SUMMARY
Maf1 is a conserved repressor of RNA polymerase (pol) III transcription; however, its physiological role in the context of a multicellular organism is not well understood. Here, we show that C. elegans MAFR-1 is functionally orthologous to human Maf1, represses the expression of both RNA pol III and pol II transcripts, and mediates organismal fecundity and lipid homeostasis. MAFR-1 impacts lipid transport by modulating intestinal expression of the vitellogenin family of proteins, resulting in cell non-autonomous defects in the developing reproductive system. MAFR-1 levels inversely correlate with stored intestinal lipids, in part by influencing the expression of the lipogenesis enzymes fasn-1/FASN and pod-2/ACC1. Animals fed a high carbohydrate diet exhibit reduced mafr-1 expression and mutations in the insulin signaling pathway genes daf-18/PTEN and daf-16/FoxO abrogate the lipid storage defects associated with deregulated mafr-1 expression. Our results reveal physiological roles for mafr-1 in regulating organismal lipid homeostasis, which ensure reproductive success.
INTRODUCTION
Initially characterized in S. cerevisiae, Maf1 is an evolutionarily conserved transcriptional co-repressor of RNA polymerase (pol) III-dependent genes, such as tRNA and 5S rRNA, which impact the biosynthetic capacity of the cell (Upadhya et al., 2002; Vannini et al., 2010). This function of Maf1 is conserved, as human, mouse and Drosophila Maf1 also represses tRNA transcription (Boguta, 2013; Boguta and Graczyk, 2011; Marshall et al., 2012; Rideout et al., 2012). Mammalian Maf1 additionally regulates certain RNA pol II-dependent promoters, including some Elk-1-regulated genes (Johnson et al., 2007). Given that Maf1 has extended roles in higher eukaryotes, we examined its function in a physiological context.
We were keen to investigate the physiological role of Maf1 in a genetically tractable system such as C. elegans. We examined the function of the related C. elegans MAF polymerase III Regulator-1 (MAFR-1) protein and elucidated the functional consequences of altered mafr-1 expression on development, reproduction, and lipid homeostasis. In C. elegans, metabolic homeostasis is maintained by multiple evolutionarily conserved mechanisms (Barros et al., 2012; Brey et al., 2009; Brock et al., 2006, 2007; O’Rourke et al., 2009; Paek et al., 2012; Soukas et al., 2009; Walker et al., 2011; Watts, 2009; Zheng and Greenway, 2012) and C. elegans has become exceptionally useful for high-throughput screening studies of complex cellular processes relevant to human diseases (Anastassopoulou et al., 2011; Squiban et al., 2012; Wahlby et al., 2012). We have discovered that MAFR-1 negatively regulates intracellular lipid accumulation and influences reproductive capacity. Taken together, these studies define the physiological roles for Maf1 and indicate the potential for targeting of Maf1 for therapeutic strategies for the prevention and treatment of metabolic diseases with deregulated lipid phenotypes.
RESULTS
C. elegans MAFR-1 is a conserved modulator of RNA pol-III and pol-II transcript levels
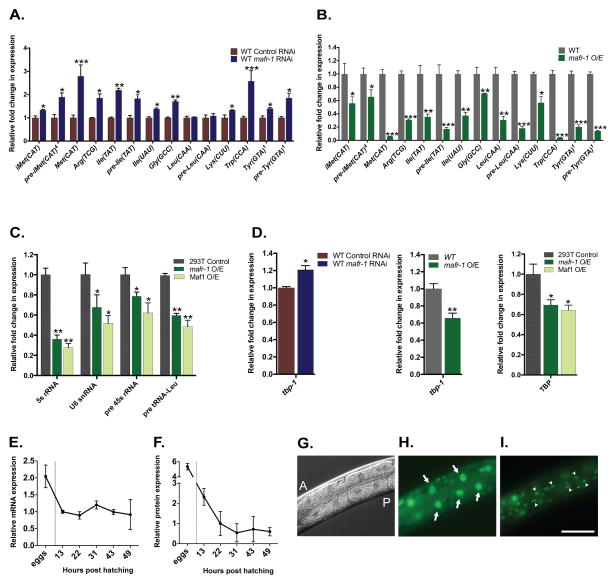
Given the conserved role of Maf1 as a negative regulator of RNA pol III in yeast, flies, and mammals, we investigated whether C. elegans MAFR-1 functions in an orthologous manner. We reduced mafr-1 expression by RNAi and measured the transcript levels of established RNA pol III transcripts, such as tRNAs. As predicted, when mafr-1 expression was reduced by approximately 50% (Figure S1A), the expression of most tRNAs were significantly increased as compared to the internal normalization control, snb-1, whose expression was stable (Figure 1A and S1A). We further examined animals harboring an additional chromosomally integrated copy of mafr-1, which results in ~80% increase in mafr-1 over expression (mafr-1 O/E) (Figure S1B) and observed a striking reduction in all tRNAs tested (Figure 1B and S1B). Furthermore, the reduction of tRNA levels observed in mafr-1 O/E animals were restored when animals were fed dsRNA targeting mafr-1, indicating that the effects on RNA pol III transcripts were specific to mafr-1 levels (Figure S1C).
Figure 1. MAFR-1 is a conserved modulator of RNA pol III and RNA pol II.
Expression of RNA pol III targets in mafr-1 RNAi treated animals (A) or animals overexpressing (O/E) mafr-1 (B). Expression of human RNA pol III targets in 293T cells transfected with mafr-1 or Maf1 (C). Expression of the RNA pol II target tbp-1/TBP (D). Developmental expression pattern of mafr-1 mRNA (E) and MAFR-1::GFP protein (F). MAFR-1 tissue expression revealed by imaging animals expressing MAFR-1::GFP DIC (G) (A, anterior and P, posterior), intestinal cells (H) (arrows), and hypodermal cells (I) (arrow heads). Data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P<0.001 versus respective controls. Scale bar = 100um. See also Figure S1.
Drosophila dMaf1 was shown to control body size and developmental timing by specifically regulating tRNAiMet synthesis (Rideout et al., 2012). In C. elegans, mafr-1 expression is inversely correlated with the synthesis of multiple tRNAs, including tRNAiMet (Figure 1A). mafr-1 RNAi increases animal body area by ~4% while mafr-1 O/E leads to a ~7% decrease in body area (Figure S1D). Unlike modulation of dMaf1 in flies, mafr-1 levels do not alter developmental timing in the worm (Figure S1E).
We tested the ability of MAFR-1 to regulate the expression of mammalian RNA pol III targets. Overexpression of either MAFR-1 or human Maf1 in human 293T cells was sufficient to reduce the expression of multiple human RNA pol III transcripts (Figure 1C). This indicates that C. elegans MAFR-1 function is conserved across metazoans. Because human Maf1 is recruited to the promoters of select RNA pol II genes, such as TBP1 (Johnson et al., 2007), we examined the ability of MAFR-1 to regulate tbp-1. Similar to mammalian Maf1, MAFR-1 is also capable of negatively regulating the expression of the RNA pol II target tbp-1 in worms as well as human TBP1 in 293T cells (Figure 1D). One model for Maf1 function is as a transcriptional repressor by interacting with components of the RNA pol III-specific TFIIIB complex, which contains RNA pol III, Tbp1, and Brf1 (Boguta, 2013; Boguta and Graczyk, 2011; Marshall et al., 2012). Consistent with previous reports in other organisms, decreased expression of tbp-1 or brf-1 in C. elegans effectively reduced the expression of RNA pol III transcripts, similar to mafr-1 O/E (Figure S1G and S1H). Importantly, RNAi of tbp-1 or brf-1 in the mafr-1 O/E strain did not further reduce the expression of tRNAs (Figure S1H), indicating that MAFR-1, TBP-1, and BRF-1 function in the same pathway to regulate RNA pol III-dependent transcription. Collectively, these results support a conserved role for MAFR-1 as a negative regulator of both RNA pol III- and RNA pol II-mediated transcription and identify MAFR-1 as a functional ortholog of mammalian Maf1.
