Abstract
Two of the central problems in biology are determining the molecular basis of adaptive evolution and understanding how cells regulate their growth. The chemostat is a device for culturing cells that provides great utility in tackling both of these problems: it enables precise control of the selective pressure under which organisms evolve and it facilitates experimental control of cell growth rate. The aim of this review is to synthesize results from studies of the functional basis of adaptive evolution in long-term chemostat selections using Escherichia coli and Saccharomyces cerevisiae. We describe the principle of the chemostat, provide a summary of studies of experimental evolution in chemostats, and use these studies to assess our current understanding of selection in the chemostat. Functional studies of adaptive evolution in chemostats provide a unique means of interrogating the genetic networks that control cell growth, which complements functional genomic approaches and quantitative trait loci (QTL) mapping in natural populations. An integrated approach to the study of adaptive evolution that accounts for both molecular function and evolutionary processes is critical to advancing our understanding of evolution. By renewing efforts to integrate these two research programs, experimental evolution in chemostats is ideally suited to extending the functional synthesis to the study of genetic networks.
Keywords: nutrient limitation, selection, copy number variation, chemostats, cell growth, adaptive evolution
Understanding how microorganisms evolve under conditions of continuous culturing using chemostats provides insight into the genetic networks that regulate cell growth and how those networks are rewired during evolution.
Understanding how microorganisms evolve under conditions of continuous culturing using chemostats provides insight into the genetic networks that regulate cell growth and how those networks are rewired during evolution.
INTRODUCTION
In 1950, Jacques Monod introduced a method for culturing microorganisms in which cells in a liquid medium of fixed volume grow continuously by constant replenishment of the medium. In the same year, Leo Szilard and Aaron Novick reported a device for culturing cells using the same approach, which they named the ‘chemostat’ (Novick and Szilard, 1950a,b) – a name that has since endured. The central principle of the chemostat is that through the continuous addition of medium containing a single growth-limiting nutrient and simultaneous removal of culture, a stable equilibrium is achieved. In this steady state, the rate at which the population of cells grows is equal to the rate at which the culture is diluted. The key advantages of the chemostat are that a steady state in a constant, defined environment is attained and the growth rate of the cells can be experimentally controlled by modulating the rate of culture dilution. The inventors of the chemostat argued that these unique properties had great utility for two areas of investigation: the study of cell growth and the study of evolution by mutation and selection. More than 60 years later, the advent of genome-scale experimental methods and a reinvigoration of a quantitative approach to biology (i.e. systems biology) have stimulated renewed interest in the use of chemostats (Hoskisson and Hobbs, 2005; Bull, 2010; Ziv et al., 2013a). The primary research applications of chemostats remain the study of cell growth and adaptive evolution, which owing to their central importance in cell and evolutionary biology, remain vital areas of investigation.
Rather than treating the study of cell growth and adaptive evolution as separate research enterprises, much stands to be gained from their integrated study. Here, we argue that many of the important questions in evolution including the role of different types of genetic variation, the predictability and repeatability of evolution, the causes and consequences of antagonistic pleiotropy, the distribution of fitness effects, and the role of epistasis are best addressed on the foundation of an understanding of the genetic networks that are the targets of selection. This requires a priori knowledge, and precise experimental control, of the selective pressure experienced by the organism. The chemostat is ideally suited to fulfilling these criteria. Moreover, as a chemostat environment usually represents a novel environment for the organism, fitness increases in evolving lineages, and populations, are typically large providing increased statistical power when dissecting multigenic and epistatic effects. Finally, the chemostat provides unparalleled control for systematically varying important parameters in adaptive evolution including population size and the strength of selection.
At the same time, identifying the targets of selection in a chemostat provides insight into how a cell regulates its growth across a range of environmental conditions. Identifying the functional basis of increased fitness in a chemostat is informative about the regulatory mechanisms that control cell growth in different environments. In principle, there may be multiple ways in which fitness can increase in a particular environment. However, only a subset of these options may be evolutionarily viable, and determining how genetic networks that control growth evolve provides insight into their structure and plasticity. As many of the principles of cell growth control are conserved across orders of life and many of the proteins and processes required for cell growth are evolutionarily, conserved, insight into the evolution of the genetic networks controlling cell growth has the potential to be broadly informative.
Several reviews of experimental evolution in microorganisms have been published in recent years (Elena and Lenski, 2003; Zeyl, 2006; Bennett and Hughes, 2009; Buckling et al., 2009; Conrad et al., 2011; Dettman et al., 2012; Desai, 2013). Reviews focused on experimental evolution in chemostats have appeared less frequently. Dykhuizen and Hartl (1983) published a comprehensive review of early microbial evolution studies using chemostats, and subsequent publications have provided updates on progress in the field (Dykhuizen, 1990; Adams, 2004; Ferenci, 2008). In addition, studies of selection in chemostats have been featured in collected volumes (Garland and Rose, 2009) and treatises on adaptive evolution (Bell, 2008). In this review, we introduce the concept of the chemostat and discuss the theoretical basis for adaptive evolution in chemostats. Although experimental evolution in chemostats has been performed in a range of microorganisms, we focus here on studies in Escherichia coli and Saccharomyces cerevisiae, which currently have the best functionally annotated genomes and therefore are ideally suited to enabling the integrated approach that we advocate. We discuss recent progress in understanding the molecular basis of adaptive evolution in chemostats in these two organisms and conclude by outlining some of the future goals that are now attainable. Our aim is to synthesize these studies to define the state of the field and to argue that selection in chemostats is an ideal system for extending the functional synthesis – the reunification of molecular biology and evolution (Dean and Thornton, 2007) – to the study of genetic networks.
THE PRINCIPLE OF THE CHEMOSTAT
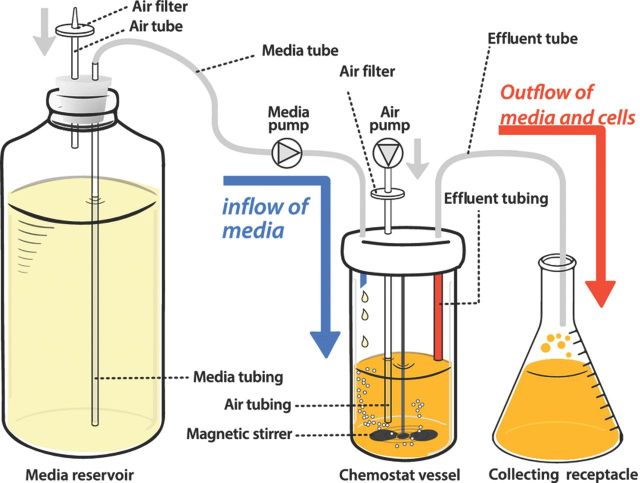
The culturing of microorganisms in a chemostat differs in several ways from the method of batch culture growth typically employed by microbiologists. In a batch culture, a small number of individual cells are inoculated into fresh medium, and following a physiological and metabolic adjustment, cell growth and division commences. Initially, the rate at which the population grows is unconstrained by nutrient abundance, and population expansion proceeds at a maximal rate. Once a population reaches a sufficiently high density, nutrients become increasingly scarce, and exhaustion of essential nutrients ultimately leads to the cessation of cell growth and initiation of a quiescent state. In a chemostat, population growth is initially qualitatively similar to batch culture growth. However, as the population grows, the medium in the culture is continuously replenished by addition of fresh medium (Fig. 1). Simultaneously, an equal volume containing both cells and spent medium is removed from the culture vessel. Thus, a chemostat is said to be ‘diluted’ through continuous addition of fresh medium and removal of culture. A single essential nutrient is present at a growth-limiting concentration in the chemostat vessel (analogous to the nutrient that is first exhausted in batch cultures), while all other nutrients are present in excess. The growth-limiting nutrient is predetermined by the experimenter and can be any nutrient essential for cell growth. For example, if glucose is the limiting nutrient, a chemostat is said to be ‘glucose-limited’.
Figure 1.
Design of a chemostat. Typically, a chemostat comprises a culture vessel in which the population grows under continuous agitation and aeration. New media flows into the vessel at a defined rate. At the same rate, culture containing cells and medium is removed from the chemostat. The flow of media and culture is maintained using a pumping apparatus and holding the chemostat vessel under positive pressure by means of a constant air flow.
In a chemostat, there are two important dynamic variables: the number of cells (x) and the concentration of the growth-limiting nutrient (s). The number of cells increases with time due to growth and division and decreases with time due to culture dilution. The concentration of the growth-limiting nutrient increases with time through addition of fresh medium and decreases through both dilution and consumption by the cells in the chemostat vessel. The temporal dynamics with which the number of cells (dx/dt) and the nutrient concentration (ds/dt) change can be modeled using the coupled ordinary differential equations:
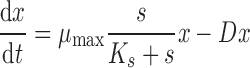 |
(1) |
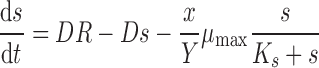 |
(2) |
In these equations, the growth rate of the cells (μ) is related to the concentration (s) of the limiting nutrient with saturating kinetics as described by the hyperbolic function, μ = μmax⋅s/(Ks + s), where Ks is the substrate concentration at half-maximal μ. This relationship between growth rate and nutrient concentration was proposed by Monod on the basis of empirical measurements of E. coli growth rates in different glucose concentrations (Monod, 1949) and also observed by Novick and Szilard (1950b) when an E. coli strain auxotrophic for tryptophan was grown in the presence of different concentrations of tryptophan. Recently, this model has been shown to agree with sensitive measurements of S. cerevisiae cells growing across a range of glucose concentrations (Ziv et al., 2013b).
The number of cells produced per mole of the limiting nutrient is defined by the parameter Y. Thus, μmax, Ks, and Y are properties of the cell and therefore potentially modified by genetic variation. By contrast, the experimenter controls the parameters R, the concentration of the limiting nutrient in the feed medium and D, the culture dilution rate, by modulating the flow rate (F) of media addition, and removal for a given culture volume (V) according to the relationship, D = F/V.
This system of equations predicts the establishment of a single nonzero stable steady state in which the cell number and the concentration of the growth-limiting nutrient remain constant per unit time (i.e. dx/dt = 0 and ds/dt = 0) (Fig. 2). At steady state, growth rate (μ) is submaximal and exponential. The remarkable implication of this model is that in steady-state conditions, the exponential growth rate constant (i.e. the ‘specific growth rate’) is equal to the dilution rate (D). Thus, the doubling time of the exponentially growing population (i.e. the generation time) is simply ln(2)/D. By varying the dilution rate, a variety of steady-state conditions can be established all of which have the property of growth rate being equal to the dilution rate so long as the dilution rate is less than the maximal growth rate of the cells. In principle, these steady states can be maintained indefinitely, enabling long-term selection in a constant environment. Although the experimental reality of some of the parameters of this model (Ferenci, 1999a) and the existence of a true steady state in the chemostat have been questioned (Ferenci, 2006), the model provides a useful conceptual framework for understanding selection in a chemostat. For further background on the principle of the chemostat, the reader is referred to the introductory book by Kubitschek (1970) and a comprehensive mathematical treatment of the chemostat (Smith and Waltman, 1995).
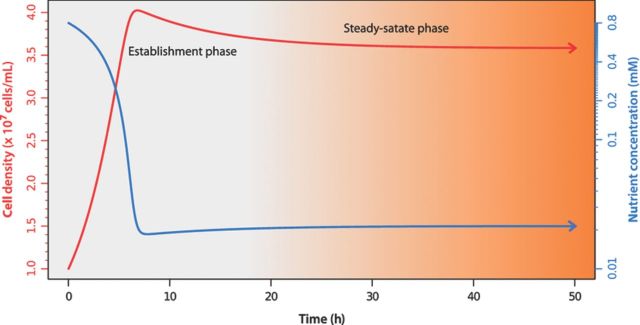
Figure 2.
