Abstract
H3N2 human influenza A virus causes epidemics of influenza mainly in the winter season in temperate regions. Since the antigenicity of this virus evolves rapidly, several attempts have been made to predict the major amino acid sequence of hemagglutinin 1 (HA1) in the target season of vaccination. However, the usefulness of predicted sequence was unclear because its relationship to the antigenicity was unknown. Here the antigenic model for estimating the degree of antigenic difference (antigenic distance) between amino acid sequences of HA1 was integrated into the process of selecting vaccine strains for H3N2 human influenza A virus. When the effectiveness of a potential vaccine strain for a target season was evaluated retrospectively using the average antigenic distance between the strain and the epidemic viruses sampled in the target season, the most effective vaccine strain was identified mostly in the season one year before the target season (pre-target season). Effectiveness of actual vaccines appeared to be lower than that of the strains randomly chosen in the pre-target season on average. It was recommended to replace the vaccine strain for every target season with the strain having the smallest average antigenic distance to the others in the pre-target season. The procedure of selecting vaccine strains for future epidemic seasons described in the present study was implemented in the influenza virus forecasting system (INFLUCAST) (http://www.nsc.nagoya-cu.ac.jp/~yossuzuk/influcast.html).
Keywords: Influenza virus, Hemagglutinin, Antigenic distance, Thermodynamic stability, Vaccine
Introduction
H3N2 influenza A virus appeared and caused a pandemic in the human population in 1968 (Coleman et al., 1968). Subsequently, this virus has been circulating in humans causing epidemics every winter season in temperate regions (Nelson and Holmes, 2007, World Health Organization, 2009). Vaccines are available for prophylaxis against human influenza viruses (Henle et al., 1943, Francis et al., 1944). The main target of vaccination is hemagglutinin (HA), which is the envelope protein most abundantly exposed on the virion surface (Mitnaul et al., 1996). HA is a homotrimeric type I transmembrane protein glycosylated at the N-linked glycosylation sites (NGS), and the HA1 domain of HA contains the sialic-acid receptor binding sites (RBS) and the neutralization epitopes (Skehel and Wiley, 2000).
The antigenicity of human influenza viruses evolves rapidly due to the high mutation rate, the short generation time, the host immune responses, and the relatively weak functional constraint operating on surface amino acids of HA1 (Suzuki and Nei, 2002, Smith et al., 2004, Suzuki, 2006, Suzuki, 2013a, Bedford et al., 2012). Consequently, vaccines are re-formulated biannually by the Global Influenza Surveillance and Response System of the World Health Organization under the assumption that the dominant strain in a year may dominate in the next year (Treanor, 2004, Russell et al., 2008a). However, this assumption does not always hold and the antigenicity of vaccines sometimes fails to match that of epidemic viruses (Centers for Disease Control and Prevention, 2004, Centers for Disease Control and Prevention, 2010).
To improve the effectiveness of vaccines, several attempts have been made to predict the major amino acid sequence of HA1 in the target season of vaccination (Bush et al., 1999, Xia et al., 2009, He and Deem, 2010, Ito et al., 2011, Suzuki, 2013b, Luksza and Lassig, 2014). However, the precision of prediction was limited by the random genetic drift, the incomplete sampling of epidemic viruses, the occurrence of de novo mutations, and the lack of knowledge on the mechanism of sequence evolution. In addition, the usefulness of predicted sequence was unclear because its relationship to the antigenicity was unknown.
The antigenicity of influenza viruses may be determined by the physical and chemical properties of surface amino acids in HA1 (Hensley et al., 2009, Lees et al., 2010), as well as their relative positions to RBS (Whittle et al., 2011) and NGS (Kobayashi and Suzuki, 2012b). Several methods have been developed for estimating the degree of antigenic difference (antigenic distance) between amino acid sequences of HA1 (Lee and Chen, 2004, Gupta et al., 2006, Lee et al., 2007, Liao et al., 2008, Huang et al., 2009, Lees et al., 2010, Suzuki, 2013b). The purpose of the present study was to integrate the information on antigenicity into the process of selecting vaccine strains for H3N2 human influenza A virus.