MAFR-1 expression in C. elegans fat storage tissues alters the expression of metabolism and reproduction genes
To identify the physiological roles of MAFR-1 in a multicellular organism, we first examined the temporal expression of mafr-1 transcripts and MAFR-1::GFP protein throughout development and documented the tissue expression patterns of MAFR-1::GFP. mafr-1 expression is highest in embryos but is reduced post-hatching and remains relatively constant throughout larval development and into adulthood (Figure 1E). After embryogenesis, the levels of MAFR-1 protein are reduced and stabilize (31 hours post hatching) in the L3/L4 larval stage (Figure 1F). We next examined the tissue expression pattern of MAFR-1 using transgenic animals expressing a MAFR-1::GFP fusion construct. We observed the strongest expression of MAFR-1::GFP in the intestine and hypodermis, which are the two major sites of fat storage in C. elegans (Mak, 2012; O’Rourke et al., 2009) (Figure 1G–I).
Our discovery that MAFR-1 can modulate the expression of RNA pol II genes, such as tbp-1, fits previous reports of the localization of human Maf1 at select RNA pol II promoters (Johnson et al., 2007). However, it remains unclear if worm MAFR-1 directly regulates these genes and the extent to which Maf1 can impact the expression of RNA pol II transcriptional targets. We therefore compared the steady state gene expression profiles of a single copy mafr-1 O/E model as compared to control animals. The canonical role of MAFR-1 as a negative regulator of transcription drove our analysis of genes whose expression was reduced when mafr-1 was overexpressed (Table 1). A bioinformatic examination of these genes identified enrichment for gene ontology (GO) terms related to reproduction and metabolism (Table 1). Enrichment for factors associated with the negative regulation of translation and lipid transport were also identified. GO-terms linked to the regulation of RNA pol III target expression (4.17% of genes of this GO-term were represented) and RNA polymerase activity (5.26% of genes of this GO-term were represented) were also identified (data not shown). We also identified transcripts that were increased in the mafr-1 O/E animals (Table S1), which may represent homeostatic responses to deregulated mafr-1 expression. These findings support a role for MAFR-1 in modulating RNA pol III, but also reveal its ability to influence RNA pol II targets with biological functions beyond protein synthesis capacity. We investigated a selection of transcripts whose expression was deregulated when mafr-1 levels are altered. These transcripts were selected based on the degree and significance of the change and the identification of a representative portion of the genes within the gene family or GO-term group.
Table 1.
GO-term analysis of 1495 genes downregulated in mafr-1O/E animals
| Gene ontology (GO) term1 | Enrichment2 |
|---|---|
| Developmental process involved in reproduction | Score = 24.51, p-value = 10E-11 |
| % genes in group | |
| Reproductive structure development | 18.74 |
| Germ cell development | 19.44 |
| Primary metabolic process | Score = 5.46, p-value = 0.004 |
| % genes in group | |
| Amino acid metabolism | 9.65 |
| Lipid metabolism | 9.09 |
| Carbohydrate metabolism | 8.29 |
| Negative regulation of translation | Score = 5.18, p-value = 0.005 |
| Lipid transport | Score = 4.82, p-value = 0.01 |
Partek Genomic suite used for microarray dataset and GO enrichment analysis fold change +/− 2, false discovery rate <= 0.001
Fishers exact test was used for GO-term enrichment.
mafr-1 negatively regulates lipid transport and fecundity
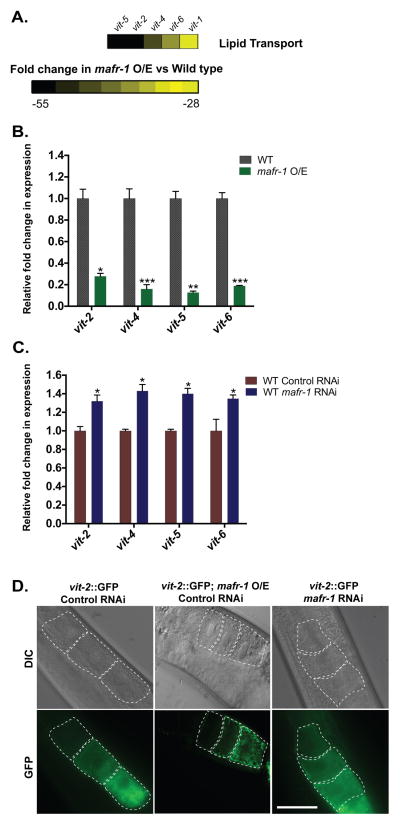
Our analysis of the transcripts sensitive to mafr-1 expression identified components of the C. elegans vitellogenin (vit) lipid transport system, which were significantly down regulated (~20- to 50-fold as compared to wild type) in our microarray analysis (Figure 2A). Vitellogenins are lipid-binding proteins that are produced in the intestine, secreted as yolk into the body cavity, and diffuse into the gonad where they are endocytosed by developing oocytes (Grant and Hirsh, 1999; Kimble and Sharrock, 1983). We measured the expression of vit genes in mafr-1 O/E (Figure 2B) and mafr-1 RNAi treated animals (Figure 2C) and discovered MAFR-1 negatively regulated the expression vit-2, vit-4, vit-5, and vit-6. The reduction in vit expression in mafr-1 O/E animals was specific to mafr-1 expression levels as RNAi targeting mafr-1 could reverse this reduction (Figure S2A). We next tested if vit gene expression was tied to the change in organismal tRNA abundance when mafr-1 levels are altered (Figure 1). As reported above, RNAi of tbp-1 or brf-1 reduced the expression of tRNAs, similar to mafr-1 O/E. However, unlike mafr-1 O/E, brf-1 RNAi treated animals in general have normal levels of vit expression (Figure S2B). Surprisingly, animals with reduced expression of tbp-1 actually increase vit gene expression (Figure S2B). Taken together these findings implicate a role for MAFR-1 in the expression of vit genes that is not simply a result of altered transcription of tRNAs.
Figure 2. Impact of mafr-1 expression on lipid transport and oocyte development.
The vitellogenin lipid transport system was significantly repressed in microarrays of mafr-1 O/E animals as compared to non-transgenic siblings (A); numbers represent fold change in expression as compared to wild type animals. qPCR analysis reveals the lipid transport genes vit-2, -4, -5, and -6 are differentially regulated by mafr-1 levels in mafr-1 O/E (B) or RNAi treated (C) animals. (D) Altered vitellogenin gene expression correlates with localization and abundance of VIT-2::GFP in developing oocytes. Scale bar = 50um. See also Figure S2.
To further examine the effects of mafr-1 expression on VIT-2 function we utilized a transgenic strain expressing a VIT-2 fusion to GFP to allow visualization of lipid transport to the reproductive system (Grant and Hirsh, 1999). Consistent with our gene expression analysis, oocytes in mafr-1 O/E animals contained less intracellular VIT-2::GFP when compared to controls (Figure 2D). Similarly, RNAi of mafr-1 led to a modest increase in VIT-2::GFP in oocytes. These data suggest that mafr-1 regulates the expression of the vitellogenin yolk precursor proteins in the intestine, which function in a cell non-autonomous manner in the developing germline of the reproductive system.