Establishment of a steady state in the chemostat. Following inoculation and initiation of culture dilution, the chemostat is characterized by a period during which the population increases and nutrient abundance declines. Eventually, a steady state is established in which the cell population remains high and the concentration of the limiting nutrient remains low. The steady state is predicted by the fundamental equations of the chemostat and depends on the parameter values used in the simulation. In this simulation, μmax = 0.4 h−1, Ks = 0.05 mM, Y = 4.6 × 107 cells mmol−1, R = 0.8 mM, and D = 0.12 h−1. The simulation was initialized with x = 1 × 107 cells mL−1 and s = 0.8 mM.
EXPERIMENTAL EVOLUTION IN CHEMOSTATS
The environment in a chemostat differs in several ways from that of a batch culture with important implications for understanding the relevant selective pressures. In a batch culture, cells initially experience nutrient-rich conditions, but as the population grows nutrients continuously decline, and are ultimately exhausted leading to arrest in cell growth and division. For the purposes of experimental evolution in batch cultures, these cycles of boom and bust are repeated using serial dilution (Elena and Lenski, 2003). During each cycle, population size change dramatically (typically, by several orders of magnitude), and in addition to a decline in nutrients, the environment is altered in other ways including the pH, oxygenation, and presence of waste products. As a consequence, this experimentally simple method of propagating microorganisms results in a complex and dynamic environment making it difficult to define the evolutionarily relevant selective force(s). By contrast, in a steady-state chemostat, cell growth is continuously constrained by the abundance of a single essential nutrient ensuring that cells are maintained in a ‘hungry’ (Ferenci, 2001) or ‘poor, not starving’ (Saldanha et al., 2004) state. The population size in a steady-state chemostat remains constant, and all environmental factors are essentially invariant. Thus, despite the increased experimental complexity of chemostats, their use greatly simplifies the selection for the purposes of experimental evolution.
Additional factors of central importance to evolutionary processes also differ between the two methods of long-term selection. Serial dilution results in regular population bottleneck events when a subset of the population is used to found the subsequent cycle of growth. This results in random purging of genetic diversity increasing the role of genetic drift (random sampling of alleles between generations) in the evolution experiments. The extent of random genetic drift is a function of the effective population size, which in a serial dilution regime is determined by the number of cells passaged with each dilution. In a chemostat, the population size remains constant; however, the dilution process introduces an element of chance dictating which genotypes are maintained in the population. For example, at a dilution rate of 0.1 h−1, there is a 10% chance that a cell will be removed from the population each hour regardless of its genotype. More generally, a cell containing a newly introduced mutation has a 50% chance of being removed by the dilution process before the population doubles.
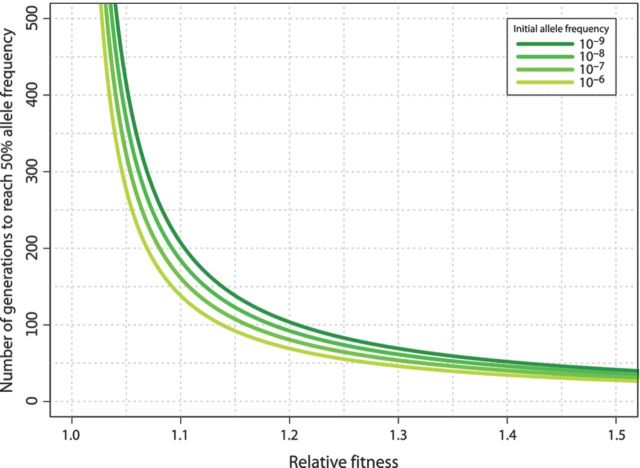
As with most evolutionary scenarios, population size (N) and mutation rate (μ) are critical parameters that dictate the speed, outcome, and reproducibility of adaptive evolution in a chemostat. The product of population size and the mutation rate, Nμ, determines the mutation supply rate. In chemostats containing E. coli or S. cerevisiae, population sizes are typically on the order of 108–1010 individuals. Whole-genome sequencing of mutation accumulation lines results in estimates of 1 × 10−3 nucleotide substitutions per genome per generation in E. coli (Lee et al., 2012) and 4 × 10−3 nucleotide substitutions per genome per generation in S. cerevisiae (Lynch et al., 2008). Thus, the mutation supply rate in a chemostat containing 108–1010 cells is around 106-108 mutations per generation. As this is equal to, or exceeds, the number of nucleotides in E. coli (4.6 × 106 bases) and S. cerevisiae (1.2 × 107 bases), on average, every possible one-step mutation is introduced into the population each generation. Moreover, the rate at which other types of genetic variation occur such as copy number variants (CNVs) and transposition events may exceed the rate of nucleotide substitutions. As a result, depending on mutation rate and population size (Chao and Cox, 1983), adaptation in a chemostat is typically not limited by mutation supply, and multiple adaptive mutations are expected to coexist in the population. Depending on the fitness effect of a particular mutation, new adaptive mutations are expected to reach 50% frequency in the population in less than a few hundred generations (Fig. 3). If an adaptive mutation was already present at more than one copy in the population, this time will be further reduced.
Figure 3.
Expected waiting time for beneficial mutations. The time until a beneficial mutations rises to a 50%, allele frequency in the population depends on the fitness effect of the mutation. Increased initial frequencies of a mutant genotype due to the occurrence of mutations during the establishment phase of the chemostat further reduce the waiting time.
The chemostat facilitates exploration of important factors that govern the outcome and dynamics of adaptive evolution. For example, the intensity of selection can be systematically varied, as increasing the dilution rate will increase the steady-state concentration of the limiting nutrient (Fig. 4a). The model of the chemostat predicts that as the dilution rate increases, the steady-state population size will decrease. However, it is possible to alter the population size in the chemostat independently of the growth rate by varying the concentration of the limiting nutrient (R) in the feed media. At an identical dilution rate, steady-state cell concentrations will increase linearly with increasing values of R, but the steady-state concentration of the limiting nutrient in the culture vessel will be identical (Fig. 4b). Thus, different population sizes can be maintained in otherwise identical selective conditions. The control of nutrient concentration and population size that is uniquely possible in a chemostat enables exploration of the effect of these parameters on adaptive evolution with unparalleled precision.
Figure 4.
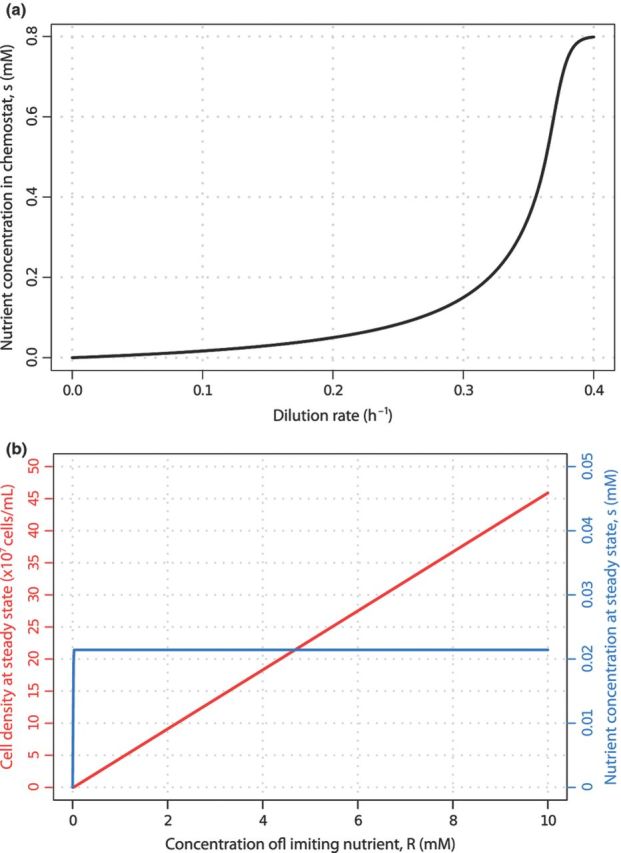
Systematic variation of selection intensity and population size in a chemostat. (a) Steady-state nutrient concentration increases with increasing dilution rates. (b) Population size (red) in the chemostat can be controlled by varying the concentration of the limiting nutrient in the feed media (R) and maintaining a constant dilution rate. In this case, the steady-state concentration of the limiting nutrient in the chemostat (blue) remains constant.
THEORETICAL ROUTES TO INCREASED FITNESS IN A CHEMOSTAT
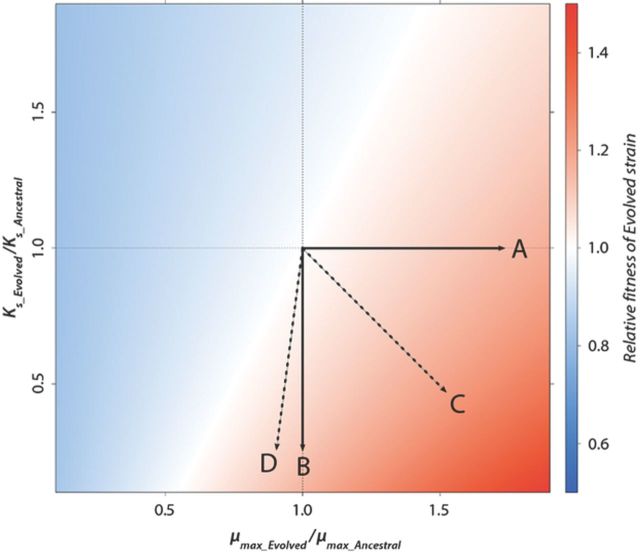
The mathematical model of the chemostat provides insight into the mechanisms by which fitness increases in the chemostat. Selection in the chemostat acts on those parameters that affect cell growth rate namely, the maximal growth rate (μmax) and the nutrient concentration required for half-maximal growth rate (Ks). Changes in these parameters due to new mutations will effect the fitness of a lineage relative to the ancestral lineage (Fig. 5). Acquisition of a genetic variant that increases only maximal growth rate (μmax) relative to the ancestral lineage results in increased fitness (mutation A in Fig. 5) as does a mutation that only decreases the nutrient concentration required for half-maximal growth rate (Ks) (mutation B in Fig. 5).
Figure 5.
The fitness landscape in a chemostat is determined by changes in Ks and μmax due to adaptive mutations. A mutation that (A) increases μmax or (B) decreases Ks will result in increased fitness. Pleiotropic mutations that (C) synergistically increase μmax and decrease Ks will be adaptive. Pleiotropic mutations that act (D) antagonistically by decreasing μmax and decreasing Ks can still be beneficial depending on the relative effects on the two parameters. Data were simulated using s = 10 mM, ancestral Ks = 10 μM, and ancestral μmax = 0.4 h−1.
A single mutation may exert pleiotropic effects and result in changes in both μmax and Ks. For example, a mutation that decreases Ks may also increase μmax and therefore act synergistically to increase the fitness of a lineage in the chemostat (mutation C in Fig. 5). Alternatively, the effect of a particular mutation may be antagonistic with respect to these two parameters. For example, a mutation that decreases Ks may also result in decreased μmax. Pleiotropic mutations that have antagonistic effects on both parameters can still result in a net increase in fitness so long as the magnitude of the effects is sufficiently different (mutation D in Fig. 5) and therefore may be selected.
Although yield (Y) is a property of the cell that is modeled in the fundamental equations of the chemostat, alterations in yield do not directly affect fitness in the chemostat as a change in Y does not impact growth rate. However, an increase in Y could increase fitness indirectly by reducing the steady-state concentration of the limiting nutrient, s.
