Abstract
Pulmonary arterial hypertension (PAH) is characterized by increased pulmonary vascular smooth muscle contraction and proliferation. Here, we analyze genome-wide mRNA expression in human pulmonary arterial smooth muscle cells (HPASMC) isolated from three control, three hereditary (HPAH), and three idiopathic PAH (IPAH) subjects using the Affymetrix Human Gene ST 1.0 chip. The microarray analysis reveals the expression of 537 genes in HPAH and 1024 genes in IPAH changed compared with control HPASMC. Among those genes, 227 genes show similar directionality of expression in both HPAH and IPAH HPASMC. Ingenuity™ Pathway Analysis (IPA) suggests that many of those genes are involved in cellular growth/proliferation and cell cycle regulation and that signaling pathways such as the mitotic activators, polo-like kinases, ATM signaling are activated under PAH conditions. Furthermore, the analysis demonstrates downregulated mRNA expression of certain vasoactive receptors such as bradykinin receptor B2 (BKB2R). Using real time PCR, we verified the downregulated BKB2R expression in the PAH cells. Bradykinin-stimulated calcium influx is also decreased in PAH PASMC. IPA also identified transcriptional factors such p53 and Rb as downregulated, and FoxM1 and Myc as upregulated in both HPAH and IPAH HPASMC. The decreased level of phospho-p53 in PAH cells was confirmed with a phospho-protein array; and we experimentally show a dysregulated proliferation of both HPAH and IPAH PASMC. Together, the microarray experiments and bioinformatics analysis highlight an aberrant proliferation and cell cycle regulation in HPASMC from PAH subjects. These newly identified pathways may provide new targets for the treatment of both hereditary and idiopathic PAH.
Keywords: PULMONARY ARTERIAL HYPERTENSION, HUMAN PULMONARY ARTERY SMOOTH MUSCLE CELLS (HPASMC), DNA MICROARRAY, GENE EXPRESSION, SMOOTH MUSCLE CELL PROLIFERATION, CELL CYCLE, INGENUITY PATHWAY ANALYSIS
Pulmonary arterial hypertension (PAH) is a rare disease characterized by elevated mean pulmonary arterial pressures and subsequent right ventricular hypertrophy and failure. The prognosis is poor with a survival rate of ~60% three years after diagnosis [Humbert et al., 2010]. While a number of PAH types exist, the two major types are idiopathic PAH (IPAH) and hereditable PAH (HPAH) [Farber and Loscalzo, 2004]. Although of variable etiology, the histological appearance of the lung tissue in all PAH is similar involving increased medial thickness, increased smooth muscle and endothelial cell proliferation, and resting constriction of smooth muscle cells [Farber and Loscalzo, 2004].
To date, available treatments have been only partially effective in stabilizing the disease [Rydell-Törmänen et al., 2013]. Meaningful development of treatments which reverse PAH and result in improved quality of life for the patient depends on identifying and targeting the as yet unidentified signaling mechanisms responsible for this pulmonary vascular pathobiology. As the pulmonary vascular smooth muscle plays a crucial role in the etiology of PAH [Humbert et al., 2004], we focused our studies on human pulmonary artery smooth muscle cells (HPASMC) derived from normal control, HPAH, and IPAH lungs. We utilized genome-wide microarray analysis to determine mRNA expression profiles from these diseased HPASMC as compared to the control cells. We also used bioinformatics analysis to develop aberrant signaling pathways in both disease conditions. We focused on genes and pathways similarly dysregulated in both types of PAH and related physiologic changes occurring during disease pathogenesis. Finally, we validated our findings in vitro using cellular, biochemical, and molecular techniques to understand the relationship between changes in gene mRNA expression and protein signaling cascades and how these may lead to abnormal cell function.
MATERIALS AND METHODS
ISOLATION AND CULTURE OF HUMAN PULMONARY ARTERY SMOOTH MUSCLE CELLS (HPASMC)
Normal, HPAH, and IPAH HPASMC were isolated at the Cleveland Clinic Foundation [Aldred et al., 2010]. The cells used herein consisted of HPASMC from three donor control subjects (CONTROL-1, CONTROL-2, and CONTROL-3), three samples from IPAH subjects (IPAH-1, IPAH-2, and IPAH-3), and three samples from patients with BMPR2 mutation (HPAH-1, HPAH-2, and HPAH-3) [Aldred et al., 2010; Yu et al., 2013]. The patients with PAH were identified based on the National Institutes of Health (NIH) registry diagnostic criteria for pulmonary hypertension. The healthy controls were individuals with no history of pulmonary or cardiac disease or symptoms. HPASMC were isolated from elastic pulmonary arteries (>500-μm diameter) dissected from lungs obtained at explantation during lung transplant. Briefly, after removal of endothelial cells, HPASMC were dissociated by digestion with collagenase type II/DNase I solution overnight at 37 °C. Cells were cultured in 15 mM HEPES buffered DMEM/F12 (50:50) media (Mediatech, Manassas, VA) containing 10% fetal bovine serum (FBS) (Lonza), and 2.5% Antibiotic-Antimycotic from GIBCO (cat. no. 15240). The smooth muscle phenotype of cultured cells was confirmed (>97% purity) by immunohistochemistry and flow cytometric analysis with antibodies against smooth muscle α-actin (Supplementary Fig. S1) and calponin. Those cells also displayed dose dependent constriction in response to endothelin-1 [Wilson et al., 2012]. Primary cultures up to passage 6 were used in the experiments. Approval to use these human cells was granted by the Boston University Institutional Review Board.
MICROARRAY ANALYSIS
The Microarray analyses were performed on three non-PAH (Normal Control), three HPAH, and three IPAH HPASMC samples. Total RNA was extracted from HPASMC using the RNeasy kit (Qiagen, Valencia, CA). Briefly, RNA quantity was determined on an Agilent Nanodrop and quality assessed on an Agilent 2100 Bioanalyzer, according to the manufacturer's instructions. Microarray experiments were carried out using the Affymetrix Human Gene ST 1.0 chip as described in the Afftymetrix standard protocol. Raw CEL files were normalized to produce Entrez Gene-identifier-specific expression values using the implementation of the Robust Multiarray Average (RMA) [Irizarry et al., 2003] in the affy package [Gautier et al., 2004] in the Bioconductor software suite [Gentleman et al., 2004] and the BrainArray Entrez Gene-specific probeset mapping (version 14.0.0) [Dai et al., 2005]. All computations were performed using the R environment for statistical computing (http://www.R-project.org/). The box plot of the normalized data was shown in Supplementary Figure S2, which shows that the expression was normalized.
Using “multtest” package in R, we selected genes that are differentially expressed in HPAH or IPAH samples using the t-test with equal variances. 443 Genes in HPAH and 785 genes in IPAH were selected with fold > 1.5 and P-value < 0.01. In addition to P < 0.01, a 1.5-fold difference in normalized expression value was used to estimate differential expression. The reason to choose 1.5 instead of 2 is that the expression of many important genes changes less than 50% but has important biological consequence. We try to avoid to miss those important genes if chose higher fold change cutoff value such as 2. We next used permutation tests to select probes with false discovery rate (FDR) <0.5% and fold change cutoff 1.5. 112 Genes in HPAH and 166 genes in IPAH were selected. Finally, the genes chosen in the previous t-test and permutation test were combined in HPAH (537 genes) and IPAH samples (1024 genes) (Supplementary Table 1 SA for HPAH and 1B for IPAH). Since the interest of this study is to probe the common gene expression signature of HPAH and IPAH HPASMC, among those genes, 227 genes were chosen for further bioinformatics analysis. The expression of those genes changed in the same direction in both HPAH and IPAH cells in comparison with the non-PAH control HPASMC (Supplementary Table S2).
SIGNAL PATHWAY ANALYSIS
Functional and canonical pathway analysis were performed using Ingenuity Pathway Analysis software (IPA) (Ingenuity1 Systems, www.ingenuity.com). From the microarray analysis of HPAH and IPAH PASMC, 227 selected differentially expressed genes were uploaded into Ingenuity website. The Ingenuity Pathways Knowledge Base (IPKB) provided all the published known functions and interactions. Fischer's exact test was used to calculate a P-value to determine the probability that each biologic function or canonical pathway assigned to the dataset was due to chance alone. We also conducted upstream regulator analysis. Ingenuity's Upstream Regulator Analysis is a tool that predicts upstream regulators from gene expression data based on the literature and compiled in the Ingenuity™ Knowledge Base. A Fisher's exact test P-value is calculated to assess the significance of enrichment of the gene expression data for the genes downstream of an upstream regulator. The goal of the IPA upstream regulator analysis was to identify the cascade of upstream regulators that can explain the observed gene expression changes in a user's dataset, which can help illuminate the biological activities occurring in the tissues or cells being studied. The DAVID Bioinformatics Resources 6.7 [Huang et al., 2008] was used to perform functional annotation on the selected genes.
