Abstract
We report a 3.1 Mb de novo deletion of 3p21.31 in a 3 ½ year old female with cortical blindness, cleft lip, CNS abnormalities, and gross developmental delays. Examination of the region showed ~80 genes to be involved in the deletion. Functional analysis of the deleted genes suggests that several of them may be important in normal neuronal maturation and function. Thus, haploinsufficiency of one or more of these genes could potentially contribute to the observed phenotype. Our patient does not have clinical features that overlap completely with either proximal or distal 3p deletions, suggesting that the deletion seen in our patient leads to a distinct clinical phenotype not described previously.
Keywords: cleft lip, cortical blindness, developmental delay, microarray, microdeletion
1. Methods of Detection
Affymetrix 50k Xba array. SNP-based microarray containing 50k probes (Affymetrix, Inc., Santa Clara, CA), as previously described [9].
Illumina HumanHap550 array. SNP-based microarray containing 550k probes (Illumina, Inc., San Diego, CA), with an average resolution of ~3 kb per the manufacturer.
2. Chromosomal Anomaly
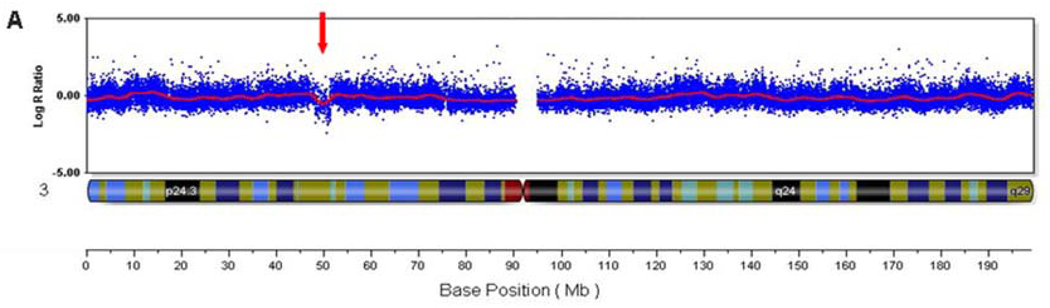
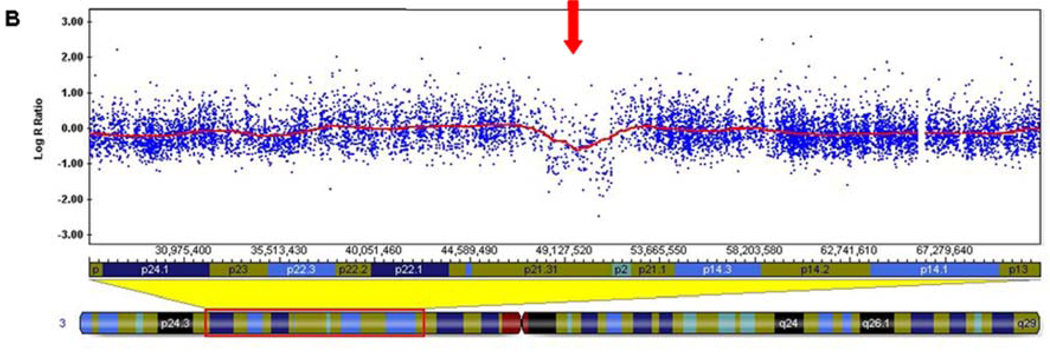
The Illumina results showed a minimal deletion size of 3.06 Mb from chromosome 3p21.31 (chr3:48,301,437-51,364,610, hg18 build) (Figure 1). The distal deleted SNP is rs6801801 (48,301,437) and the proximal deleted SNP is rs12487468 (51,364,610). The first non-deleted distal SNP is rs2889854 (48,243,513), and the first non-deleted proximal SNP is rs4974096 (51,368,086). Therefore, the maximum deletion size is 3.12 Mb.
SNP analysis using the microarray genotype results showed that the deletion was from the paternally-inherited chromosome 3.
Figure 1. Illumina 550k Human HapMap microarray results.
