S, Iwado E, et al. A novel inhibitor of the STAT3 pathway induces apoptosis in malignant glioma cells both in vitro and in vivo. ONCOGENE. 2007;26:2435–2444. doi: 10.1038/sj.onc.1210031. [DOI] [PubMed] [Google Scholar]
- 50.Smilowitz HM, Weissenberger J, Weis J, et al. Orthotopic transplantation of v-src-expressing glioma cell lines into immunocompetent mice: establishment of a new transplantable in vivo model for malignant glioma. J NEUROSURG. 2007;106:652–659. doi: 10.3171/jns.2007.106.4.652. [DOI] [PubMed] [Google Scholar]
- 51.Su Y, Li G, Zhang X, et al. JSI-124 Inhibits Glioblastoma Multiforme Cell Proliferation through G2/M Cell Cycle Arrest and Apoptosis Augment. CANCER BIOL THER. 2008:7. doi: 10.4161/cbt.7.8.6263. [DOI] [PubMed] [Google Scholar]
- 52.Ghosh MK, Sharma P, Harbor PC, et al. PI3K-AKT pathway negatively controls EGFR-dependent DNA-binding activity of Stat3 in glioblastoma multiforme cells. ONCOGENE. 2005;24:7290–7300. doi: 10.1038/sj.onc.1208894. [DOI] [PubMed] [Google Scholar]
- 53.de la Iglesia N, Konopka G, Puram SV, et al. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. GENES DEV. 2008;22:449–462. doi: 10.1101/gad.1606508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de la Iglesia N, Konopka G, Lim KL, et al. Deregulation of a STAT3-interleukin 8 signaling pathway promotes human glioblastoma cell proliferation and invasiveness. J NEUROSCI. 2008;28:5870–5878. doi: 10.1523/JNEUROSCI.5385-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sen A, Kallos MS, Behie LA. New tissue dissociation protocol for scaled-up production of neural stem cells in suspension bioreactors. TISSUE ENG. 2004;10:904–913. doi: 10.1089/1076327041348554. [DOI] [PubMed] [Google Scholar]
- 56.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. METHODS. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.