REAL TIME PCR
The mRNA used for real time PCR was isolated by the same procedure. However, to confirm the microarray results the isolation was done separately from that for the DNA array experiment. The total RNA was then subjected to reverse transcriptase-PCR using the Superscript III First Strand Synthesis System (Invitrogen, Carlsbad, CA). Negative controls were performed without the reverse transcriptase. The resultant cDNA was used to conduct Taqman real time PCR on the Applied Biosystems 7300 Real time PCR System (Applied Biosystems, Foster City, CA). Taqman reagents for detecting mRNA expression of human bradykinin receptor 2 (BDKRD2: Hs00176121_m1), human angiotensin II receptor type I (AGTR1: Hs99999095_m1), and human GAPDH (Hs99999905_m1) were purchased from Applied Biosystems (Foster City, CA). Cycling parameters were as follows: 50 °C for 2 min, 95 °C for 10 min, 45 cycles of 95 °C for 15 s, and 60 °C for 1 min. Results are presented as relative expression normalized to GAPDH and were calculated using the ΔΔCt method as described in the Applied Biosystems publication, “Guide to Performing Relative Quantitation of Gene Expression Using Real-Time Quantitative PCR” (4371095 Rev B).
CALCIUM INFLUX
The intracellular Ca2+ concentrations were determined as previously reported [Prado et al., 1998]. The HPASMC were trypsinized and washed two times in physiological buffer solution (140 mM NaCl, 5 mM KCl, 1 mM MgCl2, 10 mM glucose, 0.9 mM CaCl2, 15 mM HEPES, 0.1% BSA). The cells were resuspended at 1.5 × 107 cells/ml and incubated with 2 μM Fura-2/AM for 30 min. The cell suspension was then diluted fivefold with physiological buffer and incubated for another 15 min. Cells were pelleted and resuspended at 1 × 106 cells/ml. Intracellular Ca2+ influx was determined with a Hitachi F-2500 Fluorescence Spectrophotometer using a FL solutions 2.0 program (Hitachi Inc., Tokyo, Japan). 100 nM bradykinin was added after the basal level of calcium was detected.
PHOSPHO-ANTIBODY ARRAY ANALYSIS
The phospho-antibody array analysis was performed using the Proteome Profiler Human Phospho-Kinase Array Kit ARY003 from R&D Systems according to the manufacturer's instructions. Briefly, HPASMC were lysed with Lysis Buffer 6 (R&D Systems) and agitated for 30 min at 4 °C. Cell lysates were clarified by microcentrifugation at 14,000 × g for 5 min, and the supernatants were subjected to protein assay. Preblocked nitrocellulose membranes of the human phospho-kinase array were incubated with 500 mg of cellular extract overnight at 4 °C on a rocking platform. The membranes were washed three times with 1 × Wash Buffer (R&D Systems) to remove unbound proteins and then were incubated with a mixture of biotinylated detection antibodies and streptavidin-HRP antibodies. Chemiluminescent detection re-agents were applied to detect spot densities. Array images were analyzed using the NIH ImageJ image analysis software. The averaged density of duplicate spots representing each phosphorylated kinase protein was determined and used for the relative changes in phosphorylated kinase proteins. The list of target capture antibodies and their positions on the arrays can be found at http://www.rndsystems.com/pdf/ARY003.pdf
CELL PROLIFERATION ANALYSIS
HPASMC at 80–90% confluence were trypsinized and seeded onto 24 well culture plates at a density of 20,000 cells per well. Cells were allowed to attach overnight in growth medium (DMEM/F12K 50:50 with 10% FBS) at 37 °C. The next day triplicate wells from each cell strain were counted and the remaining wells were changed to 0.2% FBS medium. The cells were maintained under these conditions for eight more days with the medium refreshed once. On the last day cells were collected from each well and counted using a Coulter counter. Cell proliferation was determined after 5 days using the MTT Cell Proliferation Assay (ATCC, Manassas, VA).
RESULTS
PHENOTYPE RETENTION OF HPASMC IN INITIAL CULTURES
The smooth muscle phenotype of the HPASMC used for this study was confirmed by several procedures. Stain for smooth muscle actin demonstrated that the HPASMC retained their phenotype in culture (Supplementary Fig. S1).
DETERMINATION OF DIFFERENTIALLY EXPRESSED GENES
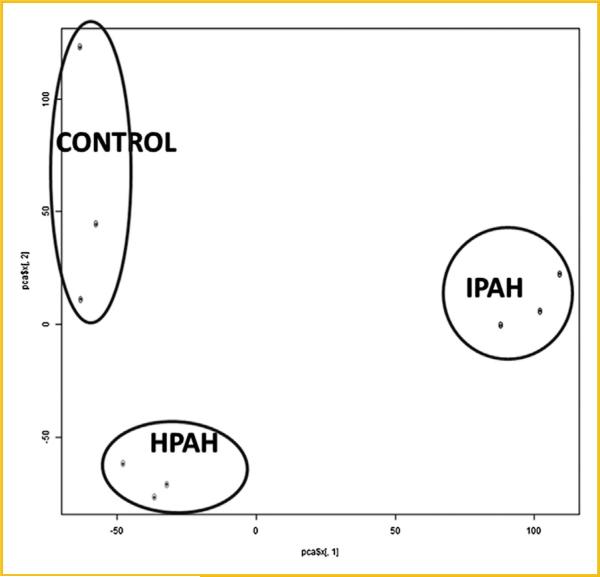
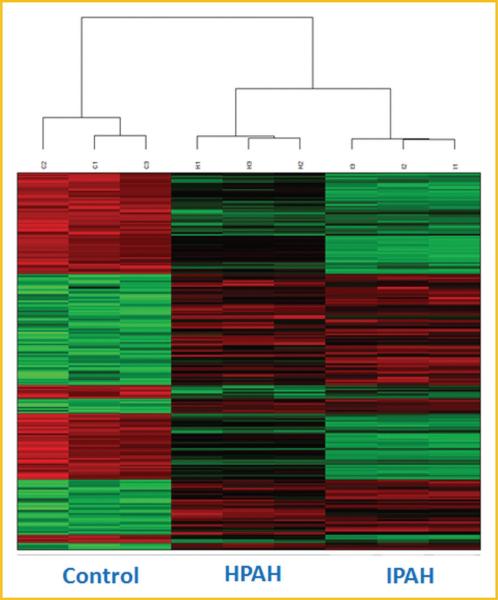
Microarray experiments were performed on three samples of non-PAH (Normal Control), HPAH, and IPAH HPASMC. Principal component analysis (PCA) showed sample clustering by disease (Fig. 1). This plot also shows that there are no extreme outliners. We performed pairwise comparisons between disease and normal conditions, and found the expression of 537 genes in HPAH (Supplementary Table S1A) and 1024 genes in IPAH (Supplementary Table S1B) changed compared with control HPASMC. Among those genes, 227 genes showed same trended expression changes in both PAH and IPAH HPASMC (Supplementary Table S2). Fifteen of the most upregulated and downregulated genes are presented in Table I. These genes show the same trend of expression in HPAH and IPAH (either increase or decrease) as is illustrated in the heatmap (Fig. 2).
Fig. 1.
Principal component analysis (PCA) plot of the mRNA expression data that characterizes the trends exhibited by the expression profiles of control, HPAH, and IPAH. Each dot represents a sample. The first component represent 49% of variance, the second component represent 26% of variance. The PCA plot was plotted using prcomp function in R program.
TABLE I.
