Abstract
Caspase-2 is an atypical caspase that regulates apoptosis, cell cycle arrest and genome maintenance, although the mechanisms are not well understood. Caspase-2 has also been implicated in chemotherapy response in lung cancer, but this function has not been addressed in vivo. Here we show that Caspase-2 functions as a tumor suppressor in Kras-driven lung cancer in vivo. Loss of Caspase-2 leads to enhanced tumor proliferation and progression. Despite being more histologically advanced, Caspase-2-deficient tumors are sensitive to chemotherapy and exhibit a significant reduction in tumor volume following repeated treatment. However, Caspase-2-deficient tumors rapidly rebound from chemotherapy with enhanced proliferation, ultimately hindering long-term therapeutic benefit. In response to DNA damage, Caspase-2 cleaves and inhibits Mdm2 and thereby promotes the stability of the tumor-suppressor p53. Caspase-2 expression levels are significantly reduced in human lung tumors with wild-type p53, in agreement with the model whereby Caspase-2 functions through Mdm2/p53 regulation. Consistently, p53 target genes including p21, cyclin G1 and Msh2 are reduced in Caspase-2-deficient tumors. Finally, we show that phosphorylation of p53-induced protein with a death domain 1 leads to Caspase-2-mediated cleavage of Mdm2, directly impacting p53 levels, activity and chemotherapy response. Together, these studies elucidate a Caspase-2-p53 signaling network that impacts lung tumorigenesis and chemotherapy response in vivo.
Caspase-2 is an atypical member of the cysteine-aspartic acid protease family. Although Caspase-2 is among the first caspases identified and is highly conserved across species, its function is not fully understood.1,2 Studies have focused on the role of Caspase-2 in apoptosis; however, mice lacking Caspase-2 are viable, fertile and undergo normal levels of apoptosis, with the exception of a limited number of cell types.3 Few endogenous Caspase-2 cleavage targets have been identified making it difficult to fully understand Caspase-2 function.4 More recently, it has become appreciated that Caspase-2 has non-apoptotic roles in cell cycle arrest, genome stability and tumor suppression,5, 6, 7 yet the mechanisms behind these processes are incompletely understood.
Loss-of-function mutations and downregulation of Caspase-2 have been observed in a variety of hematological and solid malignancies.8,9 Caspase-2 functions as a tumor suppressor in mouse models of Eμ-Myc-driven lymphoma, c-Neu-driven mammary carcinoma and in SV40/Kras-transformed mouse embryonic fibroblasts.6,9,10 Given that Myc, c-Neu and Kras function in diverse signaling pathways, Caspase-2 may act as a general tumor suppressor that cooperates non-specifically with oncogenes much like the tumor-suppressor p53. The tumor suppressor function of Caspase-2 is dependent on p53, and p53 induction is impaired in Caspase-2-deficient cells, suggesting Caspase-2 and p53 act in the same pathway.5,11
p53 is a transcription factor that mediates the response to a wide variety of cellular stresses and is the most commonly mutated gene across all cancer types, including lung cancer.12,13 Loss of p53 accelerates tumor development, promotes genomic instability and is associated with poor outcome.14,15 Although the role of p53 in chemotherapy response is controversial, in both mice and humans, p53 loss can improve response to chemotherapy in multiple tissues.16, 17, 18 Our previous work in Kras-driven lung tumors suggests that cancer cells utilize wild-type p53 as a resistance mechanism to chronic cisplatin treatment.19
Upon DNA damage, p53 induces expression of p53-induced protein with a death domain 1 (PIDD1).20 PIDD1 promotes the assembly of a complex called the Caspase-2-PIDDosome, leading to the activation of Caspase-2.21 We previously showed that Mdm2, a master regulator and inhibitor of p53, is a cleavage target of Caspase-2.20 Caspase-2 cleaves the C-terminal E3-ubiquitin ligase domain of Mdm2 preventing Mdm2 from targeting p53 for proteasomal degradation.22 Cleavage of Mdm2 increases protein levels of p53, elevates expression of p53 target genes including the cyclin-dependent kinase inhibitor p21, and induces cell cycle arrest.22 It is predicted that Caspase-2 loss leads to increased Mdm2-mediated degradation of p53, ultimately serving as a direct mechanism to explain how Caspase-2 acts as a p53-dependent tumor suppressor. Interestingly, in response to DNA damage, Pidd1 is upregulated in chemoresistant Kras-driven lung tumors, presumably leading to higher Caspase-2 activity and p53-mediated cell cycle arrest.19 However, the role of Caspase-2 in Kras-driven lung tumor development and chemotherapy response has not yet been addressed.
Here, we investigate the role of Caspase-2 in KrasG12D-driven lung tumorigenesis and chemotherapy response in vivo.
Results
Caspase-2 levels are significantly reduced in human lung adenocarcinoma with wild-type p53
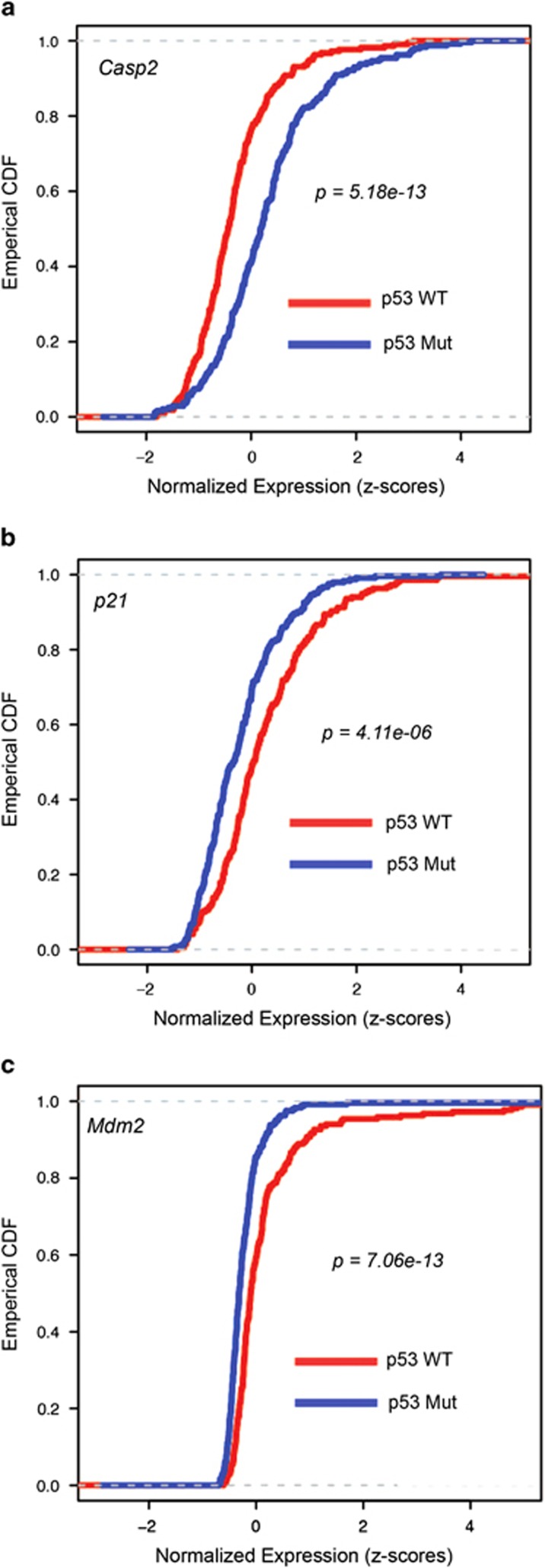
Our previous work in lung cancer demonstrated that Caspase-2 directly enhances p53 function through Mdm2 cleavage.22 Based on this model, we reasoned that selective pressure to lose Caspase-2 may occur preferentially in p53 wild-type lung tumors. To address this, we analyzed human lung adenocarcinoma data from The Cancer Genome Atlas (TCGA) publicly available data portal, containing both somatic mutation calls and gene expression profiles. Protein-altering mutations in Caspase-2 and p53 were observed in 2% (11/538 tumors with somatic mutations) and 53% of human lung adenocarcinomas, respectively. Caspase-2 mutant tumors were marginally enriched for p53 mutations (9/11 tumors, P=0.0398, hypergeometric test). In addition, global downregulation of Caspase-2 mRNA has been observed in human cancers.9 To probe the association between Caspase-2 expression levels and p53 mutation status, lung adenocarcinomas (459 tumors with mutation and gene expression data) were stratified based on p53 mutation status. We observed that 47% of tumors (217 of 459) were p53 wild-type versus 53% of tumors (242 of 459) harbored p53 mutations. Caspase-2 levels were significantly lower in p53 wild-type tumors compared with p53 mutant tumors (Figure 1a). p53 target genes, p21 and Mdm2, however, were significantly enriched in p53 wild-type tumors (Figures 1b and c). We also investigated expression of components of the Caspase-2-PIDDosome, Pidd1 and Raidd/Cradd. Pidd1 expression was not associated with p53 status, whereas Raidd expression was enriched in p53 wild-type tumors (Supplementary Figure S1). These data suggest that Caspase-2 mRNA downregulation is selectively enriched in p53 wild-type tumors, consistent with the model that Caspase-2 functions as a p53-dependent tumor suppressor through Mdm2 cleavage.