LONG-TERM SELECTION EXPERIMENTS IN CHEMOSTATS
Undertaking long-term selection experiments in chemostats is conceptually straightforward. Chemostat cultures can be established using a limiting concentration of any essential nutrient required for cell growth including carbon, nitrogen, phosphorous, and sulfur. The spectrum of possible limiting agents can be expanded by the use of auxotrophic strains. Indeed, early studies by Novick and Szilard (1950b) of mutation rates in chemostats made extensive use of E. coli auxotrophic for tryptophan. However, the fitness defects of auxotrophic strains (Baganz et al., 1997; Pronk, 2002) and the potential for unanticipated effects (Boer et al., 2008) associated with ‘unnatural limitation’ (Saldanha et al., 2004) for the auxotrophic requirement suggest caution be employed with the use of auxotrophs in studies of cell growth.
Once a steady-state culture has been established, a chemostat can be maintained indefinitely with minimal maintenance aside from regular measurement of the dilution rate. The main practical challenges entail maintaining an adequate supply of fresh media, ensuring the integrity of the tubing and pump systems and preventing contamination (Fig. 1). These challenges are readily met with experience and care. For example, improved methods for sterile assembly of chemostats have largely overcome the problems of contamination that plagued earlier efforts of long-term selection in chemostats.
As with all experimental evolution studies, regular sampling from the chemostat allows curation of a ‘fossil record’ of the evolving population (Elena and Lenski, 2003). This record comprises unbiased samples of the population obtained periodically during the selection and archived at −80 °C. These samples can be used for reconstructing allele frequency dynamics during the populations' histories and for studying the emergence of adaptation-associated traits. In a chemostat, it is possible to obtain these samples without disturbing the population by temporarily diverting the outflow (Fig. 1) to a sample tube.
PHENOTYPIC OUTCOMES OF LONG-TERM SELECTION IN CHEMOSTATS
Typically, the expected (and desired) outcome of long-term selection in chemostats is the acquisition of improved growth capabilities in the low-nutrient environment of the chemostat. This is most rigorously quantified by performing competition assays in a chemostat in which the evolved strain and the ancestral strain are co-cultured. The relative abundance of the two strains is determined over a few generations (typically < 20) in steady-state chemostats. As both lineages grow exponentially, the growth advantage per unit time, which is defined as fitness, is determined by linear regression of the natural log of the ratio of two strains against time (Dykhuizen and Hartl, 1983). To distinguish the two strains and estimate their relative abundances, they are typically marked by neutral markers such as a drug (Paquin and Adams, 1983a, b; Gresham et al., 2008) or phage resistance (Novick and Szilard, 1950b) allele, which facilitates genotype estimation by plating. The use of strains labeled with a constitutively expressed fluorescent protein facilitates genotype estimation using flow cytometry (Wenger et al., 2011; Hong and Gresham, 2014), which greatly increases the precision of fitness measurements.
Typically, fitness increases of lineages selected within a few hundred generations in chemostats are large, often exceeding 10% (relative fitness, w = 1.1) per generation, and occasionally as much as 50% (w = 1.5) per generation. This is consistent with the notion that populations introduced into a chemostat start far from the fitness optima. Importantly, the increased fitness of chemostat-adapted lineages is specific to the low-nutrient environment of the chemostat. In fact, many lineages selected in chemostats appear to have reduced growth capabilities when growing in nutrient-rich conditions, suggesting that ‘antagonistic pleiotropy’ is frequently associated with beneficial mutations selected in the chemostat (Wenger et al., 2011; Hong and Gresham, 2014).
As growth rate in a chemostat is constrained by nutrient abundance, one anticipated outcome of mutation and selection is enhanced nutrient uptake rates, which can be measured using radioactively labeled nutrients. Escherichia coli (Rosenzweig et al., 1994; Manche et al., 1999; Notley-McRobb and Ferenci, 1999b; Maharjan et al., 2007) and S. cerevisiae (Brown et al., 1998) lineages evolved in glucose-limited chemostats and E. coli lineages evolved in phosphate-limiting chemostats (Wang et al., 2010) have greatly enhanced substrate transport velocities. Most likely, increased nutrient transport velocities have the effect of increasing growth rates by decreasing Ks. Enhanced transport capabilities have been reported to be associated with both increased (Wenger et al., 2011) and decreased (Maharjan et al., 2007) biomass yields.
Selection in chemostats also appears to result in altered metabolic strategies. A number of studies have accessed genome-wide changes in gene expression in yeast (Ferea et al., 1999; Gresham et al., 2008; Kao and Sherlock, 2008) and E. coli (Kinnersley et al., 2009) lineages adapted to chemostats. These studies provide evidence for newly evolved metabolic strategies in chemostat-adapted lineages including increased oxidation of glucose in S. cerevisiae strains evolved in glucose-limited chemostats (Ferea et al., 1999). Consistent with increased metabolic efficiency, residual ethanol concentrations are typically decreased in populations evolved in glucose-limited chemostats (Gresham et al., 2008).
In a chemostat, the organism lives on the edge of starvation. However, mounting a full scale starvation response, in which the cell enters a quiescent, nonproliferative state is an evolutionary dead end as these cells will be removed by dilution without contributing to subsequent generation. There is evidence that one outcome associated with increased fitness in chemostat is loss of the ability to appropriately mount a stress response. For example, strains of E. coli adapted to glucose-limited chemostats exhibit reduced survival when subjected to temperature or oxidative stress (Notley-McRobb et al., 2002a). The loss of stress response pathways in E. coli can be qualitatively assessed by determining glycogen content in cells, using iodine staining, and catalase activity, using a ‘bubbling assay’ in hydrogen peroxide. Many E. coli lineages selected in glucose-limited chemostats show evidence for decreased glycogen levels and catalase activity, which has lead to the proposal of an antagonistic relationship between the capacity to mount a robust stress response and growth ability in nutrient-poor environments (Ferenci, 2003, 2005).
In addition to phenotypes that can be explained as a result of adaptive evolution in nutrient-limited environments, there are potential undesirable outcomes of long-term culturing in chemostats that may confound connecting genotype to phenotype. These include wall growth and flocculation, which are cases of niche specialization in the culture vessel that lead to heterogeneity within the population. However, selection for this class of mutants can be avoided by proper agitation of the culture to maintain all cells in suspension and the use of treated glass, which prevents microbial growth on vessel walls. In addition, an exception to the constant environment of a chemostat can occur if yeast cells synchronize cycles of oxidative and reductive metabolism in very slow-growing glucose-limited chemostats (Tu and McKnight, 2007). The phenotypic outcome to adaptation in this particular scenario is likely to be more complex.
REGULATION OF CELL GROWTH IN NUTRIENT-LIMITED ENVIRONMENTS
The design, analysis and interpretation of adaptive evolution studies in chemostats is informed by an understanding of how cells regulate their growth. Escherichia coli and S. cerevisiae can modulate their rate of growth over a range of suboptimal nutrient concentrations. How cells regulate their growth in response to variation in environmental nutrient status and how this is coordinated with the myriad cellular processes required for cell growth is an enduring question of central importance in biology. Classic studies of cell growth in bacteria using chemostats showed that cell physiology changes with variation in growth rate (Kjeldgaard et al., 1958; Schaechter et al., 1958). These studies demonstrated that as cell growth rates decrease as a result of reduced environmental nutrients, the mass of cells, their RNA content, and their protein content decrease. More recent studies have revisited the question of how cellular processes vary with variation in cell growth. Using chemostats, it was found that a large fraction of E. coli (Ishii et al., 2007) and S. cerevisiae (Regenberg et al., 2006; Castrillo et al., 2007; Brauer et al., 2008) mRNAs vary in relative abundance as a function of growth rate as do many intracellular metabolites (Boer et al., 2010). The gene expression and physiological changes associated with slowed growth rates in both E. coli and S. cerevisiae have important consequences for cells including increased resistance to stress (Elliott and Futcher, 1993; Notley and Ferenci, 1996; Ihssen and Egli, 2004; Lu et al., 2009).
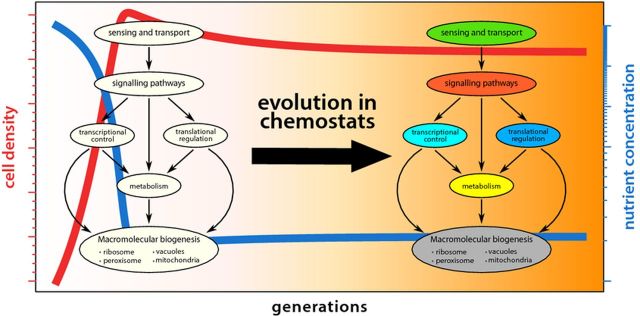
Conceptually, the regulation of cell growth in response to environmental nutrient status can be divided into distinct, but interconnected, modular processes (Fig. 6): (1) the sensing and transport of essential nutrients, (2) intracellular signaling pathways that converge on (3) transcriptional and (4) translational regulation, and (5) metabolic processes to coordinately regulate (6) the biogenesis of protein complexes and cellular organelles. In a chemostat, in which cell growth is constrained by the abundance of an essential nutrient, modification of any of these processes could potentially result in increased fitness and therefore may be targets of selection. The similarities in the principles underlying cell growth regulation in E. coli and S. cerevisiae present the possibility that common strategies may underlie adaptive evolution in chemostats.
Figure 6.
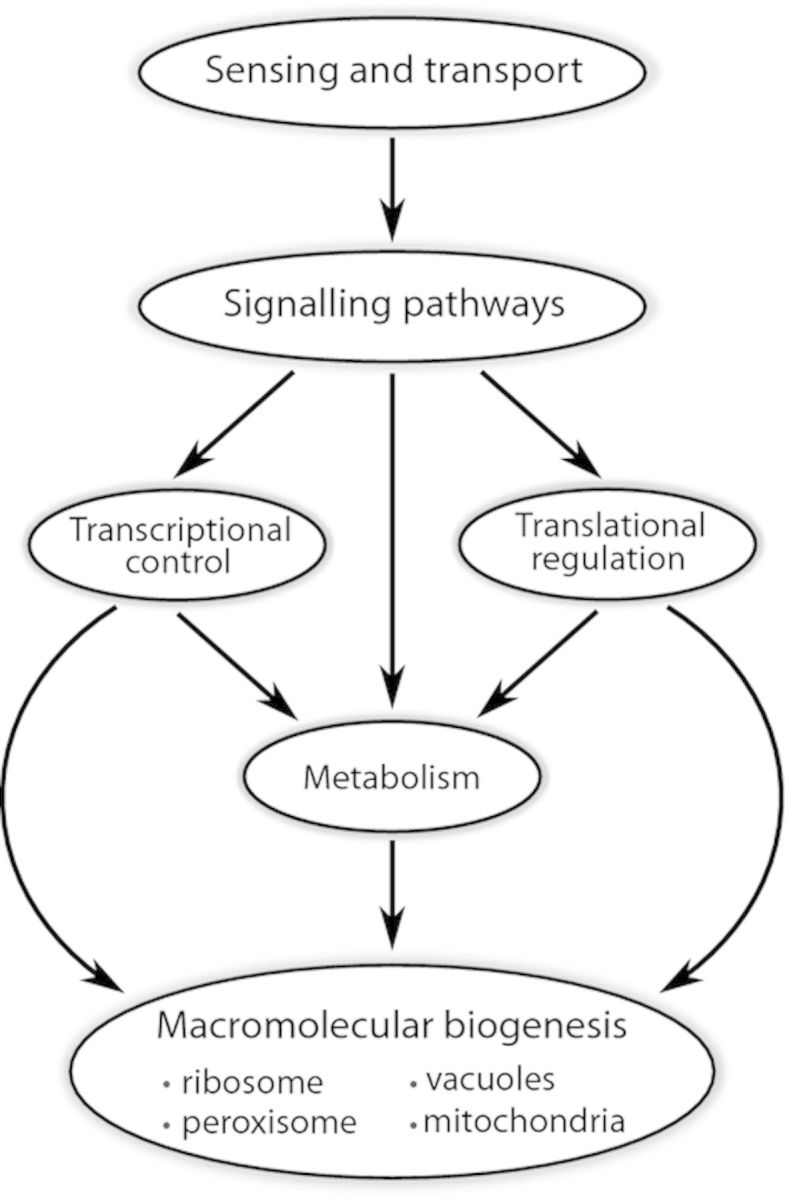
Modularity of cell growth regulating processes. A hierarchy of cellular processes control cell growth and are potential targets of selection in the chemostat.
