Abstract
Background
“Evolution Canyon” (ECI) at Lower Nahal Oren, Mount Carmel, Israel, is an optimal natural microscale model for unraveling evolution in action highlighting the basic evolutionary processes of adaptation and speciation. A major model organism in ECI is wild emmer, Triticum dicoccoides, the progenitor of cultivated wheat, which displays dramatic interslope adaptive and speciational divergence on the tropical-xeric “African” slope (AS) and the temperate-mesic “European” slope (ES), separated on average by 250 m.
Methods
We examined 278 single sequence repeats (SSRs) and the phenotype diversity of the resistance to powdery mildew between the opposite slopes. Furthermore, 18 phenotypes on the AS and 20 phenotypes on the ES, were inoculated by both Bgt E09 and a mixture of powdery mildew races.
Results
In the experiment of genetic diversity, very little polymorphism was identified intra-slope in the accessions from both the AS or ES. By contrast, 148 pairs of SSR primers (53.23%) amplified polymorphic products between the phenotypes of AS and ES. There are some differences between the two wild emmer wheat genomes and the inter-slope SSR polymorphic products between genome A and B. Interestingly, all wild emmer types growing on the south-facing slope (SFS=AS) were susceptible to a composite of Blumeria graminis, while the ones growing on the north-facing slope (NFS=ES) were highly resistant to Blumeria graminis at both seedling and adult stages.
Conclusion/Significance
Remarkable inter-slope evolutionary divergent processes occur in wild emmer wheat, T. dicoccoides at EC I, despite the shot average distance of 250 meters. The AS, a dry and hot slope, did not develop resistance to powdery mildew, whereas the ES, a cool and humid slope, did develop resistance since the disease stress was strong there. This is a remarkable demonstration in host-pathogen interaction on how resistance develops when stress causes an adaptive result at a micro-scale distance.
Introduction
The "Evolution Canyon" (EC I) model is the subject of a long-term research program that started in 1990 at Lower Nahal Oren, Mount Carmel, Israel. EC I is an optimal natural micro-scale model for unraveling evolution in action highlighting the basic evolutionary processes of adaptation and speciation [1] (Fig 1 and Fig 2). It was expanded in three additional evolution canyons: EC II at Lower Nahal Keziv, westerm Upper Galilee; EC III at Nahal Shaharut, southern Negev Desert, and EC IV Mezar, southern Golan Heights) [2]. EC I, at Mount Carmel, studied here, (32°43′N; 34°58′E), consists of Upper Cenomanian limestones. Two thpusands and five hundred species have been identified in EC I, from bacteria to mammals in an area of 7,000 m. Solar radiation (up to 800% more) on the "African" (AS) south-facing slope (SFS) causes its tropical microclimate contrasting the "European" north-facing slope[2] (Fig 2). Higher terrestrial species richness was found on the more stressful tropical “African” slope (AS). Aquatic species richness was higher on the milder ES. By analyzing the genetic diversity between the AS and ES at Mount Carmel, Nevo et al [3] draw the following conclusions. First, microclimatic selection is the major evolutionary inter-slope, fast-acting, diverging force on genotypes and phenotypes, over-riding migration and genetic drift. Secondly, ecological stress can generate global-scale, adaptive evolutionary genome and phenome strategies at micro- and macro-scales, reinforcing homeostasis and fitness, and suggesting continuity between micro-evolution and macro-evolution.
Fig 1. The “Evolution Canyon” I at Lower Nahal Oren, Mount Carmel, Isarel.

Note the "African" savannah AS = SFS slope versus the forested ES = NFS slope. Higher terrestrial species richness occurs on the more stressful tropical AS. Aquatic species richness is higher on the milder, temperate cool and humid “European” slope (ES).
Fig 2. The EC model.