Figure 1.
Caspase-2 levels are significantly reduced in human lung cancers with wild-type p53. Empirical cumulative density functions (ECDFs) of normalized and standardized expression values for Caspase-2 (a), p21 (b) and Mdm2 (c) are shown for TCGA lung adenocarcinoma tumors with protein-altering p53 mutations (blue) or tumors with wild-type p53 (red). In general, ECDF (y axis) estimates the fraction of tumors at or below a given value of gene expression. X axis represents normalized expression of indicated gene. P-values indicated in the figures. Left shift in a indicates lower expression of Caspase-2 in p53 wild-type tumors. Right shift in b and c indicates higher expression levels of p21 and Mdm2 in p53 wild-type tumors
Caspase-2 is a tumor suppressor in Kras-driven lung cancer
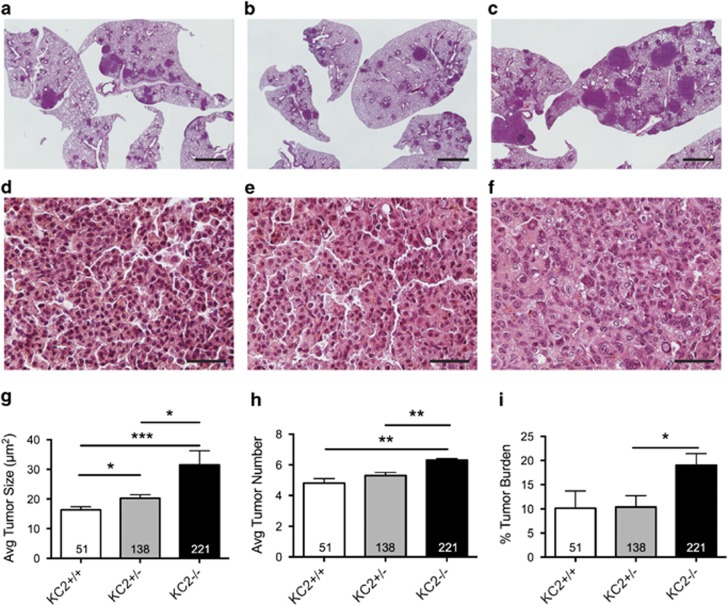
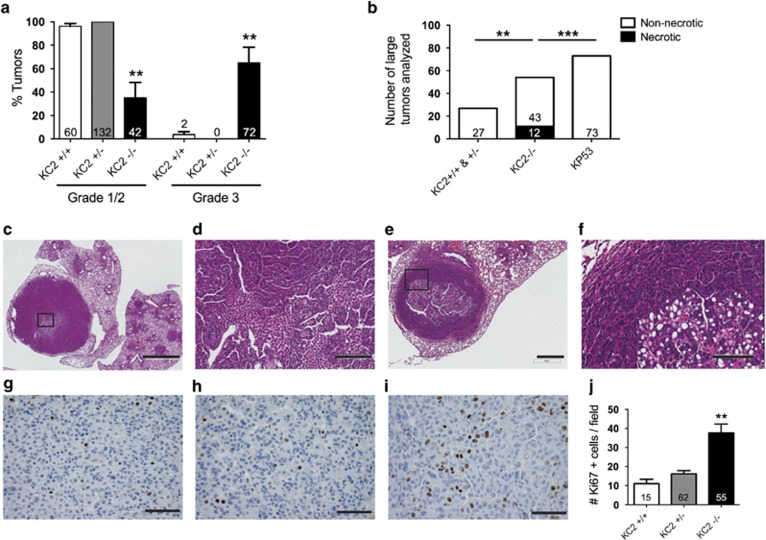
Next, we sought to determine the impact of Caspase-2 loss using a mouse model of lung adenocarcinoma, the Lox-Stop-Lox (LSL)-KrasG12D/+ mice.23 Lung tumors were generated in LSL-KrasG12D/+ mice that were Casp2−/− (KC2−/−), Casp2+/− (KC2+/−) or Casp2+/+ (KC2+/+) by intranasal administration of adenovirus carrying Cre recombinase (AdCre). Mice were killed 12 weeks after tumor initiation and the average tumor size, number and percent tumor burden were quantified from hematoxylin and eosin (H&E)-stained lung sections. KC2−/− mice harbored more lung tumors with an increased average size, contributing to a significantly higher tumor burden compared with KC2+/− and KC2+/+ littermates (Figures 2a–i). In addition, tumors in KC2−/− mice were histologically more advanced compared with KC2+/− and KC2+/+ tumors (Figures 2d–f). KC2−/− tumors were mostly high-grade (>60% grade 3) as opposed to low-grade KC2+/− and KC2+/+ tumors (100% and >96% grade 1/2, respectively; Figure 3a). Interestingly, we observed large tumors with necrotic centers specifically in KC2−/− lungs, which we have not previously observed in KrasG12D-only lungs. We compared tumors of similar size from each genotype as well as KrasG12D/p53-null lungs that produce higher grade tumors, and Caspase-2 null tumors had significantly more necrotic centers than either genotype (Figures 3b–f). In total, nine tumors with necrotic centers were identified in 20 KC2−/− animals (45% frequency). To determine why tumors were larger and more advanced in the absence of Caspase-2, we quantified tumors for cellular proliferation using Ki67 and BrdU antibodies. Tumors in KC2−/− mice exhibited increased proliferation compared with KC2+/− and KC2+/+ tumors (Figures 3g–j, Supplementary Figure S2A). To test whether Caspase-2 loss impacts basal apoptotic rates, we stained tumor sections with antibodies to cleaved Caspase-3 (CC3) or via TUNEL. We observed low basal apoptotic rates that were not significantly altered upon Caspase-2 loss (Supplementary Figure S2BC). High lung tumor burden in KC2−/− mice was associated with a reduction in overall survival compared with heterozygous but not wild-type littermates (Supplementary Figure S2D). Taken together, these data suggest that Caspase-2 functions as a tumor suppressor in lung cancer by regulating cellular proliferation.
Figure 2.
Caspase-2 is a tumor suppressor in Kras-driven lung cancer. (a–f) Representative H&E histology of KC2+/+ (a, d), KC2+/− (b, e) and KC2−/− (c, f) lungs 12 weeks after induction of tumorigenesis at x1 and x40 magnification, respectively. Scale bar in a–c represents 2 mm. Scale bar in d–f represents 50 μm. (g) Average lung tumor size in KC2+/+, KC2+/− and KC2−/− mice, *P<0.01, ***P<0.0001. (h) Average tumor number per lung area in cross-section in KC2+/+, KC2+/− and KC2−/− mice, **P<0.001. (i) Tumor area/total lung area (% tumor burden) in KC2+/+, KC2+/− and KC2−/− mice, *P<0.04. Error bars represent standard error of the mean (SEM). Numbers in each bar indicate the number of tumors quantified per genotype
Figure 3.