A) Log R ratio of all SNP probes on chromosome 3, with the 3.1 Mb deletion noted by a decrease in the Log R ratio (red arrow). Each blue dot represents a single SNP-based probe and the red line is a smoothing curve over a 200 Kb genomic window. The Log R ratio for diploid probes are distributed around 0.0. The Log R ratio for hemizygously deleted probes are distributed around −0.5. B) Higher magnification view of the 3p21.31 deletion (red arrow).
3. Methods of Confirmation
The deletion was confirmed by standard fluorescence in situ hybridization (FISH) techniques with a probe located in the deleted region (RP11-3B7).
Parental samples were also analyzed by the Illumina microarray platform as well as by standard FISH methods. These results were normal, suggesting the deletion is de novo and pathogenic in the patient.
4. Clinical Description
The patient was originally seen at 3 months of age for hypotonia, cleft lip, and gastroesophageal reflux. She was the product of a dizygotic twin pregnancy, and was the second pregnancy to a non-consanguineous couple of Ashkenazi Jewish ancestry. She was born at 38 weeks gestation to a 23 year old mother and 24 year old father by vaginal delivery. There were no difficulties during the pregnancy except for a twisted umbilical cord and oligohydramnios. The delivery was uncomplicated. There was no history of significant resuscitation efforts required at birth, but the Apgar scores were unknown. Her weight was 2.2 kg (5th centile). The fraternal twin sister was 2.6 kg (25th centile) and experienced no considerable neonatal problems. Our patient, however, was treated in the hospital for one week following birth due to feeding issues related to her cleft lip. An older brother, 14 months of age, was healthy without medical or developmental difficulties. There is no history of any family members with a cleft lip or other congenital defects.
At three months of age, the patient was not smiling or tracking, and was noticed to have abnormal eye movements with poor visual focusing due to cortical blindness. Her height was at the 3rd centile and head circumference at the 60th centile. Dysmorphic features included mild hypertelorism with upslanting palpebral fissures, cleft lip, and widely-spaced nipples. Her dermatoglyphic pattern was normal. She had generalized hypotonia, but no other focal neurological abnormalities. High-resolution chromosomes, subtelomere analysis, and FISH for a 22q11.2 deletion were all normal.
At seven months of age the patient was seen in follow-up, with medical issues including cleft lip, cortical blindness with pale fundi, short stature (50th centile for 3 months old), and gastroesophageal reflux. An MRI showed partial agenesis of the corpus callosum, underoperculized Sylvian fissures, prominence of the extra axial fluid spaces over the frontal lobes bilaterally, and no areas of abnormal signal intensity or atrophy. Developmental delays were present, where she was unable to lift her head from a prone position due to significant hypotonia. She was cooing but not babbling. Studies included a buccal swab for tetrasomy 12p (Pallister-Killian) syndrome by FISH, BAC-based array comparative genomic hybridization (aCGH) of 831 clones covering 230 loci (Signature Genomic Laboratories, LLC, Spokane, WA), transferrin isoelectric focusing, creatine kinase levels, spinal muscular atrophy deletion analysis, dysferlin protein levels, FKRP, CAPN3, and SGCA gene sequencing for limb-girdle muscular dystrophy, plasma acylcarnitine profile, plasma amino acids, very long chain fatty acids, and sweat testing for cystic fibrosis, all of which were normal.
At 17 months of age, her vision was improving, but she still suffered from gastroesophageal reflux and developmental delays. She was thought to have an intestinal malrotation at this age, but further radiographic studies excluded the diagnosis. Dysmorphic features included hypotonic-appearing facies, broad forehead, upslanting palpebral fissures, bulbous nasal tip, and long philtrum (Figure 2). A sample obtained during this visit was used in a research study to test the efficacy of the Affymetrix 50k Xba array for the detection of submicroscopic rearrangements not found by standard cytogenetic techniques [9]. A follow-up appointment at 3 years and 8 months of age demonstrated continued developmental delays, where the patient began pulling to a stand and crawling at 2 years, walking at 3 years and 4 months, but was not speaking any words. She also developed a prolonged seizure episode with an abnormal EEG. Her physical exam showed short stature (3rd centile) with a head circumference at the 50th centile. She was evaluated by a neurologist and was found to have central and peripheral hypotonia, a wide-based gait, and mildly decreased reflexes.