A) Schematic diagram, B) Cross section view of "Evolution Canyon" I (EC I), Lower Nahal Oren, Mount Carmel, and C) Air view of EC I. Note the distinct divergent plant formations on the opposite slopes. The green, lush, European, temperate, cool-mesic, ES = NFS, sharply contrasts with the open park forest, warm-xeric, tropical, “African-Asian” savanna on the AS = SFS. In EC I, as in other three "Evolution Canyons" studied in Israel, seven sampling stations are designated: three on the AS = SFS (nos. 1–3), one at the valley bottom (no. 4) and three on the NFS (nos. 5–7). Source: Reference 2.
Local microcosmic natural laboratories are dubbed “Evolution Canyon” (EC) models, where genomic, proteomic, and phenomic studies were carried out and focused on speciation and adaptation at a micro-scale. Speciation and adaptation are two important processes in studying evolution [2]. Viruses, bacteria, fungi, plants, and animals have been studied at the micro-scale involving biodiversity divergence, adaptation, and incipient adaptive ecological sympatric speciation to reveal evolution in action. Extensive studies have been conducted in "Evolution Canyon" in wild barley and other model organisms listed in Nevo "Evolution Canyon" publications at http://evolution.haifa.ac.il and in references [1–17]. These studies displayed dramatic interslope adaptive genomic divergence, slope-specific fitness components, and incipient adaptive ecological sympatric speciation on the opposite slopes.
The grass family Gramineae evolved 50–70 million years ago (Mya) [18, 19] and the sub-family Pooideae including wheat, barley, and oats had diverged around 20 Mya [20]. Wild diploid wheat (T. urartu, 2n = 2X = 14, AA) hybridized with goat grass (Aegilops speltoides, 2n = 2X = 14, BB) 300,000–500,000 BP to produce wild emmer wheat (T. dicoccoides, 2n = 2X = 28, AABB) [21]. Wild emmer occurs in Israel, Syria, Lebanon, southeast Turkey, Jordan, northern Iraq, and western Iran. Natural populations of wild emmer have wide genotypic variations in agronomic, amino acid composition, protein quality and quantity, micronutrient contents, abiotic stress tolerances, herbicide resistances, and biotic stress tolerances [22–24].
Powdery mildew, caused by Blumeria graminis f. sp. tritici (Bgt), is one of the devastating wheat diseases in areas with temperate climates. To date, several powdery mildew resistant genes (Pm16, Pm26, Pm30, Pm36, MlZec 1, PmG3M, PmG16, Pm41, and Pm42) are mapped using molecular markers in wild emmer [25–34].
Here we exhibited the patterns of 278 nuclear microsatellites and phenotypes on resistance to powdery mildew of wild emmer in “Evolution Canyon” (ECI), Lower Nahal Oren, Mt. Carmel, Israel. SSR sequences can serve functional roles as regulatory elements [35–40]. We revealed adaptive inter-slope divergence of the phenotype on resistance to powdery mildew between the tropical-xeric “African” slope and the temperate-mesic “European” slope.
Materials and Methods
Plant Materials
A total of 38 accessions of wild emmer were obtained from Eviatar Nevo from two stations (Stations 2 and 7) at the “Evolution Canyon” 1 (EC 1) in Mount Carmel. We can confirm that the field studies did not involve endangered or protected species. Israel Nature Reserve Authority responsible for Evolution Canyon. Israel Nature Reserve Authority issued permission for our study. Station 2 is on the south-facing slope, SFS, or “African” slope, AS, and station 7 is on the north-facing slope, NFS, or “European” slope, ES (Fig 2). The AS is richer in species of drought-tolerant taxa in the northern canyons, and the ES is richer in “humid” taxa, reflecting locally global patterns [1].