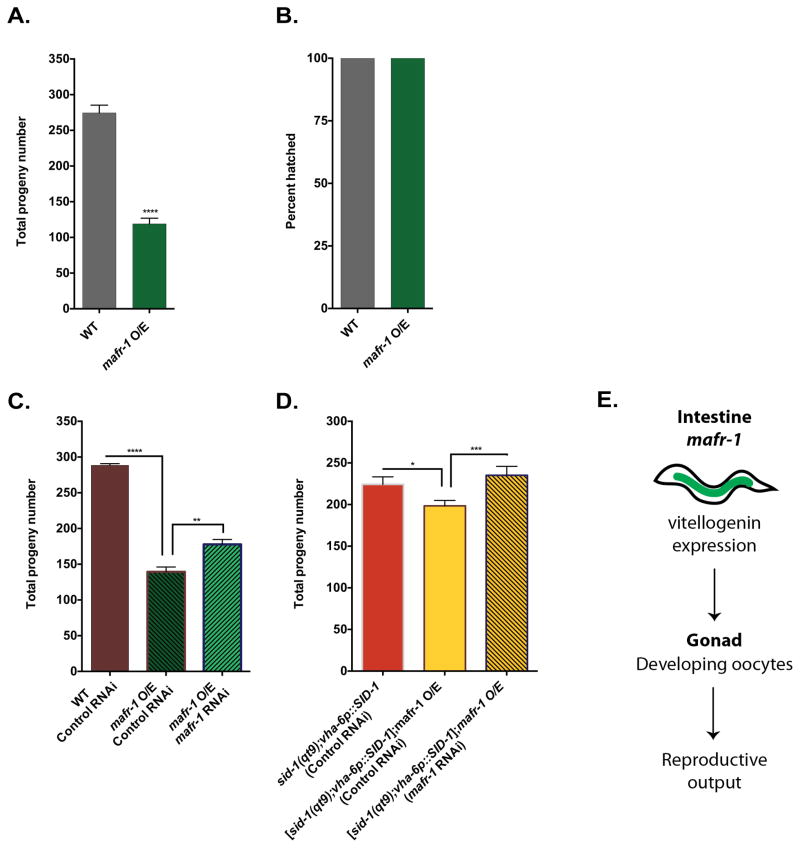
We hypothesized that the diminished abundance of VIT-2 lipid particles in the developing oocytes of mafr-1 O/E animals would result in a strong reproduction phenotype. As predicted, mafr-1 O/E animals display a >50% reduction in fecundity as compared to wild type controls (Figure 3A). In addition, mafr-1 O/E had no measurable effect on embryo viability as all eggs hatched (Figure 3B). The diminished reproduction phenotype is tied to MAFR-1 overexpression as RNAi of mafr-1 in the mafr-1 O/E strain could partially restore progeny production (Figure 3C). Intriguingly, in wild type animals, RNAi of mafr-1 was not capable of increasing total brood size (Figure S3A). brf-1 RNAi animals have a small reduction in fecundity that is synergistically reduced when combined with mafr-1 O/E, which suggests two parallel pathways converging upon reproductive output (Figure S3B). This finding is consistent with our observation that the altered expression of vit genes in the mafr-1 O/E strain could not be phenocopied by simply reducing the expression of RNA pol III transcripts following tbp-1 or brf-1 RNAi. We could not test the relationship with tbp-1 in this assay as tbp-1 RNAi treatment in worms leads to embryonic lethality (Gonczy et al., 2000; Piano et al., 2000).
Figure 3. Altered mafr-1 expression in the intestine has cell non-autonomous effects on reproductive output.
Total brood size (A) and embryo viability (B) of animals with mafr-1 O/E. Restoration of fecundity by tissue general (C) or intestine specific (D) RNAi of mafr-1. Model for cell non-autonomous role of mafr-1 expression on fecundity (E). Data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P<0.001 versus respective controls. See also Figure S3.
Since vitellogenesis occurs in the intestine we next tested whether mafr-1 expression specifically in the intestine was causal for the observed decline in reproductive output. The intestinal specific RNAi strain has a reduced brood as compared to wild type animals, which is perhaps due to the sid-1(qt9) genetic background compounded with the intestinal sid-1(+) rescue array. However, mafr-1 O/E is still capable of reducing fecundity, demonstrating that reproductive output in this strain remains sensitive to mafr-1 levels. Importantly, when we lower mafr-1 expression by RNAi specifically in the intestine of animals with mafr-1 O/E, we observed an increase in fecundity (Figure 3D and S3C). Together, these results suggest that mafr-1 expression exerts a cell non-autonomous influence on reproductive success by altering lipid transport and not through pleiotropic effects on zygote formation or embryonic development (Figure 3E).
mafr-1 negatively regulates lipid homeostasis
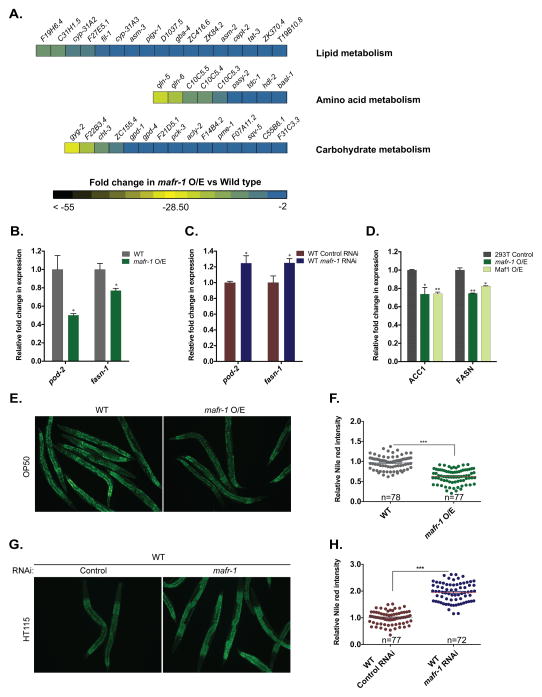
Our microarray analysis of mafr-1 overexpression animals identified a significant enrichment for lipid, amino acid, and carbohydrate metabolism genes (Figure 4A). We examined the expression of established lipid metabolism genes in mafr-1 RNAi treated and mafr-1 single copy overexpression animals (Figure 4B and 4C). We increased the specificity of our mafr-1 sensitive gene targets by imposing a requirement for reciprocal effects on transcript levels: increased expression in mafr-1 RNAi animals and decreased expression in mafr-1 O/E animals.
Figure 4. mafr-1 expression levels alter organismal lipid homeostasis.
(A), Multiple GO-terms tied to cellular metabolism were significantly repressed in microarrays of mafr-1 O/E animals as compared to non-transgenic siblings. Expression of the lipid biosynthesis genes pod-2/ACC1 and fasn-1/FASN in animals overexpressing mafr-1 (B) or with RNAi-reduction of mafr-1 (C). Expression of human ACC and FASN in human 293T cells transfected with either MAFR-1 or hMaf1 constructs (D). (E–F) Nile red staining of OP50 fed wild type and mafr-1 O/E animals. The representative animals are shown in (E) and quantitative data are shown in (F). (G–H) Nile red staining of wild type animals fed control or mafr-1 RNAi bacteria. The representative images are shown in (G) and quantitative data are shown in (H). Data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P<0.001 versus respective controls, (n) number of animals used for statistical analysis in each condition. Scale bar = 200um. See also Figure S4.
The enrichment for lipid metabolism genes was intriguing, but maintenance of lipid homeostasis requires the coordination of hundreds of lipid pathway genes. As such, we first focused on lipid biosynthesis. The expression of the C. elegans lipid biosynthesis genes pod-2/ACC1 and fasn-1/FASN, were repressed in mafr-1 O/E animals (Figure 4B) and higher when mafr-1 expression was reduced by RNAi (Figure 4C). The repression in mafr-1 O/E could be reversed by mafr-1 RNAi (Figure S4A). Similar to our findings for the expression of the vit genes, pod-2 and fasn-1 transcript levels were not altered in brf-1 RNAi animals and were increased in tbp-1 RNAi treated worms (Figure S4B). This is suggestive that the changes in the expression of lipid biosynthesis genes in mafr-1 O/E animals are not simply a response to global reduction in tRNA biosynthesis or tbp-1 expression. We also examined the expression of multiple lipid homeostasis genes that function in fatty acid oxidation and lipolysis, but could not detect any significant change when mafr-1 was reduced by RNAi or overexpressed (Figure S4C and S4D). Together these results reveal that MAFR-1 regulates intracellular lipids in part through its ability to regulate lipogenic enzyme gene expression.
