Abstract
Interaction of serum proteins and nanoparticles leads to a nanoparticle-protein complex formation that defines the rational strategy for a clinically relevant formulation for drug delivery, hyperthermia, and magnetic resonance imaging (MRI) applications in cancer nanomedicine. Given this perspective, we have examined the pattern of human serum protein corona formation with our recently engineered magnetic nanoparticles (MNPs). The alteration in particle size, zeta potential, hemotoxicity, cellular uptake/cancer cells targeting potential, and MRI properties of the MNPs after formation of human serum (HS) protein corona were studied. Our results indicated no significant change in particle size of our MNPs upon incubation with 0.5-50 wt/v% human serum, while zeta potential of MNPs turned negative due to human serum adsorption. When incubated with an increased serum and particle concentration, apolipoprotein E was adsorbed on the surface of MNPs apart from serum albumin and transferrin. However, there was no significant primary or secondary structural alterations observed in serum proteins through Fourier transform infrared spectroscopy, X-ray diffraction, and circular dichroism. Hemolysis assay suggests almost no hemolysis at the tested concentrations (up to 1 mg/mL) for MNPs compared to the sodium dodecyl sulphate (positive control). Additionally, improved internalization and uptake of MNPs by C4-2B and Panc-1 cancer cells were observed upon incubation with human serum (HS). After serum protein adsorption to the surface of MNPs, the close vicinity within T1 (~1.33-1.73 s) and T2 (~ 12.35-13.43 ms) relaxation times suggest our MNPs retained inherent MRI potential even after biomolecular protein adsorption. All these superior clinical parameters potentially enable clinical translation and use of this formulation for next generation nanomedicine for drug delivery, cancer-targeting, imaging and theranostic applications.
Keywords: Magnetic nanoparticles, protein corona, magnetic resonance imaging, hyperthermia, cancer therapeutics, drug delivery
1. Introduction
Recent advances in nanomedicine have prompted the use of various nanoparticle (NP) formulations for basic and clinical cancer therapeutics. Although 92,000 studies involving NPs are present in the literature, very few NP formulation(s) are in human clinical trials or involve human use. The primary hurdle is a lack of clear indicative synthesis and chemical modification approaches for in vivo human applications. Additionally, there is a remarkable gap and limited understanding of the physico-chemical properties of NPs in the physiological system [1]. Physiological conditions influence the interaction of biological systems with NPs, which can describe the fate and biosafety of NPs [2-4]. This information particularly reveals the sustained circulation phenomenon of NPs or possible clearance mechanism by the immune system. Thus, clinical translation and success of NPs will depend on key interactions with human proteins [2, 3, 5].
Magnetic nanoparticles (MNPs) have been widely used for theranostic applications due to their multi-functionality, i.e., drug delivery, cell labelling, hyperthermia and magnetic resonance imaging (MRI) properties [6, 7]. However, most of the MNPs are rapidly engulfed due to their aggregation by the mononuclear phagocyte system, which quickly processes them for clearing and degrading upon intravenous administration. This process not only lowers the therapeutic dose at a disease site but also induces inflammation, hampers the host defense mechanism, and sheds MNPs to un-targeted areas in the body [8-10]. These events eventually lead to accumulation at un-targeted areas and impose possible toxicity concerns [11]. Thus, synthesis of highly stable and dispersible MNPs for systemic administration is highly desirable [6, 12].
Custom made, multi-layered, and multi-functional MNPs can improve the potential impact of treatment and diagnosis at the tumor site [13-15]. Our recent studies demonstrated that dual layered drug loaded MNP formulations have shown superior anti-cancer, imaging and targeting capabilities [16-19]. Viewing the translation potential, we engineered a dual layer MNP formulation which showed improved bioavailability, biocompatibility and therapeutic potential to effectively load therapeutics in cancer cells [18]. The first cyclodextrin layer depot loaded therapeutic drugs [20, 21] and the second pluronic polymer (F127) layer acted as a repelling hydrophilic polymer to enrich biological performance [22, 23]. F127 polymer (outer layer, polyethylene glycol chains) coating on nanoparticles has been thought to decrease recognition by phagocytic cells of the reticulo endothelial system (RES). Additionally, binding of F127 pluronic polymer(s)/PEG chain-coated nanoparticles led to less adsorption of plasma proteins and their significant role on the protein corona has been elucidated [24]. The adsorption of blood serum protein components on MNPs and interactions with phagocytes can modify the size, aggregation state, and interfacial composition and thus offer a distinct “biological identity” [25]. In addition, a clear picture of the interaction mechanisms between dual layered coated MNPs and human serum proteins is currently missing. Identification of this nanomaterial–protein complex is crucial to understanding the uptake mechanisms, biodistribution, and clearance of nanoparticles. Therefore, the aim of this study is to identify the interaction of serum proteins and MNPs; and the subsequent influence of their complex on the inherent physico-chemical properties of MNPs, such as, their uptake by cancer cells, cyto-compatibility, and influence on clinically relevant properties for drug delivery applications.
2. Materials and Methods
2.1. Materials and methods
All the chemicals and reagents used in this study were purchased from Sigma Chemical Co. (St. Louis, MO) unless otherwise stated. Magnetic nanoparticles were prepared using our previous protocol [18]. Whole human serum (HS) and red blood cells of a healthy male were purchased from Biological Specialty Corporation (Colmar, PA) and used within a month.
2.2. Human serum protein binding to MNPs
In a typical experiment, 300 μg MNPs were incubated in 0.5-50 wt./v.% HS (total volume 600 μL) for 24 h at 37°C. The solutions were centrifuged at 12,000 rpm for 15 min (twice) to recover human serum protein bound MNPs (HS@MNPs) samples as pellet. The recovered human serum proteins bound MNPs in the pellet were denoted as HS0.5@MNPs, HS1@MNPs, HS2.5@MNPs, HS5@MNPs, HS10@MNPs, HS20@MNPs, HS30@MNPs, HS40@MNPs, and HS50@MNPs when MNPs were incubated with 0.5, 1, 2.5, 5, 10, 20, 30, 40, and 50% human serum, respectively. The HS@MNPs samples were immediately lyophilized using the Labconco Freeze Dry System (−48°C, 133 × 10−3mBar; Labconco, Kansas City, MO). The lyophilized HS@MNPs samples were stored at 4°C until analyzed. To examine the HS protein adsorption on MNPs, we performed Fourier transform infrared (FTIR) spectroscopy, X-ray diffraction (XRD), and thermo gravimetric (TG) analyses [3].
2.2.1. XRD
The XRD spectral acquisition of vertically mounted HS@MNP samples on carbon tape was recorded using a Rigaku D/Max-B diffractometer (Rigaku Americas Corp, Woodlands, TX) with cobalt-alpha radiation at λ = 0.1546 nm operating at 40 kV and 40 mA [16-19].
2.2.2. FTIR
FTIR spectra of HS@MNPs samples were obtained using a Smiths Detection IlluminatIR FTIR microscope (Danbury, CT) equipped with a zinc selenide crystal ATR accessory. The spectra were collected from an average of 32 scans for each HS@MNP sample from 500 to 4000 cm-1 with a resolution of 4 cm−1 [16-19]. However, FTIR data was presented from 600 to 1800 cm−1.
2.2.3. TGA
Thermogravimetric analysis (TGA) of HS@MNPs samples was performed using a Perkin Elmer Simultaneous Thermal Analyzer STA6000 (Waltham, PA). Weight loss of MNPs, human serum and HS@MNPs samples was recorded from 25°C to 700°C at a heating rate of 10°C, under a constant nitrogen gas flow (100 mL/min) [18].
2.3. Dynamic light scattering
To determine the HS protein adsorption, protein corona formation on the surface of MNPs, and stability of MNPs, we used dynamic light scattering (DLS) [26]. For this study, HS@MNPs were dispersed in PBS/different concentrations of human serum solution, and the particle size and zeta potential (ζ) (surface charge) of MNPs or HS@MNPs were measured using a Delsa™Nano C Particle Size Analyzer (Beckman Coulter, Miami, FL). Size and distribution of samples were measured for 3 min, and the zeta potential was measured for 90 runs (9 min). The data represents an average of three independent readings.
2.4. Protein adsorption and composition
HS protein binding onto MNPs was determined by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis [26]. For this experiment, various concentrations of whole human serum and MNP solutions in 1X PBS were used. Briefly, Protocol 1: MNPs (300 μg) were incubated in 1, 10, and 40% human serum for 0.5, 1, 2, 3 and 24 h at 37°C; Protocol 2: 30, 60, 120, 180, and 300 μg MNPs were incubated in 10% human serum for 24 h at 37°C; Protocol 3: 300 μg MNPs were incubated in 1, 2.5, 5, 10, 20, 30, 40, and 50% human serum for 24 h at 37°C. In all three incubation protocols, the total volume of HS@MNPs solution was maintained at 600 μL. After completion of the incubation period, the unbound HS proteins (supernatant) and the HS protein bound MNPs (palette) were separated in the centrifugation process (twice, 12,000 rpm for 15 min). The HS@MNP samples were solubilized in an electrophoresis 2X SDS sample buffer (Santa Cruz Biotechnology, Santa Cruz, CA). The HS proteins bound on MNPs were separated and denatured by heating to 95°C for 5 min. The bound HS protein samples were further separated by size in the 4–20% 1D SDS-PAGE in an electric field using electrophoresis at a constant voltage of 150 V for 60 min. All the samples/gels were run in triplicate. These SDS-PAGE gels were processed for SimplyBlue™ SafeStain (Coomassie® G-250 stain, Invitrogen, Carlsbad, CA), and images were scanned using a BioRad Gel Doc (BioRad, Hercules, CA) [26].