In E. coli, many of the key components that contribute to regulation of cell growth are known. Specialized transporters import essential nutrients such as carbon, nitrogen, phosphorous, and sulfur, and many transporters are optimized for transporting specific forms of these nutrients. Many nutrient transporters are transcriptionally regulated, and as nutrient abundance declines, classes of higher affinity transporters are expressed. A key transcriptional regulator of the cellular response to decreased nutrient abundance is the sigma factor, αS, encoded by RpoS (Notley and Ferenci, 1996), the activity of which is regulated by decreased glucose, nitrogen, and phosphorous by different mechanisms (Peterson et al., 2005). As nutrient abundance declines, E. coli cells increase expression of porins, which results in increased permeability of the outer membrane (Ferenci, 1999b). Interestingly, the environmental abundance of different essential nutrients appears to converge on the regulated increased production of important intracellular signaling molecules including cyclic adenosine monophosphate (cAMP) and guanosine tetraphosphate (ppGpp; Ferenci, 2001; Srivatsan and Wang, 2008). Production of ppGpp is controlled by the enzymes RelA and SpoT in response to distinct signals. ppGpp and the protein DksA exert growth inhibitory effects through a variety of mechanisms including inhibition of rRNA transcription (Potrykus and Cashel, 2008; Jin et al., 2011).
In S. cerevisiae, the genetic networks that regulate cell growth have been intensively studied, in part due to their conservation in humans where the same networks regulate cell growth and are frequently dysregulated in disease such as cancer (Zaman et al., 2008; Lippman and Broach, 2010; Loewith and Hall, 2011; Broach, 2012). The yeast genome contains an abundance of nutrient transporters with greatly varying affinities and specificities. The expression of specific nutrient transporters is controlled both transcriptionally and through a variety of post-transcriptional mechanisms. Environmental nutrient status is transmitted intracellularly by conserved signaling pathways including the TORC1 and Protein Kinase A/Ras pathways (Zaman et al., 2008; Broach, 2012). Although the integration of diverse environmental signals that control cell growth is not well understood in yeast cells, there is evidence that signals from these pathways are integrated by converging on signaling molecules including the protein kinase RIM15 (Pedruzzi et al., 2003). Consistent with the importance of coordinating protein translation with cell growth, the ultimate target of these signaling pathways are factors involved in ribosome biogenesis and protein translation (Jorgensen and Tyers, 2004; Loewith and Hall, 2011; Broach, 2012).
THE MECHANISTIC BASIS OF INCREASED FITNESS IN NUTRIENT-LIMITED ENVIRONMENTS
Identifying the alleles that increase fitness in chemostats, and understanding the mechanistic basis by which the encoded products improve cell growth in nutrient-limited conditions, provides a path to understanding which of the myriad growth-related processes (Fig. 6) are limiting under conditions of suboptimal resources. However, until the advent of genome-scale methods, identifying the underlying genetic basis of increased fitness in chemostats was laborious and, with some exceptions, not typically amenable to standard methods for genetic cloning. Starting at the turn of the 21st century, the development of methods for comprehensive identification of genomic variation solved the long-standing problem of characterizing the genetic variation associated with adaptive evolution in chemostats. The aggregate of these studies provides insight into the types of mutations that contribute to adaptive evolution, the genes that are targeted by selection, and the mechanistic bases of increased fitness in chemostats.
THE ROLE OF COPY NUMBER VARIATION IN ADAPTATION IN CHEMOSTATS
The adaptive role of gene amplification was an early finding from experimental evolution studies in chemostats. In some of the first studies of adaptive evolution of E. coli in lactose-limited chemostats, it was observed that evolved lineages produced unusually high amounts of β-galactosidase (Horiuchi et al., 1962), which is the result of increased copies of the lac operon (Horiuchi et al., 1963). In S. cerevisiae, amplification of the gene encoding acid phosphatase, which hydrolyzes organic phosphates, was identified in lineages selected in chemostats limited for phosphate (Hansche, 1975; Hansche et al., 1978). A seminal study of Salmonella typhimurium lineages evolved in different carbon-limited chemostats identified amplification alleles containing transporters specific to the limiting carbon source (Sonti and Roth, 1989).
Subsequent studies have shown that CNVs are an important class of adaptive variation in chemostat-adapted lineages. Some of these CNVs are amplification alleles that contain specific nutrient transporter genes that import the limiting nutrient (Table 1). Targeted analysis of the high-affinity glucose transporter genes, HXT6 and HXT7, in an S. cerevisiae strain evolved in glucose-limiting conditions identified an amplification of the HXT6-HXT7 locus (Brown et al., 1998), which has subsequently been reported in multiple independent studies of adaptive evolution in glucose-limited chemostats (Gresham et al., 2008; Kao and Sherlock, 2008). In repeated long-term selections in sulfur-limited chemostats using S. cerevisiae, multiple independent amplification alleles were identified that include SUL1, which encodes a high-affinity sulfur transporter (Gresham et al., 2008). Similarly, long-term selections in diverse nitrogen-limiting environments identified CNV alleles containing transporter genes specific to the nitrogen source in the chemostat (Gresham et al., 2010; Hong and Gresham, 2014). Although the role of CNVs has not been extensively studied in experimental evolution of E. coli, amplification of the lac operon, which includes the lacY permease gene, underlies adaptation of E. coli to lactulose (Zhong et al., 2004). The role of CNVs in adaptive evolution of E. coli lineages warrants closer attention.
Table 1.
Transporter genes within copy number variants selected in nutrient-limited chemostats.
| Gene | Function | Nutrient limitation | References | Species |
|---|---|---|---|---|
| GAP1 | General amino acid permease | Glutamine and glutamate | Gresham et al. (2010), Hong and Gresham (2014) | Saccharomyces cerevisiae |
| PUT4 | High-affinity proline transporter | Proline | ||
| DUR3 | Plasma membrane transporter for both urea and polyamines | Urea | ||
| DAL4 | Allantoin permease | Allantoin | ||
| HXT6/HXT7 | High-affinity glucose transporters (99% identical) | Glucose | Brown et al. (1998), Kao and Sherlock (2008), Gresham et al. (2008) | |
| SUL1 | High-affinity sulfate permease | Sulfur | Gresham et al. (2008) | |
| LacY | Lactose permease | Lactose | Zhong et al. (2004) | Escherichia coli |
The repeated selection of specific CNVs containing nutrient-specific transporter genes is consistent with enhanced nutrient transport capacity underlying a common adaptive strategy in chemostats. Measurement of fitness increases attributable to increased copy number of the SUL1 locus suggests that relative fitness increases as large as 50% are associated with amplification of transporter genes (Gresham et al., 2008). The critical role of CNVs in the rapid adaptive evolution observed in chemostats is consistent with their role in diverse adaptive evolution scenarios including animal domestication (Wright et al., 2009) and human evolution in response to the advent of agriculture (Perry et al., 2007). Although a subset of CNV alleles selected in chemostats contain transporter genes, many do not, and deletion alleles containing several contiguous genes are also detected (Gresham et al., 2008; Hong and Gresham, 2014). Understanding the mechanistic basis by which the many CNVs that do not include transporter genes increase fitness in the chemostat remains to be determined.
Typically, amplification alleles are the result of tandem repeat expansions at their endogenous chromosomal locus. However, the mechanisms underlying CNV generation in chemostat-evolved lineages are not well understood. Although some CNV alleles in both E. coli (Zhong et al., 2004) and S. cerevisiae (Hong and Gresham, 2014) are bounded by repetitive sequence elements, including transposon sequences, that likely facilitate intrachromosomal recombination (Mieczkowski et al., 2006), many CNV junctions show no evidence of such repeats (Gresham et al., 2008; Araya et al., 2010; Hong and Gresham, 2014). Microhomology-mediated mechanisms (Payen et al., 2008) may contribute to formation of these alleles. A notable exception to the chromosomal location of CNV alleles is the GAP1 gene in S. cerevisiae, which is unusual in being one of only two isolated genes in the yeast genome flanked by tandem long-terminal repeats. These repeats facilitate the generation of a circular DNA element containing an autonomous replication sequence and GAP1 (Gresham et al., 2010), which transports glutamine and glutamate and is selected in glutamine- and glutamate-limited chemostats.
Determining the spontaneous rate at which CNVs are generated poses a considerable technical challenge. In S. cerevisiae, estimated rates of duplications range from as low as 10−10 amplifications per cell per division (Dorsey et al., 1992; Peterson et al., 2000) to 3.4 × 10−6 amplifications per cell per division (Lynch et al., 2008). However, the rate at which amplification and deletion alleles are spontaneously generated is highly variable and dependent on local genome architecture (Romero and Palacios, 1997). Moreover, rates of CNV generation may be controlled in part by environmental factors, and the rate at which they are generated in the nutrient-poor conditions of a chemostat has not been determined. One considerable challenge to studying beneficial CNVs is that they appear to be unstable and revert at a high rate (Maharjan et al., 2013b; Hong and Gresham, 2014); if the CNV is deleterious outside of the chemostat environment, inadvertent selection for revertants may be difficult to avoid. Nonetheless, as the rate of occurrence of CNVs is likely to profoundly impact the evolutionary dynamics in chemostats, determining the rate at which these alleles are generated and the mechanistic basis by which they increase fitness is critical to understanding adaptive evolution in chemostats.
CHROMOSOMAL TRANSLOCATIONS AND ANEUPLOIDIES ARE FREQUENTLY ASSOCIATED WITH INCREASED FITNESS IN CHEMOSTATS
In addition to CNVs, large-scale chromosomal alterations associated with increased fitness in chemostats have been identified in S. cerevisiae. In lineages selected in chemostats limited for different organic phosphates, individual clones were found to contain variants in chromosome size as well as chromosomal rearrangements (Adams et al., 1992). High resolution analysis of S. cerevisiae lineages evolved in glucose-limited chemostats using array comparative genomic hybridization (aCGH) identified repeated chromosomal translocations including reciprocal and nonreciprocal translocation events (Dunham et al., 2002). In many cases, breakpoint mapping revealed that translocations are associated with transposon sequences as has been found in experimental studies of chromosomal rearrangements (Mieczkowski et al., 2006).
Although the repeated selection of chromosomal translocations points to their adaptive benefit, the underlying basis by which they result in increased fitness is not clear. In the case of clones selected from glucose-limited chemostats, some translocation breakpoints were identified immediately adjacent to CIT1, which encodes citrate synthase (Dunham et al., 2002). Although the translocation event is proposed to alter their transcriptional regulation of CIT1, this has not been formally demonstrated. More recently, a study of a diploid S. cerevisiae/Saccharomyces bayanus hybrid evolved in ammonium-limited chemostats identified reciprocal translocation events between species-specific homologous chromosomes that mapped within MEP2 (Dunn et al., 2013), which encodes a high-affinity ammonium permease. In this case, it seems likely (though untested) that the fusion protein product has enhanced transport properties compared with either of the species-specific gene products. Thus, chromosomal translocations may serve to both alter gene expression regulation and generate novel sequence variants that are beneficial.