Evaluation for Powdery Mildew Resistance
Bgt E09, a prevailing powdery mildew pathotype in China, virulent to Pm1, Pm 3a, Pm3b, Pm3c, Pm3d, Pm3e, Pm3f, Pm5, Pm6, Pm7, Pm8, Pm17, and Pm19 [34], was used to inoculate 38 accessions (16 plants in each accession) at the seedling stage under controlled biological incubator conditions, 70% humidity and 18–20°C, in the winter of 2013. Huixianhong, a kind of highly susceptible common wheat, was used for the inoculation of seedlings. Disease reactions were recorded 9–13 days after inoculation, and infection types were scored on a 0–4 scale, with 0 representing no visible symptoms, 0; for necrotic flecks and 1, 2, 3, 4 for highly resistant, resistant, susceptible and highly susceptible reactions, respectively [27]. Phenotypes were pooled into two groups, resistant (R, IT = 0–2) and susceptible (S, IT = 3, 4). Furthermore, a mixture of powdery mildew races, collected in the experimental fields, was also used to inoculate 38 accessions under controlled greenhouse conditions at the seedling and adult stages. A randomized complete block design with three replications consisting of 1 m rows, with 20 plants at 5 cm spacing, 30 cm apart in greenhouse was used in this study.
Genomic DNA Extraction and SSR Analysis
Total DNA was extracted from the healthy leaves of 38 accessions following the method of cetytrimethylammonium bromide (CTAB) [41] and there were also minor changes.
SSR markers mapped on the A and B genomes of hexaploid wheat ([42, 43]; http://www.graingenes.gov) were chosen for analysis of polymorphism between the wild emmer wheat genotypes from the AS and ES. We selected 278 SSR markers for testing (about 20 markers for each chromosome on average). 15μl volumes were taken in all PCR reactions containing 10mM Tris-Hcl, pH 8.3, 50mM Kcl, 1.5mM Mgcl2, 0.2mM dNTPs, 25 ng of each primer, 50-100ng genomic DNA and 0.75U Taq DNA polymerase. The amplification was performed at 94°C for 3 min, followed by 35 cycles at 94°C for 30s, at 55–62°C (depending on specific primers) for 30s, and at 72°C for 30s, with a final extension at 72°C for 10 min. SSR markers were performed on 8% non-denaturing polyacrylamide gels (39 acrylamide: 1 bisacrylamide). Gels were silver strained and photographed.
Results and Discussion
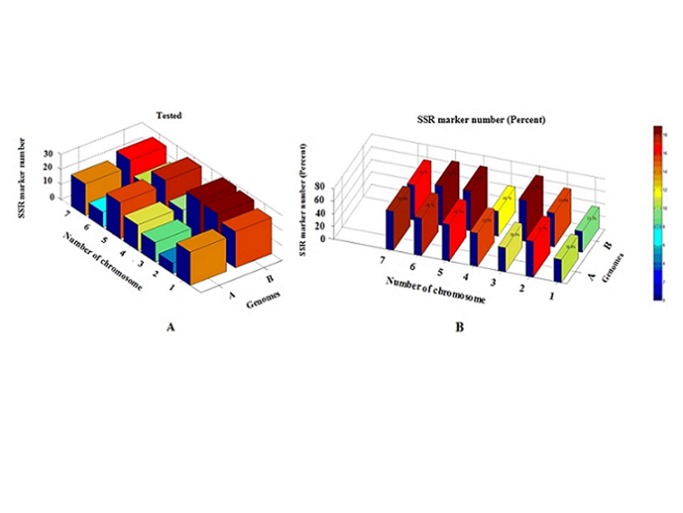
A set of 278 wheat SSR markers located separately on 14 chromosomes were used to analyze the genetic diversity between the wild emmer wheat genotypes from the AS and ES (Table 1). Out of these, 114 SSR markers belong to genome A (1A-7A), and the other 164 SSR markers were distributed on genome B (1B-7B) (Table 1, Fig 3A). Interestingly, very little polymorphism was identified on the AS and ES intra-slope. Consequently, one phenotype from each slope, i.e. AS4 (from station 2; For AS4 genotype, total 127 pairs of SSR primers were screened, no polymorphic products were amplified on the AS intra-slope.) and a second ES7 (from station 7; Total 127 pairs of SSR primers were screened, for ES7 genotype, 2 of them amplified polymorphic products on the ES intra-slope.), were selected to represent AS genotypes and ES genotypes, respectively. Overall, 148 pairs of SSR primers (53.23%) amplified polymorphic products between the AS and ES (Table 1). We also used MATLAB 8.0 and Statistics Toolbox 8.1 software to illustrate the frequency distribution of genome A and B. The frequency polymorphism of SSR markers between AS and ES were similar on the genomes A and B, which are around 50% (Table 1, Fig 3B). This illustration amplify the differences between the two wild emmer wheat genomes and the inter-slope SSR polymorphic products between genome A and B. SSR sequences can serve functional roles as regulatory elements [35–40].