Caspase-2 deficiency enhances tumor proliferation and progression. (a) Percent low-grade (grade 1/2) and advanced grade (grade 3) tumors in KC2+/+, KC2+/− and KC2−/− mice 12 weeks after tumor induction, **P<0.001 compared with KC2+/+. (b) Number of necrotic lung tumors of largest tumors present in KC2+/+ and KC2+/− (n=23), KC2−/− (n=20) and Kp53fl/fl (KP53) mice (n=7). **P<0.0069, ***P<0.0001 using Fisher's exact test, two-tailed. (c–f) Two representative tumors with necrotic centers observed exclusively in KC2−/− mice. Insets indicated by black squares in c and e. Scale bars represent 2 mm (c), 200 μm (d), 400 μm (e) and 100 μm (f). (g–i) Representative Ki67 staining in lung sections from KC2+/+ (g), KC2+/− (h) or KC2−/− (i) mice 12 weeks after tumor induction, as quantified in j. Scale bars represent 50 μm (g–i). (j) Number of Ki67-positive cells per x40 image field. **P<0.0045 using unpaired Student's t-test compared with KC2+/+. For panels a, b and j, numbers in each bar indicate the number of tumors quantified per genotype
Caspase-2-deficient tumors have reduced p53 activity but respond to chemotherapy in vivo
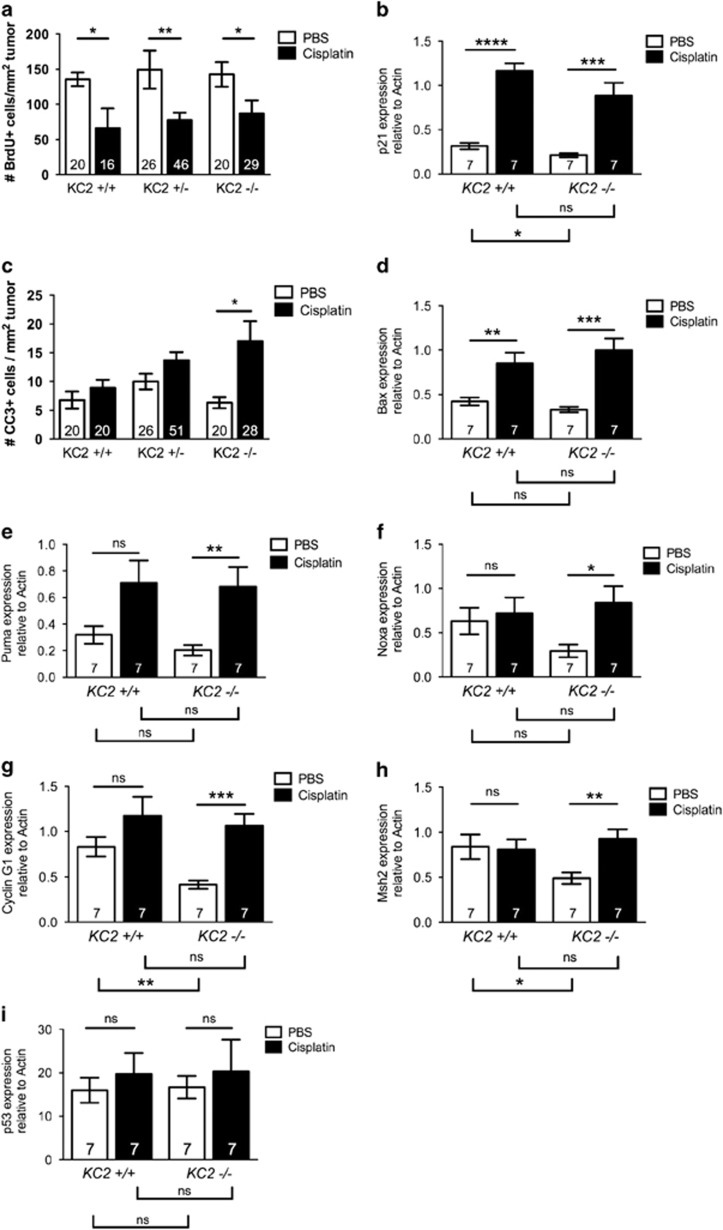
We previously demonstrated that activation of the Caspase-2-PIDDosome promotes chemotherapy resistance in human lung cancer cells, suggesting that loss of Caspase-2 may enhance chemosensitivity. To test whether Caspase-2 impacts the response to acute genotoxic stress, mice were treated with a single dose of cisplatin (7 mg/kg) 12 weeks after tumor initiation. We have observed that compared with Ki67 staining, which is retained in non-proliferating cells for a considerable time period,24 BrdU incorporation is a more sensitive measure of cell cycle arrest following chemotherapy treatment. Ki67 staining, however, is more sensitive at detecting basal changes in proliferation, likely because BrdU labels considerably fewer cells in this protocol. To assess proliferation changes following chemotherapy, we analyzed BrdU incorporation and found that cisplatin treatment significantly reduced cellular proliferation regardless of Caspase-2 status (Figure 4a). Whereas BrdU analysis did not detect significant differences in basal tumor proliferation based on Caspase-2 genotype, Ki67 staining of tumors from the same animals revealed a significant increase in tumor proliferation in KC2−/− mice (Supplementary Figure S3A). Consistently, we observed a significant induction of p21 in tumors from mice treated with cisplatin regardless of genotype (Figure 4b). In response to cisplatin, we also observed an increase in CC3-positive cells in KC2−/− mice that was not observed in KC2+/+ or KC2+/− mice at this time point (Figure 4c). Expression of the pro-apoptotic gene Bax was significantly induced in both KC2+/+ and KC2−/− tumors (Figure 4d). These data demonstrate that Caspase-2 deficiency does not hinder initial cell cycle arrest and apoptosis in response to chemotherapy, and may even sensitize tumors to apoptosis.
Figure 4.
Caspase-2-deficient tumors respond to chemotherapy treatment and exhibit reduced p53 activity. (a) Number of BrdU-positive cells per mm2 tumor area in KC2+/+, KC2+/− and KC2−/− mice treated with PBS control (white bars) or cisplatin (7 mg/kg; black bars) and analyzed after 72 h. (b) Expression of p21 in KC2+/+ and KC2−/− mice treated with PBS or cisplatin and analyzed by real-time RT-PCR after 72 h. (c) Number of CC3-positive cells per mm2 tumor area in KC2+/+, KC2+/− and KC2−/− mice treated with PBS or cisplatin and analyzed after 72 h. (d–i) Expression of Bax (d), Puma (e), Noxa (f), Cyclin G1 (g) Msh2 (h) and p53 (i) in KC2+/+ (n=6 mice each) and KC2−/− mice (n=5 mice each) treated with PBS or cisplatin and analyzed by real-time RT-PCR after 72 h. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 using Student's unpaired t-test. NS=not significant. Numbers in each bar indicate the number of tumors quantified per genotype
Our previous work demonstrated that the Caspase-2-PIDDosome promotes p53 activity in a positive feedback loop.22 Therefore, we predicted that loss of Caspase-2 may be associated with a reduction in p53 activity. To address this, we analyzed expression of additional p53 target genes in KC2+/+ and KC2−/− tumors treated with or without a single dose of cisplatin by real-time RT-PCR. Apoptotic genes including Puma and Noxa were induced by cisplatin, and to a significant level in KC2−/− tumors. Although basal levels of apoptotic genes tended to be lower in KC2−/− tumors, these data were not significant (Figures 4e and f). Genes involved in cell cycle arrest and DNA damage repair, including p21, cyclin G1 and Msh2, were significantly lower in KC2−/− tumors at the basal level (Figures 4b, g and h). However, p53 target genes controlling cell cycle arrest and DNA repair were induced upon cisplatin treatment in KC2−/− tumors. These data suggest that basal p53 activity is significantly dampened in the absence of Caspase-2, but that upon DNA damage, KC2−/− tumors are capable of inducing p53 target genes to levels similar to KC2+/+ tumors.
These data imply that Caspase-2-deficient tumors are histogically advanced but maintain p53. To test this directly, we examined levels of p53 mRNA, which did not change upon cisplatin treatment or by Caspase-2 genotype (Figure 4i). To determine whether p53 had acquired point mutations, we sequenced p53 mRNA from macro-dissected lung tumors (n=9-11 tumors per genotype) from KC2+/+ and KC2−/− mice or normal lung control. We did not detect p53 mutations in tumors regardless of Caspase-2 genotype suggesting that Caspase-2-deficient tumors maintain wild-type p53 (Supplementary Figure S3B).
Caspase-2-deficient tumors rapidly resume proliferation following chemotherapy
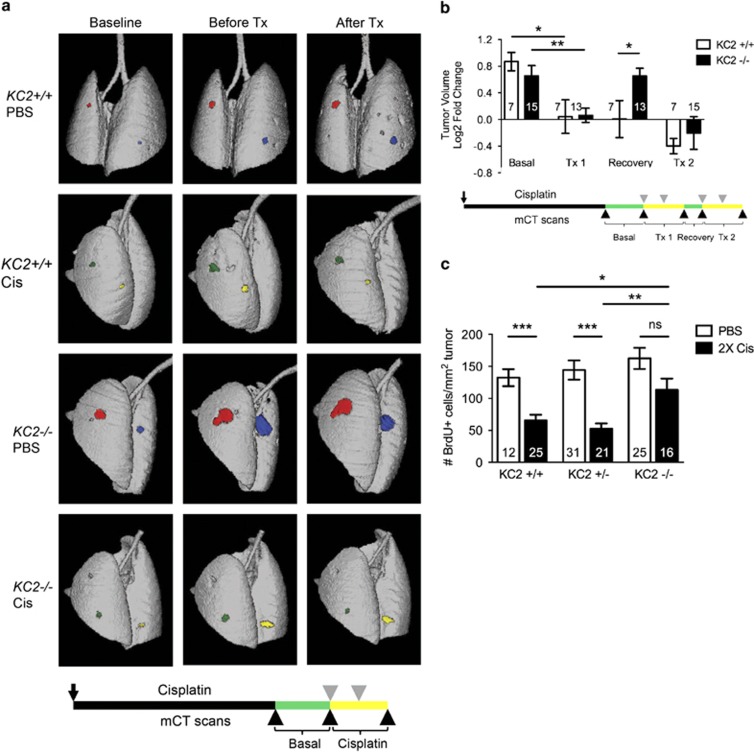
To further investigate cisplatin response in KC2−/− mice, we used micro-computed tomography (microCT) imaging to analyze the response of individual tumors longitudinally. Mice were imaged before and after two doses of cisplatin (one dose per week) and individual tumor volumes were quantified (Figure 5a). Following two doses of cisplatin treatment, lung tumors demonstrated significant growth inhibition irrespective of genotype (Figure 5b). Thus, although loss of Caspase-2 accelerates tumorigenesis, Caspase-2-deficient tumors maintain chemosensitivity similar to mice lacking p53 or p21.17,19
Figure 5.