2.5. Steady-state fluorescence spectroscopy
Steady-state fluorescence spectroscopy was used to monitor the instantaneous adsorption and binding efficiency of serum protein on MNPs [19, 27]. These measurements were conducted using a SpectraMax Plus 384 plate reader (Molecular Devices, Sunnyvale, CA). Intrinsic fluorescence of 1 and 2% human serum solution in 1X PBS was measured by titrating with 0-100 μg/mL MNPs. Fluorescence decay profiles were obtained by excited human serum solution at 295 nm, and the emission was set from 280-420 nm. Fluorescence quenching occurred due to interaction between MNPs and tryptophan units of human serum proteins. The binding constant and number of binding sites (n) were derived from the following equation:
Where Fo, F and Fs are the fluorescence area under the curve of emission of human serum, human serum titrated with various concentrations of MNPs, and human serum saturated with MNPs, respectively. The binding constant and number of binding sites were calculated following our previous reports [19]. The final values are reported as an average of three independent measurements.
2.6. CD spectra
Circular dichroism (CD) spectroscopy was used to elucidate secondary structural changes that may have occurred in human serum proteins during their instantaneous interaction with MNPs [27]. For this study, CD measurements were performed using a Jasco 815 CD spectrometer (JASCO International Inc., Ltd. Tokyo, Japan). All the differential absorption measurements were acquired between 200 and 260 nm wavelength in a standard 1 cm × 1 cm quartz cuvette at a slit width of 1 m with a scanning speed of 0.1 nm. This study was conducted at 0.5, 1, and 2% human serum with 0-100 μg/mL MNPs. During this titration, the total volume of the solution was maintained so as not to exceed 10% by volume. All CD measurements were performed at 25°C.
2.7. Magnetization saturation
A vibration sample magnetometer (Lake Shore 7400 Series, Lake Shore Cryotronics, Westerville, OH) was employed to determine the superparamagnetic property of MNPs and HS@MNPs. The hysteresis loop of samples was obtained by applying a maximum magnetic field strength of 150 Oe. The saturation magnetization (Ms) was determined from Ms versus applied field plots and extrapolated to infinite fields.
2.8. Magnetic resonance imaging
Magnetic resonance imaging (MRI) was performed using an 89 mm vertical bore MRI scanner (Agilent, Santa Clara, CA). This 9.4 T (400 MHz H1) scanner was equipped with triple axis gradients (100 G/cm) and a 4 cm Millipede transmit/receive radiofrequency coil. MRI measurements were carried out for 50 g iron/mL MNPs, HS2.5@MNPs, HS10@MNPs, and HS40@MNPs, which were diluted in 3% agar solution. T1 and T2 relaxation times were acquired using a multi-echo multi slice (MEMS) sequence. T1 relaxation times were calculated by acquisitions of 12 images with repetition time (TR) exponentially arrayed between 100 ms and 8000 ms; effective echo time, TE=10 ms; number of echoes, NE=1; FOV=10 mm2; number of excitations, NEX=1; slice thickness=1 mm; matrix size of 128 × 128; acquisition time ≈ 51 min. T2 relaxation data were generated using the same sequence with the following parameters: TR=4000 ms; TE=8.7 ms; NE=32; FOV=10 mm2; NEX=2; slice thickness=1 mm; matrix size of 128 × 128; acquisition time ≈ 17 min. The T1 and T2 relaxation times were computed using a nonlinear regression applied by a MATLAB based program. T1 and T2 measures are presented as mean ± standard deviations, which are calculated within a region of interest drawn around the sample.
2.9. Cell culture and red blood cells
Human pancreatic carcinoma (Panc-1) cell line (American Type Cell Culture, Manassas, VA) and human bone metastatic LNCaP-derivative, C4-2B prostate cancer cell line (kind gift from Dr. Sanjay Gupta, Case Western Reserve University, Cleveland, OH) were used to study the proliferation and uptake/intracellular localization of MNPs and HS@MNPs. These cells were maintained in Roswell Park Memorial Institute (RPMI)-1640 medium or Dulbecco's Modified Eagle Medium (DMEM) (HyClone Laboratories, Inc., Logan, UT) by supplementing with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA) and 1% penicillin/streptomycin (Gibco BRL, Grand Island, NY). The cells were grown as monolayers in a humidified atmosphere (5% CO2) at 37°C in a cell culture incubator. Freshly collected sodium citrate anticoagulant human whole blood of a healthy donor (Biological Specialty Corporation) was used to examine the hemolysis potential, a quantitative measure of the hemoglobin release from red blood cells (RBCs) of MNPs/HS@MNPs. The hemolysis study was performed within a month after receiving the sample.
2.10. Hemolysis Assay
The RBC suspension (500 μL) was incubated with 0.05, 0.10, 0.25, 0.50, 0.75, and 1.00 mg of MNPs, HS2.5@MNPs, HS10@MNPs, and HS40@MNPs in 1.75 mL Eppendorf tubes in an incubator for 3 h. Sodium dodecyl sulphate (SDS, 1 mg/mL) was used as a positive (100% lysis) control while 1X PBS was used as a negative (0%) control in this experiment. The sample tubes were centrifuged at 1000 rpm for 10 min and the supernatant (200 μL) was transferred to a 96-well plate to measure the concentration of hemoglobin that was released from RBCs. Hemoglobin release was measured using absorbance at 590 nm by using a plate reader (Model 680, BioRad Laboratories). The hemolysis experiment was done in triplicate and the percentage of hemolysis was calculated as follows:
2.11. Scanning electron microscopy
MNPs and HS@MNPs were incubated with RBCs and fixed with 4% paraformaldehyde for 30 min at room temperature. 5-10 μL RBC suspension was placed on a metal stage and dried at room temperature for 6 h in a chemical hood. The samples were gold-sputtered and examined using a Quanta 450 scanning electron microscope (S. No. D9234, FEI™, Hillsboro, OR) at an accelerating voltage of 5 kV.
2.12. Cellular uptake
In this study, C4-2B and Panc-1 cells (1 × 105 per well in 2 mL medium) were seeded in 6-well plates. After incubating MNPs or HS@MNPs with cells for 6 h, the cells were washed with PBS and fixed in ethanol for 5 min. The cells were then stained with Prussian blue and Nuclear Fast Red according to our previously reported standard protocol [17]. Images were obtained on an Olympus BX 41 phase contrast microscope (Olympus, Center Valley, PA). Using the same protocol, after treatment the cells were lysed in hydrochloric acid (HCl) and a calorimetric method was used to determine the overall iron content internalized in the cancer cells. This protocol specifically represents only internalized nanoparticles in cells not just bound nanoparticles. The average result was calculated from 3 samples.
2.13. Proliferation
Cell proliferation of cancer cells in the presence of MNPs and HS@MNPs was examined using a standard MTS assay. In this study, C4-2B cancer cells (5 × 104 in 100 μL in each well) were placed into a 96-well plate. After 24 h, cells were incubated with 2.5-200 μg/mL of MNPs, HS2.5@MNPs, HS10@MNPs, or HS40@MNPs for 24 h. The cells were then washed twice with PBS that was subsequently replaced with 100 μL fresh medium containing 20 μL MTS solution and incubated for 3 h. The absorbance of each well was recorded at 492 nm using a microplate reader (BioMate 3 UV-Vis spectrophotometer, Thermo Scientific, Waltham, MA). The percentage of viable cells was calculated by comparing absorbance of MNPs/HS@MNP-treated cells against the untreated cells. Data presented are from five replicates. Similarly, anti-cancer activity of curcumin and curcumin loaded HS@MNPs were evaluated in C4-2 and HPAFII cancer cells [16-19]. Curcumin loaded HS@MNPs was prepared according to our previous reports [16-19].
2.14. Animal model
Animal experiments were conducted in accordance with local Institutional Animal Care and Use Committee (IACUC) guidelines. To evaluate the toxicity profile of MNPs, we used approximately 6- to 8-week-old athymic nude (nu/nu) mice (Harlan Laboratories, Madison, WI). The mice were inoculated subcutaneously in their left flank with 5 × 106 HPAF-II human pancreatic cancer cells dispersed in PBS and Matrigel (1:1 ratio; BD Biosciences, San Jose, CA). After mice developed tumors (100 mm), 3 mice per group were injected with MNPs at a dose of 20 mg/kg (400 μg per mouse) via intraperitoneal or intravenous route using a 30G needle. No treatment mice served as control. After 24 hours, animals were euthanized and tumor, liver, spleen, and brain tissues were collected and stored at −80°C. Toxicity profiles were evaluated using H&E staining. Note: we have not employed HS@MNPs because the aim of this experiment is to examine how the bare MNPs behave after binding serum protein in vivo.
2.15. Statistical data analysis
DLS, fluorescence, CD, SDS-PAGE, cellular uptake, hemolysis, and proliferation experiments were conducted at least in triplicate. All experimental results were calculated using Microsoft Excel 2013 software and expressed as mean ± standard error of the mean (S.E.M.). The statistical significance of the data was analyzed using an unpaired, two tailed student t-test. When p < 0.05, the differences between data sets was considered statistically significant. All graphs were plotted using Origin 6.1 software.