Previous studies on Maf1 function have primarily focused on single cell models (Boguta, 2013; Johnson et al., 2007; Michels et al., 2010; Rohira et al., 2013; Shor et al., 2010). The observed changes in lipid homeostasis resulting from altering mafr-1 in the entire organism can result from cell autonomous and/or cell non-autonomous effects on physiology. The GO terms enriched in animals with altered mafr-1 expression included genes expressed in a variety of tissues, which may act in an autonomous manner. Because our microarray analysis is a snapshot of expression of all tissues in the animal combined, we took advantage of the conserved activity of MAFR-1 in mammalian cells and asked if MAFR-1 could also alter the expression of de novo lipogenesis genes in this model. Similar to our observations for RNA pol III gene targets, 293T cells overexpressing either MAFR-1 or human Maf1 have lower levels of human ACC1 and FASN mRNAs (Figure 4D).
The enrichment for metabolism genes in our microarrays prompted an investigation of the biological relevance of mafr-1 expression on metabolic homeostasis. Similar to most animals, C. elegans fat homeostasis is influenced by multiple metabolic pathways (Mak, 2012). As such, we first examined the levels of stored fat by fixed Nile red and Oil red O staining in mafr-1 O/E and mafr-1 RNAi animals. Overexpression of mafr-1 led to a significant 35% reduction of intracellular lipids (Figure 4E and 4F and Figure S4F), while RNAi of mafr-1 resulted in a striking 94% increase in stored intestinal fat (Figure 4G, 4H and S4E). Similar to previous reports, we found the steady state levels of stored lipids to be lower in worms fed RNAi bacteria HT115 as compared to OP50 (Figure 4E, 4G, S4G and S4H). Importantly, the changes in stored fat were not a result of altered food intake, as measured by pharyngeal pumping rates (Figure S4I). These results indicate that organismal lipid homeostasis is sensitive to even single copy variations in mafr-1 expression.
MAFR-1 is regulated by glucose and insulin signaling
The measured reduction in stored lipids observed in animals overexpressing mafr-1 led us to test if mafr-1 O/E could counteract diet-induced obesity in the C. elegans model. To this end, we challenged mafr-1 O/E animals on a high carbohydrate diet (HCD), which is capable of inducing increased storage of intracellular lipids (Lee et al., 2009; Pang and Curran, 2014; Pang et al., 2014; Schulz et al., 2007) (Figure 5A and 5B). Consistent with the steady state reduction in stored lipids on a normal diet, mafr-1 O/E could partially reduce the accumulation of lipids on the HCD by ~10%, albeit these animals were still significantly more fat than animals raised on a normal diet. In fact, because the mafr-1 O/E animals are less fat than wild type on a regular diet, the increase (166%) when mafr-1 O/E animals are fed a HCD compared to a regular diet is much larger than the increase (112%) observed in wild type animals on similar diets (Figure 5B).
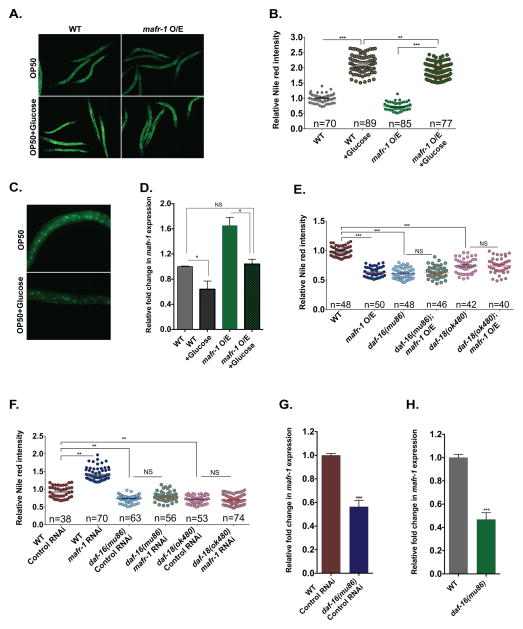
Figure 5. Glucose and insulin signaling pathways regulate the expression of mafr-1.
(A–B) Nile red staining of worms with indicated genotypes fed OP50 or OP50 plus 2% glucose. The representative images are shown in (A) and quantitative data are shown in (B). Dietary glucose reduces the steady state levels of MAFR-1 protein (C) and mafr-1 transcripts (D). Quantification of Nile red staining of animals overexpressing mafr-1 (E) or RNAi treated for mafr-1 (F) in daf-18/PTEN or daf-16/FoxO mutant backgrounds. Expression of mafr-1 in daf-16/FoxO RNAi treated (G) or genetic null mutant (H) animals. Data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P<0.001 versus respective controls, (n) number of animals used for statistical analysis in each condition. Scale bar = 600um. See also Figure S5.
In the presence of ample dietary sugars, excess carbohydrates are converted to triglycerides (Hillgartner et al., 1995). The ability of mafr-1 levels to alter de novo lipogenesis, but not effectively abrogate the total lipid increase on a HCD suggested mafr-1 itself could be sensitive to nutrient availability. To test this, we first examined the levels of MAFR-1::GFP in animals fed a HCD. MAFR-1 protein was reduced in animals raised in the presence of glucose (Figure 5C). The reduction in MAFR-1 was in part the result of reduced expression of mafr-1 on the HCD, both in wild type and MAFR-1::GFP expressing animals (Figure 5D). This diet-induced repression effectively abrogates the increased expression of mafr-1 in mafr-1 O/E animals when glucose is present. Previous biochemical characterization of the E. coli strains OP50 and HT115 have revealed that the HT115 diet has a higher carbohydrate composition than OP50 (Brooks et al., 2009). Consistent with these findings, a comparison of the mafr-1 levels in animals raised on these two diets reveals lower expression of mafr-1 when fed the HT115 diet (Figure S5A). These data indicate mafr-1 expression and MAFR-1 protein levels are sensitive to the abundance of dietary carbohydrates.
In C. elegans, the presence of glucose represses the FoxO transcription factor DAF-16 (Lee et al., 2009) through a conserved insulin/IGF-like signaling pathway. We tested whether inhibition of DAF-16/FoxO activity was sufficient to suppress the effects of mafr-1 expression levels using animals lacking the DAF-16 regulator daf-18/PTEN. In the absence of daf-18/PTEN, the insulin signaling pathway is de-repressed and DAF-16 activity is inhibited (Ogg and Ruvkun, 1998). In this genetic background, the induced changes in stored lipids mediated by mafr-1 overexpression (Figure 5E, S5B) or RNAi reduction (Figure 5F, S5C) were abrogated. In addition, we tested the ability of mafr-1 expression levels to alter intracellular lipid homeostasis in the absence of daf-16. The mafr-1-dependent change in steady state levels of intracellular lipids was abolished in the absence of DAF-16/FoxO (Figure 5E, 5F, S5B, and S5C). These data reveal a requirement for the insulin-like signaling pathway in MAFR-1 mediated regulation of stored lipids. This finding is consistent with the reported role for the insulin receptor acting down stream of dMaf1 in regulating growth in flies (Rideout et al., 2012).
We further investigated why these daf-16 null-mutants were insensitive to mafr-1 O/E. The requirement of DAF-16 in the MAFR-1 lipid phenotype could result from transcriptional regulation of mafr-1. We measured mafr-1 expression in animals lacking daf-16 and found a 50% reduction in expression as compared to wild type animals (Figure 5G and 5H). Next, we looked at the requirement of DAF-16 in the induction of tRNAs, vit, and lipid biosynthesis genes when mafr-1 expression was reduced by RNAi. In the absence of daf-16, mafr-1 RNAi was still able to induce the expression of most tRNAs, although the basal expression of multiple tRNAs was initially lower in the daf-16(mu86) null mutant background as compared to wild type controls. mafr-1 RNAi was similarly able to increase the expression of pod-2/ACC1, vit-2, -4, and -5, but intriguingly, not fasn-1 and vit-6 (Figure S5D).