Caspase-2-deficient tumors rapidly resume proliferation following cisplatin treatment. (a) Representative microCT 3D reconstructions of lungs 12 days before (baseline), 1 day before (before Tx) and 5 days after two doses of PBS or cisplatin (Cis; After Tx). Individual tumors on the lung surface are pseudo-colored for visualization. Below is schematic of longitudinal mCT analysis during acute cisplatin treatment. Black arrow indicates AdCre infection. Gray triangles indicate Cis treatments; black arrowheads indicate mCT scans. 'Basal' (green) corresponds to period of baseline to before Tx. 'Cisplatin' (yellow) corresponds to period of before Tx to after Tx. (b) Log2 tumor volume fold-change of individual tumors in KC2+/+ (white bars) and KC2−/− mice (black bars). Individual tumor volumes were quantified before treatment (basal), before and after doses 1–2 (Tx1), during the recovery period (recovery), and before and after doses 3–4 (Tx2), *P<0.01. **P<0.005. Below is schematic of long-term cisplatin treatment and longitudinal mCT analysis. Mice were infected with AdCre at day 0 (black arrow). Lung tumors were quantified by mCT 2 weeks prior or 1 day before treatment and then 5 days after treatments 1–2 and 3–4 (black arrowheads). Mice were treated with PBS control or cisplatin at weeks 12, 13, 15 and 16 (gray arrowheads). (c) Number of BrdU-positive cells per tumor area in KC2+/+, KC2+/− and KC2−/− mice treated with two doses of PBS (white bars) or two doses of cisplatin (black bars) and analyzed after 10 days, equivalent to the recovery period, ***P<0.0002, **P<0.002, *P<0.012, NS=not significant. Error bars represent SEM. Numbers in each bar graph indicate the number of tumors quantified per genotype
To determine the effect of Caspase-2 on long-term cisplatin response, we used a previously established regimen of cisplatin treatment whereby mice are treated with two doses of cisplatin followed by a 2-week recovery period before receiving two additional doses of cisplatin (four total doses).19 We analyzed tumor volume change before and after the second cycle of cisplatin (doses 3 and 4) by microCT imaging (Figure 5b). Cisplatin-treated tumors in KC2−/− mice displayed a significant reduction in tumor volume in response to the second cycle of cisplatin similar to KC2+/+ tumors, demonstrating that KC2−/− tumors maintain sensitivity to chemotherapy over repeated treatment (Figure 5b). Next, we analyzed tumor growth during the 2-week drug-free recovery period in between doses 2 and 3 (Figure 5b). This period is similar to the 'drug holiday' given to cancer patients to alleviate the negative side effects associated with therapy. Surprisingly, KC2−/− tumors had a significant increase in tumor volume during the recovery phase, whereas no change was detected during this time period in KC2+/+ littermates (Figure 5b). Consistently, lung tumors in cisplatin-treated KC2−/− mice proliferated significantly more than KC2+/+ and KC2+/− tumors during the recovery period as determined by BrdU incorporation (Figure 5c). These findings demonstrate that tumors in KC2−/− mice rapidly re-enter the cell cycle following cisplatin treatment. Loss of Caspase-2, therefore, may limit long-term therapeutic benefit because of enhanced proliferation and cellular recovery following chemotherapy.
Loss of Caspase-2 impacts long-term chemotherapy treatment
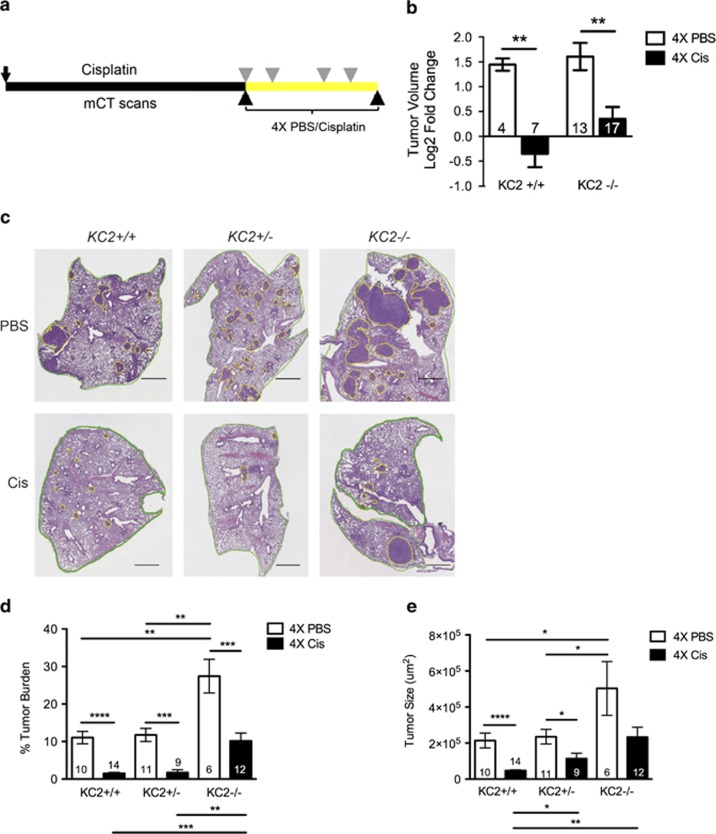
As Caspase-2 loss promoted a rapid rebound following chemotherapy response, we sought to determine the long-term impact of Caspase-2 loss on tumor burden. We treated KC2+/+ and KC2−/− mice with four doses of PBS control or cisplatin. Lung tumor volume was quantified by microCT imaging before the first treatment and 5 days after the fourth treatment (Figure 6a). Untreated tumors from both KC2+/+ and KC2−/− mice grew over time, and cisplatin treatment caused a significant reduction in tumor growth in both genotypes (Figure 6b). To take into consideration the total tumor burden, H&E-stained sections were analyzed for total tumor burden in an independent cohort of mice killed after the fourth dose of cisplatin. Cisplatin caused a reduction in tumor burden irrespective of genotype (Figures 6c and d). However, KC2−/− mice exhibited significantly greater tumor burden following four doses of cisplatin compared with KC2+/+ and KC2+/− littermates (10% versus 2%, respectively; Figure 6d). In addition, tumors remaining in KC2−/− mice following cisplatin treatment were significantly larger than KC2+/+ tumors (Figure 6e). Together, these data demonstrate that Caspase-2 loss impedes the long-term benefit of cisplatin treatment in Kras-driven lung tumors.
Figure 6.