3. Results
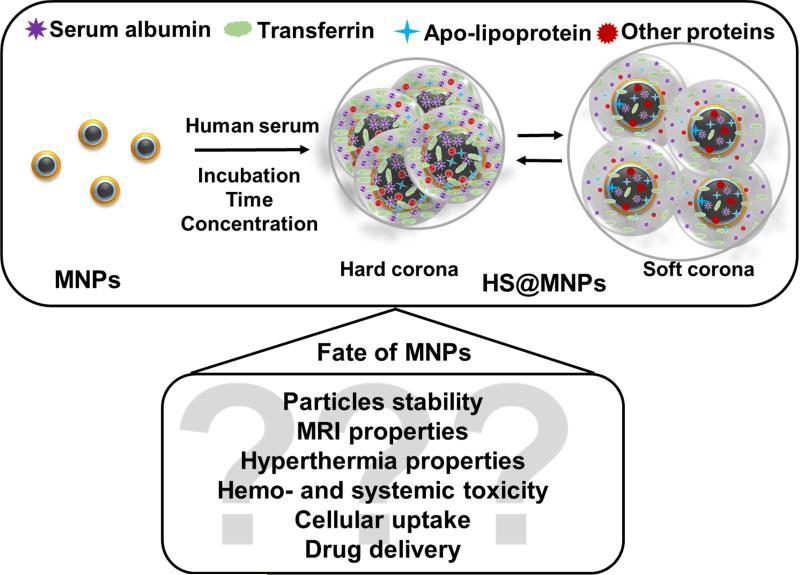
Magnetic nanoparticles have shown a good tolerance profile with drug delivery and diagnostic applications [15, 28]. To achieve superior properties with better targeting features a number of custom made MNPs are readily synthesized. All these modifications may or may not affect the human serum binding profile, which dictates the fate of NPs in vivo. Therefore, it is important to study the MNP-serum protein interaction, which will allow examination of the delivery of medicine in preclinical and clinical settings. Many studies have reported that incubation of NPs with human serum leads to the formation of a soft (loosely bound) or hard (permanent) corona on NPs or their equilibrium. A putative mechanism is proposed for binding of the serum proteins with MNPs and the possible influence on the physico-chemical properties of MNPs (Figure 1).
Figure 1. A putative MNPs-protein corona formation and influence on the physico-chemical properties of MNPs.
Schematic illustration of double layer magnetic nanoparticles incubated in whole human serum resulted in the formation of human serum protein corona on MNPs. The formed human serum protein corona is either soft or hard and under equilibrium state.
3.1. HS@MNPs formation
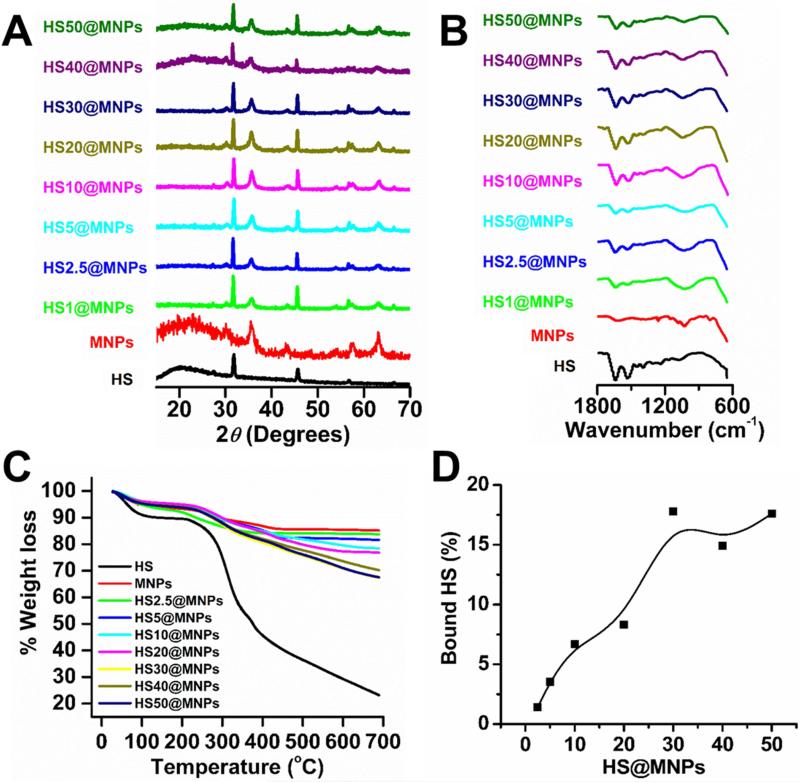
It is expected that a small number of human serum proteins bind to MNPs upon incubation with whole human serum due to hydrophilic polymer/groups (PEG) of pluronic F127 polymer on MNPs. To confirm the presence and extent of human serum proteins on MNPs, XRD (Figure 2A), FTIR (Figure 2B), and TG (Figure 2C-D) analyses were performed. Six diffraction peaks at 2θ (30.2, 35.5, 43.2, 53.5, 57.2, and 62.7°) were observed for MNPs that can be indexed to the 220, 311, 400, 422, 511, and 440 planes, and support the face centered cubic spinel phase of MNPs (Figure 2A, red diffraction spectrum). In all HS@MNPs (Figure 2A, green to olive diffraction spectra), the peak positions of MNPs were unchanged but additional peaks (32.4 and 46.3°) resulted from adsorption of HS proteins (Figure 2A, black diffraction spectra) on MNPs. The FTIR spectrum of MNPs exhibited a broad absorption peak between 3500 and 3000 cm−1 due to the presence of hydroxyl/amino groups (data not shown), an intense band at 1010 cm−1 due to glycosidic (C–O–C) vibration, CH2 rocking and the coupled (C–C/C–O) stretch vibrations of cyclodextrin and F127 polymer layers on MNPs (Figure 2B, red spectrum). The appearance of HS protein peaks in the spectra of HS@MNPs at 1651 cm−1 and 1548 cm−1 are assigned to the amide bond stretch region (the amide I and amide II bands) (Figure 2B, black and green to olive spectra). This confirms adsorption of HS proteins on the surface of the MNPs and possible MNPs-protein corona formation. However, due to saturation adsorption of HS proteins, the FTIR spectra of HS@MNPs showed no dependence on the HS concentration. To confirm the presence of human serum proteins in HS@MNPs, thermogravimetric analyses were performed. Thermograms at 700°C revealed that MNPs (Figure 2C, red thermogram) exhibit lowest weight loss due to an iron oxide core while human serum (Figure 2C, black thermogram) displayed maximum weight loss due to excessive protein degradation at higher temperatures. The weight loss of all HS@MNPs (Figure 2C, green to olive thermogram) was inbetween the MNPs and HS samples. This demonstrates that all HS@MNPs contain various amounts of HS due to protein corona formation. Figure 2D discloses the percent of HS proteins bound on each HS@MNP. The amount of HS binding gradually increases up to HS30@MNPs and then a slight decrease was observed.
Figure 2. Physico-chemical analysis of HS@MNPs confirms the presence of human serum proteins bound on MNPs.
(A) X-ray diffraction patterns of human serum (HS), magnetic nanoparticles (MNPs) and human serum protein bound on MNPs (HS@MNPs). HS exhibits peak positions at 32.4 and 46.3°; MNPs exhibit peak positions at 30.2, 35.5, 43.2, 53.5, 57.2, and 62.7°; and all HS@MNPs exhibit a total of eight peak positions at 32.4 and 46.3° (resulted from adsorption of HS proteins), and 30.2, 35.5, 43.2, 53.5, 57.2, and 62.7° (belongs to MNPs). (B) FTIR spectra of HS, MNPs and HS@MNPs. Appearance of characteristic peaks at 1651 cm−1 and 1548 cm−1 indicate the presence of human serum proteins on MNPs. (C) Weight loss thermograms of HS, MNPs, HS@MNPs. Excessive weight loss of HS@MNPs specifies human serum protein degradation in thermograms. (D) The percent weight loss of HS@MNPs in thermograms was converted to bound human serum proteins.
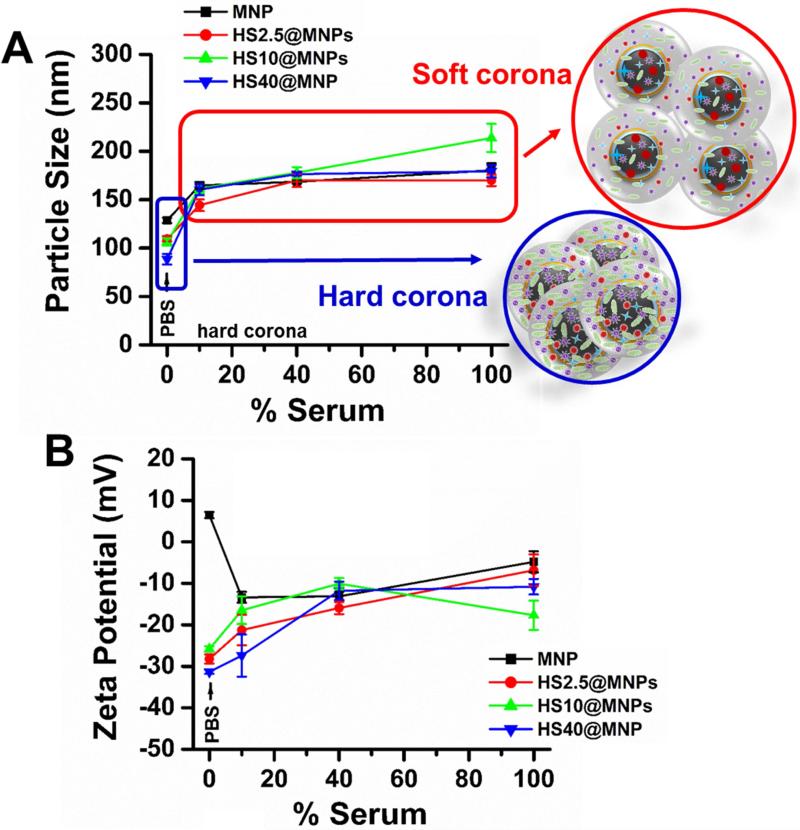
3.2. Particle size and zeta potential
MNPs have to be stable in buffer and human serum for medical applications. Serum albumin can stabilize or de-stabilize the NPs depending on the composition, surface charge, and density of NPs [29]. Therefore, the stability of MNPs and HS@MNPs was investigated in PBS and 2.5-100% human serum solutions by using a dynamic light scattering instrument to measure particle size and zeta potential (Figure 3). The particle size of MNPs, HS2.5@MNPs, HS10@MNPs, and HS40@MNPs in PBS solution ranged between 88 and 128 nm (Figure 3A) in the following order: MNPs > HS2.5@MNPs > HS10@MNPs > HS40@MNPs. In HS@MNPs, the bound HS protein chains on the MNPs give a hard corona (Figure 3A, indicated in the black circle) after incubation for 24 h, which reduces the aggregation of MNPs by forming a steric layer on the surface. This provides stabilization to HS@MNPs in PBS solution. On the other hand, a clear increase in particle size was observed for MNPs, HS2.5@MNPs, HS10@MNPs, and HS40@MNPs in 10-100% human serum (HS) solution (Figure 3A). The reason for this may be the HS solution provides instantaneous binding onto MNPs or HS@MNPs, which establishes a soft corona (Figure 3A, indicated in red circle). In this condition, HS proteins loosely bind to the MNPs or HS@MNPs, which leads to slight increase in particle size. Zeta potential of MNPs was observed to be 6.4 mV, but establishment of HS hard corona on HS2.5@MNPs, HS10@MNPs, and HS40@MNPs leads to negative zeta potentials (-28.2 to -31.35 mV) (Figure 3B). This is highly acceptable because HS is negatively charged at pH 7.4. Similarly, after dispersion of the MNPs in 10-100% HS solutions, zeta potential decreased from 6.4 mV to -13.1 mV. However, a slight increase in zeta potential of HS2.5@MNPs, HS10@MNPs, and HS40@MNPs was observed in 10-100% HS solutions compared to PBS solution. The reason could be instantaneous adsorption/dis-adsorption of HS proteins on MNPs.