Figure 2. Photograph of patient at 18 months of age.
Note the hypotonic-appearing facies, upslanting palpebral fissures, bulbous nasal tip, and long philtrum.
5. Discussion
In this report, we present a patient with multiple congenital anomalies including a cleft lip, dysmorphic features, and motor and language delays. She had many previously normal genetic studies including a high-resolution karyotype, subtelomere in situ hybridization, and BAC-based aCGH. She was found to have a 3.1 Mb deletion of 3p21.31 by higher-density oligonucleotide microarray analysis. Review of the literature and the DECIPHER database (https://decipher.sanger.ac.uk/) did not identify any patients with this specific deletion. There was one patient in DECIPHER (patient 00004666) with joint laxity, mental retardation, speech delay, and a long/thin face, who had a partially overlapping deletion from 49,290,812–54,276,939 Mbp. However, most patients described in the literature with interstitial 3p deletions are more proximal and involve band 3p12 [7,11,15], which is not included in our patient’s deletion. These patients have variable phenotypic findings, but are thought to commonly share psychomotor delay, cardiovascular anomalies, early growth retardation, and dysmorphic facial features such as dysplastic and low-set ears. A cleft lip or cleft palate has been seen in two patients [7,11]. Another patient was described with an interstitial deletion distal to our patient (3p26.2-p25.3), with clinical findings consisting of ptosis, microcephaly, growth retardation, and developmental delay [3]. Patients with terminal 3p deletion syndrome typically have breakpoints in 3p25 resulting in haploinsufficiency of the CALL gene, and the clinical characteristics include growth retardation, developmental delays, microcephaly, hypertelorism, ptosis, abnormal nose, low-set ears, and a long philtrum [2,10]. Our patient does have some clinical features that overlap with known proximal and distal 3p deletion syndromes, but the variability of the observed phenotype suggests this particular deletion is a distinct clinical entity.
The 3.1 Mb region deleted in our patient contains over 80 genes. The genes with known functions that may be relevant to this patient’s phenotype are listed in Table I. Although it is premature to pinpoint which one of these genes is involved in the patient’s phenotype, the genes that are expressed in the brain are presumed to explain many of the observed neurological features, such as the developmental delays, cortical blindness, and hypotonia. A review of the deleted genes identified several possible candidates associated with the clinical findings. The PLXNB1 gene encodes for the semaphorin 4D receptor and is important in axon guidance in the developing nervous system, and can either induce extension or retraction of axons [8]. Mice deficient for this gene show abnormal axon collapsing; however they do not have specific developmental defects [6]. CELSR3 is a protocadherin gene involved in axonal development and neuronal cell polarity, as evidenced by Celsr3 deficient mice showing anomalies of several axonal fascicles [14]. Studies of rat neuron cultures showed Celsr3 suppresses neurite growth whereas Celsr2 enhances growth, and the balance of these two genes is important for normal neurite growth regulation [12]. The protein product of another deleted gene, bassoon (BSN), has been shown to be involved in presynaptic transmission of excitatory neurons, the loss of which decreases the transmission of the excitatory neurotransmitter glutamine in the brain and retina [1,5]. GNAT1 is involved in autosomal dominant congenital stationary night blindness, but this typically is due to a dominant negative effect of a constitutively active mutant protein and not to haploinsufficiency [13]. DOCK3, also expressed in the brain, was found to be interrupted in a patient with a 3p inversion and attention deficit hyperactivity disorder-like behaviors, suggesting this gene may be associated with normal brain development and function [4].
Table 1.