Loss of Caspase-2 impacts long-term chemotherapy treatment. (a) Schematic of long-term cisplatin treatment and longitudinal microCT analysis. Individual tumors were quantified by mCT 1 day before, and 5 days after, four doses of PBS or cisplatin (black arrowheads). Mice were infected with AdCre at day 0 (black arrow) and treated with PBS control or cisplatin (7 mg/kg) at weeks 12, 13, 15 and 16 (gray arrowheads). (b) Log2 tumor volume fold change of individual tumors in KC2+/+ and KC2−/− mice treated with PBS control (white bars) or cisplatin (black bars), as indicated in a, **P<0.001. (c) Representative images of indicated mice treated with four doses of PBS or cisplatin (Cis). Green outlines lung area; yellow outlines tumor. Scale bar represents 1 mm. (d) Quantification of tumor area/total lung area (% tumor burden) of H&E-stained sections in KC2+/+, KC2+/− and KC2−/− mice treated with four doses of PBS or cisplatin, **P<0.003, ***P<0.001, ****P<0.0001. (e) Average tumor size in cross-section from KC2+/+, KC2+/− and KC2−/− mice treated with four doses of PBS or cisplatin. *P<0.04, **P<0.001, ****P<0.0001. Error bars represent SEM. Numbers in each bar graph indicate the number of tumors (b) or mice (d, e) analyzed per genotype
PIDD1 phosphorylation regulates Mdm2 cleavage and p53 activity
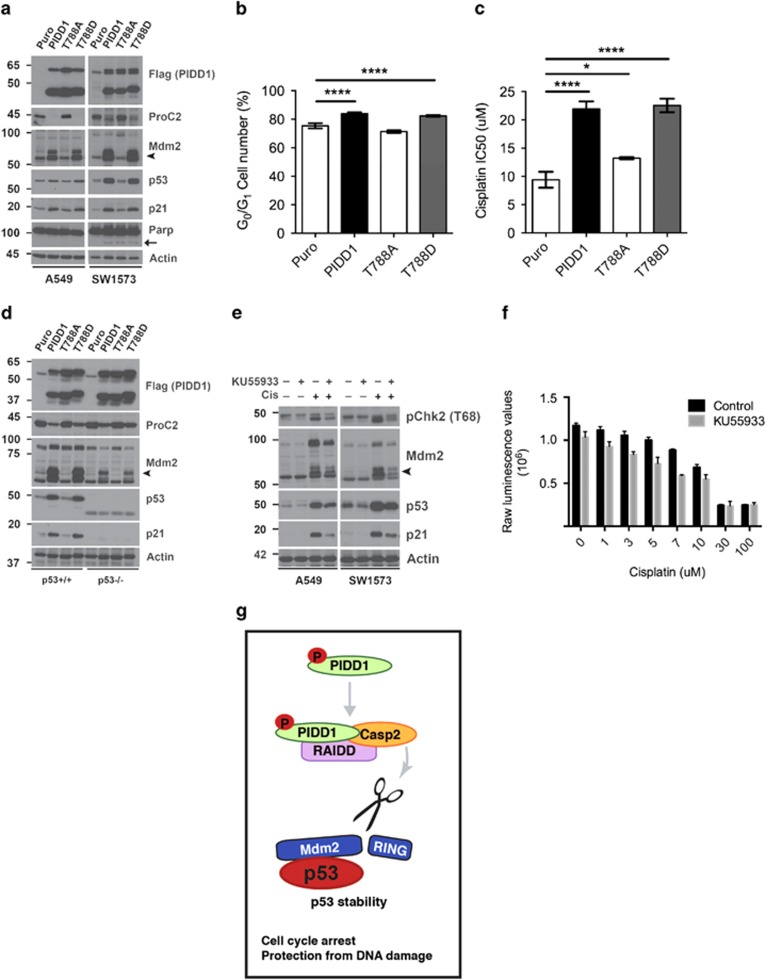
Our previous work demonstrated that high basal phosphorylation of the checkpoint kinase Chk2 and genomic instability were associated with cisplatin-resistant lung tumors in this model.19 High basal phospho-Chk2 suggests that resistant tumors may have increased ATM activity, potentially as a result of genomic instability. Recently, Ando et al. showed that ATM phosphorylates PIDD1 on Thr788 and this phosphorylation is critical for Caspase-2-PIDDosome formation.25 Activation of the Caspase-2-PIDDosome promotes Mdm2 cleavage and resistance to a variety of genotoxic agents in a p53-dependent manner.22 Together, this suggests that an ATM-Caspase-2 signaling pathway may regulate chemoresistance in p53 wild-type lung tumor cells. Whether PIDD1 phosphorylation impacts Caspase-2 cleavage targets and the biological response of p53 wild-type cells has not been tested. To investigate the impact of PIDD1 phosphorylation on Mdm2 cleavage, we stably expressed PIDD1 mutants that either mimic or prevent T788 phosphorylation in p53 wild-type lung cancer cell lines A549 and SW1573 using retroviral infection (Figure 7a). Wild-type and phospho-mimetic PIDD1 T788D, but not phospho-mutant PIDD1 T788A, were sufficient to activate Caspase-2 as demonstrated by the absence of pro-Caspase-2 (Figure 7a). Importantly, only wild-type PIDD1 and PIDD T788D led to Mdm2 cleavage product, p60 (Figure 7a). Mdm2 cleavage was associated with increased p53 and p21 protein levels as predicted (Figure 7a). Upon PIDD1 expression, cells exhibited undetectable or low levels of PARP cleavage (Figure 7a), but this did not lead to apoptosis as assessed by subG1 cell cycle analysis (Supplementary Figure S4A). Consistent with p21 induction, expression of wild-type and T788D PIDD1, but not T788A, led to G1-cell cycle arrest as measured by propidium iodide staining and flow cytometry (Figure 7b). Finally, expression of PIDD1 T788D led to enhanced cell viability in the presence of cisplatin similar to wild-type PIDD1 (Figure 7c). These data demonstrate that PIDD1 phosphorylation directly impacts Mdm2 cleavage, p53 activity and chemotherapy response.
Figure 7.
PIDD1 phosphorylation regulates Mdm2 cleavage and p53 activity. (a) Immunoblot for Flag-tag (PIDD1), pro-Caspase-2 (proC2), Mdm2, p53, p21 and Parp in A549 and SW1573 cells with retroviral overexpression of control (Puro) or indicated PIDD1 constructs. Actin serves as a loading control. Arrowhead indicates cleaved Mdm2, p60; arrow indicates cleaved Parp. (b) Percent of A549 cells in G0/G1 phase of the cell cycle as analyzed by propidium iodide staining by flow cytometry in three independent experiments, ****P <0.0001. (c) Cisplatin IC50s of A549 cells overexpressing indicated constructs in triplicate in three independent experiments, *P <0.05 and ****P <0.0001. (d) Immunoblot for Flag-tag (PIDD1), proC2, Mdm2, p53 and p21 in HCT116 p53+/+ and p53−/− cells with retroviral overexpression of control (Puro) or indicated PIDD1 constructs. Actin serves as a loading control. Arrowhead indicates cleaved Mdm2, p60. (e) Immunoblot analysis of A549 and SW1573 cells treated +/− cisplatin (10 and 9 μM, respectively) +/− KU55933 (10 μM) for 72 h. Whole-cell lysates were blotted for Mdm2, p53 and p21. Actin serves as a loading control and pChk2 serves as functional readout of KU55933 activity. Arrowhead indicates cleaved Mdm2, p60. (f) Raw luminescent values of SW1573 cells treated in triplicate with increasing doses of cisplatin +/− KU55933 or vehicle control, analyzed for cell viability by CTG assay after 72 h. Statistical significance determined using Student's unpaired t-test. (g) PIDD1 phosphorylation regulates Caspase-2-PIDDosome cleavage of Mdm2 and p53 activity. Under conditions of DNA damage, PIDD1 phosphorylation promotes assembly of the Caspase-2-PIDDosome, leading to Caspase-2 activation. Activated Caspase-2 cleaves and inhibits Mdm2, promoting p53 stability and activity. In p53 wild-type cells, our data suggest that p53 activity promotes cell cycle arrest and resistance to DNA damage
To investigate the requirement for p53 in chemoresistance induced by Caspase-2-PIDDosome activation, we performed the same experiments in the p53 isogenic cell lines, HCT116 p53+/+ and p53−/−. Wild-type and T788D PIDD1 promoted Caspase-2 activation and Mdm2 cleavage regardless of p53 status. In the absence of p53, cleaved Mdm2 cannot promote p53 activation and thus, p21 induction is abolished (Figure 7d). These findings support a critical role for PIDD1 T788 phosphorylation in Caspase-2-mediated Mdm2 cleavage and p53-dependent drug resistance.
DNA damage activates p53 leading to PIDD1 expression, activation of the Caspase-2-PIDDosome and Mdm2 cleavage.21,22 We hypothesized that ATM inhibition should block endogenous PIDD1-induced activation of Caspase-2 and reduce Mdm2 cleavage and p53 levels. To test this, we treated cell lines with the ATM inhibitor KU55933 in the presence of a sublethal dose of cisplatin. KU55933 treatment attenuated Caspase-2 activity as demonstrated by reduced accumulation of Mdm2 cleavage product (Figure 7e). Furthermore, KU55933 treatment led to corresponding reductions in p53 and p21 levels (Figure 7e). KU55933 treatment did not effect Mdm2 cleavage in the context of forced overexpression of PIDD1, possibly due to the high levels of PIDD1 expression (Supplementary Figure S4B). Combining KU55933 with cisplatin significantly increased cell death compared with cisplatin alone at multiple concentrations of cisplatin (Figure 7f). Together, these results suggest a model whereby PIDD1 phosphorylation promotes Caspase-2-PIDDosome-mediated cleavage of Mdm2, leading to enhanced p53 activity, cell cycle arrest and drug resistance (Figure 7g).