Figure 3. Particle size and zeta potential of human serum protein corona on MNPs.
(A) Hydrodynamic diameter of MNPs and HS@MNPs in PBS and various concentrations of human serum solutions. (B) Zeta potential of MNPs and HS@MNPs in PBS and various concentrations of human serum solutions. Size and distribution of samples were measured for 3 min, and the zeta potential was measured for 90 runs (9 min). Data represents the average of 3 readings.
3.3. Adsorption behavior and composition of serum proteins
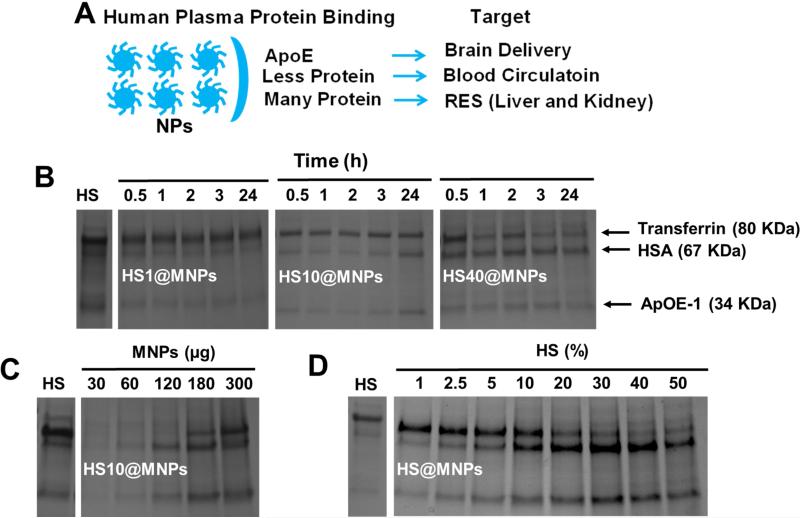
The NP-protein interaction determines how NPs can best be utilized in therapeutics [1, 3]. Different adsorption patterns dictate the intended target (Figure 4A). Using SDS-PAGE, we found that human serum proteins were heterogeneously adsorbed to the surface of MNPs (Figure 4A-D). It was noticed in all the adsorption studies that major binding proteins are: transferrin, serum albumin and apo-lipoprotein E-1 (Figure 4B-D). The adsorption capacity of proteins is dependent upon protein concentration and time (Figure 4B). At a low concentration of HS there is no significant difference in binding of various serum proteins on MNPs during incubation. At 24 h, a considerable increase was observed in adsorption pattern of proteins, i.e., increased HSA and ApOE-1. A higher concentration of serum HS40 resulted in the instantaneous binding of all proteins (within 30 min). Interestingly, we also observed that the adsorption of proteins on MNPs depends on the concentration of MNPs. With an increased concentration of MNPs there is an increase of bound proteins on the surface (Figure 4C). Quite interestingly, a clear change of protein binding was observed when the concentration of serum was increased (HS1 to HS50) (Figure 4D). Transferrin adsorption remains the same up to HS10, followed by a decrease in binding capacity (Figure 4D). HSA adsorption increased continuously up to HS40 and then had a slight downfall in adsorption (Figure 4C). Of note, in all conditions, ApOE-1 seemed to either maintain a constant adsorption pattern or increase adsorption (Figure 4B-D) at various incubation durations, serum concentrations, and MNPs concentrations. From this data, it is expected that a small number of human serum proteins bind to MNPs upon incubation with whole human serum due to hydrophilic polymer/groups (PEG) of pluronic F127 polymer on MNPs. The protein corona on MNPs gives rise to a change in overall dispersion and particle aggregation pattern that was determined by TEM (Support Information I and II). However, from our DLS data it is evident that nanoparticles maintain their integrity and size range, demonstrating nanoparticles do not precipitate in the presence of serum.
Figure 4. Evaluation of human serum protein adsorption on MNPs.
(A) Schematic representation of MNPs after binding with human serum proteins and their intended target based on the extent of MNPs and HS proteins interactions. (B-D) Resolved SDS-PAGE of adsorbed human plasma proteins on MNPs. Data represents that human serum proteins bind to MNPs depending on the (B) duration of time of indication, (C) concentration of MNPs, and (D) concentration of human serum. L indicates ladder and HS indicates control human serum as control. The represented SDS-PAGE gels represent 3 independent runs.
3.4. Rate of binding
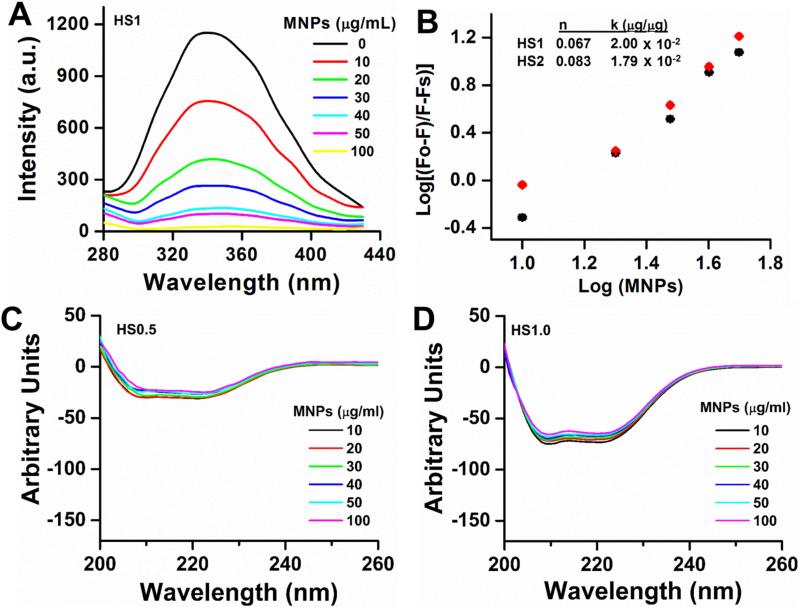
It is widely accepted that NP and serum protein interactions depend on the surface functional groups, surface charge and size of NPs [3, 30, 31]. The type of serum protein plays a greater role. The rate of association and dissociation of serum proteins are competitive with other plasma proteins. To determine overall protein binding effects on MNPs, we chose a steady-state fluorescence quenching method. A gradual increase in fluorescence quenching was observed due to part of the tryptophan residues of HS proteins that are involved in the binding process on MNPs (Figure 5A). The initially rapid decrease in fluorescence intensity was due to the fast adsorption of 1% HS proteins, which then decelerated due to the gradual complexation. From the plots of Fo/(Fo−Fs) vs. log [MNPs] (slope and intercept), the number of binding sites and binding constants were calculated (Figure 5B). From this study, it is clear that the average number of binding sites per particles varies from 0.067 to 0.083 with binding constants 2.00 × 10−2 to 1.79 × 10−2 μg/μg, with an increase of HS concentration.
Figure 5. Human serum binding on MNPs.
(A) Representative fluorescence spectral profiles of 1% human serum with increased concentration of MNPs (0-100 μg/mL) at room temperature (λex =295 nm). (B) Number of binding sites (n) and binding constant (k) calculation graph from fluorescence spectral data. F0, F and Fs are the relative fluorescence emission intensity of human serum alone, in the presence of MNPs, and infinity saturated MNPs, respectively. The solid black and red points are used to fit curves. Data is a mean of three replicates. (C-D) Circular dichroic spectral profiles of 0.5-1% human serum with increased concentration of MNPs (0-100 μg/mL). Note: No significance changes occur in the α-helical content of the protein with increasing concentration of MNPs.
Interaction of HS with MNPs is a direct quantitative measurement that can also be quantified by CD spectral analysis. CD spectral signals arise from electronic transitions of secondary protein structures. Abnormal changes in secondary structures of HS may indicate aggressive binding behavior of MNPs to HS. No significant HS alterations in secondary chemical structure were observed as a function of the incubation with MNPs at 208 nm degree of molar ellipticity (θ) (Figure 5C-D). Overall, serum albumin before and after incubation with MNPs that contains predominantly α-helices (~70%) as secondary structure indicates that HS proteins retain their original protein structure. A slight decrease of spectra at 208 nm is due to the HS adsorption on MNPs, not the loss of the α-helix secondary structure.