Materials and methods
Influenza seasons
H3N2 human influenza A virus causes epidemics of influenza mainly in the winter season in temperate regions. Therefore, in the present study, the influenza seasons are defined such that season 2000.0 indicates the period between October of year 1999 and March of year 2000, corresponding to the winter season in the northern hemisphere, and season 2000.5 indicates the period between April and September of year 2000, corresponding to the winter season in the southern hemisphere (Luksza and Lassig, 2014). The recommended composition of vaccine strains for human influenza viruses has been publicized such that the vaccine strains for season 2001.0 were announced in February of year 2000, and those for season 2001.5 in September of year 2000 (World Health Organization, 2014a, World Health Organization, 2014b). In the present study, the vaccine strain for target season 2001.0 was attempted to be identified from the strains isolated up to the end of season 2000.0, and the vaccine strain for target season 2001.5 from the strains isolated up to the end of season 2000.5, and so forth. Here, seasons 2000.0 and 2000.5 are called the pre-target season for target seasons 2001.0 and 2001.5, respectively.
Antigenic distance
Difference in antigenicity between influenza virus strains can be quantified using the hemagglutination inhibition (HI) titer. HI titer tij is defined as the magnitude of the maximum dilution of anti-serum raised against strain i that inhibits hemagglutination of strain j. It is considered that vaccination by strain i effectively prevents infection by strain j if tij/tii ≤ 4 (antigenic escape threshold). The antigenic distance between strains i and j (dij) is defined as (Lees et al., 2010). Models have been developed for estimating the antigenic distance from the difference in amino acid sequences of HA1 (Lee and Chen, 2004, Gupta et al., 2006, Lee et al., 2007, Liao et al., 2008, Huang et al., 2009, Lees et al., 2010, Suzuki, 2013b).
In the model of Suzuki (2013b), dij is estimated by
where s denotes the amino acid site, vol and pI denote the volume (Grantham, 1974) and the isoelectric point (Nelson and Cox, 2008) of amino acid representing the physical and chemical properties, respectively, and wvol and wpI denote the weights for them. The dRBS and dNGS indicate the Euclidean distances of Cα atom in amino acid from the closest Cα atom in RBS and NGS, respectively, in the three-dimensional structure of HA trimer for A/Hong Kong/19/1968 (Protein Data Bank [PDB] ID: 2HMG). The functions f(x) and g(y) are given by f(x) = e− kx (k > 0) and g(y) = 1 − e− ly (l > 0). The δ is 1 or 0 according to whether the relative solvent accessibility of amino acid is > 0 or 0, respectively, which is obtained by computer program ASAVIEW (Ahmad et al., 2004) using 2HMG.
A total of 1673 antigenic distances experimentally determined between the strains of H3N2 human influenza A virus, for which the nucleotide sequences encoding HA1 were available in the Influenza Virus Resource at the National Center for Biotechnology Information (Bao et al., 2008), were retrieved from the literature (Supplementary Table S1). After eliminating the distances associated with the sequences containing minor gaps, ambiguous nucleotides, or premature termination codons and averaging the distances for the same pairs of strains, 382 distances between 88 strains were available for optimizing model parameters wvol, wpI, k, and l (Supplementary Table S2).
In the present study, the antigenic model was integrated into the process of selecting vaccine strains for H3N2 human influenza A virus. To imitate the real situation of selecting vaccine strains, the model parameters were re-optimized for each target season using only the antigenic distances between the strains that were isolated up to the end of pre-target season. Optimization was performed by minimizing the square sum of residual errors in the estimates of antigenic distances using the genetic algorithm (Tomita et al., 2000). Three sets of random real numbers were assigned as the initial parameter values to confirm the convergence of the results (Suzuki, 2013b).
Sequence data
Effectiveness of a potential vaccine strain for a target season of H3N2 human influenza A virus may be evaluated retrospectively using the average antigenic distance between the strain and the epidemic viruses circulating in the target season; strains with smaller average antigenic distances may be considered as more effective (Luksza and Lassig, 2014). To obtain reliable estimates for the average antigenic distance, it is required to sample a relatively large number of epidemic viruses in the target season, so that they could reflect the composition of epidemic viruses in nature.