Discussion
We show that Caspase-2 acts as a tumor suppressor in lung cancer by blocking tumor cell proliferation and progression. Loss of Caspase-2 does not hinder the initial response of lung tumors to chemotherapy. However, Caspase-2-deficient tumors rapidly re-enter the cell cycle following chemotherapy-induced growth arrest, ultimately impeding long-term therapeutic benefit. Caspase-2 thus acts similar to p53 in lung cancer, in that loss of p53 accelerates cell proliferation and tumor progression but is not required for chemosensitivity.19 Our previous work demonstrated that loss of Caspase-2 leads to increased levels of its cleavage target, Mdm2, with a corresponding decrease in p53, ultimately establishing a direct mechanistic link between loss of Caspase-2 and decreased p53 activity.22 Consistently, Caspase-2-deficient tumors have reduced expression of p53 target genes involved in cell cycle arrest and DNA damage repair.
Caspase-2 acts as a tumor suppressor in lymphoma and mammary tumor models driven by expression of Myc and Neu oncogenes, respectively.6,10 Our studies show that loss of Caspase-2 also enhances tumorigenesis in the context of oncogenic Kras in the lung. Caspase-2 mutations have been found in lung cancer and decreased Caspase-2 expression is found in multiple cancer types.9,26 Consistent with the model that Caspase-2 acts as a p53-dependent tumor suppressor, we observe that Caspase-2 levels are significantly reduced in p53 wild-type human adenocarcinomas compared with tumors with p53 mutations. An alternative explanation for this association is that p53 can repress Caspase-2 expression.27 Despite the mechanism by which Caspase-2 expression is reduced in p53 wild-type tumors, this reduction may contribute to reduced p53 activity by abolishing the Caspase-2-Mdm2-p53-positive feedback loop.22
Together, this suggests that Caspase-2 acts as a general tumor suppressor, reminiscent of p53, which cooperates with numerous oncogenes to prevent tumorigenesis in multiple tissues. Our data suggest that the primary mechanism of tumorigenesis upon Caspase-2 loss in vivo is increased cellular proliferation, consistent with in vitro studies where knockdown of Caspase-2 accelerates cell growth.6,28 We observed significant reductions in the expression of basal p53 target genes including p21, cyclin G1 and Msh2, providing a potential mechanistic link to the loss of growth control and advanced histopathology observed in Caspase-2 null tumors. Loss of expression of these genes may be involved in the genome maintenance functions of Caspase-2 as well.5,7
Our previous work implicated the Caspase-2-PIDDosome in drug resistance in p53 wild-type lung tumors, suggesting that loss of Caspase-2 may confer drug sensitivity. Although loss of Caspase-2 did not dramatically enhance chemosensitivity per se, it did not hamper initial drug sensitivity or drug sensitivity following repeated doses, despite the fact that Caspase-2-deficient tumors were more advanced than wild-type tumors. MicroCT imaging and tumor proliferation studies indicated that Caspase-2 loss inhibits the long-term response to chemotherapy by accelerating tumor recovery following DNA damage. Similar to our lung tumor model, mammary tumors with wild-type p53 also arrest in response to chemotherapy, limiting their therapeutic benefit compared with p53-null tumors that continuously respond to treatment.17 Furthermore, a p53-like gene expression signature is associated with platinum-based chemotherapy resistance in human bladder cancer.18
Our previous studies demonstrated that chemoresistant Kras-driven lung tumors have high basal phospho-Chk2, suggesting elevated ATM activity.19 Recent studies demonstrated that ATM phosphorylates PIDD1 and this modification is required for the formation of the Caspase-2-PIDDosome. However, the impact of PIDD1 phosphorylation on Caspase-2 cleavage targets was not investigated.25 We show that phosphorylation of PIDD1 is sufficient to induce Mdm2 cleavage, and enhance p53 activity, cell cycle arrest and drug resistance in human lung cancer cells. Together with our previous report of increased Pidd1 expression in resistant tumors responding to DNA damage, these findings suggest that ATM-induced PIDD1 activity may contribute to cell cycle arrest and chemoresistance in vivo. Although these data suggest that PIDD1 phosphorylation serves as one mechanism of Caspase-2 activation, PIDD1-independent regulation of Caspase-2 has also been reported.11 Future studies should address whether pharmacological modulation of ATM or Caspase-2 in an acute manner enhance chemotherapy response in p53 wild-type tumors.
In contrast to previous studies, our results implicate the Caspase-2-PIDDosome in cell cycle arrest as opposed to apoptosis.25 An important difference between these studies is that PIDD1-induced apoptosis was observed in the context of p53-deficient cells with Chk1 inhibition.25 We observe that PIDD1 phosphorylation dictates Caspase-2 activity and Mdm2 cleavage in p53 wild-type cells in the absence of Chk1 inhibition. This suggests that activation of the Caspase-2-PIDDosome is potentially relevant to a much broader cellular context than previously described and may serve to protect cells from reparable amounts of DNA damage.
Materials and Methods
Mouse breeding and drug treatment
Mice were housed in an environmentally controlled room according to the Committee of Animal Care. LSL-KrasG12D/+ mice were kindly provided by Tyler Jacks and Casp2tm1Yuan-deficient mice were purchased from Jackson Labs (Bar Harbor, ME, USA). Mice were backcrossed over six generations and bred onto a 129svJae background. Mice were infected with 6.47 × 107 plaque-forming units of AdCre (University of Iowa) by intranasal instillation as described previously15 and allowed to develop tumors for 8–12 weeks before cisplatin treatment. Mice were given freshly prepared cisplatin (7 mg/kg body weight in PBS, Sigma, St. Louis, MO, USA) or PBS by intraperitoneal (i.p.) injection. For BrdU labeling experiments, BrdU (Sigma) was injected i.p. (30 mg/kg) 24 h before killing.
Antibodies
Flag-Tag (F3165, 1:1000) and actin (A2066, 1:1000) were obtained from Sigma. BrdU (B44, 1:200) and Ki67 antibodies (556528, 1:300) were obtained from BD Pharmingen (San Jose, CA, USA). Caspase-2 (11B4, 1:500) was obtained from Alexis (Farmingdale, NY, USA). Mdm2 N-terminal (IF2, 1:200) and p53 (DO-1, 1:2000) were obtained from Calbiochem (Billerica, MA, USA). p21 (F-5, 1:100), p53 (FL393, 1:1000) and Mdm2 (SMP14, 1:1000) were obtained from Santa Cruz (Dallas, TX, USA). Cleaved Caspase-3 (9661, 1:500), phospho-Chk2 (T68) (2661, 1:1000), and Parp (9532, 1:1000) were obtained from Cell Signaling Technology (Danvers, MA, USA).
Cell culture and viral preparation
Cell lines 293T, A549, SW1573 and HCT116 p53 isogenic lines were maintained in DMEM. For generating viral supernatants, 293T cells were transfected using Mirus LT-1 transfection reagent with gene plasmids as indicated along with Gag/pol and Env plasmids. Supernatants were harvested after 48–72 h and human cell lines were infected twice with viral supernatant at 1:1 (media/supernatant) with 1000 × polybrene (8 μg/ml). Cells were then selected for 2–3 days in puromycin.
Cell viability assays
Cells were seeded in triplicate (5000/well) in opaque 96-well plates. The next day, cells were treated with increasing doses of cisplatin (0–100 μM). After 48 h of treatment, cell viability was measured using Cell Titer Glo (Promega, Madison, WI, USA) on a luminometer. Normalized, transformed dose response curves were generated and analyzed using GraphPad Prism (GraphPad, La Jolla, CA, USA) to determine IC50. KU55933 was obtained from Selleck Chemicals (Houston, TX, USA) and used at 10 μM by addition 24 h before cisplatin treatment in immunoblot and cell viability assays.
Cell cycle analysis
Cells were harvested with trypsin, washed twice in PBS and 1 × 106 cells were fixed in ice-cold 70% ethanol overnight. Cells were washed 2 × in PBS/1% BSA, treated with 500 μg/ml RNase for 30 min at 37 C, then stained with 50 μg/ml propidium iodide overnight at 4 C. Cells were analyzed on a BD FACScan flow cytometer and cell cycle analysis was performed using FlowJo software (FlowJo LLC, Ashland, OR, USA).