3.5. Effect of HS protein corona
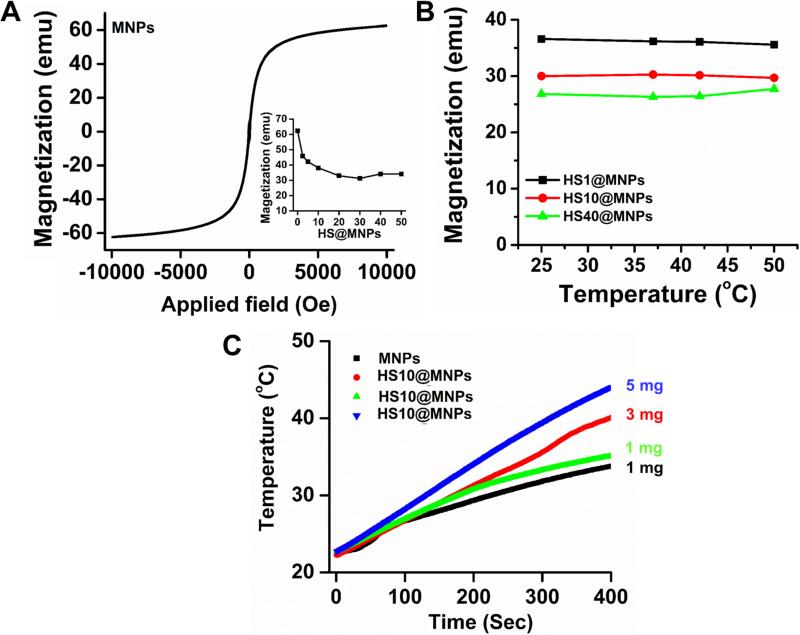
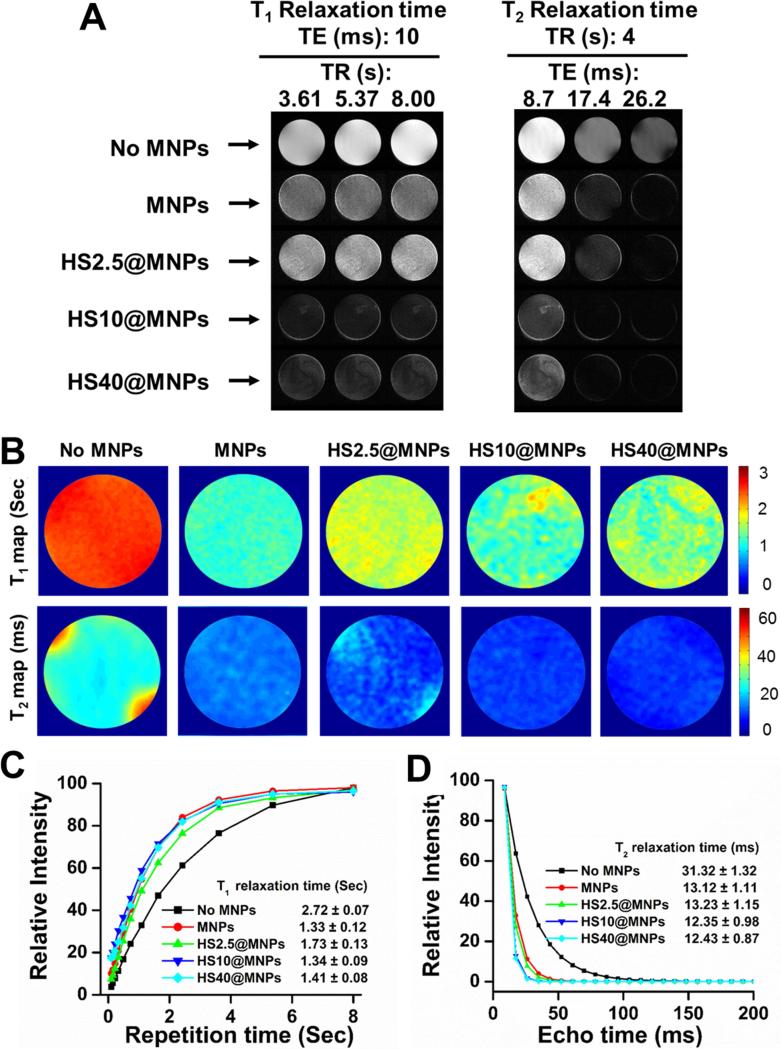
Protein corona can alter the existing inherent properties of MNPs [32], including magnetization, magnetic hyperthermia and magnetic resonance imaging properties. All these properties depend on the magnetization saturation (Ms) of MNPs. The MNPs exhibit superparamagnetic properties, Ms = 64 emu (Figure 6A). It was readily observed that the Ms decreased with an increase of serum used (HS2.5 to HS50) in HS@MNPs (Figure 6A, inset), which is due to the surface disorder and modified HS protein corona. The decrease in Ms is attributed to the HS corona, which prevents the core spins from aligning along the field direction. No change was observed in the magnetization saturation values of HS1@MNPs, HS10@MNPs, and HS40@MNPs at different temperatures (Figure 6B), indicating a significant application in MRI. However, a different trend was observed in hyperthermia (increase of temperature in alternating current) properties of HS10@MNPs (green line) over MNPs (black line) (Figure 6C). It was noted that MNPs (1 mg) reached 33.6°C while HS10@MNPs (1, 3, and 5 mg) achieved a temperature of 39.8, 34.9, and 43.7°C, respectively, within 400 s under applied field current. This may be due to the fact that HS protein corona stabilizes MNPs and is more uniformly distributed in aqueous medium, thus the core of MNPs are readily available for its heating action. Both MNPs and HS@MNPs exhibit superior MRI properties (Figure 7A-B). T1 relaxation times were increased for HS2.5@MNPs (1.73 ± 0.13 s), HS10@MNPs (1.34 ± 0.09 s) and HS40@MNPs (1.41 ± 0.09 s) compared to MNPs (1.33 ± 0.12 s). This pattern is clearly seen in Figure 7C. In addition, it was also noticed that the T2 relaxation times were slightly altered upon HS protein formation on MNPs (Figure 7D). The order of T1 relaxation times (s) is: HS2.5@MNPs (1.73 ± 0.13) > HS40@MNPs (1.41 ± 0.08) > HS10@MNPs (1.34 ± 0.09) > MNPs (1.33 ± 0.12). In contrast, no significant variation was observed in T2 relaxation times (ms): HS2.5@MNPs (13.23 ± 1.15) ≥ MNPs (13.12 ± 1.11) > HS40@MNPs (12.43 ± 0.87) ≥ HS10@MNPs (12.35 ± 0.98). This data indicates T2 relaxation times characteristic of MNPs and HS@MNPs are very similar while T1 relaxation times of HS2.5@MNPs and MNPs differ by 30%.
Figure 6. Influence of human serum protein corona on magnetization and hyperthermia properties of MNPs.
(A) Magnetization saturation of MNPs. Inset represents decrease of magnetization saturation upon formation of human serum protein coronas. This data obtained from the plots of Magnetization saturation versus applied field. (B) Magnetization saturation of HS1@MNPs, HS10@MNPs, and HS40@MNPs at different temperatures (25, 35, 42 and 50°C). Data indicates no significant change in magnetization saturation. (C) Temperature increase profile under applied current in hyperthermia instrument. The increased temperature profile indicates that MNPs have superior specific absorption rate which can be used for the magnetic hyperthermia application.
Figure 7. Human serum protein corona influence MR Imaging properties of MNPs.
(A) Gray scale T1 and T2 relaxation maps of phantom gels of MNPs, HS2.5@MNPs, HS10@MNPs, and HS40@MNPs. (B) Representative color T1 and T2 relaxation of phantom gels of MNPs, HS2.5@MNPs, HS10@MNPs, and HS40@MNPs. (C-D) T1 and T2 relaxation curves of phantom gels of MNPs, HS2.5@MNPs, HS10@MNPs, and HS40@MNPs.
3.6. Toxicity evaluation
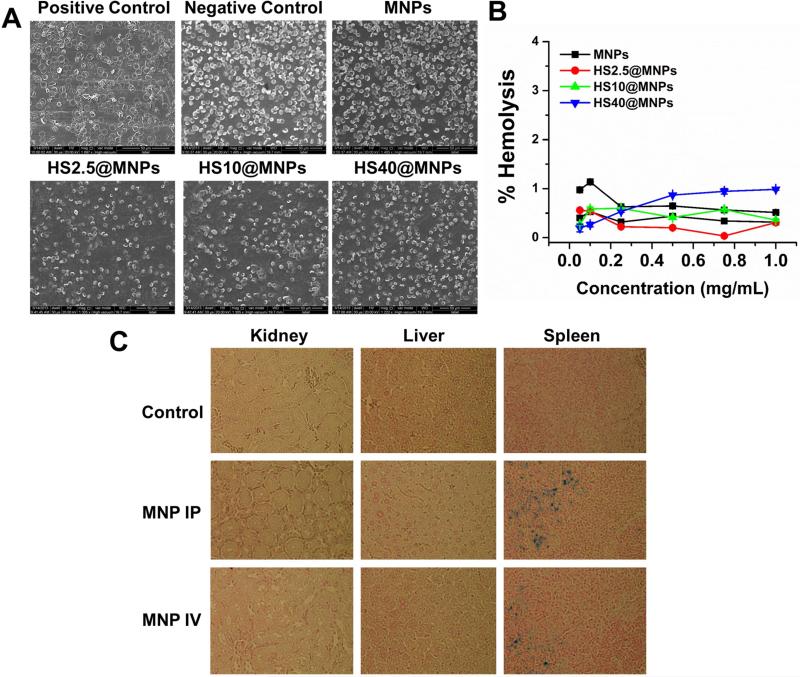
The improved uptake, accumulation and localization of magnetic nanoparticles in cancer cells lead to efficient drug delivery [33]. After administration, NPs immediately interact with blood cells; if those particles are toxic to these cells, unknown side effects in the body occur. To delineate this phenomenon for MNPs, we evaluated the toxicity profile of MNPs and HS@MNPs with human red blood cells (Figure 8A-B). Scanning electron microscopic (SEM) images depict that the positive control (sodium dodecyl sulphate) significantly causes toxicity to RBCs, which is evident by the rupture of RBC membrane and a de-morphed oval biconcave disk shape (Figure 8A). However, no sign of toxicity was observed in the case of MNPs and HS@MNPs as RBCs maintained a healthy smooth oval bioconcave disc shape structure like a negative control treatment (Figure 8A). Furthermore, this hemo-compatible behavior was confirmed by hemolysis assay (Figure 8B). Similarly, no hemolysis was observed even in the presence of serum media instead of PBS (Supporting Information III). Negligible hemolysis values were observed for HS@MNPs and MNPs upon incubation with RBCs (less than 1.25%), which indicate the formulations are hemo-compatible. This data is consistent with other reported magnetic nanoparticle formulations [34-36].