Top Genes Which Are Expressed Differently in HPAH and IPAH HPASMC Compared With Control Cells
| Symbol | Entrez gene name | Fold change (HPAH) | P-value (HPAH) | Fold change (IPAH) | P-value (IPAH) |
|---|---|---|---|---|---|
| Mostly upregulated genes with related biological functions | |||||
| CDC20 | Cell division cycle 20 | 2.717 | 4.02E-03 | 7.059 | 3.05E-04 |
| PLK1 | Polo-like kinase 1 | 2.517 | 5.73E-03 | 6.442 | 4.78E-04 |
| ERG | v-ets avian erythroblastosis virus E26 oncogene homolog | 8.529 | 3.40E-03 | 6.278 | 5.79E-03 |
| CDKN3 | Cyclin-dependent kinase inhibitor 3 | 2.134 | 2.51E-02 | 5.987 | 1.50E-03 |
| ANLN | Anillin, actin binding protein | 2.356 | 8.74E-03 | 5.827 | 6.49E-04 |
| CCNB2 | Cyclin B2 | 1.986 | 9.20E-03 | 5.419 | 4.72E-04 |
| CDK1 | Cyclin-dependent kinase 1 | 1.981 | 3.75E-03 | 4.634 | 1.86E-04 |
| PLXDC1 | Plexin domain containing 1 | 2.774 | 2.04E-05 | 3.712 | 1.69E-06 |
| BMP4 | Bone morphogenetic protein 4 | 2.405 | 3.17E-05 | 3.323 | 1.20E-05 |
| SMOC1 | SPARC related modular calcium binding 1 | 3.172 | 2.09E-03 | 3.304 | 1.94E-03 |
| PDPN | Podoplanin | 2.781 | 4.19E-04 | 2.998 | 2.77E-04 |
| WISP3 | WNT1 inducible signaling pathway protein 3 | 2.664 | 8.05E-05 | 2.659 | 2.04E-04 |
| CTSH | Cathepsin H | 3.57 | 1.48E-04 | 2.252 | 7.90E-04 |
| ADRB2 | Adrenoceptor beta 2, surface | 2.089 | 2.18E-04 | 2.191 | 1.48E-04 |
| DAPK1 | Death-associated protein kinase 1 | 3.125 | 1.55E-04 | 1.532 | 2.49E-03 |
| Mostly downregulated genes with related biological functions | |||||
| EDNRB | Endothelin receptor type B | –7.616 | 4.90E-04 | –2.321 | 6.53E-03 |
| CCL20 | Chemokine (C-C motif) ligand 20 | –2.526 | 9.66E-04 | –2.356 | 1.16E-03 |
| KSR1 | Kinase suppressor of ras 1 | –1.594 | 6.41E-04 | –2.452 | 1.23E-04 |
| ADAMTS5 | ADAM metallopeptidase with thrombospondin type 1 motif, 5 | –1.683 | 3.55E-04 | –2.554 | 1.02E-04 |
| MFAP4 | Microfibrillar-associated protein 4 | –2.299 | 3.96E-03 | –2.589 | 2.51E-03 |
| FOXE1 | Forkhead box E1 (thyroid transcription factor 2) | –2.693 | 2.54E-03 | –2.788 | 2.76E-03 |
| PTN | Pleiotrophin | –2.089 | 2.81E-03 | –2.951 | 6.16E-04 |
| RGS7 | Regulator of G-protein signaling 7 | –2.527 | 1.79E-04 | –3.068 | 2.07E-04 |
| TRPC4 | Transient receptor potential cation channel, subfamily C, member 4 | –3.557 | 6.01E-04 | –3.28 | 5.75E-04 |
| PCDHGA3 | Protocadherin gamma subfamily A, 3 | –2.642 | 2.05E-04 | –3.344 | 6.09E-05 |
| LPPR4 | Lipid phosphate phosphatase-related protein type 4 | –6.696 | 3.04E-03 | –4.406 | 7.10E-03 |
| PCDHB16 | Protocadherin beta 16 | –1.949 | 5.45E-03 | –5.14 | 1.83E-04 |
| EREG | Epiregulin | –6.881 | 6.81E-04 | –6.292 | 4.23E-04 |
| DSG2 | Desmoglein 2 | –5.663 | 3.97E-06 | –6.511 | 7.42E-06 |
| MXRA5 | Matrix-remodeling associated 5 | –1.815 | 2.07E-03 | –7.209 | 4.11E-05 |
The table showed the entrez gene ID, gene symbol, the fold change of mRNA expression in HPAH and IPAH samples compared with normal HPASMC, and the normalized expression value of control, HPAH, and IPAH samples of the 15 mostly upregulated and 15 mostly downregulated genes with related biological functions.
Fig. 2.
Heat map the selected genes that changed in both HPAH and IPAH HPASMC. 227 Genes were selected as differentially expressed in both HPAH and IPAH HPASMC. Color represents the log intensities. Red: upregulated genes; green: downregulated genes. The dendogram demonstrates clustering of gene expression of control, HPAH, and IPAH samples. The heatmap was plotted using “Heatplus” package in R program.
BIOLOGICAL FUNCTIONS AND SIGNALING PATHWAY ANALYSIS
The 227 selected genes in Supplementary Table S2 were imported into the IPA software to obtain insight on the biological function of these genes. The most cogent disease and biological functions predicted by the IPA core analysis are shown in Table II. The full list of functions are listed in Supplementary Table S3. The related functions include changes in cellular growth and proliferation, cell cycle, cellular movement, cellular maintenance, and cell death and survival. The most cogent predicted canonical signal transduction pathways activated in both HPAH and IPAH are shown in Table III. All the pathways are listed in Supplementary Table S4. Interestingly, many affected signaling pathways appear to involve cell cycle regulation. These include mitotic roles of polo-like kinase, DNA damage-induced 14-3-3δ signaling, ATM signaling, cyclins and cell cycle regulation, G2/M DNA damage checkpoint regulation, role of CHK proteins in cell cycle checkpoint control, cell cycle regulation by BTG family proteins. Other signaling pathways illustrated in Supplementary Table S4 include IL-6 signaling, IL-22 signaling, IL-1 signaling, BMP signaling pathway, superoxide radicals degradation, inhibition of matrix metalloproteases, ErbB signaling, prostanoid biosynthesis, Cdc42 signaling, HIF1α signaling, and endothelin-1 signaling.
TABLE II.
Most Significantly Pertinent Diseases and Biological Functions Predicted by IPA Based on the Differentially Expressed Genes in HPAH and IPAH vs Control
| Category | P-value |
|---|---|
| Cancer | 3.08E-17-8.72E-03 |
| Cell cycle | 9.67E-09-8.72E-03 |
| Cellular assembly and organization | 9.67E-09-8.09E-03 |
| DNA replication, recombination, and repair | 9.67E-09-8.51E-03 |
| Cell death and survival | 3.28E-08-7.9E-03 |
| Cellular movement | 1.61E-06-8.76E-03 |
| Cellular growth and proliferation | 2.78E-06-8.99E-03 |
| Cell-to-cell signaling and interaction | 5.06E-06-8.31E-03 |
| Cardiovascular system development and function | 7.91E-06-7.9E-03 |
| Metabolic disease | 1.05E-05-7.9E-03 |
| Inflammatory response | 1.33E-04-8.75E-03 |
| Cellular compromise | 1.57E-04-6.09E-03 |
| Post-translational modification | 2.73E-04-2.73E-04 |
| Cell signaling | 5.12E-04-8.09E-03 |
| Respiratory system development and function | 8.03E-04-1.19E-03 |
| Molecular transport | 1.35E-03-8.5E-03 |
| Carbohydrate metabolism | 1.66E-03-4.56E-03 |
| Cardiovascular disease | 1.88E-03-4.41E-03 |
| Respiratory disease | 2.13E-03-7.89E-03 |
The Biological Functional Analysis™ identified the biological functions and/or diseases that were most significant to the dataset. The 227 selected genes in Supplementary Table S2 were associated with a biological function in the ingenuity knowledge base were considered for analysis. Fischer's exact test was used to calculate a P-value determining the probability that each biological function and/or disease assigned to that data set is due to chance alone. The full list of the predicted diseases and biological functions, and the genes involved in those functions were shown in Supplementary Table S3.
TABLE III.