Plasmids
MSCV-Puro and MSCV-PIDD1 were kindly provided by A Tinel and J Tschopp. MSCV-PIDD1 point mutants (T788A and T788D) were cloned by site-directed mutagenesis using the following primers from (Stratagene, Quikchange, La Jolla, CA, USA): Pidd_T788A_For 5-AGACCGGCTTTCTGGCGCAGAGCAACCTG-3; Pidd_T788A_Rev 5-CAGGTTGCTCTGCGCCAGAAAGCCGGTCT-3; Pidd_T788D_For 5-CCGAGACCGGCTTTCTGGATCAGAGCAACCTGCTGAG-3; and Pidd_T788D_Rev 5-CTCAGCAGGTTGCTCTGATCCAGAAAGCCGGTCTCGG-3. Cloning was performed according to the manufacturer's instructions and verified by DNA sequencing.
Histology and Immunohistochemistry
Mice were killed by carbon dioxide asphyxiation and lungs were inflated with PBS, fixed overnight in normalized buffered formalin, and transferred to 70% ethanol. Lung lobes were separated and embedded in paraffin according to the standard procedures. Grossly cutting the lungs served to increase the lung area examined microscopically. Lungs were sectioned at 4 μm and stained with H&E for tumor pathology. Percent tumor burden and all subsequent staining indices per tumor area were quantified using whole slide imaging. H&E-stained slides were digitally scanned using Aperio XT instrument (Aperio Inc., Vista, CA, USA). Image analysis was performed using ImageScope software (Aperio) or AxioVision software (Zeiss, Oberkochen, Germany). For staining with BrdU, CC3 or Ki67 antibodies, 4% normalized buffered formalin-fixed lungs were paraffin embedded. 4 μm sections were dewaxed, rehydrated and subjected to high temperature antigen retrieval, 10 min boiling in a pressure cooker in 0.01 M citrate buffer, pH 6.0. Slides were blocked in 3% H2O2 for 15 min, blocked in 5% goat serum in PBS/0.1% Tween-20 for 1 h, and stained overnight in 5% goat serum in PBS/0.1%Tween-20 with BrdU (1:100), CC3 (1:100 in SignalStain Diluent, Cell Signaling Technology) or Ki67 (1:300) antibodies. A goat anti-mouse or anti-rabbit HRP-conjugated secondary antibody (Vector Laboratories, Burlingame, CA, USA) was used at 1:200 dilution in 5% goat serum in PBS-Triton, incubated for 45 min at room temperature, followed by DAB staining (Vector Laboratories). For CC3 staining, secondary antibody staining was performed using the MOM kit (Vector Laboratories). All staining was performed with Sequenza coverplate technology. IHC slides were visualized using Zeiss Axio Scope.A1 microscope and analyzed using AxioVision SE64 software.
Statistical analysis
All statistical analyses were performed using Graphpad Prism 5.0. For column statistics to determine P-values, unpaired two-tailed Student's t-tests were performed. For IC50 analysis, nonlinear fit-log (inhibitor) versus normalized response (variable slope) was performed. Error bars represent standard error of the mean, unless otherwise noted.
MicroCT imaging
At indicated time points, mice were scanned for 2 min under isoflurane anesthesia using a small animal Quantum FX microCT (Perkin Elmer, Waltham, MA, USA) at 45-μm resolution, 90 kV, with 160-μA current. Images were acquired using Perkin Elmer Quantum FX software and processed with Analyze 11.0 (AnalyzeDirect, Overland Park, KS, USA).
Real-time RT-PCR
For gene expression analysis by real-time RT-PCR, RNA was isolated by TRIzol (Invitrogen, Grand Island, NY, USA) as described19 and 1 μg of total RNA was converted to cDNA using iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). Real-time RT-PCR was performed using gene-specific primers and Sybr Green Supermix (Bio-Rad) in a 20 μl reaction in triplicate on a Bio-Rad CFX96 Real-Time PCR machine. Analysis was performed using Bio-Rad CFX Manager software and expression values were based on 10-fold serial dilutions of standards and normalized to Actin levels. Primers for real-time RT-PCR are given in Table 1.
Table 1. Table 1List of mouse primers used.
| Primer | Sequence (5'–3') | Size (bp) |
|---|---|---|
| Actin B2 Ex5 | GGCATAGAGGTCTTTACGGATGTC | 139 |
| Actin F2 Ex4 | TATTGGCAACGAGCGGTTCC | 139 |
| Bax-F1 | TGCAGAGGATGATTGCTGAC | 173 |
| Bax-R1 | GATCAGCTCGGGCACTTTAG | 173 |
| CCNG1 (Cyc G1) F3 | CTCAGTTCTTTGGCTTTGACACG | 160 |
| CCNG1 (Cyc G1) B14 | TGGGACATTCCTTTCCTCTTCAG | 160 |
| Msh2-F1 | CAGGTGGAAAACCACGAGTT | 226 |
| Msh2-R1 | TGTTGTTGCGAAGCACTTTC | 226 |
| Noxa (Pmaip1) F1 | CCCACTCCTGGGAAAGTACA | 139 |
| Noxa (Pmaip1) R1 | AATCCCTTCAGCCCTTGATT | 139 |
| p21-F | TCCAGACATTCAGAGCCACA | 184 |
| p21-R | ACGAAGTCAAAGTTCCACCG | 184 |
| Puma (bbc3) F | GCCCAGCAGCACTTAGAGTC | 191 |
| Puma (bbc3) R | TGTCGATGCTGCTCTTCTTG | 191 |
| p53 F | TATCCTGCCATCACCTCACTGC | 220 |
| p53 R | GAAGCCATAGTTGCCCTGGTAAG | 220 |
p53 mRNA sequencing
The p53 DNA-binding domain was sequenced from cDNA. RNA was isolated by TRIzol (Invitrogen) from individual tumors as described,19 DNase-1-treated and 1 μg of total RNA was converted to cDNA using iScript cDNA synthesis kit (Bio-Rad). Two PCR reactions covering p53 exons 4–8 and exons 6–11 were purified using QIAquick gel extraction kit (Qiagen, Venlo, Netherlands) and sequenced using Sanger sequencing. Base call and mutation detection analysis was performed manually with ApE software (M.Wayne Davis, Salt Lake City, UT, USA). NCBI Reference Sequence XP_006533220.1 was used as the reference sequence for mutation analysis, and tumors were also compared with normal lung.
Primers used to amplify the p53 DNA-binding domain were: p53 Exon 4F 5-ATCTGTTGCTGCCCCAGGATGTTG-3, p53 Exon 8R 5-AGGCACAAACACGAACCTC-3, p53 Exon 6F 5-AAGACAGGCAGACTTTTCG-3 and p53 Exon 11R 5-AAAAAGGCAGCAGAAGGGACCG-3. Primers used for sequencing were: p53 Exon 4F Seq 5-GCCCCTGTCATCTTTTGTC-3, p53 Exon 7R Seq 5-GAGTCTTCCAGTGTGATGATG-3, p53 Exon 7F Seq 5-AACCGCCGACCTATCCTTAC-3 and p53 Exon 11R Seq 5-CCTGAAGTCATAAGACAGCAAG-3.
Western blotting
Protein lysates were prepared as previously described.19 Protein samples were separated via SDS-PAGE and transferred to a PVDF membrane. Membranes were blocked for 1 h, followed by overnight incubation with primary antibodies at 4 °C. Membranes were washed 6 × 5 min at room temperature in PBS-T. HRP-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA, USA, 1:10 000) were incubated for 1 h at room temperature. For detection, membranes were exposed to WesternBright HRP Quantum substrate (Advansta, Menlo Park, CA, USA) and detected on Hyblot CL film (Denville Scientific Inc., South Plainfield, NJ, USA).
Human lung adenocarcinoma bioinformatics
Gene expression profiles and somatic mutation calls for a large collection of lung adenocarcinoma tumors (n=459 primary tumors from the RNASeqV2 collection) were downloaded from “The Cancer Genome Atlas” (TCGA) data portal (https://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp). Expression levels of selected genes were analyzed between tumors with protein-altering mutations in the P53 gene (n=242 tumors) and tumors with wild-type P53 (n=217). Briefly, RNA-seq gene expression levels were normalized to reads per kilobase exon per million mapped reads (RPKM). Normalized reads per kilobase exon per million mapped expression values for the selected gene were standardized into z-scores (mean=0, s.d.=1) and the nonparametric Kolmogorov–Smirnov test was used to assess significance between the empirical distribution functions of the p53 mutant and wild-type sample sets.
Acknowledgments
Thanks to members of the Oliver Lab for technical support, especially C Lin, U Kc, P Hale, J Clegg, S Pop and J Hidalgo. Thanks to HCI and ARUP for histological services. MRT was funded in part by a NIH Genetics Training grant T32 #GM07464. TGO was supported in part by a Research Scholar Award #RSG-13-300-01-TBG from the American Cancer Society and the National Cancer Institute of the National Institutes of Health #R01CA187457. TGO is a Damon Runyon-Rachleff Innovator and V Foundation Scholar. Thanks to Etienne Meylan and Alejandro Sweet-Cordero for advice and critical reading of the manuscript.