Figure 8. MNPs exhibit hemo- and bio-compatibility.
(A) Scanning electron microscopic images of RBCs treated with positive (sodium dodecyl sulphate), negative control (1X PBS), MNPs, HS2.5@MNPs, HS10@MNPs, and HS40@MNPs. Positive control shows visual evidence of membrane rupture and destruction of flexible and oval biconcave disk shape. MNPs and HS@MNPs samples exhibit healthy smooth oval bioconcave disc shape as control RBCs. (B) Hemolysis of RBCs upon incubation with 0.05-1 mg/mL MNPs, HS2.5@MNPs, HS10@MNPs, and HS40@MNPs. Data represents triplicates. (C) In vivo safety profile of MNPs. Hematoxylin-eosin staining of liver and spleen organs of mice treated with MNPs. No apoptotic or necrotic signs were observed. Tissue images were captured at 400X magnification.
PEGylated MNPs significantly accumulate in the liver, spleen, and intestine, while much lower accumulation is observed in the kidney, heart, and lungs [37]. Therefore, we examinedcytotoxicity of MNPs in vivo in a mice model by injecting MNPs intravenously or intraperitoneally (Figure 8C). Our results demonstrate that after treating mice with MNPs, there are no signs of apoptosis or necrosis in liver and spleen cells (hematoxylin-eosin, H&E staining method), where NPs are known to accumulate more frequently. Such compatibility was achieved even with MNP-CUR formulation in mice study (Supporting Information IV). This in vivo study concludes that these MNPs have a great potential in medical application. However, efficient and validated target moiety on MNPs can help to direct the NP formulations directly to the tumor sites for efficient delivery of therapeutics.
3.7. Cellular uptake and drug delivery
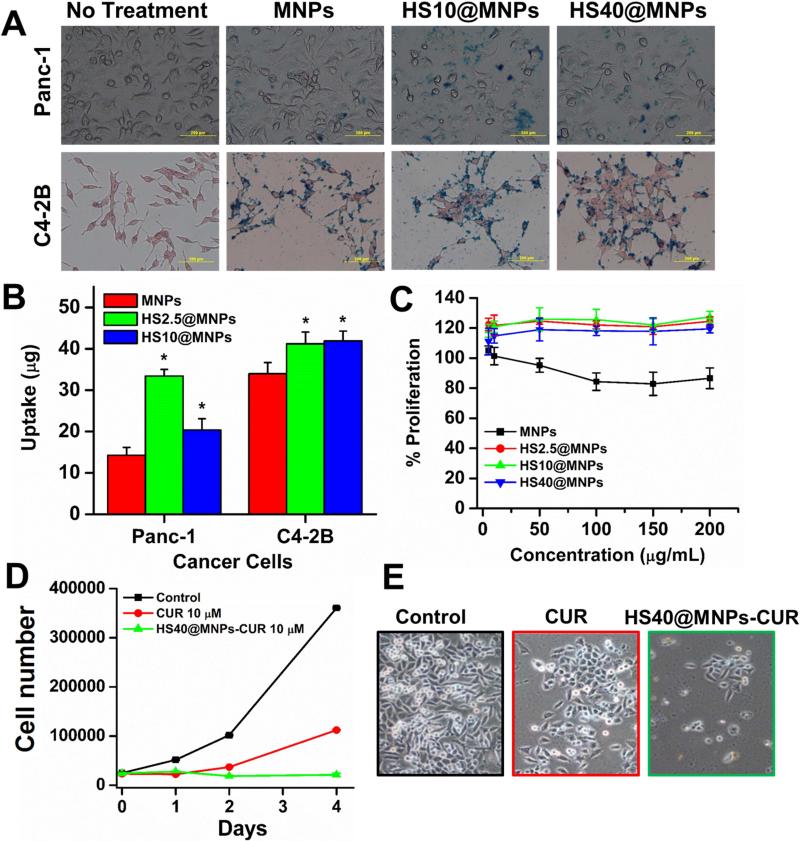
Superior biocompatibility and low cytotoxicity enable the use of MNPs in cancer therapeutics [38]. Efficient internalization of NPs in cancer cells is required for efficient drug delivery application. Figure 9A presents visual representation of the internalization patterns of MNPs in cancer cell lines. The order of uptake was determined to be: HS2.5@MNPs > HS10@MNPs > MNPs in Panc-1 cells and HS2.5@MNPs ≥ HS10@MNPs > MNPs in C4-2B cells (Figure 9B). The internalization of MNPs was calculated by Prussian blue stain and a plate reader. The uptake of HS2.5@MNPs and HS10@MNPs is significantly higher than MNPs in both cancer cell lines. This demonstrates that after the protein corona formation, MNPs undergo an uptake process that is either quicker than or interacts to a greater extent with cellular membranes, which enhances their internalization process. Further, HS@MNPs also indicate there is no cytotoxic effects on C4-2B cancer cells (Figure 9C). In fact, HS@MNPs show increased proliferation of cancer cells over MNPs, which also supports the uptake process. In this situation, cancer cells utilize the iron present in HS@MNPs in the cell cycle process and therefore, proliferation of cancer cells is slightly increased. More importantly, after loading with an anticancer model drug (curcumin) HS40@MNPs have proven to have superior anti-cancer potential in C4-2 (Figure 9D-E) and HPAF-II cancer cells (Supporting information V).
Figure 9. Human serum protein corona effects on the uptake and proliferation in cancer cells.
(A) Phase contrast images of Panc-1 and C4-2B cancer cells treated with 50 μg MNPs, HS2.5@MNPs and HS10@MNPs. Internalization of NPs can be seen as blue stain produced by Prussian blue from the iron of MNPs/HS@MNPs. (B) Quantification of Prussian blue staining of cancer cells recorded using plate reader at 510 nm. Data from triplicate samples. (C) Proliferation of C4-2B cancer cells with treatment of 2.5-200 μg/mL MNPs, HS2.5@MNPs, HS10@MNPs, and HS40@MNPs. Data represents 5 replicates. (D-E) HS40@MNPs containing curcumin exhibits superior anti-cancer activity in C4-2 cells. Cell images were captured at 200X magnification.
4. Discussion
Various NP formulations have been under pre-clinical and clinical trials [39] for improved and targeted delivery of therapeutic drugs at the tumor sites. To achieve efficient delivery of drugs, numerous strategies have been employed to modify the surface of NPs with specific functional group(s)/polymer layer(s) and targeting ligand(s) [40, 41]. Such approaches can improve the binding and internalization of NPs via binding to receptors on the target cells and facilitating the endocytosis process. Although in vitro investigations of NPs show excellent targeting efficacy in the cancer cells, low targeting and poor biodistribution profiles are found in in vivo animal models and in human studies. This is due to the influence of biological fluids where serum proteins alter the fate of NPs in vivo. Therefore, identification of such biological phenomenon at the nanoparticle-serum protein corona has received considerable attention in the nanopharmaceutics field.
NPs and serum protein interaction occurs due to the overall change in enthalpy which is small and negative; thus, Gibbs free energy becomes negative [42], making the interaction process between NPs and serum proteins feasible. This process mainly occurs via electrostatic interactions between the protein and the NP surface [1, 31, 43]. The NP–protein interaction is a complex and dynamic phenomenon, which leads to the development of a protein sheath or corona around the NPs (Figure 1). The composition and type of protein corona (soft or hard) on NPs define their appropriate use for a specific therapeutic application. The pattern of adsorption can be regulated by choosing optimal size, shape, surface moieties, surface zeta potential, and concentration of NPs. This study delineates the existence of a HS protein corona on MNPs using FTIR, XRD, TGA and DLS analyses (Figures 2-3). Dynamic light scattering (DLS) measurements confirm that hard corona produces a highly stable formulation with reduced particle size and low negative zeta potential. Instantaneous protein adsorption or soft corona tends to slightly increase the particle size and alter the zeta potential of NPs towards the positive. As expected, change of particle size is not significant due to the hard/soft corona of HS proteins, which concludes that overall, MNPs are stable formulations in biological milieu. Jansch et al. [44] also made a similar finding where the total amount of proteins adsorbed on the surface of the iron oxide nanoparticles increased after incubation with increased plasma concentration. The HS proteins binding on MNPs is continuously increased up to HS30@MNPs while after that HS protein binding lessens (Figure 4D). This indicates saturation of binding of HS proteins or sufficient hard corona is formed at HS30@MNPs.
It is also known that various serum proteins bind to NPs immediately upon intravenous administration. Therefore, in our study we chose whole human serum to evaluate such NPs and protein interactions by steady state fluorescence and CD spectral analyses. Poly(ethylene glycol) chains of a F127 pluronic polymer layer on the top of MNPs not only induce hydrophobicity but negative zeta potential of surface MNPs, and govern a lower rate of interaction with HS. This was confirmed from the number of binding sites over other hydrophobic NPs (Figure 5A-B). Furthermore, the CD analysis data confirms that MNPs do not persuade significant changes in the secondary structure of serum proteins (Figure 5C-D).