In addition to partial chromosomal events, duplications of entire chromosomes (aneuploidies) are repeatedly associated with adaptive evolution in chemostats (Dunham et al., 2002; Gresham et al., 2008; Hong and Gresham, 2014). The adaptive benefit of chromosomal aneuploidies remains to be determined; however, their beneficial role in adaptive evolution in chemostats is consistent with their reported role in suppression of engineered genetic lesions (Hughes et al., 2000; Rancati et al., 2008) and adaptive evolution in other selective regimes such as high temperature growth (Yona et al., 2012). The reason that aneuploidies are beneficial in particular circumstances is an open question with clear relevance to understanding their role in human tumors (Torres et al., 2007, 2010; Pavelka et al., 2010a,b; Tang et al., 2011).
TRANSPOSONS CONTRIBUTE TO ADAPTIVE EVOLUTION IN CHEMOSTATS
Given their pervasive role in evolution (Biemont, 2010; Fedoroff, 2012), transposable elements – including ‘cut and paste’ DNA transposons and ‘copy and paste’ retrotransposons – are likely to play a role in adaptive evolution in chemostats. However, few studies have explicitly explored the contribution of transposition events to adaptive evolution in chemostats. In E. coli, both the Tn5 (Biel and Hartl, 1983) and Tn10 (Chao et al., 1983) transposons have been shown to have beneficial effects in adaptive evolution in chemostats. In the case of Tn10, it was shown that this advantage is due to transposition events that presumably generate beneficial mutations (Chao et al., 1983). Similarly in S. cerevisiae, the retrotransposon family, Ty1, has been shown to play an important role in generating adaptive-genetic variation in chemostats (Adams and Oeller, 1986; Adams et al., 1992; Wilke and Adams, 1992).
However, retrotransposon sequences serve two distinct roles in generating genetic variation: first, their repetitive nature increases the frequency with which intra- and interchromosomal recombination events occur; second, they can generate novel alleles through retrotransposition events. The facilitation of genome rearrangements appears to be a particularly important role for transposon sequences as many rearrangements map to these repetitive elements (Dunham et al., 2002; Zhong et al., 2004; Gresham et al., 2008; Hong and Gresham, 2014). In a small number of cases, novel transposition events in chemostat-evolved lines have been demonstrated to confer fitness benefits (Blanc and Adams, 2003; Gresham et al., 2008; Gaffé et al., 2011). However, the contribution of transposons to adaptive evolution in chemostats remains poorly understood. One of the potential perils of using next generation sequencing, with its current reliance on short read sequences, is that transposition events may be missed. Therefore, employing methods that specifically identify novel insertion sites of retrotransposon (Gabriel et al., 2006; van Opijnen et al., 2009; Mularoni et al., 2012) remains an important aspect of analyzing genomes of evolved lineages.
IDENTIFICATION OF GENETIC NETWORKS UNDER SELECTION BY WHOLE-GENOME SEQUENCING
Prior to the advent of genome-scale methods for interrogating DNA sequence variation, the identification of nucleotide substitutions associated with adaptive evolution in chemostats largely relied on the identification of adaptation-associated phenotypes with known, or readily testable, genetic bases. In E. coli, the careful analysis of phenotypes in chemostat-evolved lines enabled significant progress in identifying the genetic bases of adaptation in chemostats. Loss of function mutations in rpoS results in decreased glycogen production and catalase activity in selected lineages (Notley-McRobb et al., 2002a), facilitating identification of recurrent selection for loss of rpoS in glucose-limited chemostats (Ferenci, 2003; Kinnersley et al., 2009). Similarly, on the basis of known regulation of the high-affinity glucose transporter encoded by LamB, mutations in the transcriptional repressor mlc and the transcriptional activator malT were identified in lineages adapted to glucose-limited chemostats (Notley-McRobb and Ferenci, 1999b; Kinnersley et al., 2009). The mglBAC regulon, which transports galactose, is highly expressed in glucose-limited chemostats (Hua et al., 2004) in which it also functions as a high-affinity glucose transporter. The repressor of the mglBAC genes, mglD (or galS), is repeatedly mutated in clones adapted to glucose limitation (Notley-McRobb and Ferenci, 1999a). Selection for mgl mutations appears to be specific to growth in aerobic glucose-limited chemostats: in anaerobic glucose-limited chemostat mutations in ptsG, a regulator of the PEP:glucose phosphotransferase system is repeatedly selected (Manche et al., 1999).
The advent of next generation sequencing methods has enabled complete characterization of the spectrum of mutations in E. coli strains selected in chemostats. These unbiased characterizations have confirmed the importance of some of the loci identified in earlier studies including rpoS, mglD, and malT in glucose-limited clones (Wang et al., 2010; Maharjan et al., 2012). Whole-genome sequencing of clones selected in phosphate-limited chemostats identified mutations in rpoS (Wang et al., 2010), providing evidence that loss of rpoS is not a specific adaptive response to glucose limitation.
Other stress responsive pathways may be general targets of selection. Sequencing of clones evolved in phosphate-limited chemostats also identified mutations in hfq, which encodes an RNA chaperone important for post-transcriptional regulation of many transcripts by small RNAs. Reduction of ppGpp production could potentially confer benefits in the chemostat as it inhibits cell growth. However, although ppGpp levels are high in E. coli cells growing in chemostats, whole-genome sequencing of lineages adapted to glucose limitation has not yet identified mutations in spoT (Maharjan et al., 2012), a regulator of ppGpp production, whereas mutations in spoT have been identified in lineages adapted to phosphate limitation (Wang et al., 2010). Interestingly, mutations in spoT are also frequently selected in experimental evolution using serial dilution (Herron and Doebeli, 2013).
In contrast to studies in E. coli, the absence of characteristic phenotypes of known genetic bases in S. cerevisiae strains evolved in chemostats impeded the identification of point mutations using candidate gene approaches in early studies. This was initially solved by the use of tiling DNA microarray-based methods to comprehensively identify genomic variation throughout the genome (Gresham et al., 2006; Kao and Sherlock, 2008). A notable finding from early studies of accumulated sequence variation in S. cerevisiae lineages selected in chemostat was the relatively small number of mutations (Gresham et al., 2006, 2008; Kao and Sherlock, 2008). Thus, mutator strains, which have been reported in experimental evolution studies of E. coli using chemostats (Cox and Gibson, 1974; Notley-McRobb et al., 2002b,c) and serial dilution (Arjan et al., 1999), do not seem to contribute to adaptive evolution of S. cerevisiae in chemostat environments.
More recently, next generation sequencing has been used to define the complete spectrum of mutations in many S. cerevisiae lineages selected in chemostats. Among repeated studies in glucose-limited chemostats, loci that regulate transcription of glucose-responsive genes are frequently mutated. In particular, a negative regulator of the glucose-sensing signal transduction pathway, MTH1, has been found to contain loss of function mutations in several lineages in different labs (Gresham et al., 2008; Kvitek and Sherlock, 2011). In lineages selected in ammonium-limited chemostats, missense mutations in GAT1, which encodes a positive transcriptional regulator of the high-affinity ammonium permease MEP2, are recurrently selected (Hong and Gresham, 2014). Selection in ammonium-limited chemostats also provides a rare example in which a transporter gene has been found to acquire coding sequence variants as multiple independent mutations were found in MEP2 (Hong and Gresham, 2014). A comparative analysis of loci selected in chemostats limited for different nitrogen sources identified repeated selection for variation in VAC14 (Hong and Gresham, 2014). VAC14 is a regulator of phosphatidylinositol-3,5-bisphosphate production with roles in protein trafficking and vacuole biogenesis. This study also found that loss of function mutations in nutrient transporters that are expressed but futile in a particular environment are frequent including GAP1 in urea and allantoin limitation and the proline permease gene PUT4 in arginine limitation (Gresham et al., 2010; Hong and Gresham, 2014). To date, the small number of genomes that have been characterized for S. cerevisiae strains evolved in phosphate- or sulfur-limited chemostats (Gresham et al., 2008; Araya et al., 2010) preclude the identification of common targets of selection in these lineages.
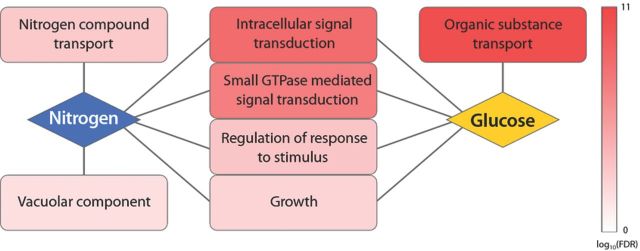
Comparison of loci that acquire adaptive variation in S. cerevisiae lineages evolving in glucose- and nitrogen-limited environments provides evidence for common themes underlying adaptive evolution in chemostats (Fig. 7). Although the repeated selection of the identical gene appears to occur infrequently, the function of genes that are mutated and selected in chemostats shows considerable coherence. Mutated loci are enriched for nutrient transport functions that are specific to the molecular form of the limiting nutrient consistent with the importance of nutrient transport for fitness in the chemostat. Genetic loci underlying adaptive evolution that are common to nitrogen- and glucose-limited environments are enriched for signaling pathways that control cell growth in response to nutrient status. Some of the same specific targets are shared and repeatedly observed to be mutated and selected in different chemostat selections (Hong and Gresham, 2014). In particular, loss of function mutations in RIM15, which encodes a protein kinase downstream of the TORC1 and PKA pathways, is selected in a variety of glucose and nitrogen-limited chemostat selections (Gresham et al., 2008; Kao and Sherlock, 2008; Kvitek and Sherlock, 2011; Hong and Gresham, 2014). RIM15 regulates initiation of a quiescent G0 state in response to different nutrient starvations (Reinders et al., 1998; Pedruzzi et al., 2003), and its loss presumably reduces the probability of a cell entering this state in response to the poor nutrient conditions in the chemostat. Thus, selection for loss of RIM15 in S. cerevisiae lineages evolving in chemostats may be analogous to loss of rpoS in E. coli lineages evolving in similar conditions. It is likely that with deeper sampling and increased replication of selection experiments, additional mechanisms of adaptation in chemostats will be identified. The integration of orthogonal methods for identifying the genetic basis of fitness increases in chemostats, such as the use of pools of gene deletion mutants (Delneri et al., 2007), with the outcomes of long-term selection is a potential way of anticipating and interpreting novel pathways that may be identified as targets of selection.
Figure 7.
Convergence of cellular processes that are targets of adaptive evolution in chemostats. We performed GO term enrichment of networks generated using genemania (Warde-Farley et al., 2010) seeded with loci containing sequence variants identified in lineages and whole populations evolved in glucose-limited and nitrogen-limited chemostats from Gresham et al. (2008), Kvitek and Sherlock (2011) and Hong and Gresham (2014).