Glossary
- SV40
simian vacuolating virus 40
- c-Neu (V-Erb-B2)
avian erythroblastic leukemia viral oncogene homolog 2
- Kras
Kirsten rat sarcoma viral oncogene homolog
- Myc
v-myc avian myelocytomatosis viral oncogene homolog
- p53
tumor protein p53
- Pidd1
p53 induced death domain protein 1
- Mdm2
mouse double minute 2
- p21 (Cdkn1a)
cyclin-dependent kinase inhibitor 1A
- TCGA
The Cancer Genome Atlas
- Raidd/Cradd
RIP-associated ICH1/CED3-homologous protein with death domain/CASP2 and RIPK1 domain containing adaptor with death domain
- LSL-KrasG12D
Lox-Stop-Lox-KrasG12D
- KC2
LSL-KrasG12D Caspase-2
- AdCre
adenovirus expressing Cre recombinase
- ECDF
empirical cumulative density functions
- KP53
LSL-KrasG12D
- tumor protein p53
CC3, cleaved Caspase 3
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labeling
- Brdu
bromodeoxyuridine
- RT-PCR
real-time quantitative polymerase chain reaction
- Msh2
mutS protein homolog 2
- Bax
bcl-2-associated X protein
- Noxa (Pmaip1)
protein encoded by phorbol-12-myristate-13-acetate-induced protein 1
- Puma
p53 upregulated modulator of apoptosis
- microCT (mCT)
micro-computed tomography
- H&E
hematoxylin and eosin
- Chk2
checkpoint kinase 2
- ATM
ataxia telangiectasia mutated
- Cis
cisplatin
- Tx
treatment
- PBS
phosphate buffered saline
- Puro
puromycin
- ProC2
Pro-Caspase-2
- Parp
Poly ADP ribose polymerase
- IC50
inhibitory concentration 50
- DMEM
Dulbecco's modified Eagle's medium
- CTG
Cell Titer Glo
- MSCV
murine stem cell virus
- H2O2
hydrogen peroxide
- HRP
horseradish peroxidase
- DAB
3,3'-diaminobenzidine
- MOM kit
mouse on mouse kit
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- PVDF
polyvinylidene fluoride
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by B Zhivotovsky
Supplementary Material
References
- Kumar S, Kinoshita M, Noda M, Copeland NG, Jenkins NA. Induction of apoptosis by the mouse Nedd2 gene, which encodes a protein similar to the product of the Caenorhabditis elegans cell death gene ced-3 and the mammalian IL-1 beta-converting enzyme. Genes Dev. 1994;8:1613–1626. doi: 10.1101/gad.8.14.1613. [DOI] [PubMed] [Google Scholar]
- Bouchier-Hayes L, Green DR. Caspase-2: the orphan caspase. Cell Death Differ. 2012;19:51–57. doi: 10.1038/cdd.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron L, Perez GI, Macdonald G, Shi L, Sun Y, Jurisicova A, et al. Defects in regulation of apoptosis in caspase-2-deficient mice. Genes Dev. 1998;12:1304–1314. doi: 10.1101/gad.12.9.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumschnabel G, Sohm B, Bock F, Manzl C, Villunger A. The enigma of caspase-2: the laymen's view. Cell Death Differ. 2009;16:195–207. doi: 10.1038/cdd.2008.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorstyn L, Puccini J, Wilson CH, Shalini S, Nicola M, Moore S, et al. Caspase-2 deficiency promotes aberrant DNA-damage response and genetic instability. Cell Death Differ. 2012;19:1288–1298. doi: 10.1038/cdd.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho LH, Taylor R, Dorstyn L, Cakouros D, Bouillet P, Kumar S. A tumor suppressor function for caspase-2. Proc Natl Acad Sci USA. 2009;106:5336–5341. doi: 10.1073/pnas.0811928106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puccini J, Shalini S, Voss AK, Gatei M, Wilson CH, Hiwase DK, et al. Loss of caspase-2 augments lymphomagenesis and enhances genomic instability in Atm-deficient mice. Proc Natl Acad Sci USA. 2013;110:19920–19925. doi: 10.1073/pnas.1311947110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Kim HS, Jeong EG, Soung YH, Yoo NJ, Lee SH. Somatic mutations of caspase-2 gene in gastric and colorectal cancers. Pathol Res Pract. 2011;207:640–644. doi: 10.1016/j.prp.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Ren K, Lu J, Porollo A, Du C. Tumor-suppressing function of caspase-2 requires catalytic site Cys-320 and site Ser-139 in mice. J Biol Chem. 2012;287:14792–14802. doi: 10.1074/jbc.M112.347625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons MJ, McCormick L, Janke L, Howard A, Bouchier-Hayes L, Green DR. Genetic deletion of caspase-2 accelerates MMTV/c-neu-driven mammary carcinogenesis in mice. Cell Death Differ. 2013;20:1174–1182. doi: 10.1038/cdd.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzl C, Peintner L, Krumschnabel G, Bock F, Labi V, Drach M, et al. PIDDosome-independent tumor suppression by Caspase-2. Cell Death Differ. 2012;19:1722–1732. doi: 10.1038/cdd.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical Lung Cancer Genome Project A genomics-based classification of human lung tumors. Sci Transl Med. 2013;5:209ra153. doi: 10.1126/scitranslmed.3006802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek DW. Tumour suppression by p53: a role for the DNA damage response? Nat Rev Cancer. 2009;9:714–723. doi: 10.1038/nrc2716. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- Bertheau P, Turpin E, Rickman DS, Espie M, de Reynies A, Feugeas JP, et al. Exquisite sensitivity of TP53 mutant and basal breast cancers to a dose-dense epirubicin-cyclophosphamide regimen. PLoS Med. 2007;4:e90. doi: 10.1371/journal.pmed.0040090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JG, Pant V, Li Q, Chang LL, Quintas-Cardama A, Garza D, et al. p53-mediated senescence impairs the apoptotic response to chemotherapy and clinical outcome in breast cancer. Cancer Cell. 2012;21:793–806. doi: 10.1016/j.ccr.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W, Porten S, Suengchan K, Willis D. Identification of Distinct Basal and Luminal Subtypes of Muscle-Invasive Bladder Cancer with Different Sensitivities to Frontline Chemotherapy. Cancer Cell. 2014;25:152–165. doi: 10.1016/j.ccr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver TG, Mercer KL, Sayles LC, Burke JR, Mendus D, Lovejoy KS, et al. Chronic cisplatin treatment promotes enhanced damage repair and tumor progression in a mouse model of lung cancer. Genes Dev. 2010;24:837–852. doi: 10.1101/gad.1897010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Ma W, Benchimol S. Pidd a new death-domain-containing protein, is induced by p53 and promotes apoptosis. Nat Genet. 2000;26:122–127. doi: 10.1038/79102. [DOI] [PubMed] [Google Scholar]
- Tinel A, Tschopp J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science. 2004;304:843–846. doi: 10.1126/science.1095432. [DOI] [PubMed] [Google Scholar]
- Oliver TG, Meylan E, Chang GP, Xue W, Burke JR, Humpton TJ, et al. Caspase-2-mediated cleavage of Mdm2 creates a p53-induced positive feedback loop. Mol Cell. 2011;43:57–71. doi: 10.1016/j.molcel.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuveson DA, Shaw AT, Willis NA, Silver DP, Jackson EL, Chang S, et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- van Dierendonck JH, Keijzer R, van de Velde CJ, Cornelisse CJ. Nuclear distribution of the Ki-67 antigen during the cell cycle: comparison with growth fraction in human breast cancer cells. Cancer Res. 1989;49:2999–3006. [PubMed] [Google Scholar]
- Ando K, Kernan JL, Liu PH, Sanda T, Logette E, Tschopp J, et al. PIDD death-domain phosphorylation by ATM controls prodeath versus prosurvival PIDDosome signaling. Mol Cell. 2012;47:681–693. doi: 10.1016/j.molcel.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- Baptiste-Okoh N, Barsotti AM, Prives C. Caspase 2 is both required for p53-mediated apoptosis and downregulated by p53 in a p21-dependent manner. Cell Cycle. 2008;7:1133–1138. doi: 10.4161/cc.7.9.5805. [DOI] [PubMed] [Google Scholar]
- Puccini J, Dorstyn L, Kumar S. Caspase-2 as a tumour suppressor. Cell Death Differ. 2013;20:1133–1139. doi: 10.1038/cdd.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.