Table 1. Polymorphism investigated by SSR markers between AS and ES on 14 chromosomes.
| SSR markers | SSR markers | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Chromosome | Tested | Polymorphism between AS and ES | Chromosome | Tested | Polymorphism between AS and ES | ||||
| N | N | % | N | N | % | ||||
| 1A | 22 | 8 | 36.4 | 1B | 24 | 8 | 33.3 | ||
| 2A | 7 | 4 | 57.1 | 2B | 28 | 15 | 53.6 | ||
| 3A | 13 | 5 | 38.5 | 3B | 29 | 19 | 65.5 | ||
| 4A | 17 | 9 | 52.9 | 4B | 15 | 6 | 40 | ||
| 5A | 24 | 14 | 58.3 | 5B | 27 | 17 | 63 | ||
| 6A | 10 | 6 | 60 | 6B | 16 | 10 | 62.5 | ||
| 7A | 21 | 13 | 61.9 | 7B | 25 | 14 | 56 | ||
| Total | 114 | 59 | 51.2 | Total | 164 | 89 | 54.3 | ||
| Mean | 16 | 8 | 50 | Mean | 23 | 13 | 56.5 | ||
Fig 3. Interslope SSR polymorphism divergence in the A and B genomes of wild emmer wheat Triticum dicoccoides.
A) Tested SSR markers between AS and ES on genomes A and B, B) SSR frequency polymorphism between AS and ES on the two genomes. Red color indicates high frequency and blue color low frequency.
Remarkably, biodiversity evolution including functional ecological genomics diverges drastically on the abutting slopes across life from bacteria to mammals in biodiversity, adaptations and speciation patterns [1–17]. Microsites divergent sharply ecologically abiotically (by climate, rocks, soils, or chemicals), or biotically (parasites and pathogens) are optimal for identifying genetic resources for crop improvement.
The reactions of the 38 accessions to Bgt E09 are listed in Table 2. Remarkably, there were 18 AS genotypes susceptible to powdery mildew isolates of Bgt E09, while another 20 of ES genotypes were highly resistant at the seedling stage under the same controlled biological incubator conditions (Table 2). The same results were obtained under the controlled greenhouse conditions using a mixture of powdery mildew races at both seedling and adult stages. This is the first research on the phenotype and genotype adaptability of the resistance to powdery mildew of wild emmer between the abutting “African” slope and “European” slope in EC I. We have shown inter-slope adaptive and speciational patterns at ECII across phylogeny from bacteria, through soil fungi, plants and animals including mammals (Nevo list of "Evolution Canyon" publications at http://evolution.haifa.ac.il, including studies on wild emmer wheat and wild barley [1–17]). Our study on genetic diversity and the phenotype/genotype adaptability of the resistance to powdery mildew of wild emmer wheat locally at EC followed our regional analysis across Israel [44]. These regional and local studies are complementary. The uniqueness of the current study is its unfolding resistance locally across very close abutting slopes, separate, on average, by only 250 meters. Future studies could map the resistant genes, clone and transform them to cultivated crops thereby increasing world food production. We suggest that the EC microscale model offers an optimal opportunity to study the influence of microclimatic ecological divergence on the population genetics and genome evolution of natural populations. These studies could substantially support crop improvement and food production in a world whose population will reach 10 billions in 2050.