Nucleotide sequences encoding HA1 of H3N2 human influenza A virus without minor gaps, ambiguous nucleotides, or premature termination codons, for which the information on the time (year and month) and the place (country) of isolation was available, were retrieved from the Influenza Virus Resource (Bao et al., 2008) on July 25, 2014. The sequences were classified according to the season and the continent (Africa, Asia, Europe, North America, Oceania, or South America) of isolation, and the identical sequences classified in the same category were collapsed into one (Luksza and Lassig, 2014). Since the sample size was relatively large (≥ 5) for the seasons of 1992.0 and later, 6297 sequences were regarded as representatives of epidemic viruses in seasons 1992.0 to 2014.5 (Supplementary Table S3).
Fitness model
A model for predicting the composition of epidemic viruses in season t + 1 from that observed in the sample obtained in season t has been proposed for H3N2 human influenza A virus (Luksza and Lassig, 2014). Given the relative frequency of strain i observed in season t (xi(t)), its relative frequency in season t + 1 (xi(t + 1)) was estimated by
where fi denotes the fitness of strain i determined by the cross-immunity to the former epidemic viruses and the thermodynamic stability of HA1 (Luksza and Lassig, 2014). In the present study, fi was defined as
where ci denotes the sum of cross-immunity to strain j isolated in season τ of 1992.0 ≤ τ ≤ t; that is . The gi indicates the thermodynamic stability of HA1 measured as the difference in the ∆G value from that for A/Hong Kong/19/1968 (PDB ID: 2HMG); that is gi = ∑sΔΔGs, where ∆∆Gs is the difference at amino acid site s estimated by FOLDX (version 3.0 beta 5.1) (Guerois et al., 2002, Schymkowitz et al., 2005). It was assumed that the stability of HA1 is optimum at go and the fitness declines linearly to the difference from it (Bloom and Glassman, 2009). The wc and wg are the weights for cross-immunity and thermodynamic stability, respectively, and f0 is the constant to set .
Similar to the case for the antigenic model, model parameters wc, wg, and go were re-optimized for each target season t as follows. First, in each of season τ, the strain with the smallest average antigenic distance to those sampled in season τ + 1 was identified using the antigenic model. Then, wc, wg, and go were optimized by minimizing the square sum of the estimates of average antigenic distances between the identified strains in season τ and the predicted compositions of epidemic viruses in season τ + 1, using the genetic algorithm (Tomita et al., 2000) with three sets of random real numbers assigned as the initial parameter values to confirm the convergence of the results (Suzuki, 2013b).
Results and discussion
Effectiveness of actual vaccines and the most effective vaccine strains
The average antigenic distances were first computed between the actual vaccines for H3N2 human influenza A virus that have been administered in practice (Table 1) and the epidemic viruses sampled in their target seasons of 2001.0 to 2014.5, to evaluate their effectiveness retrospectively. Here it was assumed that vaccination did not affect the composition of epidemic viruses to any large extent because worldwide coverage of vaccination is small (Osterholm et al., 2012). The average antigenic distances for actual vaccines were found to be relatively large, ranging from 0.892 to 3.170 with the overall average of 1.558 (Table 1). In particular, for 5 out of 28 seasons examined, the average antigenic distance exceeded the antigenic escape threshold of 2. Indeed, the occurrence of antigenic drift and the discrepancy in antigenicity between actual vaccines and epidemic viruses have been reported in some of these seasons (Centers for Disease Control and Prevention, 2004, Centers for Disease Control and Prevention, 2010).
Table 1.