DYNAMICS AND CONSTRAINTS OF SELECTION IN CHEMOSTATS
Identification of the targets of selection in the chemostat facilitates the analysis of many questions important for our understanding of the dynamics and constraints of adaptive evolution. Early studies in the chemostat found evidence supporting periodic selection as the dominant mode of adaptation dynamics (Novick and Szilard, 1950b; Paquin and Adams, 1983a). In this regime, genotypes of increasing fitness are sequentially replaced by filter genotypes leading to selective sweeps, which were monitored using a neutral genetic marker. However, the role of periodic selection in the chemostat was questioned on the basis of the expected mutation supply rate (Dykhuizen, 1990), and more recent studies have shown that clonal interference - competition between multiple lineages carrying beneficial mutations - characterizes the evolutionary dynamics in a chemostat (Maharjan et al., 2006; Kao and Sherlock, 2008). Clonal interference prevents genotypes from sweeping to fixation, and as a result, lineages are rarely found at fixation in chemostats (Gresham et al., 2008; Kao and Sherlock, 2008). However, population level sequencing has shown that the extent of genetic heterogeneity is limited, and evolving populations contain only a small number of dominant lineages even when more complex limitations are used (Hong and Gresham, 2014). This points to an important role for chance and luck in the evolutionary dynamics in the chemostat. Once a lineage containing adaptive mutations has risen to an appreciable frequency, it will limit the probability of a similarly fit lineage increasing in frequency even in large populations. New adaptive mutations are then likely to occur in the background of the dominant lineages, which results in the sequential accumulation of adaptive mutations within lineages (Hong and Gresham, 2014). Studies in E. coli suggest a more complex scenario in which multiple lineages with different genotypes, likely corresponding to distinct adaptive strategies, coexist within a population (Maharjan et al., 2012).
The order in which adaptive mutations accumulate is likely to be constrained by epistatic interactions, which impacts both the pace of adaptive evolution and the topology of adaptive landscapes. Analysis of the fitness effect of individual mutations and their combinations from S. cerevisiae lineages selected in glucose-limited chemostats showed that combinations of adaptive mutations are less than the sum of their individuals effects (Kvitek and Sherlock, 2011). This phenomenon, termed ‘diminishing returns epistasis’, has also been observed in E. coli lineages selected in glucose-limited chemostats (Maharjan and Ferenci, 2013) as well as long-term serial dilution selections (Chou et al., 2011; Khan et al., 2011) and may be a general phenomenon of adaptive evolution (Kryazhimskiy et al., 2014).
Although negative epistasis between beneficial mutations may be a general phenomenon, other types of nonadditive genetic interactions may depend more specifically on the functional relationship between the encoded gene products. Sign epistatic interactions occur when individually beneficial mutations are deleterious in the same genetic background (Weinreich et al., 2005). One such example was found for loss of function mutations in MTH1 and amplification of the HXT6/7 locus, which are both adaptive in glucose-limited chemostats. It was observed that both mutations are never found in the same lineage and consistent with a sign epistatic relationship; when both mutations were engineered into the same strain, the strain was less fit than the ancestral lineage (Kvitek and Sherlock, 2011). Analysis of a recurrently selected three-locus genotype comprised of variation in MEP2, GAT1, and LST4 found that variation in LST4 is only beneficial in the background of a mutation in MEP2 or GAT1 consistent with positive epistasis (Hong and Gresham, 2014). Sign epistatic relationships have also been found between rpoS and hfq and between mglD and malT in E. coli (Maharjan and Ferenci, 2013). Interestingly, the negative interaction between mglD and malT mutations is alleviated by the presence of an rpoS loss of function mutation, suggesting that higher order epistasis may be important in chemostat evolution experiments.
The reproducibility of adaptive evolution outcomes in the chemostat has been addressed by performing parallel selection experiments in the same environment. It is clear that there is a finite number of solutions to the selection imposed by a chemostat. However, the extent to which convergent solutions are observed depends on the precise selection. For example, the diversity of outcomes at both the genetic and phenotypic level is far greater for selection in glucose- and phosphate-limited chemostats than sulfur-limited chemostats (Gresham et al., 2008). Similar comparative studies have not yet been performed in E. coli; however, it seems likely that the reproducibility of evolution depends on the distribution of fitness effects associated with adaptive mutations, which will likely differ for different environments.
The beneficial effect of mutations selected in chemostats is uniquely specific to the environment. In fact, many mutations that are beneficial in the chemostat exhibit antagonistic pleiotropy in that they are deleterious in nutrient-rich conditions (Wenger et al., 2011; Hong and Gresham, 2014). Mutations in hfq are beneficial at low dilution rates in which the mutations result in increased expression of glucose transporters; however, the same mutations are deleterious when cells are grown at a higher dilution rate for reasons that remain unclear (Maharjan et al., 2013a). In general, the basis of antagonistic pleiotropy is not well understood and could result from specific functional effects that are deleterious in alternative environments or from the increased load imposed by increased production of particular proteins that are not required in the alternative environment. Understanding the ‘cost’ of protein production requires careful experimentation and interpretation (Stoebel et al., 2008).
CONCLUDING REMARKS AND OPEN QUESTIONS
Several general themes emerge from the accumulated studies of adaptive evolution in chemostats. First, it is clear that selection for improved nutrient transport capabilities in both E. coli and S. cerevisiae underlies adaptive evolution in chemostats. The specificity with which CNVs containing nutrient transporters are selected is consistent with increased nutrient transport production underlying increased growth rates in nutrient-limited environments. Whereas amplification alleles result in increased production of nutrient transporter mRNAs (Hong and Gresham, 2014), adaptive point mutations preferentially target regulators of these same genes. This occurs through a variety of mechanisms including loss of function mutations in negative regulators (e.g. galS and MTH1 mutations) and possible gain of function mutations in positive regulators (e.g. malT and GAT1), which are expected to have the net effect of increasing transporter abundance. Second, signaling pathways, and their downstream targets, that regulate cellular responses to environmental nutritional status are targets of selection in the chemostat in both E. coli and S. cerevisiae. The repeated loss of function mutations in rpoS in E. coli lineages and RIM15 in S. cerevisiae lineages in different nutrient limitations indicates that loss of the capacity to fully induce a stress response in nutrient-limited conditions is beneficial in the chemostat. Interestingly, there is little evidence in either species for selection of enhanced or new enzymatic functions in metabolic genes despite the centrality of metabolic processes for optimized growth in nutrient-limited environments. In addition, there is little evidence that translational regulation of nutrient transporters is a target of selection.
On the foundation of the precise control provided by the chemostat and our accumulated understanding of adaptive evolution in the chemostat from over 60 years of investigation, there are several evolutionary questions that are now readily tackled. It will be important to continue to identify the pathways that are targets of selection constraint and to determine which of these are nutrient-specific responses as opposed to generic solutions to growth by any nutrient. This will allow us to determine how many routes to increased fitness exist within a single selection. These studies will provide insight into the distribution of fitness effects attributable to acquired mutations and how this differs in different selective environments and different genetic backgrounds. The chemostat is uniquely suited to asking how the adaptive response changes with the intensity of selection and variation in population size. Furthermore, the maintenance of constant populations sizes provides the ideal scenario for characterizing the allele frequency dynamics of evolving populations. Finally, precise modulation of the environment enables analysis of the mechanistic basis of antagonistic pleiotropy.
Addressing these questions using chemostats is empowered by an understanding of the structure and function of the genetic networks and cellular processes that control cell growth. The study of protein evolution is making great strides by embracing the ‘functional synthesis’ (Dean and Thornton, 2007) in which molecular biology, biochemistry, and evolutionary biology are integrated. Experimental evolution in chemostats is an ideally suited to extending the functional synthesis to the study of gene networks. The convergence of DNA sequencing methods, a renewed appreciation for the critical importance of cell growth control, and detailed functional annotation of the genomes of E. coli and S. cerevisiae mean that experimental evolution in chemostats is entering an exciting new phase in which the study of adaptive evolution will benefit from renewing the close relationship with cell and molecular biology embraced by the founders of the field (Monod, 1950; Novick and Szilard, 1950).
Acknowledgments
We thank members of the Gresham lab for helpful discussions. Research in the Gresham lab is supported by the National Science Foundation of the USA (MCB-1244219) and the National Institute of Health (GM107466) and a Dupont Corporation Young Professor Award.
Conflict of interest statement. None declared.
REFERENCES
- Adams J. Microbial evolution in laboratory environments. Res Microbiol. 2004;155:311–8. doi: 10.1016/j.resmic.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Adams J, Oeller PW. Structure of evolving populations of Saccharomyces cerevisiae: adaptive changes are frequently associated with sequence alterations involving mobile elements belonging to the Ty family. P Natl Acad Sci USA. 1986;83:7124–7. doi: 10.1073/pnas.83.18.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams J, Puskas-Rozsa S, Simlar J, Wilke CM. Adaptation and major chromosomal changes in populations of Saccharomyces cerevisiae. Curr Genet. 1992;22:13–9. doi: 10.1007/BF00351736. [DOI] [PubMed] [Google Scholar]
- Araya CL, Payen C, Dunham MJ, Fields S. Whole-genome sequencing of a laboratory-evolved yeast strain. BMC Genomics. 2010;11:88. doi: 10.1186/1471-2164-11-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arjan JA, Visser M, Zeyl CW, Gerrish PJ, Blanchard JL, Lenski RE. Diminishing returns from mutation supply rate in asexual populations. Science. 1999;283:404–6. doi: 10.1126/science.283.5400.404. [DOI] [PubMed] [Google Scholar]
- Baganz F, Hayes A, Marren D, Gardner DC, Oliver SG. Suitability of replacement markers for functional analysis studies in Saccharomyces cerevisiae. Yeast. 1997;13:1563–73. doi: 10.1002/(SICI)1097-0061(199712)13:16<1563::AID-YEA240>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Bell G. Selection. 2nd edn. New York, NY: Oxford University Press; 2008. [Google Scholar]
- Bennett A, Hughes B. Microbial experimental evolution. Am J Physiol Regul Integr Comp Physiol. 2009;297:R17–25. doi: 10.1152/ajpregu.90562.2008. [DOI] [PubMed] [Google Scholar]