Table 2. The reactions of the 38 genotypes to Bgt E09, a prevailing powdery mildew pathotype in the Beijing area.
| Accessions | Plant reaction a | Genotypes | Plant reaction a |
|---|---|---|---|
| ES1 | R | AS1 | S |
| ES2 | R | AS3 | S |
| ES3 | R | AS4 | S |
| ES4 | R | AS5 | S |
| ES5 | R | AS6 | S |
| ES6 | R | AS7 | S |
| ES7 | R | AS8 | S |
| ES8 | R | AS9 | S |
| ES9 | R | AS10 | S |
| ES10 | R | AS11 | S |
| ES11 | R | AS12 | S |
| ES12 | R | AS13 | S |
| ES13 | R | AS14 | S |
| ES14 | R | AS15 | S |
| ES15 | R | AS16 | S |
| ES16 | R | AS17 | S |
| ES17 | R | AS18 | S |
| ES18 | R | AS19 | S |
| ES19 | R | ||
| ES20 | R |
a Plant reaction: R resistant (IT = 0–2), S susceptible (IT = 3–4)
Acknowledgments
We thank Avigdor Beiles and Robin Permut for their comments on the manuscript scientifically and linguistically, respectively.
Data Availability
All relevant data are within the paper.
Funding Statement
This study was funded by Ancell-Teicher Research Foundation of Genetics and Molecular Evolution, National Natural Science Foundation of China (NSFC) (31471488), 973 developmental program (2014CB138100) and Advanced Seed Engineering Project of Shandong Province provided financial support. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Nevo E (1995) Asian, African and European biota meet at "Evolution Canyon", Israel: local tests of global biodiversity and genetic diversity patterns. Proc Roy Soc Lond B 262: 149–155. [Google Scholar]
- 2. Nevo E (2012) “Evolution Canyon,” a potential microscale monitor of global warming across life. Proc Natl Acad Sci USA 109: 2960–2965. 10.1073/pnas.1120633109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nevo E, Lu Z, Pavliček T (2006) Global evolutionary strategies across life caused by shared ecological stress: fact or fancy? Isr J Plant Sci 54: 1–8. [Google Scholar]
- 4. Nevo E, Apelbaum-Elkaher I, Garty J, Beiles A (1997) Natural selection causes microscale allozyme diversity in wild barley and a lichen at "Evolution Canyon" Mt. Carmel, Israel. Heredity 78: 373–382. [Google Scholar]
- 5. Owuor ED, Fahima T, Beiles A, Korol A, Nevo E (1997) DNA-RAPD diversity caused by microclimatic ecological stress in wild barley, Hordeum spontaneum . Mol Ecol 6:1177–1187. [Google Scholar]
- 6. Kalendar RJ, Tanskanen S, Immonen S, Nevo E, Schulman AH (2000) Genome evolution of wild barley (Hordeum spontaneum) by Bare-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc Natl Acad Sci USA 97: 6603–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nevo E (2001) Evolution of genome-phenome diversity under environmental stress. Natl Acad Sci USA 98:6233–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nevo E (2011) Selection overrules gene flow at "Evolution Canyons", Israel In: Advances in Genetics Research Vol 5 Urbano K.V. (ed) chapter 2. Nova Science Publishers Inc; pp 67–89. [Google Scholar]
- 9. Li YC, Krugman T, Fahima T, Beiles A, Nevo E (1998) Genetic diversity of alcohol dehydrogenase 3 in wild barley population at the "Evolution Canyon" microsite, Nahal Oren, Mt. Carmel, Israel. Barley Genetics Newsletter 28: 58–60. [Google Scholar]
- 10. Gupta PK, Sharma PK, Balyan HS, Roy JK, Sharma S, et al. (2002) Polymorphism at rDNA loci in barley and its relation with climatic variables. Theor Appl Genet 104: 473–481. [DOI] [PubMed] [Google Scholar]
- 11. Nevo E (2004) Evolution of genome dynamics under ecological stress In: Parisi V., De Fonzo V. and Alluffi-Pentini F. (Eds). Dynamical Genetics 81: 7736–231–3. [Google Scholar]
- 12. Nevo E, Beharav A, Meyer RC, Hackett CA, Forster BP, et al. (2005) Genomic microsatellite adaptive divergence of wild barley by microclimatic stress in “Evolution Canyon”, Israel. Biol J Linn Soc 84: 205–224. [Google Scholar]