In flies, inhibition of dMaf1 by RNAi results in the release of insulin-like peptides and systemic insulin signaling. We therefore tested whether mafr-1 had a similar effect on insulin-like signaling in C. elegans. We equated activation of insulin-like signaling by measuring the induction of the DAF-16/FoxO target gene sod-3. Unlike the effect of the hypomorphic allele of the insulin-like receptor, daf-2(e1368), which leads to a dramatic ~60-fold increase in the expression of sod-3, overexpression of mafr-1 led to a small but significant ~2.5-fold increase in sod-3 expression (Figure S5E). Feedback and feed-forward regulation by endocrine systems are well established (Murphy and Hu, 2013; Rutter, 1999) and our data are consistent with previous reports that insulin/IGF-I signaling can potentiate organismal responses to Maf1 (Rideout et al., 2012). Future dissection of where and how MAFR-1 integrates into the insulin-like signaling pathway will be of great interest.
DISCUSSION
We have identified C. elegans MAFR-1 as the functional ortholog of mammalian Maf1 and uncovered roles for MAFR-1 in the regulation of organismal fecundity and lipid homeostasis (Figure 6). We find that MAFR-1 can function in an orthologous manner to human Maf1 (hMaf1) in cell culture. Specifically, we find that MAFR-1 is capable of repressing the expression of human RNA pol-III transcribed targets, which indicates functional conservation between invertebrates and mammals. Moreover, MAFR-1 can alter the transcript levels of RNA pol II targets such as tbp-1, similar to mammalian Maf1 (Johnson et al., 2007). tbp-1 RNAi reduces tRNA transcript levels in a manner similar to the effect observed in mafr-1 O/E. However, the reduction in tbp-1 expression is not solely responsible for all of the physiological responses we have documented, as tbp-1 RNAi fails to phenocopy the mafr-1 O/E strain. This finding is intriguing, especially in light of the increased expression of the vit genes observed in tbp-1 RNAi treated animals. The C. elegans genome encodes one TBP-related factor, tlf-1, that could possibly compensate for the absence of tbp-1. We find that although tbp-1 levels are responsive to mafr-1, tlf-1 levels are not altered when mafr-1 is reduced or overexpressed (Figure S1F). Intriguingly, tlf-1 levels are also lower in tbp-1 RNAi treated animals, and therefore, not likely compensating for tbp-1 (Figure S1F). This is consistent with previous studies that suggest TLF-1 performs a unique function in activating bulk RNA pol II transcription during embryogenesis that is distinct from TBP-1 (Kaltenbach et al., 2000).
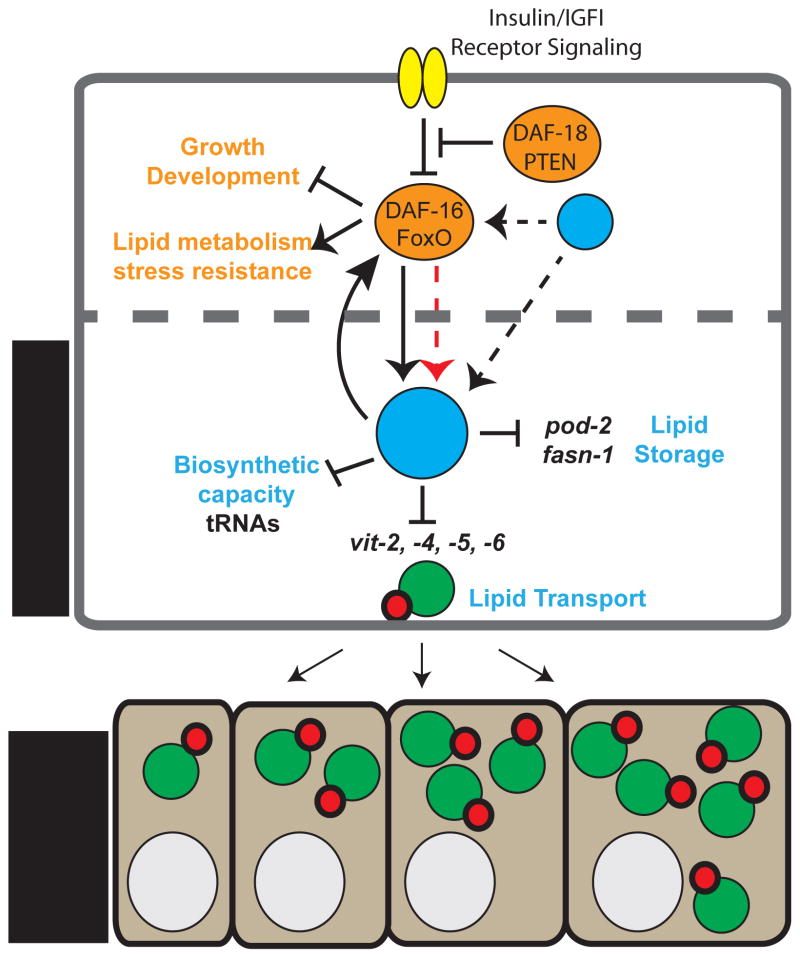
Figure 6. Model of the central role of MAFR-1 in organismal physiology.
MAFR-1 can influence animal physiology in both an insulin/FoxO-dependent (orange) and independent (blue) manner. Altered mafr-1 levels in the intestine cell non-autonomously impacts fecundity through changes in the production of the vitellogenin family of proteins that deregulates lipid transport to developing oocytes in the germ line. Black arrows and bars indicate genetic interactions. DAF-16/FoxO may directly regulate the expression of mafr-1 (dashed red arrow). Other factors that regulate DAF-16/FoxO-dependent and independent regulation of Maf1 may exist (?) and influence Maf1 cell autonomously and non-autonomously. Dashed cell boundary indicates potential auto-, para-, or endocrine signals resulting from Maf1 activity.
Our studies uncover the surprising finding that Maf1 regulates intracellular lipids. The transcriptional and physiological effects of altering mafr-1 abundance are specific to mafr-1, as we can suppress the observed changes in the mafr-1 O/E strain with RNAi knockdown of mafr-1. An examination of the mechanism by which this occurs reveals that Maf1 represses the expression of lipid biosynthesis (pod-2 and fasn-1) and transport (vit) genes. The changes observed in lipid biosynthesis genes and vitellogenins are not simply due to changes in RNA pol III transcription, as directly altering RNA pol III-dependent transcription does not change the expression of these RNA pol II-transcribed genes. Although 293T cell have abnormal metabolism, our findings are further supported by a complementary study from Deborah Johnson’s group showing hMaf1 ChIPs to the FASN promoter and regulates intracellular lipid abundance in Huh7 cells, whose lipid metabolism and physiology are more clearly defined than 293T cells (D.L.J., unpublished data). Although this doesn’t exclude the possibility that Maf1 may participate in other pathways to regulate lipid homeostasis, we identify Maf1 as an important negative regulator of lipid biogenesis.