- Biel SW, Hartl DL. Evolution of transposons: natural selection for Tn5 in Escherichia coli K12. Genetics. 1983;103:581–92. doi: 10.1093/genetics/103.4.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemont C. A brief history of the status of transposable elements: from junk DNA to major players in evolution. Genetics. 2010;186:1085–93. doi: 10.1534/genetics.110.124180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc VM, Adams J. Evolution in Saccharomyces cerevisiae: identification of mutations increasing fitness in laboratory populations. Genetics. 2003;165:975–83. doi: 10.1093/genetics/165.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer VM, Amini S, Botstein D. Influence of genotype and nutrition on survival and metabolism of starving yeast. P Natl Acad Sci USA. 2008;105:6930–5. doi: 10.1073/pnas.0802601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer VM, Crutchfield CA, Bradley PH, Botstein D, Rabinowitz JD. Growth-limiting intracellular metabolites in yeast growing under diverse nutrient limitations. Mol Biol Cell. 2010;21:198–211. doi: 10.1091/mbc.E09-07-0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, Boer VM, Troyanskaya OG, Botstein D. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell. 2008;19:352–67. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach JR. Nutritional control of growth and development in yeast. Genetics. 2012;192:73–105. doi: 10.1534/genetics.111.135731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Todd KM, Rosenzweig RF. Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Mol Biol Evol. 1998;15:931–42. doi: 10.1093/oxfordjournals.molbev.a026009. [DOI] [PubMed] [Google Scholar]
- Buckling A, Craig Maclean R, Brockhurst MA, Colegrave N. The Beagle in a bottle. Nature. 2009;457:824–9. doi: 10.1038/nature07892. [DOI] [PubMed] [Google Scholar]
- Bull AT. The renaissance of continuous culture in the post-genomics age. J Ind Microbiol Biotechnol. 2010;37:993–1021. doi: 10.1007/s10295-010-0816-4. [DOI] [PubMed] [Google Scholar]
- Castrillo JI, Zeef LA, Hayle DC, et al. Growth control of the eukaryote cell: a systems biology study in yeast. J Biol. 2007;6:4. doi: 10.1186/jbiol54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao L, Cox EC. Competition between high and low mutating strains of Escherichia coli. Evolution. 1983;37:125–34. doi: 10.1111/j.1558-5646.1983.tb05521.x. [DOI] [PubMed] [Google Scholar]
- Chao L, Vargas C, Spear BB, Cox EC. Transposable elements as mutator genes in evolution. Nature. 1983;303:633–5. doi: 10.1038/303633a0. [DOI] [PubMed] [Google Scholar]
- Chou H-H, Chiu H-C, Delaney NF, Segre D, Marx CJ. Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science. 2011;332:1190–2. doi: 10.1126/science.1203799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad TM, Lewis NE, Palsson BO. Microbial laboratory evolution in the era of genome-scale science. Mol Syst Biol. 2011;7:1–11. doi: 10.1038/msb.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox EC, Gibson TC. Selection for high mutation rates in chemostats. Genetics. 1974;77:169–84. doi: 10.1093/genetics/77.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean AM, Thornton JW. Mechanistic approaches to the study of evolution: the functional synthesis. Nat Rev Genet. 2007;8:675–88. doi: 10.1038/nrg2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delneri D, Hoyle DC, Gkargkas K, et al. Identification and characterization of high-flux-control genes of yeast through competition analyses in continuous cultures. Nat Genet. 2007;40:113–7. doi: 10.1038/ng.2007.49. [DOI] [PubMed] [Google Scholar]
- Desai MM. Statistical questions in experimental evolution. J Stat Mech. 2013;2013:P01003. [Google Scholar]
- Dettman JR, Rodrigue N, Melnyk AH, Wong A, Bailey SF, Kassen R. Evolutionary insight from whole-genome sequencing of experimentally evolved microbes. Mol Ecol. 2012;21:2058–77. doi: 10.1111/j.1365-294X.2012.05484.x. [DOI] [PubMed] [Google Scholar]
- Dorsey M, Peterson C, Bray K, Paquin CE. Spontaneous amplification of the ADH4 gene in Saccharomyces cerevisiae. Genetics. 1992;132:943–50. doi: 10.1093/genetics/132.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham MJ, Badrane H, Ferea T, Adams J, Brown PO, Rosenzweig F, Botstein D. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. P Natl Acad Sci USA. 2002;99:16144–9. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn B, Paulish T, Stanbery A, Piotrowski J, Koniges G, Kroll E, Louis EJ, Liti G, Sherlock G, Rosenzweig F. Recurrent rearrangement during adaptive evolution in an interspecific yeast hybrid suggests a model for rapid introgression Fay, JC, ed. PLoS Genet. 2013;9:e1003366. doi: 10.1371/journal.pgen.1003366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykhuizen DE. Experimental studies of natural selection in bacteria. Annu Rev Ecol Syst. 1990;21:373–98. [Google Scholar]
- Dykhuizen DE, Hartl DL. Selection in chemostats. Microbiol Rev. 1983;47:150–68. doi: 10.1128/mr.47.2.150-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena SF, Lenski RE. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat Rev Genet. 2003;4:457–69. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- Elliott B, Futcher B. Stress resistance of yeast cells is largely independent of cell cycle phase. Yeast. 1993;9:33–42. doi: 10.1002/yea.320090105. [DOI] [PubMed] [Google Scholar]
- Fedoroff NV. Presidential address. Transposable elements, epigenetics, and genome evolution. Science. 2012;338:758–67. doi: 10.1126/science.338.6108.758. [DOI] [PubMed] [Google Scholar]
- Ferea TL, Botstein D, Brown PO, Rosenzweig RF. Systematic changes in gene expression patterns following adaptive evolution in yeast. P Natl Acad Sci USA. 1999;96:9721–6. doi: 10.1073/pnas.96.17.9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenci T. “Growth of bacterial cultures” 50 years on: towards an uncertainty principle instead of constants in bacterial growth kinetics. Res Microbiol. 1999a;150:431–8. doi: 10.1016/s0923-2508(99)00114-x. [DOI] [PubMed] [Google Scholar]
- Ferenci T. Regulation by nutrient limitation. Curr Opin Microbiol. 1999b;2:208–13. doi: 10.1016/S1369-5274(99)80036-8. [DOI] [PubMed] [Google Scholar]
- Ferenci T. Hungry bacteria – definition and properties of a nutritional state 2008. Environ Microbiol. 2001;3:605–11. doi: 10.1046/j.1462-2920.2001.00238.x. [DOI] [PubMed] [Google Scholar]
- Ferenci T. What is driving the acquisition of mutS and rpoS polymorphisms in Escherichia coli? Trends Microbiol. 2003;11:457–61. doi: 10.1016/j.tim.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Ferenci T. Maintaining a healthy SPANC balance through regulatory and mutational adaptation. Mol Microbiol. 2005;57:1–8. doi: 10.1111/j.1365-2958.2005.04649.x. [DOI] [PubMed] [Google Scholar]
- Ferenci T. A cultural divide on the use of chemostats. Microbiology. 2006;152:1247–8. doi: 10.1099/mic.0.28651-0. [DOI] [PubMed] [Google Scholar]
- Ferenci T. Bacterial physiology, regulation and mutational adaptation in a chemostat environment. Adv Microb Physiol. 2008;53:169–229. doi: 10.1016/S0065-2911(07)53003-1. [DOI] [PubMed] [Google Scholar]
- Gabriel A, Dapprich J, Kunkel M, Gresham D, Pratt SC, Dunham MJ. Global mapping of transposon location. PLoS Genet. 2006;2:e212. doi: 10.1371/journal.pgen.0020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffé J, McKenzie C, Maharjan RP, Coursange E, Ferenci T, Schneider D. Insertion sequence-driven evolution of Escherichia coli in chemostats. J Mol Evol. 2011;72:398–412. doi: 10.1007/s00239-011-9439-2. [DOI] [PubMed] [Google Scholar]
- Garland T, Jr, Rose MR. Experimental Evolution. Berkeley: University of California Press; 2009. [Google Scholar]
- Gresham D, Ruderfer DM, Pratt SC, Schacherer J, Dunham MJ, Botstein D, Kruglyak L. Genome-wide detection of polymorphisms at nucleotide resolution with a single DNA microarray. Science. 2006;311:1932–6. doi: 10.1126/science.1123726. [DOI] [PubMed] [Google Scholar]
- Gresham D, Desai MM, Tucker CM, Jenq HT, Pai DA, Ward A, Desevo CG, Botstein D, Dunham MJ. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 2008;4:e1000303. doi: 10.1371/journal.pgen.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresham D, Usaite R, Germann SM, Lisby M, Botstein D, Regenberg B. Adaptation to diverse nitrogen-limited environments by deletion or extrachromosomal element formation of the GAP1 locus. P Natl Acad Sci USA. 2010;107:18551–6. doi: 10.1073/pnas.1014023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansche PE. Gene duplication as a mechanism of genetic adaptation in Saccharomyces cerevisiae. Genetics. 1975;79:661–74. doi: 10.1093/genetics/79.4.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansche PE, Beres V, Lange P. Gene duplication in Saccharomyces cerevisiae. Genetics. 1978;88:673–87. [PMC free article] [PubMed] [Google Scholar]
- Herron MD, Doebeli M. Parallel evolutionary dynamics of adaptive diversification in Escherichia coli. PLoS Biol. 2013;11:e1001490. doi: 10.1371/journal.pbio.1001490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J, Gresham D. Molecular specificity, convergence and constraint shape adaptive evolution in nutrient-poor environments. PLoS Genet. 2014;10:e1004041. doi: 10.1371/journal.pgen.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi S, Tomizawa J-I, Novick A. Isolation and properties of bacteria capable of high rates of β-galactosidase synthesis. Biochimica et Biophysica Acta (BBA) – Specialized Section on Nucleic Acids and Related Subjects. 1962;55:152–63. doi: 10.1016/0006-3002(62)90941-1. [DOI] [PubMed] [Google Scholar]
- Horiuchi T, Horiuchi S, Novick A. The genetic basis of hyper-synthesis of β-galactosidase. Genetics. 1963;48:157. doi: 10.1093/genetics/48.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskisson PA, Hobbs G. Continuous culture Ndash; making a comeback? Microbiology. 2005;151:3153–9. doi: 10.1099/mic.0.27924-0. [DOI] [PubMed] [Google Scholar]
- Hua Q, Yang C, Oshima T, Mori H, Shimizu K. Analysis of gene expression in Escherichia coli in response to changes of growth-limiting nutrient in chemostat cultures. Appl Environ Microbiol. 2004;70:2354–66. doi: 10.1128/AEM.70.4.2354-2366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Roberts CJ, Dai H, et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet. 2000;25:333–7. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- Ihssen J, Egli T. Specific growth rate and not cell density controls the general stress response in Escherichia coli. Microbiology. 2004;150:1637–48. doi: 10.1099/mic.0.26849-0. [DOI] [PubMed] [Google Scholar]
- Ishii N, Nakahigashi K, Baba T, et al. Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science. 2007;316:593–7. doi: 10.1126/science.1132067. [DOI] [PubMed] [Google Scholar]
- Jin DJ, Cagliero C, Zhou YN. Growth rate regulation in Escherichia coli. FEMS Microbiol Rev. 2011;36:269–87. doi: 10.1111/j.1574-6976.2011.00279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P, Tyers M. How cells coordinate growth and division. Curr Biol. 2004;14:R1014–27. doi: 10.1016/j.cub.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Kao KC, Sherlock G. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat Genet. 2008;40:1499–504. doi: 10.1038/ng.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AI, Dinh DM, Schneider D, Lenski RE, Cooper TF. Negative epistasis between beneficial mutations in an evolving bacterial population. Science. 2011;332:1193–6. doi: 10.1126/science.1203801. [DOI] [PubMed] [Google Scholar]
- Kinnersley MA, Holben WE, Rosenzweig F. E Unibus Plurum: genomic analysis of an experimentally evolved polymorphism in Escherichia coli Guttman, DS, ed. PLoS Genet. 2009;5:e1000713. doi: 10.1371/journal.pgen.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjeldgaard NO, Maaloe O, Schaechter M. The transition between different physiological states during balanced growth of Salmonella typhimurium. J Gen Microbiol. 1958;19:607–16. doi: 10.1099/00221287-19-3-607. [DOI] [PubMed] [Google Scholar]
- Kryazhimskiy S, Rice DP, Jerison E, Desai MM. 2014. Global epistasis makes adaptation predictable despite sequence-level stochasticity. [DOI] [PMC free article] [PubMed]
- Kubitschek HE. Introduction to Research with Continuous Cultures. Englewood Cliffs: Prentice Hall; 1970. [Google Scholar]
- Kvitek DJ, Sherlock G. Reciprocal sign epistasis between frequently experimentally evolved adaptive mutations causes a rugged fitness landscape. PLoS Genet. 2011;7:e1002056. doi: 10.1371/journal.pgen.1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Popodi E, Tang H, Foster PL. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. P Natl Acad Sci USA. 2012;109:E2774–83. doi: 10.1073/pnas.1210309109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman SI, Broach JR. Chapter 16 – Systems Biology and TOR: Past, Present, and Future. 1st edn. Academic Press; 2010. [Google Scholar]