Average antigenic distances between actual vaccines or potential vaccine strains and epidemic viruses sampled in target seasons.
| Target season | Actual vaccine | Antigenic distance | Most effective vaccine straina | Antigenic distance | Center-of-mass strain | Antigenic distance | Strain selected by fitness model | Antigenic distance |
|---|---|---|---|---|---|---|---|---|
| 2001.0 | A/Moscow/10/1999 | 1.487 | A/Malaysia/12974/1999 | 0.687 | A/Nelson Marlborough/1/2000 | 1.221 | N.A. | N.A. |
| 2001.5 | A/Moscow/10/1999 | 1.600 | A/Malaysia/12974/1999 | 0.767 | ||||
| A/Hong Kong/CUHK28038/2000 | 0.822 | A/Western Australia/5/2000 | 0.824 | N.A. | N.A. | |||
| 2002.0 | A/Moscow/10/1999 | 1.613 | A/Netherlands/118/2001 | 0.621 | A/Western Australia/9/2000 | 1.147 | N.A. | N.A. |
| 2002.5 | A/Moscow/10/1999 | 1.726 | A/Queensland/20/2001 | 0.658 | A/West Coast/55/2001 | 1.144 | N.A. | N.A. |
| 2003.0 | A/Moscow/10/1999 | 2.491 | A/Perth/201/2002 | 1.112 | A/New York/22/2002 | 1.790 | N.A. | N.A. |
| 2003.5 | A/Moscow/10/1999 | 2.943 | A/Hunan/407/2002 | 0.565 | A/Western Australia/31/2002 | 2.355 | N.A. | N.A. |
| 2004.0 | A/Moscow/10/1999 | 3.170 | A/Sydney/7/2003 | 0.411 | A/Sydney/7/2003 | 0.411 | N.A. | N.A. |
| 2004.5 | A/Fujian/411/2002 | 0.892 | A/Chanthaburi/219/2003 | 0.374 | A/Uruguay/UY-PA03-4/2003 | 0.629 | N.A. | N.A. |
| 2005.0 | A/Fujian/411/2002 | 1.022 | A/Taiwan/296/2004 | 0.387 | A/Texas/TX-AC24395/2003 | 0.783 | N.A. | N.A. |
| 2005.5 | A/Wellington/1/2004 | 0.894 | A/Jeonnam/336/2004 | 0.502 | ||||
| A/California/CA-S71051/2004 | 0.588 | A/Whanganui/386/2004 | 0.594 | N.A. | N.A. | |||
| 2006.0 | A/California/7/2004 | 1.196 | A/Tianjin/275/2005 | 0.434 | A/Western Australia/63/2004 | 0.730 | N.A. | N.A. |
| 2006.5 | A/California/7/2004 | 1.372 | A/Argentina/AG-R570-05/2005 | 0.496 | A/Argentina/AG-R570-05/2005 | 0.496 | N.A. | N.A. |
| 2007.0 | A/Wisconsin/67/2005 | 1.350 | A/Western Australia/71/2005 | 0.688 | A/Western Australia/71/2005 | 0.688 | N.A. | N.A. |
| 2007.5 | A/Wisconsin/67/2005 | 1.396 | A/Connecticut/CT-5204-7547/2006 | 0.355 | A/Victoria/503/2006 | 0.733 | N.A. | N.A. |
| 2008.0 | A/Wisconsin/67/2005 | 1.511 | A/Wisconsin/03/2007 | 0.351 | A/New York/UR06-0510/2007 | 0.862 | N.A. | N.A. |
| 2008.5 | A/Brisbane/10/2007 | 0.994 | A/LangSon/LS218/2007 | 0.181 | A/Perth/142/2007 | 0.253 | N.A. | N.A. |
| 2009.0 | A/Brisbane/10/2007 | 1.100 | A/Wisconsin/09/2008 | 0.258 | A/Wisconsin/09/2008 | 0.258 | N.A. | N.A. |
| 2009.5 | A/Brisbane/10/2007 | 2.259 | A/Wisconsin/13/2008 | 1.400 | A/Wisconsin/13/2008 | 1.400 | N.A. | N.A. |
| 2010.0 | A/Brisbane/10/2007 | 2.038 | A/Texas/WRAIR1239P/2009 | 0.961 | A/Wisconsin/05/2009 | 1.179 | N.A. | N.A. |
| 2010.5 | A/Perth/16/2009 | 1.166 | A/Australia/30/2009 | 0.512 | A/Hawaii/14/2009 | 1.002 | N.A. | N.A. |
| 2011.0 | A/Perth/16/2009 | 1.317 | A/Pennsylvania/02/2010 | 0.636 | A/Florida/26/2009 | 0.697 | A/Singapore/C2009.803bV/2009 | 1.235 |
| 2011.5 | A/Perth/16/2009 | 1.485 | A/Peru/PER379/2010 | 0.787 | A/Peru/PER379/2010 | 0.787 | A/Wyoming/04/2010 | 0.819 |