Selection of genes in the deleted region.
| RefSeq Name |
Function related to phenotype |
|---|---|
| FBXW12 | Component of E3 ubiquitin ligase complex involved in protein degradation. |
| PLXNB1 |
Receptor for SEMA4D. Plays a role in axon guidance, invasive growth and cell migration. |
| TREX1 | Required for checkpoint signaling after DNA damage. |
| UCN2 | Suppresses food intake, delays gastric emptying and decreases heatinduced edema. |
| CELSR3 |
Receptor that may have an important role in cell/cell signaling during nervous system formation. |
| SLC25A20 | Defects may lead to muscle weakness and wasting. |
| LAMB2 | May have role in maturation of neuromuscular junctions. |
| DAG1 | Forms part of the dystrophin-associated protein complex (DAPC) which may have a role in maintaining structural integrity in muscle tissues. |
| BSN |
Essential in regulated neurotransmitter release from a subset of brain glutamatergic synapses. |
| GNAT1 |
Involved as modulators or transducers in various transmembrane signaling systems. |
| SEMA3B | The semaphorin/collapsin family of molecules plays a critical role in the guidance of growth cones during neuronal development. |
| RASSF1 | This gene encodes a protein similar to the RAS effector proteins and may be involved in cell cycle regulation. |
| CACNA2D2 | A member of the alpha-2/delta subunit family of calcium channels expressed in neurons. |
| CISH | May have a role in regulation of cytokine signal transduction. |
| MAPKAPK3 | Involved in the regulation of tissue-specific gene expression and cell differentiation. |
| DOCK3 | Belongs to a family of proteins that may have a role in apoptosis. |
Information obtained from the UCSC Genome Browser: http://genome.ucsc.edu.
In summary, we present here a patient exhibiting dysmorphic features and developmental delays. Analysis using high-density oligonucleotide microarrays demonstrated a de novo 3.1 Mb deletion of 3p21.31 involving approximately 80 genes. Several of the deleted genes are expressed in the brain and may be required for normal brain development. This chromosomal region and deleted genes should be considered for analysis in patients with a similar clinical phenotype. Since the parental studies were normal, the parents were able to have 2 more unaffected children, knowing that the likelihood of germ line mosaicism was very low. This case highlights the utility of genome-wide, high-density microarrays in detecting deletions or duplications in patients with congenital anomalies and mental retardation.
Acknowledgments
We would like to thank the patient’s family for their participation in the research protocol, and the Center for Applied Genomics at the Children’s Hospital of Philadelphia for excellent technical assistance. This work is partially supported by a grant from the NIH (GM081519) to T.H.S. C.H-E. was supported by a Medical Genetics Research Training Grant, 5-T32-GM-008638-11, to the University of Pennsylvania.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Altrock WD, tom Dieck S, Sokolov M, Meyer AC, Sigler A, Brakebusch C, Fässler R, Richter K, Boeckers TM, Potschka H, Brandt C, Löscher W, Grimberg D, Dresbach T, Hempelmann A, Hassan H, Balschun D, Frey JU, Brandstätter JH, Garner CC, Rosenmund C, Gundelfinger ED. Functional inactivation of a fraction of excitatory synapses in mice deficient for the active zone protein bassoon. Neuron. 2003;37:787–800. doi: 10.1016/s0896-6273(03)00088-6. [DOI] [PubMed] [Google Scholar]
- 2.Angeloni D, Lindor NM, Pack S, Latif F, Wei M-H, Lerman MI. CALL gene is haploinsufficient in a 3p-syndrome patient. Am. J. Hum. Genet. 1999;86:482–485. doi: 10.1002/(sici)1096-8628(19991029)86:5<482::aid-ajmg15>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 3.Cargile CB, Goh DL-M, Goodman BK, Chen X-N, Korenberg JR, Semenza GL, Thomas GH. Molecular cytogenetic characterization of a subtle interstitial del(3)(p25.3p26.2) in a patient with deletion 3p syndrome. Am. J. Med. Genet. 2002;109:133–138. doi: 10.1002/ajmg.10323. [DOI] [PubMed] [Google Scholar]