- Loewith R, Hall MN. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics. 2011;189:1177–201. doi: 10.1534/genetics.111.133363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Brauer MJ, Botstein D. Slow growth induces heat-shock resistance in normal and respiratory-deficient yeast. Mol Biol Cell. 2009;20:891–903. doi: 10.1091/mbc.E08-08-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Sung W, Morris K, et al. A genome-wide view of the spectrum of spontaneous mutations in yeast. P Natl Acad Sci USA. 2008;105:9272–7. doi: 10.1073/pnas.0803466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharjan RP, Ferenci T. Epistatic interactions determine the mutational pathways and coexistence of lineages in clonal Escherichia coli populations. Evolution. 2013;67:2762–8. doi: 10.1111/evo.12137. [DOI] [PubMed] [Google Scholar]
- Maharjan R, Seeto S, Notley-McRobb L, Ferenci T. Clonal adaptive radiation in a constant environment. Science. 2006;313:514–7. doi: 10.1126/science.1129865. [DOI] [PubMed] [Google Scholar]
- Maharjan RP, Seeto S, Ferenci T. Divergence and redundancy of transport and metabolic rate-yield strategies in a single Escherichia coli population. J Bacteriol. 2007;189:2350–8. doi: 10.1128/JB.01414-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharjan RP, Ferenci T, Reeves PR, Li Y, Liu B, Wang L. The multiplicity of divergence mechanisms in a single evolving population. Genome Biol. 2012;13:R41. doi: 10.1186/gb-2012-13-6-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharjan R, McKenzie C, Yeung A, Ferenci T. The basis of antagonistic pleiotropy in hfq mutations that have opposite effects on fitness at slow and fast growth rates. Heredity (Edinb) 2013a;110:10–8. doi: 10.1038/hdy.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharjan RP, Gaff JL, Plucain J, Schliep M, Wang L, Feng L, Tenaillon O, Ferenci T, Schneider D. A case of adaptation through a mutation in a tandem duplication during experimental evolution in Escherichia coli. BMC Genomics. 2013b;14:441. doi: 10.1186/1471-2164-14-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manche K, Notley-McRobb L, Ferenci T. Mutational adaptation of Escherichia coli to glucose limitation involves distinct evolutionary pathways in aerobic and oxygen-limited environments. Genetics. 1999;153:5–12. doi: 10.1093/genetics/153.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski PA, Lemoine FJ, Petes TD. Recombination between retrotransposons as a source of chromosome rearrangements in the yeast Saccharomyces cerevisiae. DNA Repair (Amst.) 2006;5:1010–20. doi: 10.1016/j.dnarep.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Monod J. The growth of bacterial cultures. Annu Rev Microbiol. 1949;3:371–94. [Google Scholar]
- Monod J. La technique de culture continue: Théorie et applications. Paris: Annales de l'Institut Pasteur; 1950. pp. 390–410. [Google Scholar]
- Mularoni L, Zhou Y, Bowen T, Gangadharan S, Wheelan SJ, Boeke JD. Retrotransposon Ty1 integration targets specifically positioned asymmetric nucleosomal DNA segments in tRNA hotspots. Genome Res. 2012;22:693–703. doi: 10.1101/gr.129460.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notley L, Ferenci T. Induction of RpoS-dependent functions in glucose-limited continuous culture: what level of nutrient limitation induces the stationary phase of Escherichia coli? J Bacteriol. 1996;178:1465–8. doi: 10.1128/jb.178.5.1465-1468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notley-McRobb L, Ferenci T. Adaptive mgl-regulatory mutations and genetic diversity evolving in glucose-limited Escherichia coli populations. Environ Microbiol. 1999a;1:33–43. doi: 10.1046/j.1462-2920.1999.00002.x. [DOI] [PubMed] [Google Scholar]
- Notley-McRobb L, Ferenci T. The generation of multiple co-existing mal-regulatory mutations through polygenic evolution in glucose-limited populations of Escherichia coli. Environ Microbiol. 1999b;1:45–52. doi: 10.1046/j.1462-2920.1999.00003.x. [DOI] [PubMed] [Google Scholar]
- Notley-McRobb L, King T, Ferenci T. rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. J Bacteriol. 2002a;184:806–11. doi: 10.1128/JB.184.3.806-811.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notley-McRobb L, Pinto R, Seeto S, Ferenci T. Regulation of mutY and nature of mutator mutations in Escherichia coli populations under nutrient limitation. J Bacteriol. 2002b;184:739–45. doi: 10.1128/JB.184.3.739-745.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notley-McRobb L, Seeto S, Ferenci T. Enrichment and elimination of mutY mutators in Escherichia coli populations. Genetics. 2002c;162:1055–62. doi: 10.1093/genetics/162.3.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick A, Szilard L. Description of the chemostat. Science. 1950a;112:715–6. doi: 10.1126/science.112.2920.715. [DOI] [PubMed] [Google Scholar]
- Novick A, Szilard L. Experiments with the chemostat on spontaneous mutations of bacteria. P Natl Acad Sci USA. 1950b;36:708–19. doi: 10.1073/pnas.36.12.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin C, Adams J. Frequency of fixation of adaptive mutations is higher in evolving diploid than haploid yeast populations. Nature. 1983a;302:495–500. doi: 10.1038/302495a0. [DOI] [PubMed] [Google Scholar]
- Paquin CE, Adams J. Relative fitness can decrease in evolving asexual populations of S. cerevisiae. Nature. 1983b;306:368–70. doi: 10.1038/306368a0. [DOI] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Li R. Dr Jekyll and Mr Hyde: role of aneuploidy in cellular adaptation and cancer. Curr Opin Cell Biol. 2010a;22:809–15. doi: 10.1016/j.ceb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010b;468:321–5. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payen C, Koszul R, Dujon B, Fischer G. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet. 2008;4:e1000175. doi: 10.1371/journal.pgen.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedruzzi I, Dubouloz F, Cameroni E, Wanke V, Roosen J, Winderickx J, De Virgilio C. TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol Cell. 2003;12:1607–13. doi: 10.1016/s1097-2765(03)00485-4. [DOI] [PubMed] [Google Scholar]
- Perry GH, Dominy NJ, Claw KG, et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 2007;39:1256–60. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C, Kordich J, Milligan L, Bodor E, Siner A, Nagy K, Paquin CE. Mutations in RAD3, MSH2, and RAD52 affect the rate of gene amplification in the yeast Saccharomyces cerevisiae. Environ Mol Mutagen. 2000;36:325–34. doi: 10.1002/1098-2280(2000)36:4<325::aid-em8>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Peterson CN, Mandel MJ, Silhavy TJ. Escherichia coli starvation diets: essential nutrients weigh in distinctly. J Bacteriol. 2005;187:7549–53. doi: 10.1128/JB.187.22.7549-7553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrykus K, Cashel M. (p)ppGpp: still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- Pronk JT. Auxotrophic yeast strains in fundamental and applied research. Appl Environ Microbiol. 2002;68:2095–100. doi: 10.1128/AEM.68.5.2095-2100.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancati G, Pavelka N, Fleharty B, Noll A, Trimble R, Walton K, Perera A, Staehling-Hampton K, Seidel CW, Li R. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell. 2008;135:879–93. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regenberg B, Grotkjaer T, Winther O, Fausbϕll A, Akesson M, Bro C, Hansen LK, Brunak S, Nielsen J. Growth-rate regulated genes have profound impact on interpretation of transcriptome profiling in Saccharomyces cerevisiae. Genome Biol. 2006;7:R107. doi: 10.1186/gb-2006-7-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders A, Bürckert N, Boller T, Wiemken A, De Virgilio C. Saccharomyces cerevisiae cAMP-dependent protein kinase controls entry into stationary phase through the Rim15p protein kinase. Genes Dev. 1998;12:2943–55. doi: 10.1101/gad.12.18.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero D, Palacios R. Gene amplification and genomic plasticity in prokaryotes. Annu Rev Genet. 1997;31:91–111. doi: 10.1146/annurev.genet.31.1.91. [DOI] [PubMed] [Google Scholar]
- Rosenzweig RF, Sharp RR, Treves DS, Adams J. Microbial evolution in a simple unstructured environment: genetic differentiation in Escherichia coli. Genetics. 1994;137:903–17. doi: 10.1093/genetics/137.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ, Brauer MJ, Botstein D. Nutritional homeostasis in batch and steady-state culture of yeast. Mol Biol Cell. 2004;15:4089–104. doi: 10.1091/mbc.E04-04-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaechter M, Maaloe O, Kjeldgaard NO. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J Gen Microbiol. 1958;19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- Smith HL, Waltman P. The Theory of the Chemostat. Cambridge: Cambridge University Press; 1995. [Google Scholar]
- Sonti RV, Roth JR. Role of gene duplications in the adaptation of Salmonella typhimurium to growth on limiting carbon sources. Genetics. 1989;123:19–28. doi: 10.1093/genetics/123.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivatsan A, Wang JD. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr Opin Microbiol. 2008;11:100–5. doi: 10.1016/j.mib.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Stoebel DM, Dean AM, Dykhuizen DE. The cost of expression of Escherichia coli lac operon proteins is in the process, not in the products. Genetics. 2008;178:1653–60. doi: 10.1534/genetics.107.085399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y-C, Williams BR, Siegel JJ, Amon A. Identification of aneuploidy-selective antiproliferation compounds. Cell. 2011;144:499–512. doi: 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–24. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, Amon A. Identification of aneuploidy-tolerating mutations. Cell. 2010;143:71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP, McKnight SL. The yeast metabolic cycle: insights into the life of a eukaryotic cell. Cold Spring Harb Symp Quant Biol. 2007;72:339–43. doi: 10.1101/sqb.2007.72.019. [DOI] [PubMed] [Google Scholar]
- van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6:767–72. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Spira B, Zhou Z, Feng L, Maharjan RP, Li X, Li F, McKenzie C, Reeves PR, Ferenci T. Divergence involving global regulatory gene mutations in an Escherichia coli population evolving under phosphate limitation. Genome Biol Evol. 2010;2:478–87. doi: 10.1093/gbe/evq035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–20. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich DM, Watson RA, Chao L. Perspective: sign epistasis and genetic constraint on evolutionary trajectories. Evolution. 2005;59:1165–74. [PubMed] [Google Scholar]
- Wenger JW, Piotrowski J, Nagarajan S, Chiotti K, Sherlock G, Rosenzweig F. Hunger artists: yeast adapted to carbon limitation show trade-offs under carbon sufficiency. PLoS Genet. 2011;7:e1002202. doi: 10.1371/journal.pgen.1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke CM, Adams J. Fitness effects of Ty transposition in Saccharomyces cerevisiae. Genetics. 1992;131:31–42. doi: 10.1093/genetics/131.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright D, Boije H, Meadows JRS, et al. Copy number variation in intron 1 of SOX5 causes the Pea-comb phenotype in chickens. PLoS Genet. 2009;5:e1000512. doi: 10.1371/journal.pgen.1000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona AH, Manor YS, Herbst RH, Romano GH, Mitchell A, Kupiec M, Pilpel Y, Dahan O. Chromosomal duplication is a transient evolutionary solution to stress. P Natl Acad Sci USA. 2012;109:21010–5. doi: 10.1073/pnas.1211150109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annu Rev Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
- Zeyl C. Experimental evolution with yeast. FEMS Yeast Res. 2006;6:685–91. doi: 10.1111/j.1567-1364.2006.00061.x. [DOI] [PubMed] [Google Scholar]
- Zhong S, Khodursky A, Dykhuizen DE, Dean AM. Evolutionary genomics of ecological specialization. P Natl Acad Sci USA. 2004;101:11719–24. doi: 10.1073/pnas.0404397101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv N, Brandt NJ, Gresham D. The use of chemostats in microbial systems biology. J Vis Exp. 2013a doi: 10.3791/50168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv N, Siegal ML, Gresham D. Genetic and nongenetic determinants of cell growth variation assessed by high-throughput microscopy. Mol Biol Evol. 2013b;30:2568–78. doi: 10.1093/molbev/mst138. [DOI] [PMC free article] [PubMed] [Google Scholar]