| 2012.0 | A/Perth/16/2009 | 1.356 | A/Peru/PER315/2010 | 0.576 | A/Hyogo/10 K304/2011 | 0.898 | A/Sao Paulo/14805/2011 | 0.905 |
| 2012.5 | A/Perth/16/2009 | 1.482 | A/Peru/PER337/2011 | 0.702 | A/Peru/PER337/2011 | 0.702 | A/Peru/PER397/2011 | 0.967 |
| 2013.0 | A/Victoria/361/2011 | 1.404 | A/Wisconsin/07/2012 | 0.553 | A/Peru/PER358/2012 | 0.892 | A/California/2972/2012 | 1.098 |
| 2013.5 | A/Victoria/361/2011 | 1.486 | A/Alabama/17/2012 | 0.359 | A/Peru/PER331/2012 | 0.982 | A/Peru/PER353/2012 | 1.732 |
| 2014.0 | A/Victoria/361/2011 | 1.710 | A/Wyoming/03/2013 | 0.455 | A/Sao Paulo/11079/2013 | 0.615 | A/Colorado/20/2012 | 2.342 |
| 2014.5 | A/Texas/50/2012 | 1.158 | A/Texas/91/2012 | 0.635 | ||||
| A/Oklahoma/3414/2013 | 0.689 | A/Santiago/p36d5/2013 | 0.765 | A/Oklahoma/3414/2013 | 0.689 | |||
| 2015.0 | A/Texas/50/2012 | N.A.b | N.A. | N.A. | A/Guam/3712/2013 | N.A. | A/Guam/3712/2013 | N.A. |
| 2015.5 | A/Switzerland/9715293/2013 | N.A. | N.A. | N.A. | A/Virginia/10/2014 | N.A. | A/Washington/3810/2014 | N.A. |
Two strains are indicated when the most effective vaccine strain identified in seasons 1992.0 to the pre-target season (upper) was different from that identified in the pre-target season (lower).
Not available.
In human influenza viruses, the vaccine strain for target season t is identified at the end of season t − 1 (pre-target season) from the epidemic viruses isolated by then (Treanor, 2004, Russell et al., 2008a). To identify, retrospectively, the most effective vaccine strain for each target season of 2001.0 ≤ t ≤ 2014.5, the average antigenic distance was computed between each of the strains isolated in seasons 1992.0 to t − 1 and the epidemic viruses sampled in season t. The strain with the smallest average antigenic distance was considered as the most effective vaccine strain (Table 1). It was observed that the average antigenic distance for the most effective vaccine strain was always smaller than 2, ranging from 0.181 to 1.400 with the overall average of 0.587 (Table 1). These values were also always smaller than those for actual vaccines obtained above. Notably, the most effective vaccine strain was identified in the pre-target season for 25 out of 28 seasons examined (Table 1).
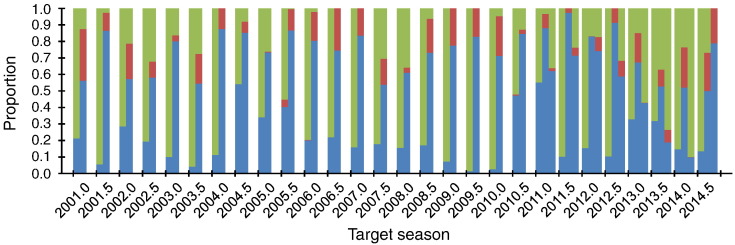
When the average antigenic distance for the actual vaccine was compared with those for the strains isolated in the pre-target season, the former was greater than the latter in 79.1% of the cases on average (same: 0.2%; smaller: 20.7%) (Fig. 1). These results suggest that a strain randomly chosen in the pre-target season was expected to be more effective than the actual vaccine. This is probably because H3N2 human influenza A virus has been continuously escaping from the host immune responses through antigenic evolution (Shih et al., 2007, Suzuki, 2008). It is therefore recommended to replace the vaccine strain for every target season by a strain isolated in the pre-target season.