Any in situ or ex situ modification to MNPs alters their inherent properties. This is also true in the case of bimolecular or serum protein binding on MNPs [3, 7]. Due to excessive protein-corona on MNPs, the magnetization saturation of MNPs decreased from 64 to 33 emu (Figure 6A). However, this corona formation helps in achieving superior hyperthermia properties (Figure 6C). Other than HS2.5@MNPs, there is no significant change in the inherent MR imaging characteristics even upon various serum protein corona formations on MNPs (Figure 7), which is an important aspect of this study.
It is apparent from the available literature that most of the anticancer drugs have side effects due to their direct interaction with blood cells or significant alteration of secondary structures of serum proteins upon intravenous administration. To protect against such adverse effects, anticancer drugs are encapsulated into NPs. Previous studies from our lab and other groups demonstrated that dual layered MNPs harness flexibility with improved circulation in mice [16, 19, 45-47]. Such improved circulation characteristic of MNPs simultaneously increases their levels in blood over time and establishes transient interactions with RBCs. If MNPs had toxicity groups on their surface or significant aggregation, damage to RBCs, and thus hemolysis, occurred. This study supports that our MNPs and HS@MNPs interact with RBCs but do not cause any toxicity (Figure 8A). This is evident from the ex vivo hemolysis assay amplitude curves (Figure 8B). This is an important parameter characterizing the safety of nanoparticles. Such safety profile is attributed to negative zeta potential and non-aggregative behavior of MNPs and HS@MNPs. Another possible reason is that overall, MNP and HS@MNPs are composed of biocompatible compounds such as cyclodextrin, F127 pluronic polymer, and HS at the surface of NPs.
In general, the discrepancy between the in vitro and in vivo results of NPs is due to absence of an immune system or serum proteins. When nanoparticles are exposed to the biological environment, their surfaces become covered with proteins and biomolecules. The majority of studies conducted in vitro assessments of MNPs toxicity using cell culture model(s). However, the transformation of such results in vivo are largely unknown. Such nanomaterials always require validation in vivo for safety. Our study suggests that no signs of toxicity exist in major organs where MNPs are known to reach upon systemic administration, thereby demonstrating in vivo experimental safety (Figure 8C).
MNPs are readily internalized via endocytosis and phagocytosis processes in cells, depending on their nature [48]. Cells view NPs and NPs-protein corona in various ways and handle them differently [49]. Thus, it is important to understand the behavior of NPs on specific cells at the molecular level, in the presence of a protein corona [50]. Although composition of protein coronas vary on MNPs, the HS@MNPs complexes are efficiently trafficked through the endocytic mechanism (Figure 9A-B). This suggests that MNPs retain their internalization capacity and indeed exhibit an improved uptake upon protein corona formation [50, 51]. Upon loading an anti-cancer drug in HS@MNPs, superior anti-cancer activity occurred in cancer cells compared to free drug (Figure 9D-E and Supporting Information V). On the other hand, we can expect that improved targeting of cancer cells by the protein corona that surrounds the MNPs can help to improve the efficiency of MNP in vivo.
5. Conclusion
This study demonstrates the dynamic interaction of human serum proteins with MNPs using a multi-instrumental approach. The interaction of proteins depends on the concentration of serum/MNPs and time. The dynamic corona formed on MNPs does not alter significantly the particle size, MR imaging, or cyto-compatibility of MNPs. However, a clear change in zeta potential, magnetization saturation, cellular uptake, and hyperthermia properties was observed. Overall, this human serum protein and nanoparticle interaction study might lead to new classification for nanotoxicology. In addition, this study indeed delineates the impact of protein corona and integrates design principles of MNPs that could be used in the future to engineer an advanced nanomedicine with precise control of properties for drug delivery, cancer-targeting, imaging and theranostics. Further investigations are needed to translate the targeting efficiency of MNPs (using specific targeting moiety) in in vivo animal models.
Supplementary Material
Acknowledgments
FTIR, TGA, CD, XRD, and magnetization saturation measurements were carried out in the Department of Chemistry and the Nebraska Center for Materials and Nanoscience at the University of Nebraska, Nebraska-Lincoln. SEM studies were conducted in the Department of Biomedical Engineering at the University of South Dakota, Sioux Falls, SD. This work was partially supported by grants from the National Institutes of Health (R01 CA142736 to SCC; U01 CA162106A to SCC and MJ; and a pilot grant through P20 GM103548 to MMY); and the College of Pharmacy 2013 Dean's Seed Grant of the University of Tennessee Health Science Center (to MJ and MMY). The authors thank Cathy Christopherson (Sanford Research) for valuable editorial suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
7. Conflict of interest
The authors declare that they have no conflict of interests in this work.
8. Disclaimer
The views, opinions and statements expressed in this research article are those of the authors and do not reflect the official policy or position of the University of Tennessee Health Science Center and the National Institutes of Health.
Supporting Information Available: TEM images of MNPs and HS@MNPs, hemolysis in the presence of serum, in vivo safety profile of MNP-CUR formulation, and anti-cancer activity of curcumin loaded HS@MNPs formulation. This material is available free of charge via the Internet.
References
- 1.Lynch I, Dawson KA. Protein-nanoparticle interactions. Nano Today. 2008;3:40–7. [Google Scholar]
- 2.Lynch I, Salvati A, Dawson KA. Protein-nanoparticle interactions: What does the cell see? Nature Nanotechnol. 2009;4:546–7. doi: 10.1038/nnano.2009.248. [DOI] [PubMed] [Google Scholar]
- 3.Mahmoudi M, Lynch I, Ejtehadi MR, Monopoli MP, Bombelli FB, Laurent S. Protein- nanoparticle interactions: opportunities and challenges. Chem Rev. 2011;111:5610–37. doi: 10.1021/cr100440g. [DOI] [PubMed] [Google Scholar]
- 4.Yang ST, Liu Y, Wang YW, Cao A. Biosafety and bioapplication of nanomaterials by designing protein-nanoparticle interactions. Small. 2013;9:1635–53. doi: 10.1002/smll.201201492. [DOI] [PubMed] [Google Scholar]
- 5.Huang R, Carney RP, Stellacci F, Lau BL. Protein-nanoparticle interactions: the effects of surface compositional and structural heterogeneity are scale dependent. Nanoscale. 2013;5:6928–35. doi: 10.1039/c3nr02117c. [DOI] [PubMed] [Google Scholar]
- 6.Yoo D, Lee JH, Shin TH, Cheon J. Theranostic magnetic nanoparticles. Acc Chem Res. 2011;44:863–74. doi: 10.1021/ar200085c. [DOI] [PubMed] [Google Scholar]
- 7.Ho D, Sun X, Sun S. Monodisperse magnetic nanoparticles for theranostic applications. Acc Chem Res. 2011;44:875–82. doi: 10.1021/ar200090c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Millward JM, Schnorr J, Taupitz M, Wagner S, Wuerfel JT, Infante-Duarte C. Iron oxide magnetic nanoparticles highlight early involvement of the choroid plexus in central nervous system inflammation. ASN neuro. 2013;5:e00110. doi: 10.1042/AN20120081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sosnovik DE, Nahrendorf M. Cells and iron oxide nanoparticles on the move: magnetic resonance imaging of monocyte homing and myocardial inflammation in patients with ST-elevation myocardial infarction. Circu Cardiovasc Imaging. 2012;5:551–4. doi: 10.1161/CIRCIMAGING.112.978932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hauptman JS, Safaee M. From the bench to the bedside: Spinal cord regeneration, niacin for stroke, magnetic nanoparticles, stimulation for epilepsy, role of galanins in epilepsy, functions of the supramarginal gyri, and the role of inflammation in postoperative cognitive disturbances. Surg Neurol Int. 2010;1:66. doi: 10.4103/2152-7806.71985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh N, Jenkins GJ, Asadi R, Doak SH. Potential toxicity of superparamagnetic iron oxide nanoparticles (SPION). Nano Rev. 2010:1. doi: 10.3402/nano.v1i0.5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He H, David A, Chertok B, Cole A, Lee K, Zhang J, et al. Magnetic nanoparticles for tumor imaging and therapy: a so-called theranostic system. Pharm Res. 2013;30:2445–58. doi: 10.1007/s11095-013-0982-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang HW, Hua MY, Liu HL, Huang CY, Wei KC. Potential of magnetic nanoparticles for targeted drug delivery. Nanotechnol Sci Appl. 2012;5:73–86. doi: 10.2147/NSA.S35506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manju S, Sreenivasan K. Enhanced drug loading on magnetic nanoparticles by layer-by- layer assembly using drug conjugates: blood compatibility evaluation and targeted drug delivery in cancer cells. Langmuir. 2011;27:14489–96. doi: 10.1021/la202470k. [DOI] [PubMed] [Google Scholar]