- 13. Cronin JK, Bundock PC, Henry RJ, Nevo E (2007) Adaptive climatic molecular evolution in wild barley at the lsa defense locus. Proc. Natl Acad Sci USA 104: 2773–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nevo E (2006) “Evolution Canyon": a microcosm of life's evolution focusing on adaptation and speciation. Isr J Ecol Evol 52: 501–506. [Google Scholar]
- 15. Nevo E (2009) Evolution in action across life at "Evolution Canyon", Israel. Trends in Evol Biol pp. e3. [Google Scholar]
- 16. Yang Z, Zhang T, Bolshoy A, Beharav A, Nevo E (2009) Adaptive microclimatic structural and expressional dehydrin 1 evolution in wild barley, Hordeum spontaneum, at "Evolution Canyon", Mount Carmel, Israel. Mol Ecol 18:2063–2075. 10.1111/j.1365-294X.2009.04140.x [DOI] [PubMed] [Google Scholar]
- 17. Nevo E (2014) Evolution in action: adaptation and incipient sympatric speciation with gene flow across life at "Evolution Canyon", Israel. Israel Journal of Ecology and Evolution (in press). [Google Scholar]
- 18. Kellogg EA (2001) Evolutionary history of the grasses. Plant Physiol 125: 1198–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang S, Sirikhachornkit A, Su X, Faris J, Gill B, et al. (2002) Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. P Natl Acad Sci USA 99: 8133–8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Inda LA, Segarra-Moragues JG, Müller J, Peterson PM, Catalán P (2008) Dated historical biogeography of the temperate Loliinae (Poaceae, Pooideae) grasses in the northern and southern hemispheres. Mol Phylogenet Evol 46: 932–957. 10.1016/j.ympev.2007.11.022 [DOI] [PubMed] [Google Scholar]
- 21. Dvorak J, Akhunov ED (2005) Tempos of gene locus deletions and duplications and their relationship to recombination rate during diploid and polyploid evolution in the Aegilops-Triticum alliance. Genetics 171: 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nevo E, Korol AB, Beiles A, Fahima T (2002) Evolution of wild emmer and wheat improvement Population Genetics, Genetic resources, and genome organization of wheat's progenitor, Triticum dicoccoides. Springer; pp. 364. [Google Scholar]
- 23. Xie W, Nevo E (2008) Wild emmer: genetic resources, gene mapping and potential for wheat improvement. Euphytica 164: 603–614. [Google Scholar]
- 24. Nevo E (2014) Evolution of wild emmer wheat and crop improvement. J Syst Evol 52: 673–696. [Google Scholar]
- 25. Rong JK, Millet E, Manisterski J, Feldman M (2000) A new powdery mildew resistance gene: Introgression from wild emmer into common wheat and RFLP-based mapping. Euphytica 115:121–126. [Google Scholar]
- 26. Reader Jk, Miller TE (1991) The introduction into bread wheat of a major gene for resistance of powdery mildew from wild emmer wheat. Euphytica 53: 57–60. [Google Scholar]
- 27. Liu ZY, Sun QX, Ni ZF, Nevo E, Yang TM (2002) Molecular characterization of a novel powdery mildew resistance gene Pm30 in wheat originating from wild emmer. Euphytica 123:21–29. [Google Scholar]
- 28. Mohler V, Zeller FJ, Wenzel G, Hsam SLK (2005) Chromosomal location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L. em Thell.). 9. Gene MlZec1from the Triticum dicoccoides-derived wheat line Zecoi-1. Euphytica 142:161–167. [Google Scholar]
- 29. Chen XM, Luo YH, Xia XC, Xia LQ, Chen X, et al. (2005) Chromosomal location of powdery mildew resistance gene Pm16in wheat using SSR marker analysis. Plant Breeding 124:225–228. [Google Scholar]
- 30.McIntosh RA, Devos KM, Dubcovsky J, Rogers WJ, Morris CF, et al. (2007) Catalogue of gene symbols for wheat: 2007 supplement. http://wheat.pw.usda.gov/ggpages/awn/53/Textfile/WGC.html. Accessed 13 Dec 2007.