- 4.de Silva MG, Elliot K, Dahl H-H, Fitzpatrick E, Wilcox S, Delatycki M, Williamson R, Efron D, Lynch M, Forrest S. Disruption of a novel member of a sodium/hydrogen exchanger family and DOCK3 is associated with an attention deficit hyperactivity disorder-like phenotype. J. Med. Genet. 2003;40:733–740. doi: 10.1136/jmg.40.10.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dick O, tom Dieck S, Altrock WD, Ammermüller J, Weiler R, Garner CC, Gundelfinger ED, Brandstätter JH. The presynaptic active zone protein bassoon is essential for photoreceptor ribbon synapse formation in the retina. Neuron. 2003;37:775–786. doi: 10.1016/s0896-6273(03)00086-2. [DOI] [PubMed] [Google Scholar]
- 6.Fazzari P, Penachioni J, Gianola S, Rossi F, Eickholt BJ, Maina F, Alexopoulou L, Sottile A, Comoglio PM, Flavell RA, Tamagnone L. Plexin-B1 plays a redundant role during mouse development and in tumor angiogenesis. BMC Dev. Biol. 2007;7:55. doi: 10.1186/1471-213X-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lalli C, Galasso C, Lo Castro AL, Nardone AM, Di Paolo A, Curatolo P. Interstitial deletion of a proximal 3p: a clinically recognizable syndrome. Brain Dev. 2007;29:312–316. doi: 10.1016/j.braindev.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 8.Lin X, Ogiya M, Takajara M, Yamaguchi W, Furuyama T, Tanaka H, Tohyama M, Inagaki S. Sema4D-plexin-B1 implicated in regulation of dendritic spine density through RhoA/ROCK pathway. Neurosci. Lett. 2007;428:1–6. doi: 10.1016/j.neulet.2007.09.045. [DOI] [PubMed] [Google Scholar]
- 9.Ming JE, Geiger E, James AC, Ciprero KL, Nimmakayalu M, Zhang Y, Huang A, Vaddi M, Rappaport E, Zackai EH, Shaikh TH. Rapid detection of submicroscopic chromosomal rearrangements in children with multiple congenital anomalies using high density oligonucleotide arrays. Hum. Mutat. 2006;27:467–473. doi: 10.1002/humu.20322. [DOI] [PubMed] [Google Scholar]
- 10.Mowrey PN, Chorney MJ, Venditti CP, Latif F, Modi WS, Lerman MI, Zbar B, Robins DB, Rogan PK, Ladda RL. Clinical and molecular analyses of deletion 3p25-pter syndrome. Am. J. Med. Genet. 1993;46:623–629. doi: 10.1002/ajmg.1320460604. [DOI] [PubMed] [Google Scholar]
- 11.Petek E, Windpassinger C, Simma B, Mueller T, Wagner K, Kroisel PM. Molecular characterization of a 15 Mb constitutional de novo interstitial deletion of a chromosome 3p in a boy with developmental delay and congenital anomalies. J. Hum. Genet. 2003;48:283–287. doi: 10.1007/s10038-003-0023-5. [DOI] [PubMed] [Google Scholar]
- 12.Shima Y, Kawaguchi S-y, Kosaka K, Nakayama M, Hoshino M, Nabeshim Y, Hirano T, Uemura T. Opposing roles in neurite growth control by two seven-pass transmembrane cadherins. Nat. Neurosci. 2007;10:963–969. doi: 10.1038/nn1933. [DOI] [PubMed] [Google Scholar]
- 13.Szabo V, Kreienkamp H-J, Rosenberg T, Gal A. p.Glu200Glu, a putative constitutively active mutant of rod alpha-transducin (GNAT1) in autosomal dominant congenital stationary night blindness. Hum. Mutat. 2007 doi: 10.1002/humu.9499. Mutation in Brief #970. [DOI] [PubMed] [Google Scholar]
- 14.Tissir F, Bar I, Jossin Y, De Backer O, Goffinet AM. Protocadherin Celsr3 is crucial in axonal tract development. Nat. Neurosci. 2005;8:451–457. doi: 10.1038/nn1428. [DOI] [PubMed] [Google Scholar]
- 15.Wieczorek D, Bolt J, Schwechheimer K, Gillessen-Kaesbach G. A patient with interstitial deletion of the short arm of chromosome 3 (pter→p21.2::p12→qter) and a CHARGE-like phenotype. Am. J. Med. Genet. 1997;69:413–417. doi: 10.1002/(sici)1096-8628(19970414)69:4<413::aid-ajmg15>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]