Excess dietary carbohydrate added to the growth medium can increase lipids and decrease survival (Lee et al., 2009). Although carbohydrates represent just one major macronutrient difference between the OP50 and HT115 diets, our data reveal a role for MAFR-1 in metabolic homeostasis and that C. elegans lipid metabolism is sensitive to even small changes in mafr-1 expression. Multiple genetic phenotypes have been shown to only manifest on either the OP50 or HT115 diet. Our understanding of the underlying molecular mechanisms regulating these differences is only beginning to emerge (Maier et al., 2010; Pang and Curran, 2014; Pang et al., 2014). Under our standard laboratory growth conditions, the expression of key lipid biosynthesis genes, but not fatty acid oxidation genes, are repressed by mafr-1, and we have found that endogenous mafr-1 levels are regulated in part by available dietary carbohydrates. These two findings could be functionally linked, as lipid biosynthesis is also regulated by carbohydrate availability. As such, MAFR-1 could play an integral role in fine-tuning this pathway, which ultimately impacts steady state levels of stored intracellular lipids. Importantly, these results define additional layers of specificity of the MAFR-1 biologically relevant pathways, as we do not observe deregulation in the expression of all genes when mafr-1 levels are deregulated.
The ability of an organism to integrate information on available nutrients to coordinate growth and metabolism is essential for survival (Mair, 2013; Pang and Curran, 2014; Pang et al., 2014). We find that the levels of MAFR-1 are influenced by the abundance of nutritional glucose and that mafr-1 expression is lower in animals fed an E. coli K-12 HT115 diet versus the standard E. coli B OP50 diet. Intriguingly, the observed reduction in expression on these two diets may involve the insulin-signaling pathway, as the expression of mafr-1 in the absence of daf-16/FoxO is similar on either food source. This finding is correlative with previous characterization of these bacterial strains, which revealed a 3 to 5-fold higher level of endogenous carbohydrates in HT115 bacteria (Brooks et al., 2009). We also noted a trend for a slightly larger brood size in wild type animals fed the HT115 diet, but the inability for mafr-1 RNAi to further increase this brood. These results are consistent with the lower expression of mafr-1 on the HT115 diet and suggest that further reduction of mafr-1 cannot synergistically enhance reproductive capacity, whose maximum is dictated by spermatogenesis (Hughes et al., 2007; Ward and Carrel, 1979). Obviously the use of bacteria as a food source increases the complexity of deciphering diet-dependent phenotypes, particularly when additives such of glucose are utilized, which can alter bacterial growth and physiology. Although animals feeding on the HT115 diet do not show an increase in stored fat, despite diminished mafr-1 expression, this could be the result of a variety of metabolic actions initiated by the host in response to this complex diet (Brooks et al., 2009). The identification of other specific macronutrient triggers will be needed to determine the extent to which MAFR-1 integrates into other metabolic pathways.
The reduction in total cellular fat accumulation when mafr-1 is overexpressed in worms is intriguing and suggests that mafr-1 could be a potential target for therapies of obesity and metabolic diseases. Although our worm model of mafr-1 overexpression when fed a HCD was statistically less fat than wild type animals fed the same diet, the percent increase in fat as compared to the same animals on a normal diet was greater for the mafr-1 O/E strain. This could be due to the fact that our overexpression system is a result of a single additional copy of mafr-1. Future studies will investigate if higher gene dosages can provide additional protection from a HCD and if manipulating mafr-1 specifically in the intestine can cell autonomously impact lipid metabolism.
We also identify DAF-16/FoxO as a genetic regulator of mafr-1 expression. However, the strong reduction in MAFR-1 protein levels on the HCD can only partially be explained by changes in mafr-1 expression. Whether DAF-16 directly regulates the transcription of mafr-1 remains to be elucidated. A bioinformatic investigation of the mafr-1 promoter reveals a canonical DAF-16 associated element (DAE) binding site at −766nt from the translational start site (Murphy et al., 2003). An unbiased ChIP Seq screen by the modENCODE project identified DAF-16::GFP associated with the mafr-1 locus, near a non-canonical DAF-16 binding element (DBE) (Gerstein et al., 2010) (Figure S5F), which suggests DAF-16/FoxO could indeed regulate mafr-1. Collectively, our studies have uncovered roles for the conserved protein MAFR-1 in the maintenance of reproduction and lipid homeostasis, which genetically engage the insulin signaling effector protein DAF-16/FoxO (Figure 6).
Lipid transport within a multicellular organism is facilitated by lipoproteins that emulsify lipids and allow these fats to move through aqueous environments. Vitellogenesis is a hormonally controlled and conserved process in birds, reptiles, fish, and many invertebrates that generate yolk as a nutrient source for early embryogenesis (Schneider, 1996). We have discovered that mafr-1 levels influence vitellogenin production. Animals overexpressing mafr-1 have reduced expression of vit-2, -4, -5, and -6 and developing oocytes contain less VIT-2 protein, as measured by a VIT-2::GFP fusion protein. As such, the change in mafr-1 expression, which leads to a change in the intestinal expression of vitellogenins, exerts a cell non-autonomous influence on oocyte maturation in the germline.
A second physiological consequence uncovered from altered mafr-1 expression is a change in organismal reproductive capacity. Fecundity is under both genetic and environmental control, and we identify mafr-1 as a central player in this essential biological process. MAFR-1 levels are continually reduced and settle to their lowest point at the peak of germ cell development. The reduction of MAFR-1 levels during the maturation of the reproductive system could be of functional relevance since animals with increased mafr-1 expression have reduced egg production. This reduced fecundity correlates with deregulated vitellogenin synthesis and localization, which is essential for oocyte maturation. Previous studies have found that the vit genes are transcribed in excess in the intestine and can be titrated to cope with reproduction demands (DePina et al., 2011). As such, increases in yolk synthesis would not necessarily increase fecundity, as we found, but in the context of low yolk availability, such as in the mafr-1 overexpression lines, the increased production of vitellogenins following mafr-1 RNAi can partially restore embryo development. The use of whole-organism microarray and qPCR analysis of gene expression can confound the extrapolation of cell autonomous versus cell nonautonomous effects. Thus, it remains possible that pod-2/ACC1 and fasn-1/FASN expression in the germline may have important roles in fecundity. Nevertheless, the fact that the vit family-of-genes are expressed specifically in the intestine and our finding that intestine specific regulation of mafr-1 levels can alter reproductive output provides evidence for mafr-1 in the intestine exerting a cell non-autonomous effect on the reproductive system. Further dissection of the cell autonomous and non-autonomous mechanisms that mediate Maf1 function on the organism level will be of future interest.
Investigating the differences in the regulation of and the physiological responses to Maf1 across species is of particular interest. Similar to dMaf1, altering the expression of mafr-1, in a tissue specific manner can have significant impact at the organism level (Rideout et al., 2012). mafr-1 expression does not appear to influence developmental timing, as dMaf1 does in flies, although it can modestly impact organismal growth. While the physiological response from altering mafr-1 expression does not appear to influence the insulin/IGF-I signaling pathway as potently as it does in flies, we nevertheless find a partial requirement for DAF-16/FoxO in mediating these mafr-1-sensitive physiological parameters. Moreover, daf-16/FoxO was dispensable for the mafr-1-dependent changes to tRNA genes. This is consistent with the idea that the insulin/IGF-I signaling pathway functions downstream of mafr-1, as it has been described in flies (Figure 6). Our results emphasize the importance of examining the role of central cellular regulators in the context of a multicellular system, which have led to the identification of physiological roles for Maf1 in reproduction and lipid metabolism. Defining the tissues and cells where insulin signaling participates, either cell autonomously and/or non-autonomously, in the mafr-1-dependent phenotypes we have described will be important to further refine our model.