Most Significantly Pertinent Canonical Signal Transduction Pathways Predicted by IPA Based on the Differentially Expressed Genes in HPAH and IPAH vs Control
| Ingenuity Canonical Pathways | –log (P-value) | Ratio | Molecules |
|---|---|---|---|
| Mitotic roles of polo-like kinase | 6.04 | 1.08E-01 | KIF23,PLK4,CDC20,CCNB2,PLK1,PPP2R1B,CDK1,KIF11 |
| Salvage pathways of pyrimidine ribonucleotides | 3.12 | 5.31E-02 | DAPK1,NEK2,PLK1,TTK,CDK1,APOBEC3G |
| DNA damage-induced 14-3-3s signaling | 2.91 | 1.36E-01 | CCNE2,CCNB2,CDK1 |
| IL-6 signaling | 2.67 | 4.84E-02 | VEGFA,TNFAIP6,MAPK10,MAPK13,IL1R1,A2M |
| ATM signaling | 2.32 | 6.06E-02 | MAPK10,CCNB2,MAPK13, CDK1 |
| Inhibition of angiogenesis by TSP1 | 2.17 | 7.14E-02 | VEGFA,MAPK10,MAPK13 |
| Cyclins and cell cycle regulation | 1.88 | 4.17E-02 | CCNE2,CCNB2,PPP2R1B, CDK1 |
| Cell cycle: G2/M DNA damage checkpoint regulation | 1.86 | 6.12E-02 | CCNB2,PLK1,CDK1 |
| Role of CHK proteins in cell cycle checkpoint control | 1.61 | 5.08E-02 | PLK1,PPP2R1B,CDK1 |
| Role of JAK family kinases in IL-6-type cytokine signaling | 1.49 | 7.14E-02 | MAPK10,MAPK13 |
| IL-17A signaling in airway cells | 1.44 | 3.95E-02 | MAPK10,CCL20,MAPK13 |
| BMP signaling pathway | 1.25 | 3.49E-02 | BMP4,MAPK10,MAPK13 |
Canonical pathways analysis identified the pathways from the Ingenuity Pathways Analysis™ library of canonical pathways that were most significant to the dataset. The 227 selected genes in Supplementary Table S2 were associated with a canonical pathway in the ingenuity knowledge base were considered for analysis. The significance of the association between the dataset and the canonical pathway was determined as a P-value calculated by Fisher's exact test, a measure of the probability that the association between the proteins in the dataset and the canonical pathway is taking place by chance alone. The ratio is calculated as follows: number ofgenes in a given pathway that meet the cutoff criteria, divided by the total number of genes that make up that pathway and that are in the reference gene set. The molecules involved in those biological pathways were also listed in the table. The identity of those molecules can be retrieved from Supplementary Table S2. The full list of the predicted pathways was shown in Supplementary Table S4.
The expression of a number of genes involved in cell proliferation is changed in PAH as illustrated in Supplementary Table S5. For example, CCNB2 (cyclin B2) increased 5.468-fold (P = 0.0005) in IPAH, and 1.974-fold (P = 0.009) in HPAH. CCNE2 (cyclin E2) increased 2.066-fold (P = 0.004) in IPAH, and 1.681-fold (P = 0.0007) in HPAH. CDC20 increased 2.717-fold (P = 0.004) in HPAH and 7.059-fold (P = 0.0003) in IPAH. CDK1 (cyclin-dependent kinase 1) increased 4.674-fold (P = 0.0002) in IPAH and 1.969-fold (P = 0.004) in HPAH. MCM10 increased in both HPAH (1.56-fold, P = 0.004)) and IPAH (2.25-fold, P = 0.0004) cells. MYBL2, a transcriptional factor involved in the regulation of cell cycle [Martinez and DiMaio, 2011] increased 1.6-fold (P = 0.001) in HPAH and 2.5-fold (P = 0.0002) in IPAH. SSTR1 (somatostatin receptor-1) which induces cell cycle arrest and inhibits tumor growth in pancreatic cancer [Li et al., 2008] decreased in HPAH (−1.75, P = 0.007) and IPAH (−2.3, P = 0.001). Uhrf1 (ubiquitin-like with PHD and ring finger domains) increased 1.6-fold (P = 0.002) in HPAH and 2.2-fold (P = 0.0002) in IPAH. Uhrf1 was shown to increase the entry into S phase of terminally differentiated cells with cyclin E and CDK2 [Bonapace et al., 2002]. Among those molecules, many are involved in cell cycle regulation.
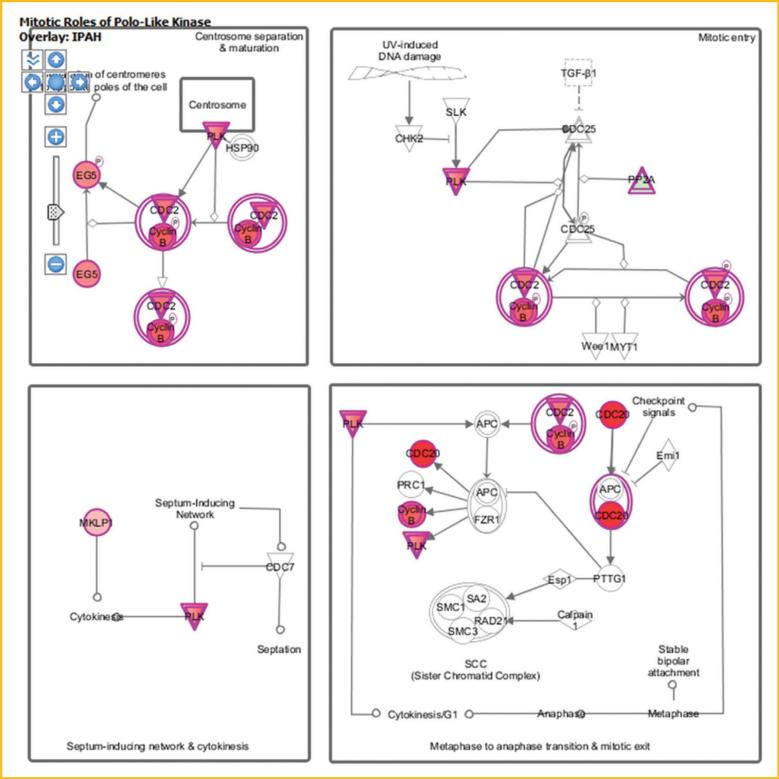
Cell cycle progression is regulated via a number of signal transduction pathways. One such pathway that is activated in PAH HPASMC is the polo-like kinase (Plks) path as illustrated in Figure 3. The activation of this pathway is enhanced in both HPAH and IPAH. Components of this pathway, CCNB1 (cyclin B1), CCNB2 (cyclin B2), CDC20, CDK1, FBXO5 (F-box protein 5), KIF11 (kinesin family member 11), KIF23 (kinesin family member 23), PKMYT1 (protein kinase, membrane associated tyrosine/threonine 1), PLK1 (polo-like kinase 1), PLK4 (polo-like kinase 4), PPP2R1B (protein phosphatase 2, regulatory subunit A, beta), PRC1 (protein regulator of cytokinesis 1), and PTTG1 (pituitary tumor-transforming 1) are upregulated in both HPAH and IPAH. The detailed information about these molecules is shown in Table IV.
Fig. 3.
The expression of the known genes involved in polo signaling in PAH HPASMC. The diagram shows the differentially expressed genes in polo like kinase signaling pathway in IPAH HPASMC. Green indicates downregulation, red indicates upregulation, and color intensity is proportional to fold change. HPAH and IPAH displayed similar changes. CCNB2: cyclin B2; CDC20: cell division cycle 20; CDK1: cyclin-dependent kinase 1; KIF11: kinesin family member 11; KIF23: kinesin family member 23; PLK1: polo-like kinase 1; PLK4: polo-like kinase 4; PPP2R1B: protein phosphatase 2, regulatory subunit A, beta.
TABLE IV.
Differentially Expressed Genes Involved in the Polo-Like Pathway
| Symbol | Entrez gene name | Entrez gene ID | Fold change (HPAH) | P-value (HPAH) | Fold change (IPAH) | P-value (IPAH) |
|---|---|---|---|---|---|---|
| CCNB2 | Cyclin B2 | 9133 | 1.986 | 9.20E-03 | 5.419 | 4.72E-04 |
| CDC20 | Cell division cycle 20 | 991 | 2.717 | 4.02E-03 | 7.059 | 3.05E-04 |
| CDK1 | Cyclin-dependent kinase 1 | 983 | 1.981 | 3.75E-03 | 4.634 | 1.86E-04 |
| KIF11 | Kinesin family member 11 | 3832 | 2.019 | 5.95E-03 | 4.051 | 5.74E-04 |
| KIF23 | Kinesin family member 23 | 9493 | 1.576 | 4.07E-04 | 2.318 | 3.79E-05 |
| PLK1 | Polo-like kinase 1 | 5347 | 2.517 | 5.73E-03 | 6.442 | 4.78E-04 |
| PLK4 | Polo-like kinase 4 | 10733 | 1.610 | 5.01E-04 | 3.550 | 1.27E-05 |
| PPP2R1B | Protein phosphatase 2, regulatory subunit A, beta | 5519 | –1.676 | 4.67E-04 | –1.747 | 1.45E-04 |
Detailed description (symbol, entrez gene name and entrez fold change, and P-value) of the differentially expressed genes involved in the polo-like pathways.