- 31. Xie WL, Ben-David R, Zeng B, Distelfeld A, Röder MS, et al. (2012) Identification and characterization of a novel powdery mildew resistance gene PmG3M derived from wild emmer wheat, Triticum dicoccoides . Theor Appl Genet 124: 911–22. 10.1007/s00122-011-1756-8 [DOI] [PubMed] [Google Scholar]
- 32. Ben-David R, Xie WL, Peleg Z, Saranga YS, Dinoor A, et al. (2010) Identification and mapping of PmG16, a powdery mildew resistance gene derived from wild emmer wheat. Theor Appl Genet 121: 499–510. 10.1007/s00122-010-1326-5 [DOI] [PubMed] [Google Scholar]
- 33. Li G, Fang T, Zhang H, Xie C, Li H, et al. (2009) Molecular identification of a new powdery mildew resistance gene Pm41 on chromosome 3BL derived from wild emmer (Triticum turgidum var. dicoccoides). Theor Appl Genet 119: 531–539. 10.1007/s00122-009-1061-y [DOI] [PubMed] [Google Scholar]
- 34. Hua W, Liu Z, Zhu J, Xie C, Yang T, et al. (2009) Identification and genetic mapping of pm42, a new recessive wheat powdery mildew resistance gene derived from wild emmer (Triticum turgidum var. dicoccoides). Theor Appl Genet 119:223–230. 10.1007/s00122-009-1031-4 [DOI] [PubMed] [Google Scholar]
- 35. Karlin S, Campbell AM, Mrazek J (1998) Comparative DNA analysis across diverse genomes. Annu Rev Genet 32: 185–225. [DOI] [PubMed] [Google Scholar]
- 36. Kashi Y, Soller M (1999) Functional roles of microsatellites and minisatellites In: Goldstein DB, Schlotterer C (eds) Microsatellites: Evolution and Application, Oxford University Press: Oxford: pp 10–23. [Google Scholar]
- 37. King AG, Soller M (1999) Variation and fidelity: the evolution of simple sequence repeats as functional elements in adjustable genes In: Wasser SP (ed) Evolutionary Theory and Processes: modern perspective, papers in honor of Eviatar Nevo, Kluwer Academic Publishers: The Netherlands: pp 65–85. [Google Scholar]
- 38. Li YC, Fahima T, Röder MS, Kirzhner VM, Beiles A, et al. (2003) Genetic effects on microsatellite diversity in wild emmer wheat (Triticum dicoccoides) at the Yehudiyya microsite, Israel. Heredity 90:150–156. [DOI] [PubMed] [Google Scholar]
- 39. Li YC, Korol AB, Fahima T, Beiles A, Nevo E (2002) Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol Ecol 11:2453–2465. [DOI] [PubMed] [Google Scholar]
- 40. Li YC, Korol AB, Fahima T, Nevo E (2004) Microsatellites within genes: structure, function and evolution. Mol Biol Evol 21:991–1007. [DOI] [PubMed] [Google Scholar]
- 41. Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. P Natl Acad Sci USA 81: 8014–8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier MH, et al. (1998) A microsatellite map of wheat. Genetics 149: 2007–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ganal MW, Röder MS (2007) Microsatellite and SNP markers in wheat breeding: Genomics-assisted crop improvement. Springer; Netherlands: pp. 1–24. [Google Scholar]
- 44. Moseman JG, Nevo E, Morshidy MAEL, Zohary D (1984) Resistance of Triticum dicoccoides to infection with Erysiphe graminis tritici. Euphytica 33:41–47. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.