Aside from developmental timing, there are intriguing differences between the biological role of Maf1 in flies and worms. In flies, dMaf1 RNAi can impact the expression of iMet, Ile, and Leu tRNAs while dMaf1 overexpression had no effect (Rideout et al., 2012). In worms, we find that increased mafr-1 expression decreased the abundance of all tRNA species tested and mafr-1 RNAi led to the increased expression of 11 out of 13 of these tRNAs (Figure 1A, 1B, S1A and S1B). Second, in flies, dMaf1 and the RNA pol III factor Brf1, which controls tRNA biosynthesis, are tied to the growth and developmental timing phenotypes (Marshall et al., 2012). In worms, brf-1 and mafr-1 can both regulate tRNA transcript levels, however the lipid homeostasis phenotypes observed in the mafr-1 O/E strains cannot be phenocopied by reduction of brf-1 expression. This data implicates a role for MAFR-1 beyond modulating RNA pol III transcript levels. Third, although dMaf1 expression in the fat body was found to be key to its function, it remains unknown if dMaf1 can influence fat accumulation or reproduction, which are two physiological process intimately linked to signaling from the fly fat body (Arrese and Soulages, 2010). While there will certainly be indirect effects resulting from homeostatic responses to the changes in RNA pol-III transcript levels and/or possible positional effects due to the chromosomal proximity of RNA pol-III and pol-II targets (Hull et al., 1994; Kinsey and Sandmeyer, 1991), defining the molecular mechanisms underlying Maf1 function will be critical to understanding the physiological differences observed in flies, worms, and mammals.
EXPERIMENTAL PROCEDURES
Standard Growth Conditions
C. elegans were raised on standard 6 cm nematode growth media plates supplemented with streptomycin and seeded with Escherichia coli OP50. For RNAi experiments, NGM plates containing 5 mM IPTG and 100 ug ml−1 carbencillin were seeded with overnight cultures of double-stranded RNAi-expressing HT115 bacteria. Plates were allowed to induce overnight, followed by transfer of age-synchronous populations of C. elegans. Animals were fed either OP50 or HT115 food sources for at least two generations before analysis to avoid diet dependent effects.
Gene Expression
Developmentally synchronous worms of indicated genotype and developmental stages were collected, washed in M9 buffer, and then homogenized in Trizol reagent (Invitrogen). RNA was extracted according to manufacturer’s protocol. DNA contamination was digested with DNase I (New England Biolabs). cDNA was generated with the SuperScript® III First-Strand Synthesis System (Life Technologies). RNA samples were also used for Affymetrix C. elegans Gene 1.0 ST Arrays. Data and statistics were analyzed with Partek Genomics Suite Software version 6.6.
Quantitative PCR was performed by using SYBR Green (BioRad). The expression of snb-1 was used to normalize C. elegans RNA samples and GAPDH for human cell culture samples. The efficiencies of all primers used were within 5% of each other. Human RNA pol III primer sequences were taken from Johnson et al (Johnson et al., 2007). Primers for C. elegans genes were designed using the Primer3Plus algorithm (Untergasser et al., 2012). Primers for C. elegans RNA pol III targets were designed using the Genomic tRNA database (Chan and Lowe, 2009).
Protein extraction
Worms were washed twice with PBS followed by once with PBS supplemented with protease inhibitors. An equal volume of 2X LDS buffer was added and worms were flash frozen and thawed at 90 degrees; the cycle was repeated three times. Crude lysates were passed through a 26G needle, centrifuged at max speed for 20 minutes and supernatants were used for protein analysis.
Staining of fat
Nile red staining was performed as previously described (Pino et al., 2013). Briefly, animals of indicated genotypes were collected, fixed in 40% isopropanol, and stained in Nile red working solution in dark for two hours. Worms were washed once with PBS/0.01% triton-X in dark for 30 minutes, mounted on slides and imaged. Fluorescent density was measured by using NIH ImageJ software. In brief, images were converted to a 32-bit format, and then the background/GFP intensity was measured using the Integrated Density(ID) method. The region of interest tool was used for normalization to body size.
For Oil Red O staining, animals were collected and fixed in 1% formaldehyde in PBS. Then samples were frozen and thawed three times in a dry ice/ethanol bath. Worms were washed with PBS three times before staining with freshly prepared Oil Red O solution. After staining for 30 minutes, worms were washed for 15 minutes, mounted on slides, and imaged under a bright field illumination.
Supplementary Material
Acknowledgments
We thank E. Griffin, M. Li Dual, and L.E. Thomas for technical assistance; H.M. Dalton, J.Y. Lo, D.A. Lynn, and S.S. Pang for critical reading of the manuscript. The intestine specific RNAi strain was provided by Dr. Gary Ruvkun. Some strains were provided by the CGC, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). S.P.C. is an Ellison Medical Foundation New Scholar in Aging; this work was funded by the NIH (GM109028 and AG032308), the American Heart Association (14GRNT20380731), the Ellison Medical Foundation (AG-NS-0748-11) and the American Federation of Aging Research (002666-00001).
Footnotes
AUTHOR CONTRIBUTIONS
S.P.C. designed the experiments. A.K. and S.P.C. conducted all experiments. D.L.J. provided the 293T cell line and human Maf1 cDNA clone. A.K. and S.P.C. analyzed the data. S.P.C. wrote the manuscript. A.K., D.L.J. and S.P.C. edited the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anastassopoulou CG, Fuchs BB, Mylonakis E. Caenorhabditis elegans-based model systems for antifungal drug discovery. Curr Pharm Des. 2011;17:1225–1233. doi: 10.2174/138161211795703753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrese EL, Soulages JL. Insect fat body: energy, metabolism, and regulation. Annual review of entomology. 2010;55:207–225. doi: 10.1146/annurev-ento-112408-085356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros AG, Liu J, Lemieux GA, Mullaney BC, Ashrafi K. Analyses of C. elegans fat metabolic pathways. Methods Cell Biol. 2012;107:383–407. doi: 10.1016/B978-0-12-394620-1.00013-8. [DOI] [PubMed] [Google Scholar]
- Boguta M. Maf1, a general negative regulator of RNA polymerase III in yeast. Biochim Biophys Acta. 2013;1829:376–384. doi: 10.1016/j.bbagrm.2012.11.004. [DOI] [PubMed] [Google Scholar]
- Boguta M, Graczyk D. RNA polymerase III under control: repression and de-repression. Trends Biochem Sci. 2011;36:451–456. doi: 10.1016/j.tibs.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Brey CW, Nelder MP, Hailemariam T, Gaugler R, Hashmi S. Kruppel-like family of transcription factors: an emerging new frontier in fat biology. Int J Biol Sci. 2009;5:622–636. doi: 10.7150/ijbs.5.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock TJ, Browse J, Watts JL. Genetic regulation of unsaturated fatty acid composition in C. elegans. PLoS Genet. 2006;2:e108. doi: 10.1371/journal.pgen.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock TJ, Browse J, Watts JL. Fatty acid desaturation and the regulation of adiposity in Caenorhabditis elegans. Genetics. 2007;176:865–875. doi: 10.1534/genetics.107.071860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks KK, Liang B, Watts JL. The influence of bacterial diet on fat storage in C. elegans. PLoS One. 2009;4:e7545. doi: 10.1371/journal.pone.0007545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePina AS, Iser WB, Park SS, Maudsley S, Wilson MA, Wolkow CA. Regulation of Caenorhabditis elegans vitellogenesis by DAF-2/IIS through separable transcriptional and posttranscriptional mechanisms. BMC Physiol. 2011;11:11. doi: 10.1186/1472-6793-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P, Echeverri C, Oegema K, Coulson A, Jones SJ, Copley RR, Duperon J, Oegema J, Brehm M, Cassin E, et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- Grant B, Hirsh D. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol Biol Cell. 1999;10:4311–4326. doi: 10.1091/mbc.10.12.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillgartner FB, Salati LM, Goodridge AG. Physiological and molecular mechanisms involved in nutritional regulation of fatty acid synthesis. Physiol Rev. 1995;75:47–76. doi: 10.1152/physrev.1995.75.1.47. [DOI] [PubMed] [Google Scholar]