UPSTREAM REGULATORS/EXPRESSION OF TUMOR SUPPRESSOR TRANSCRIPTION FACTORS
We also conducted upstream regulator analysis using IPA. Interestingly, the activity of two tumor suppressor genes, p53 and Rb, decreased in HPAH and IPAH. There are 43 proteins in the dataset, which illustrate regulatory effects by p53. Among them, 24 genes have expression direction consistent with the inhibition of p53 (z-score = −2.173, overlap P-value = 1.08E-12). Eight of fifteen genes have expression direction consistent with the inhibition of Rb (z-score = −2.80, overlap P-value = 3.13E-08). Eight molecules are regulated by both p53 and Rb. On the other hand, the activity of the upstream regulators such as Myc and FOXM1 (Forkhead box M1) is predicted to be increased. Eight of ten genes have expression direction consistent with the activation of FoxM1 (z-score = 2.436, overlap P-value 7.45E-09). Nine of thirteen genes have expression direction consistent with the activation of Myc (z-score = 2.218, overlap P-value = 0.0678). Those four transcriptional factors regulate many common downstream target genes as shown in Supplementary Table S6, and illustrated in Figure 4. These genes are actively involved in the regulation of cell growth and proliferation.
Fig. 4.
The downstream target genes of the transcriptional factors p53, Rb, FoxM1, and Myc. The network displays the relationship between the four upstream regulators and their target molecules in PAH cells. The colors indicate the level of mRNA expression: upregulated genes are represented in red and downregulated genes in green. HPAH and IPAH displayed similar changes in RNA expression. The details of the genes in the diagram are described in Supplementary Table S6.
OTHER GENES RELATED TO THE PHYSIOLOGIC FUNCTIONS OF PAH
To further identify candidate genes related to PAH biology, we reviewed the available information for each gene as illustrated in Supplementary Table S2. IPA, PubMed, and other public available databases such as ExPASy (the SIB Bioinformatics Resource Portal) and DAVID were used in this analysis. The section lists additional examples of sizable expression changes among the 227 genes which may be associated with PAH.
ANLN (anillin), an actin binding protein increased in HPAH (2.356 ×, P = 0.009) and in IPAH (5.827 ×, P = 0.0006). Anillin is a scaffold protein that links RhoA, actin, and myosin during cytokinesis, which occurs during the cell cycle [Piekny and Glotzer, 2008].
The expression of ANGPT1 (angiopoietin 1) decreased in both HPAH and IPAH. Angiopoietins are proteins with important role in vascular development and angiogenesis. The expression of ANGPT1 decreased by about 50% in both HPAH and IPAH patients. Angiopoietin-1 has powerful vascular protective effects: suppressing plasma leakage, inhibiting vascular inflammation, and preventing cell death [Brindle et al., 2006]. Thus, the decreased expression of ANGPT1 in HPAH and IPAH HPASMC might be important in the vascular pathology of PAH.
DAPK1 (Death associated protein kinase 1) increased 3.1-fold (P = 0.0002) in HPAH and 1.53-fold (P = 0.002) in IPAH. DAPK1 is a Ca2+/calmodulin-regulated serine/threonine kinase that mediates cell death. Interestingly, DAPK3 has recently been shown to mediate vascular inflammation and development of hypertension in spontaneously hypertensive rats.
The expressions of a group of protocadherin (PCDHB16, PCDHGA12, PCDHGA2, PCDHGA3, PCDHGA6) decreased in both HPAH and IPAH cells. Protocadherin-12 deficiency leads to modifications in the structure and function of arteries in mice [Philibert et al., 2012].
SOD3 (superoxide dismutase 3) decreased 1.8-fold in HPAH and 1.6-fold in IPAH. SOD3 involved in superoxide radicals degradation. This suggests an increased level of oxide radicals in PAH HPASMC.
EXPRESSION OF THE BRADYKININ B2 AND ANGIOTENSIN AT1 RECEPTOR 1 mRNAs AS DETERMINED BY PCR
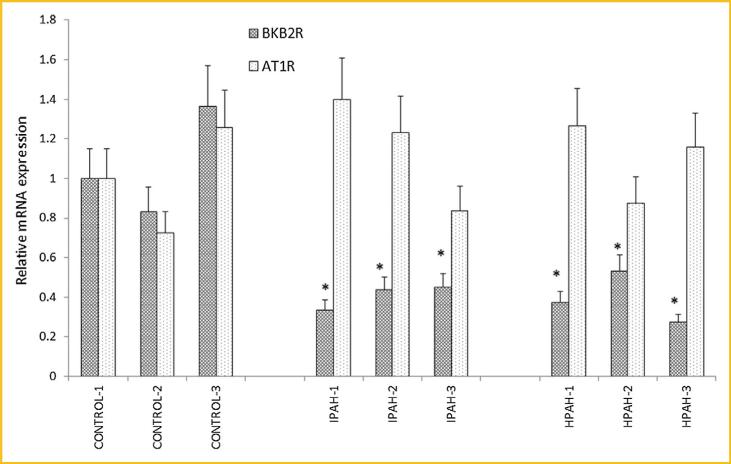
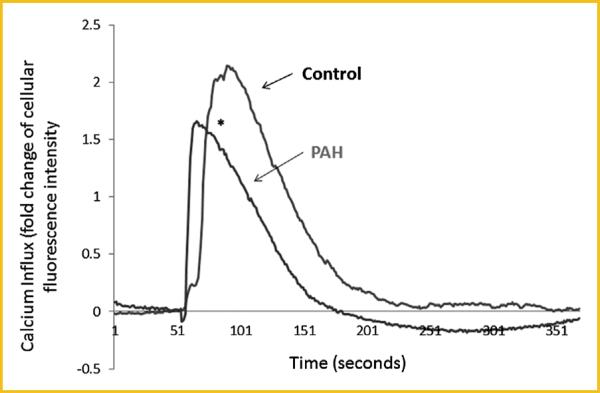
The microarray data demonstrated that the bradykinin B2 receptor mRNA expression decreased in both HPAH (61% of control, P = 0.009) and IPAH (59% of control, P = 0.005) HPASMC. We validated this result by real time PCR, using angiotensin type I receptor, which the microarray showed did not change as compared to control (Fig. 5), as a negative control for expression changes. We found significantly decreased expression of BKB2R in HPAH (38% of control, P = 0.015) and IPAH HPASMC (37% of control, P = 0.018) compared to normal control. The expression of the AT1 receptor was not altered (P > 0.05). Next, we determined bradykinin (100 nM) induced calcium influx. In the PAH cells, bradykinin induced a considerably weaker influx than in normal HPASMC (Fig. 6).
Fig. 5.
mRNA levels of bradykinin B2 receptor (BKB2R) and angiotensin II type I receptor (AT1R) in the normal control and PAH HPASMC. Results are presented as relative expression normalized to GAPDH and were calculated using the ΔΔCt method. The mRNA level from CONTROL-1 samples was set as 1. *P < 0.05 compared with the BKB2R expression level of CONTROL-1. #P < 0.05 compared with AT1R expression level from CONTROL-1 HPASMC.
Fig. 6.
Bradykinin induced Ca2+ mobilization in the HPASMC from normal control and PAH subjects. The bradykinin (100 nM) induced calcium mobilization were measured in the control, PAH HPASMC as described in Materials and Methods. *The maximal Ca2+ response was significantly weaker in the HPASMC from PAH subject compared with control cells (P < 0.05).