- 15.Lee JH, Kim JW, Cheon J. Magnetic nanoparticles for multi-imaging and drug delivery. Mol Cells. 2013;35:274–84. doi: 10.1007/s10059-013-0103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yallapu MM, Ebeling MC, Khan S, Sundram V, Chauhan N, Gupta BK, et al. Novel curcumin-loaded magnetic nanoparticles for pancreatic cancer treatment. Mol Cancer Ther. 2013;12:1471–80. doi: 10.1158/1535-7163.MCT-12-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yallapu MM, Othman SF, Curtis ET, Bauer NA, Chauhan N, Kumar D, et al. Curcumin- loaded magnetic nanoparticles for breast cancer therapeutics and imaging applications. Inter J Nanomed. 2012;7:1761–79. doi: 10.2147/IJN.S29290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yallapu MM, Othman SF, Curtis ET, Gupta BK, Jaggi M, Chauhan SC. Multi-functional magnetic nanoparticles for magnetic resonance imaging and cancer therapy. Biomaterials. 2011;32:1890–905. doi: 10.1016/j.biomaterials.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yallapu MM, Foy SP, Jain TK, Labhasetwar V. PEG-functionalized magnetic nanoparticles for drug delivery and magnetic resonance imaging applications. Pharm Res. 2010;27:2283–95. doi: 10.1007/s11095-010-0260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar S, Bhargava D, Thakkar A, Arora S. Drug carrier systems for solubility enhancement of BCS class II drugs: a critical review. Crit Rev Ther Drug Carrier Syst. 2013;30:217–56. doi: 10.1615/critrevtherdrugcarriersyst.2013005964. [DOI] [PubMed] [Google Scholar]
- 21.Loftsson T, Brewster ME. Cyclodextrins as functional excipients: methods to enhance complexation efficiency. J Pharm Sci. 2012;101:3019–32. doi: 10.1002/jps.23077. [DOI] [PubMed] [Google Scholar]
- 22.Batrakova EV, Kabanov AV. Pluronic block copolymers: evolution of drug delivery concept from inert nanocarriers to biological response modifiers. J Control Release : official journal of the Controlled Release Society. 2008;130:98–106. doi: 10.1016/j.jconrel.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alakhova D, Kabanov AV. Pluronics and MDR reversal - an update. Mol Pharm. 2014 doi: 10.1021/mp500298q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Owens DE, 3rd, Peppas NA. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int J Pharm. 2006;307:93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 25.Sakulkhu U, Mahmoudi M, Maurizi L, Salaklang J, Hofmann H. Protein corona composition of superparamagnetic iron oxide nanoparticles with various physico-chemical properties and coatings. Sci Rep. 2014;4:5020. doi: 10.1038/srep05020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yallapu MM, Ebeling MC, Chauhan N, Jaggi M, Chauhan SC. Interaction of curcumin nanoformulations with human plasma proteins and erythrocytes. Int J Nanomedicine. 2011;6:2779–90. doi: 10.2147/IJN.S25534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Q, Liang J, Han H. Probing the interaction of magnetic iron oxide nanoparticles with bovine serum albumin by spectroscopic techniques. J Phys Chem B. 2009;113:10454–8. doi: 10.1021/jp904004w. [DOI] [PubMed] [Google Scholar]
- 28.Niemirowicz K, Markiewicz KH, Wilczewska AZ, Car H. Magnetic nanoparticles as new diagnostic tools in medicine. Adv Med Sci. 2012;57:196–207. doi: 10.2478/v10039-012-0031-9. [DOI] [PubMed] [Google Scholar]
- 29.Sperling RA, Parak WJ. Surface modification, functionalization and bioconjugation of colloidal inorganic nanoparticles. Philos Trans A Math Phys Eng Sci. 2010;368:1333–83. doi: 10.1098/rsta.2009.0273. [DOI] [PubMed] [Google Scholar]
- 30.Podila R, Chen R, Ke PC, Brown JM, Rao AM. Effects of surface functional groups on the formation of nanoparticle-protein corona. Appl Phys Lett. 2012;101:263701. doi: 10.1063/1.4772509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rahman M, Laurent S, Tawil N, Yahia L, Mahmoudi M. Nanoparticle and protein corona. In: Martinac B, editor. Protein-nanoparticle interactions. Springer; 2013. pp. 21–44. [Google Scholar]
- 32.Amiri H, Bordonali L, Lascialfari A, Wan S, Monopoli MP, Lynch I, et al. Protein corona affects the relaxivity and MRI contrast efficiency of magnetic nanoparticles. Nanoscale. 2013;5:8656–65. doi: 10.1039/c3nr00345k. [DOI] [PubMed] [Google Scholar]
- 33.Gupta AK, Naregalkar RR, Vaidya VD, Gupta M. Recent advances on surface engineering of magnetic iron oxide nanoparticles and their biomedical applications. Nanomedicine. 2007;2:23–39. doi: 10.2217/17435889.2.1.23. [DOI] [PubMed] [Google Scholar]
- 34.Soler MA, Bao SN, Alcantara GB, Tiburcio VH, Paludo GR, Santana JF, et al. Interaction of erythrocytes with magnetic nanoparticles. J Nanosci Nanotechnol. 2007;7:1069–71. doi: 10.1166/jnn.2007.423. [DOI] [PubMed] [Google Scholar]
- 35.Mai TD, d'Orlye F, Menager C, Varenne A, Siaugue JM. Red blood cells decorated with functionalized core-shell magnetic nanoparticles: elucidation of the adsorption mechanism. Chem Commun. 2013;49:5393–5. doi: 10.1039/c3cc41513a. [DOI] [PubMed] [Google Scholar]
- 36.Rothen-Rutishauser BM, Schurch S, Haenni B, Kapp N, Gehr P. Interaction of fine particles and nanoparticles with red blood cells visualized with advanced microscopic techniques. Environ Sci Technol. 2006;40:4353–9. doi: 10.1021/es0522635. [DOI] [PubMed] [Google Scholar]
- 37.Dai L, Liu Y, Wang Z, Guo F, Shi D, Zhang B. One-pot facile synthesis of PEGylated superparamagnetic iron oxide nanoparticles for MRI contrast enhancement. Mater Sci Eng C Mater Biol Appl. 2014;41:161–7. doi: 10.1016/j.msec.2014.04.041. [DOI] [PubMed] [Google Scholar]
- 38.Pan Y, Du X, Zhao F, Xu B. Magnetic nanoparticles for the manipulation of proteins and cells. Chem Soc Rev. 2012;41:2912–42. doi: 10.1039/c2cs15315g. [DOI] [PubMed] [Google Scholar]
- 39.Swami A, Shi J, Gadde S, Votruba AR, Kolishetti N, Farokhzad OC. Nanoparticles for Targeted and Temporally Controlled Drug Delivery. In: Svenson A, Prud'homme RK, editors. Multifunctional Nanoparticles for Drug Delivery Applications: Imaging, Targeting, and Delivery, Nanostructure Science and Technology. Spinger; 2012. pp. 9–29. [Google Scholar]
- 40.Walkey CD, Olsen JB, Song F, Liu R, Guo H, Olsen DW, et al. Protein corona fingerprinting predicts the cellular interaction of gold and silver nanoparticles. ACS nano. 2014;8:2439–55. doi: 10.1021/nn406018q. [DOI] [PubMed] [Google Scholar]
- 41.Beech JR, Shin SJ, Smith JA, Kelly KA. Mechanisms for targeted delivery of nanoparticles in cancer. Curr Pharm Des. 2013;19:6560–74. doi: 10.2174/1381612811319370002. [DOI] [PubMed] [Google Scholar]
- 42.Paul BK, Bhattacharjee K, Bose S, Guchhait N. A spectroscopic investigation on the interaction of a magnetic ferrofluid with a model plasma protein: effect on the conformation and activity of the protein. Phys Chem Chem Phys. 2012;14:15482–93. doi: 10.1039/c2cp42415k. [DOI] [PubMed] [Google Scholar]
- 43.Jiang W, Lai K, Wu Y, Gu Z. Protein corona on magnetite nanoparticles and internalization of nanoparticle-protein complexes into healthy and cancer cells. Arch Pharm Res. 2014;37:129–41. doi: 10.1007/s12272-013-0292-2. [DOI] [PubMed] [Google Scholar]
- 44.Jansch M, Stumpf P, Graf C, Ruhl E, Muller RH. Adsorption kinetics of plasma proteins on ultrasmall superparamagnetic iron oxide (USPIO) nanoparticles. Int J Pharm. 2012;428:125–33. doi: 10.1016/j.ijpharm.2012.01.060. [DOI] [PubMed] [Google Scholar]
- 45.Sadhukha T, Niu L, Wiedmann TS, Panyam J. Effective elimination of cancer stem cells by magnetic hyperthermia. Mol Pharm. 2013;10:1432–41. doi: 10.1021/mp400015b. [DOI] [PubMed] [Google Scholar]
- 46.Foy SP, Manthe RL, Foy ST, Dimitrijevic S, Krishnamurthy N, Labhasetwar V. Optical imaging and magnetic field targeting of magnetic nanoparticles in tumors. ACS nano. 2010;4:5217–24. doi: 10.1021/nn101427t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jain TK, Reddy MK, Morales MA, Leslie-Pelecky DL, Labhasetwar V. Biodistribution, clearance, and biocompatibility of iron oxide magnetic nanoparticles in rats. Mol Pharm. 2008;5:316–27. doi: 10.1021/mp7001285. [DOI] [PubMed] [Google Scholar]
- 48.Wiogo HT, Lim M, Bulmus V, Gutierrez L, Woodward RC, Amal R. Insight into serum protein interactions with functionalized magnetic nanoparticles in biological media. Langmuir. 2012;28:4346–56. doi: 10.1021/la204740t. [DOI] [PubMed] [Google Scholar]
- 49.Mahmoudi M, Saeedi-Eslami SN, Shokrgozar MA, Azadmanesh K, Hassanlou M, Kalhor HR, et al. Cell “vision”: complementary factor of protein corona in nanotoxicology. Nanoscale. 2012;4:5461–8. doi: 10.1039/c2nr31185b. [DOI] [PubMed] [Google Scholar]
- 50.Treuel L, Jiang X, Nienhaus GU. New views on cellular uptake and trafficking of manufactured nanoparticles. J R Soc Interface. 2013;10:20120939. doi: 10.1098/rsif.2012.0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang X, Weise S, Hafner M, Rocker C, Zhang F, Parak WJ, et al. Quantitative analysis of the protein corona on FePt nanoparticles formed by transferrin binding. J R Soc Interface. 2010;7(Suppl 1):S5–S13. doi: 10.1098/rsif.2009.0272.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.