- Hughes SE, Evason K, Xiong C, Kornfeld K. Genetic and pharmacological factors that influence reproductive aging in nematodes. PLoS Genet. 2007;3:e25. doi: 10.1371/journal.pgen.0030025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull MW, Erickson J, Johnston M, Engelke DR. tRNA genes as transcriptional repressor elements. Mol Cell Biol. 1994;14:1266–1277. doi: 10.1128/mcb.14.2.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SS, Zhang C, Fromm J, Willis IM, Johnson DL. Mammalian Maf1 is a negative regulator of transcription by all three nuclear RNA polymerases. Mol Cell. 2007;26:367–379. doi: 10.1016/j.molcel.2007.03.021. [DOI] [PubMed] [Google Scholar]
- Kaltenbach L, Horner MA, Rothman JH, Mango SE. The TBP-like factor CeTLF is required to activate RNA polymerase II transcription during C. elegans embryogenesis. Mol Cell. 2000;6:705–713. doi: 10.1016/s1097-2765(00)00068-x. [DOI] [PubMed] [Google Scholar]
- Kimble J, Sharrock WJ. Tissue-specific synthesis of yolk proteins in Caenorhabditis elegans. Dev Biol. 1983;96:189–196. doi: 10.1016/0012-1606(83)90322-6. [DOI] [PubMed] [Google Scholar]
- Kinsey PT, Sandmeyer SB. Adjacent pol II and pol III promoters: transcription of the yeast retrotransposon Ty3 and a target tRNA gene. Nucleic Acids Res. 1991;19:1317–1324. doi: 10.1093/nar/19.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Murphy CT, Kenyon C. Glucose shortens the life span of C. elegans by downregulating DAF-16/FOXO activity and aquaporin gene expression. Cell Metab. 2009;10:379–391. doi: 10.1016/j.cmet.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier W, Adilov B, Regenass M, Alcedo J. A neuromedin U receptor acts with the sensory system to modulate food type-dependent effects on C. elegans lifespan. PLoS Biol. 2010;8:e1000376. doi: 10.1371/journal.pbio.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W. Tipping the energy balance toward longevity. Cell Metab. 2013;17:5–6. doi: 10.1016/j.cmet.2012.11.011. [DOI] [PubMed] [Google Scholar]
- Mak HY. Lipid droplets as fat storage organelles in Caenorhabditis elegans: Thematic Review Series: Lipid Droplet Synthesis and Metabolism: from Yeast to Man. J Lipid Res. 2012;53:28–33. doi: 10.1194/jlr.R021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall L, Rideout EJ, Grewal SS. Nutrient/TOR-dependent regulation of RNA polymerase III controls tissue and organismal growth in Drosophila. Embo J. 2012;31:1916–1930. doi: 10.1038/emboj.2012.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels AA, Robitaille AM, Buczynski-Ruchonnet D, Hodroj W, Reina JH, Hall MN, Hernandez N. mTORC1 directly phosphorylates and regulates human MAF1. Mol Cell Biol. 2010;30:3749–3757. doi: 10.1128/MCB.00319-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, Hu PJ. Insulin/insulin-like growth factor signaling in C. elegans. WormBook. 2013:1–43. doi: 10.1895/wormbook.1.164.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- O’Rourke EJ, Soukas AA, Carr CE, Ruvkun G. C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab. 2009;10:430–435. doi: 10.1016/j.cmet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S, Ruvkun G. The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol Cell. 1998;2:887–893. doi: 10.1016/s1097-2765(00)80303-2. [DOI] [PubMed] [Google Scholar]
- Paek J, Lo JY, Narasimhan SD, Nguyen TN, Glover-Cutter K, Robida-Stubbs S, Suzuki T, Yamamoto M, Blackwell TK, Curran SP. Mitochondrial SKN-1/Nrf mediates a conserved starvation response. Cell Metab. 2012;16:526–537. doi: 10.1016/j.cmet.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang S, Curran SP. Adaptive Capacity to Bacterial Diet Modulates Aging in C. elegans. Cell Metab. 2014;19:221–231. doi: 10.1016/j.cmet.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang S, Lynn DA, Lo JY, Paek J, Curran SP. SKN-1 and Nrf2 couples proline catabolism with lipid metabolism during nutrient deprivation. Nature communications. 2014;5:5048. doi: 10.1038/ncomms6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piano F, Schetter AJ, Mangone M, Stein L, Kemphues KJ. RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr Biol. 2000;10:1619–1622. doi: 10.1016/s0960-9822(00)00869-1. [DOI] [PubMed] [Google Scholar]
- Pino EC, Webster CM, Carr CE, Soukas AA. Biochemical and high throughput microscopic assessment of fat mass in Caenorhabditis elegans. J Vis Exp. 2013 doi: 10.3791/50180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout EJ, Marshall L, Grewal SS. Drosophila RNA polymerase III repressor Maf1 controls body size and developmental timing by modulating tRNAiMet synthesis and systemic insulin signaling. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1139–1144. doi: 10.1073/pnas.1113311109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohira AD, Chen CY, Allen JR, Johnson DL. Covalent small ubiquitin-like modifier (SUMO) modification of Maf1 protein controls RNA polymerase III-dependent transcription repression. J Biol Chem. 2013;288:19288–19295. doi: 10.1074/jbc.M113.473744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter GA. Insulin secretion: feed-forward control of insulin biosynthesis? Curr Biol. 1999;9:R443–445. doi: 10.1016/s0960-9822(99)80277-2. [DOI] [PubMed] [Google Scholar]
- Schneider WJ. Vitellogenin receptors: oocyte-specific members of the low-density lipoprotein receptor supergene family. Int Rev Cytol. 1996;166:103–137. doi: 10.1016/s0074-7696(08)62507-3. [DOI] [PubMed] [Google Scholar]
- Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Shor B, Wu J, Shakey Q, Toral-Barza L, Shi C, Follettie M, Yu K. Requirement of the mTOR kinase for the regulation of Maf1 phosphorylation and control of RNA polymerase III-dependent transcription in cancer cells. J Biol Chem. 2010;285:15380–15392. doi: 10.1074/jbc.M109.071639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukas AA, Kane EA, Carr CE, Melo JA, Ruvkun G. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes & development. 2009;23:496–511. doi: 10.1101/gad.1775409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squiban B, Belougne J, Ewbank J, Zugasti O. Quantitative and automated high-throughput genome-wide RNAi screens in C. elegans. J Vis Exp. 2012 doi: 10.3791/3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhya R, Lee J, Willis IM. Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol Cell. 2002;10:1489–1494. doi: 10.1016/s1097-2765(02)00787-6. [DOI] [PubMed] [Google Scholar]
- Vannini A, Ringel R, Kusser AG, Berninghausen O, Kassavetis GA, Cramer P. Molecular basis of RNA polymerase III transcription repression by Maf1. Cell. 2010;143:59–70. doi: 10.1016/j.cell.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Wahlby C, Kamentsky L, Liu ZH, Riklin-Raviv T, Conery AL, O’Rourke EJ, Sokolnicki KL, Visvikis O, Ljosa V, Irazoqui JE, et al. An image analysis toolbox for high-throughput C. elegans assays. Nat Methods. 2012;9:714–716. doi: 10.1038/nmeth.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AK, Jacobs RL, Watts JL, Rottiers V, Jiang K, Finnegan DM, Shioda T, Hansen M, Yang F, Niebergall LJ, et al. A conserved SREBP-1/phosphatidylcholine feedback circuit regulates lipogenesis in metazoans. Cell. 2011;147:840–852. doi: 10.1016/j.cell.2011.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S, Carrel JS. Fertilization and sperm competition in the nematode Caenorhabditis elegans. Dev Biol. 1979;73:304–321. doi: 10.1016/0012-1606(79)90069-1. [DOI] [PubMed] [Google Scholar]
- Watts JL. Fat synthesis and adiposity regulation in Caenorhabditis elegans. Trends Endocrinol Metab. 2009;20:58–65. doi: 10.1016/j.tem.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Greenway FL. Caenorhabditis elegans as a model for obesity research. Int J Obes (Lond) 2012;36:186–194. doi: 10.1038/ijo.2011.93. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.