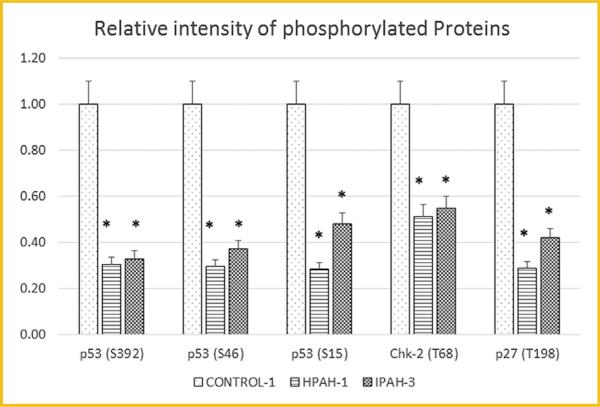
PHOSPHO-ANTIBODY ARRAY ANALYSIS OF CELL CYCLE REGULATING PROTEINS IN CONTROL AND PAH HPASMC
IPA predicted the decreased activity of the tumor suppressor gene p53 in HPAH, and IPAH HPASMC compared with control. We used an human phospho-antibody array to probe the phosphorylation of a panel of proteins, including p53. The phospho-protein array data were collected in one non-PAH, one HPAH, and one IPAH sample as demonstrated in Figure 7. The phosphorylation of all three sites, Serine392, Serine46, and Serine15 of p53 decreased in both HPAH and IPAH HPASMC compared with normal control. This fits with the prediction by IPA that the p53 activity is decreased in PAH samples. Other cell cycle related proteins also showed changes in phospho levels. For example, the phosphorylation of the CHK-2 (T68) and p27 (T198) were decreased in both HPAH and IPAH cells (Fig. 7). As a positive control, we found that the phospho-p38/MAPK level increased in both HPAH (3.9-fold) and IPAH (1.8-fold) HPASMC. The microarray data showed that MAPK13 (a p38 MAPK isoform), increased 1.84-fold (P = 0.0001) in HPAH and 1.86-fold (P = 0.0001) in IPAH.
Fig. 7.
Phosphorylation protein level of p53, chk-2, and p27 in control, HPAH, and IPAH HPASMC. The human phospho-antibody array detected phosphorylated proteins in cell lysates from one donor control, one HPAH, and one IPAH HPASMC. Phosphorylation profiles were created by quantifying the mean spot pixel densities normalized by positive control dots. Array signals from scanned X-ray film images were analyzed using image analysis software NIH ImageJ. The intensity of the dots were normalized by the positive control. The statistical analysis was performed using t-test. *P < 0.05 compared with control.
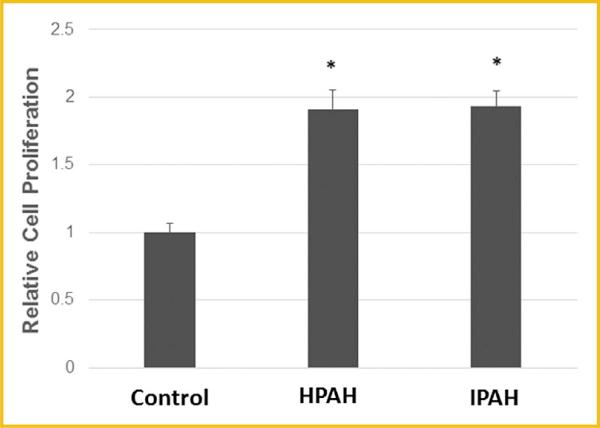
PROLIFERATION OF CONTROL AND PAH HPASMC
Function analysis predicted an enhanced proliferation in HPASMC derived from PAH patients. To confirm this prediction, cell growth of normal and PAH cells was examined in the absence of a growth stimulant under quiescent growth conditions. The cell proliferation experiments confirmed increased, dysregulated proliferation of both HPAH and IPAH cells while the normal control cell growth was negligible (Fig. 8).
Fig. 8.
Typical cell proliferation of PAH and control HPASMC in serum deprived medium. Cells were plated in 0.2% FBS medium on day 1. Cell number was determined at day 9. *P < 0.05 compared with control.
DISCUSSION
Statistical methods including t-test and FDR analysis were used to identify genes which are differentially expressed in HPASMC obtained from HPAH and IPAH patients compared to non-PAH controls. We were particularly interested in genes whose physiologic function relate to PAH symptoms and whose expression is changed similarly in the two types of PAH. Using the bioinformatics IPA tool and other available databases, we found altered biological functions and signaling pathways in the PAH cells. These pathways reflect alterations taking place in the physiology of PAH and are likely involved in the etiology and progress of the disease. Furthermore, our data suggest a change in the phosphorylation level of upstream transcription factors in PAH cells, which may promote the global transcriptional changes observed in the microarray. The array results provide an insight into the biology of PAH. Furthermore, the cell proliferation experiments with HPASMC obtained from transplanted lungs of PAH subjects confirm the development of dysregulated growth in the disease.
Our functional assay was based on the 227 commonly changed genes found in both HPAH and IPAH HPASMC. This approach simplified information processing and allowed identification of the common pathway changes in HPAH and IPAH populations. Also this approach allowed for investigation of the commonly shared pathways in HPAH and IPAH HPASMC. We used IPA core analysis to predict the biological functions and signal transduction pathways that are affected by these genes with altered expression. The IPA results point to the regulation of cell cycle progression and proliferation as a primary area of altered gene expression in the PAH cells. Related functions that were also altered include cancer and cell death and survival pathways.
Our results show that human polo-like kinase 1 (PLK1) increased 2.5-fold in HPAH and 6.4-fold in IPAH. Also, human polo-like kinase 4 (PLK4) increased 1.61-fold in HPAH and 3.55-fold in IPAH. PLK1 is essential during mitosis and in the maintenance of genomic stability. PLK1 is overexpressed in human tumors and has prognostic potential in cancer, indicating its involvement in carcinogenesis and its potential as a therapeutic target. Inhibitors of Plk1 activity target all rapidly dividing cells irrespective of being tumor cells or non-transformed normal proliferating cells [Cholewa et al., 2013]. To our knowledge, the role of polo-like kinases in pulmonary hypertension has not been investigated. PLK1 has been investigated as a target for cancer therapy for several years [Yim, 2013]. Inhibiting PLK1 action could provide an option for the treatment of pulmonary hypertension.
IPA analysis of our data suggests a reduction in activated of p53 and Rb in both HPAH and IPAH HPASMC. The phospho-protein array determinations confirmed decreased phospho-p53 level in both PAH HPASMC. p53 is an important tumor suppressor, it plays essential roles in cell cycle regulation and cancer development [Vousden and Prives, 2009]. The gene is also involved in atherosclerosis and has been shown to regulate apoptosis of the vascular smooth muscle cells under these disease conditions [Mercer and Bennett, 2006]. Furthermore, p53 has been shown to play a role in pulmonary hypertension. Specifically, p53 gene deficiency promotes hypoxia-induced pulmonary hypertension and vascular remodeling in mice [Mizuno et al., 2011]. Additionally, a recent publication showed that activating p53 prevents and reverses experimental pulmonary hypertension [Mouraret et al., 2013]. Interestingly, p53 appears to interact with polo like kinases, in that it represses the promoter of polo-like kinase 1. Polo-like kinase 1 inhibits the expression of p53 and its family members p63 and p73 [Cholewa et al., 2013]. A very recent proteomic study identified a novel association between Plk1 and p53 through heterogeneous ribonucleoprotein C1/C2. The communication illustrated that plk1 inhibition causes a marked increase in p53 protein expression and activation in melanoma cells [Cholewa et al., 2014].
IPA predicted increased activation of the transcription factors Myc and FoxM1. The two factors are involved in the promotion of cell cycle progression and proliferation. In pulmonary artery smooth muscle cells decreased c-myc expression has been associated with cell cycle arrest [Cantoni et al., 2013]. Increased c-myc level was related with HPASMC proliferation in hypoxia induced pulmonary hypertension via an ERK1/2/p27/c-fos/c-myc pathway [Li et al., 2012]. FOXM1 (Forkhead box M1) is a tumor-related transcription factor which stimulates cell proliferation and cell cycle progression by promoting the entry into S-phase and M-phase [Wierstra, 2013]. Foxm1 is essential for the development of lung vasculature [Kim et al., 2005]. It is critical for proliferation of smooth muscle cells. Depletion of Foxm1 in cultured SMC causes G2 arrest and a decreased number of cells undergoing mitosis [Ustiyan et al., 2009]. Foxm1-deficiency in vitro and in vivo is associated with reduced expression of cell cycle regulatory genes, including cyclin B1, Cdk1-activator Cdc25b phosphatase, polo-like 1 and JNK1 kinases, and cMyc transcription factor [Ustiyan et al., 2009]. Thus, our microarray data point to roles of the upstream factors p53, Rb, Myc, and FoxM1 in the dysregulated proliferation of PAH cells.
The prevailing changes in mRNA expression strongly suggest dysregulation of proliferation in the PAH cells as Supplementary Table S5 displays. These results are strengthened by the observation of increases in kinases and docking sites involved in cell cycle progression. In fact, the phospho-protein array experiment demonstrates the phosphorylation of check point protein CHK2 and cell cycle regulating protein p27 decreased in both HPAH and IPAH cells. CHK2 is a CAMK protein kinase of the Rad53 family. It is a cell cycle checkpoint regulator and putative tumor suppressor [Reinhardt and Yaffe, 2009]. DNA damage leads to the activation of CHK2 by phosphorylation, and the phosphor-CHK2 inhibits CDC25C phosphatase, preventing entry into mitosis, and has been shown to stabilize p53, leading to cell cycle arrest in G1 phase [Chehab et al., 2000]. The phosphorylation of p27 Kip (T198) was also found to be decreased in both HPAH and IPAH.p27Kip1 (cyclin-dependentkinase inhibitor 1B) binds to cyclin D to inhibit the catalytic activity of Cdk4, thus, preventing the phosphorylation of Rb. Thus, increased levels of p27 typically cause cells to arrest in the G1 phase of the cell cycle. Lowered levels of activated p27 leads to cell cycle progression [Blain, 2008]. The phosphorylation of p27Kip at T198 leads to the stabilization of p27 and growth arrest [De Vita et al., 2012]. We found the phosphop27Kip level was decreased in all PAH samples, suggesting an uncontrolled proliferation. Indeed our data on cell growth substantiates a dysregulated proliferation of both HPAH and IPAH cells. Together, these results point to the cancer paradigm of PAH vascular cells [Rai et al., 2008]. Interestingly, a very recent publication reports a case of complete remission of idiopathic pulmonary arterial hyper-tension following chemotherapy [Harbaum et al., 2014].
Paradoxically, the expression of PDE5A mRNA decreased in both HPAH and IPAH. This may indicate that some compensational mechanisms are taking place in the PAH smooth muscle cells to counter the disturbed homeopathic, diseased state. Phosphodiesterase type 5 A (PDE5A) selectively hydrolyzes cyclic GMP. To retain higher cGMP levels inhibitors of PDE5A such as sildenafil have been used for the treatment of cardiovascular disease. PDE5A plays an important role in the pulmonary vasculature where its inhibition appears to benefit patients with pulmonary hypertension [Kass et al., 2007].
Finally, the microarray results show interesting expression changes of vasoactive G-protein coupled receptors (GPCR). For example, the expression of the bradykinin B2 receptor (BKB2R) decreases in both HPAH and IPAH cells. Real time PCR confirmed this finding. Bradykinin is a nine amino acid peptide with vasodilatory, cardiovascular protective function [Heitsch, 2003; Veeravalli and Akula, 2004]. A BKB2R agonist (B9972) has been illustrated to reduce severe pulmonary hypertension and right ventricular hypertrophy in a rat model of PAH [Taraseviciene-Stewart et al., 2005]. Thus, the decreased expression of BKB2R is potentially an important factor in the pathogenesis of PAH. Another GPCR, endothelin-1 receptor B (ETB), mRNA expression also decreases in HPAH (12% of CONTROL) and IPAH (51% of CONTROL) HPASMC. Using radio-ligand binding assays we also found that the receptor number of ETB decreased in both HPAH and IPAH HPASMC [Yu et al., 2013]. Interestingly, while the ETB receptor displays some beneficial effects within the cardiovascular system [Schneider et al., 2007] limiting some of its signaling capacity improved the physiology of PAH in a hypoxic rat model [Green et al., 2013].
Together, the differential expression analysis, bioinformatics analysis, and biological validation highlight an aberrant proliferation and cell cycle regulation in the PAH affected HPASMC. These newly identified pathways such as polo-like kinases, may provide new targets for the treatment of both hereditary and idiopathic PAH.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH Grant number HL25776. The microarray experiments with Affymetrix Human Gene ST 1.0 chips were carried out at microarray core at Boston University Medical Center with Funds subsidized by Boston University Clinical and Translational Science Institute (CTSI grant # UL1 RR025771). We thank Dr. Suzy Comhair and Dr. Serpil Erzurum from the Cleveland Clinic Pathobiology Tissue Sample and Cell Culture Core (supported by NIH PO1 HL081064 and RC37 HL60917) and Dr. Marlene Rabinovitch, Stanford University under the Pulmonary Hyper-tension Breakthrough Initiative (PHBI, supported by the Cardiovascular Medical Research and Education Fund) for pulmonary smooth muscle cell samples. We also thank Dr. Michael Nagle from the Department of Neurology, Boston University School of Medicine for the constructive discussions and proofreading of the manuscript.
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of this article at the publisher's web-site.
REFERENCES
- Aldred MA, Comhair SA, Varella-Garcia M, Asosingh K, Xu W, Noon GP, Thistlethwaite PA, Tuder RM, Erzurum SC, Geraci MW, et al. Somatic chromosome abnormalities in the lungs of patients with pulmonary arterial hypertension. Am J Respir Crit Care Med. 2010;182:1153–1160. doi: 10.1164/rccm.201003-0491OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin ED, Menon S, Hemnes AR, Robinson LR, Talati M, Fox KL, Cogan JD, Hamid R, Hedges LK, Robbins I, et al. Idiopathic and heritable PAH perturb common molecular pathways, correlated with increased MSX1 expression. Pulm Circ. 2011;1:389–398. doi: 10.4103/2045-8932.87308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blain S. Switching cyclin D-Cdk4 kinase activity on and off. Cell Cycle. 2008;7:892–898. doi: 10.4161/cc.7.7.5637. [DOI] [PubMed] [Google Scholar]
- Bonapace IM, Latella L, Papait R, Nicassio F, Sacco A, Muto M, Crescenzi M, Di Fiore PP. Np95 is regulated by E1A during mitotic reactivation of terminally differentiated cells and is essential for S phase entry. J Cell Biol. 2002;157:909–914. doi: 10.1083/jcb.200201025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindle NPJ, Saharinen P, Alitalo K. Signaling and functions of angiopoietin-1 in vascular protection. Circ Res. 2006;98:1014–1023. doi: 10.1161/01.RES.0000218275.54089.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantoni S, Galletti M, Zambelli F, Valente S, Ponti F, Tassinari R, Pasquinelli G, Galiè N, Ventura C. Sodium butyrate inhibits platelet-derived growth factor-induced proliferation and migration in pulmonary artery smooth muscle cells through Akt inhibition. FEBS J. 2013;280:2042–2055. doi: 10.1111/febs.12227. [DOI] [PubMed] [Google Scholar]
- Chehab NH, Malikzay A, Appel M, Halazonetis TD. Chk2/hCds1 functions as a DNA damage checkpoint in G1 by stabilizing p53. Genes Dev. 2000;14:278–288. [PMC free article] [PubMed] [Google Scholar]
- Cholewa BD, Liu X, Ahmad N. The role of polo-like kinase 1 in carcinogenesis: cause or consequence. Cancer Res. 2013;73:6848–6855. doi: 10.1158/0008-5472.CAN-13-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholewa BD, Pellitteri-Hahn MC, Scarlett CO, Ahmad N. Large-scale label-free comparative proteomics analysis of polo-like kinase 1 inhibition via the small-molecule inhibitor BI 6727 (Volasertib) in BRAFV600E mutant melanoma cells. J Proteome Res. 2014 doi: 10.1021/pr5002516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vita F, Riccardi M, Malanga D, Scrima M, De Marco C, Viglietto G. PKC-dependent phosphorylation of p27 at T198 contributes to p27 stabilization and cell cycle arrest. Cell Cycle. 2012;11:1583–1592. doi: 10.4161/cc.20003. [DOI] [PubMed] [Google Scholar]
- Farber HW, Loscalzo J. Pulmonary arterial hypertension. New Engl J Med. 2004;351:1655–1665. doi: 10.1056/NEJMra035488. [DOI] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Gentleman R, Carey V, Bates D, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DS, Rupasinghe C, Warburton R, Wilson JL, Sallum CO, Taylor L, Yatawara A, Mierke D, Polgar P, Hill N. A cell permeable peptide targeting the intracellular loop 2 of endothelin B receptor reduces pulmonary hypertension in a hypoxic rat model. PLoS One. 2013;8:81309. doi: 10.1371/journal.pone.0081309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbaum L, Hennigs JK, Baumann HJ, Bokemeyer C, Olschewski H, Klose H. Complete resolution of idiopathic pulmonary arterial hypertension following chemotherapy. Eur Respir J. 2014;43:1513–1515. doi: 10.1183/09031936.00185713. [DOI] [PubMed] [Google Scholar]
- Heitsch H. The therapeutic potential of bradykinin B2 receptor agonists in the treatment of cardiovascular disease. Expert Opin Invest Drugs. 2003;12:759–770. doi: 10.1517/13543784.12.5.759. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protocols. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Humbert M, Morrell NW, Archer SL, Stenmark KR, MacLean MR, Lang IM, Christman BW, Weir EK, Eickelberg O, Voelkel NF, et al. Cellular and molecular pathobiology of pulmonary arterial hypertension. J Am Coll Cardiol. 2004;43:S13. doi: 10.1016/j.jacc.2004.02.029. [DOI] [PubMed] [Google Scholar]
- Humbert M, Sitbon O, Chaouat A, Bertocchi M, Habib G, Gressin V, Yaïci A, Weitzenblum E, Cordier J-F, Chabot F, et al. Survival in patients with idiopathic, familial, and anorexigen-associated pulmonary arterial hypertension in the modern management era. Circulation. 2010;122:156–163. doi: 10.1161/CIRCULATIONAHA.109.911818. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Kass DA, Champion HC, Beavo JA. Phosphodiesterase type 5: expanding roles in cardiovascular regulation. Circ Res. 2007;101:1084–1095. doi: 10.1161/CIRCRESAHA.107.162511. [DOI] [PubMed] [Google Scholar]
- Kim I-M, Ramakrishna S, Gusarova GA, Yoder HM, Costa RH, Kalinichenko VV. The forkhead box M1 transcription factor is essential for embryonic development of pulmonary vasculature. J Biol Chem. 2005;280:22278–22286. doi: 10.1074/jbc.M500936200. [DOI] [PubMed] [Google Scholar]
- Li X-W, Hu C-P, Wu W-H, Zhang W-F, Zou X-Z, Li Y-J. Inhibitory effect of calcitonin gene-related peptide on hypoxia-induced rat pulmonary artery smooth muscle cells proliferation: Role of ERK1/2 and p27. Eur J Pharm. 2012;679:117–126. doi: 10.1016/j.ejphar.2012.01.015. [DOI] [PubMed] [Google Scholar]
- Li M, Wang X, Li W, Li F, Yang H, Wang H, Brunicardi FC, Chen C, Yao Q, Fisher WE. Somatostatin receptor-1 induces cell cycle arrest and inhibits tumor growth in pancreatic cancer. Cancer Sci. 2008;99:2218–2223. doi: 10.1111/j.1349-7006.2008.00940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez I, DiMaio D. B-Myb, cancer, senescence, and MicroRNAs. Cancer Res. 2011;71:5370–5373. doi: 10.1158/0008-5472.CAN-11-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer J, Bennett M. The role of p53 in atherosclerosis. Cell Cycle. 2006;5:1907. doi: 10.4161/cc.5.17.3166. [DOI] [PubMed] [Google Scholar]
- Mizuno S, Bogaard HJ, Kraskauskas D, Alhussaini A, Gomez-Arroyo J, Voelkel NF, Ishizaki T. p53 Gene deficiency promotes hypoxia-induced pulmonary hypertension and vascular remodeling in mice. AJP–Lung Cell Mol Physiol. 2011;300 doi: 10.1152/ajplung.00286.2010. [DOI] [PubMed] [Google Scholar]
- Mouraret N, Marcos E, Abid S, Gary-Bobo G, Saker M, Houssaini A, Dubois-Rande J-L, Boyer L, Boczkowski J, Derumeaux G, et al. Activation of lung p53 by nutlin-3a prevents and reverses experimental pulmonary hypertension. Circulation. 2013;127:1664–1676. doi: 10.1161/CIRCULATIONAHA.113.002434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert C, Bouillot S, Huber P, Faury G. Protocadherin-12 deficiency leads to modifications in the structure and function of arteries in mice. Pathol Biol. 2012;60:34–40. doi: 10.1016/j.patbio.2011.11.005. [DOI] [PubMed] [Google Scholar]
- Piekny AJ, Glotzer M. Anillin is a scaffold protein that links rhoa, actin, and myosin during cytokinesis. Curr Biol. 2008;18:30–36. doi: 10.1016/j.cub.2007.11.068. [DOI] [PubMed] [Google Scholar]
- Prado GN, Mierke DF, Pellegrini M, Taylor L, Polgar P. Motif mutation of bradykinin B2 receptor second intracellular loop and proximal C terminus is critical for signal transduction, internalization, and resensitization. J Biol Chem. 1998;273:33548–33555. doi: 10.1074/jbc.273.50.33548. [DOI] [PubMed] [Google Scholar]
- Rai PR, Cool CD, King JAC, Stevens T, Burns N, Winn RA, Kasper M, Voelkel NF. The cancer paradigm of severe pulmonary arterial hypertension. Am J Respir Criti Care Med. 2008;178:558–564. doi: 10.1164/rccm.200709-1369PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt HC, Yaffe MB. Kinases that control the cell cycle in response to DNA damage: Chk1, Chk2, and MK2. Curr Opin Cell Biol. 2009;21:245–255. doi: 10.1016/j.ceb.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydell-Törmänen K, Risse P-A, Kanabar V, Bagchi R, Czubryt MP, Johnson JR. Smooth muscle in tissue remodeling and hyper-reactivity: airways and arteries. Pulm Pharmacol Therapeut. 2013;26:13–23. doi: 10.1016/j.pupt.2012.04.003. [DOI] [PubMed] [Google Scholar]
- Schneider MP, Boesen EI, Pollock DM. Contrasting actions of endothelin ETA and ETB receptors in cardiovascular disease. Ann Rev Pharm Toxicol. 2007;47:731–759. doi: 10.1146/annurev.pharmtox.47.120505.105134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraseviciene-Stewart L, Scerbavicius R, Stewart JM, Gera L, Demura Y, Cool C, Kasper M, Voelkel NF. Treatment of severe pulmonary hypertension: a bradykinin receptor 2 agonist B9972 causes reduction of pulmonary artery pressure and right ventricular hypertrophy. Peptides. 2005;26:1292–1300. doi: 10.1016/j.peptides.2005.03.050. [DOI] [PubMed] [Google Scholar]
- Ustiyan V, Wang IC, Ren X, Zhang Y, Snyder J, Xu Y, Wert SE, Lessard JL, Kalin TV, Kalinichenko VV. Forkhead box M1 transcriptional factor is required for smooth muscle cells during embryonic development of blood vessels and esophagus. Dev Biol. 2009;336:266–279. doi: 10.1016/j.ydbio.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeravalli KK, Akula A. Involvement of nitric oxide and prostaglandin pathways in the cardioprotective actions of bradykinin in rats with experimental myocardial infarction. Pharmacol Res. 2004;49:23–29. doi: 10.1016/j.phrs.2003.07.010. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Wierstra I. Chapter six – FOXM1 (Forkhead box M1) in tumorigenesis: overexpression in human cancer, implication in tumorigenesis, oncogenic functions, tumor-suppressive properties, and target of anticancer therapy. In: Kenneth DT, Paul BF, editors. Advances in Cancer Research. Academic Press; 2013. pp. 191–419. [DOI] [PubMed] [Google Scholar]
- Wilson JL, Taylor L, Polgar P. Endothelin-1 activation of ETB receptors leads to a reduced cellular proliferative rate and an increased cellular footprint. Exp Cell Res. 2012;318:1125–1133. doi: 10.1016/j.yexcr.2012.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim H. Current clinical trials with polo-like kinase 1 inhibitors in solid tumors. Anti-cancer Drugs. 2013;24:999–1006. doi: 10.1097/CAD.0000000000000007. [DOI] [PubMed] [Google Scholar]
- Yu J, Taylor L, Wilson J, Comhair S, Erzurum S, Polgar P. Altered expression and signal transduction of endothelin-1 receptors in heritable and idiopathic pulmonary arterial hypertension. J Cell Physiol. 2013;228:322–329. doi: 10.1002/jcp.24132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.