Fig. 1.
Proportions of epidemic viruses sampled in the pre-target seasons with greater (blue), same (red), and smaller (green) average antigenic distances to those sampled in the target seasons of 2001.0 to 2014.5, in comparison to the actual vaccines (left), the center-of-mass strains (middle), and the strains identified by the fitness model (right). Note that the results of the fitness model could be obtained only for the target seasons of 2011.0 to 2014.5.
Selecting vaccine strains
In replacing the vaccine strain for every target season of H3N2 human influenza A virus, it is preferable to identify the most effective vaccine strain in the pre-target season. For this purpose, it may be helpful to understand the evolutionary dynamics of the virus. It has been proposed that H3N2 human influenza A virus circulates continuously in East and Southeast (E-SE) Asia, and epidemics in temperate regions are seeded from E-SE Asia every winter season (Rambaut et al., 2008, Russell et al., 2008b). Indeed, the phylogenetic tree of HA is known to comprise trunk branches and side clusters, which apparently represent evolutionary pathways of this virus in E-SE Asia and temperate regions, respectively. Furthermore, positive and negative selection has been detected mainly on trunk branches and side clusters, respectively, suggesting that antigenic evolution occurs mainly in E-SE Asia (Fitch et al., 1991, Suzuki, 2008). According to this evolutionary scenario, it would be expected that the viruses circulating in E-SE Asia are not only positioned near the antigenic center of epidemic viruses in each season, but also most closely related to those in the following seasons. Then, the center-of-mass strain, which is defined as the strain with the smallest average antigenic distance to the others, in the pre-target season, may correspond to the most effective vaccine strain for the target season.
The center-of-mass strains identified for the target seasons of 2001.0 to 2014.5 are listed in Table 1. The average antigenic distances of these strains to the epidemic viruses sampled in their target seasons ranged from 0.253 to 2.355 with the overall average of 0.887, which were always smaller than those for actual vaccines (Table 1). The center-of-mass strain turned out to be the most effective vaccine strain for 7 out of 28 seasons examined (Table 1). In each pre-target season, the average antigenic distance for the center-of-mass strain was greater than those for the other strains only in 13.0% of the cases on average (same: 13.9%; smaller: 73.1%), suggesting that the center-of-mass strain was expected to be more effective than a strain randomly chosen in the pre-target season (Fig. 1). Vaccine strains for the future epidemic seasons of 2015.0 and 2015.5 could also be identified as the center-of-mass strains, which were A/Guam/3712/2013 and A/Virginia/10/2014, respectively (Table 1).
It has been proposed that the relative frequency of a strain in season t + 1 may be predicted from that observed in season t using the fitness model, where the fitness of a strain was assumed to be determined by the cross-immunity to the former epidemic viruses and the thermodynamic stability of HA1 (Luksza and Lassig, 2014). Using this model, the vaccine strain may be identified in the pre-target season as the strain with the smallest average antigenic distance to the predicted composition of epidemic viruses in the target season (Luksza and Lassig, 2014). Note that identifying the center-of-mass strain in the pre-target season is equivalent to assuming the same composition of epidemic viruses in the target and pre-target seasons for selecting vaccine strains by the fitness model (Luksza and Lassig, 2014).
In the retrospective analysis, vaccine strains could be identified by the fitness model only for the target seasons of 2011.0 to 2014.5, because of a failure in optimizing the model parameters due to relatively small numbers of data points available for computation (Table 1). In addition, for seven out of eight seasons examined, the average antigenic distance for the vaccine strain identified by the fitness model was greater than that for the center-of-mass strain obtained above (Fig. 1). The vaccine strain for the future epidemic season of 2015.0 identified by the fitness model (A/Guam/3712/2013) was the same as the center-of-mass strain, supporting the reliability. For season 2015.5, however, a different strain (A/Washington/3810/2014) was identified by the fitness model, which was likely to be less reliable than the center-of-mass strain as suggested from the results of the retrospective analysis (Table 1). Similar results were observed when either the cross-immunity or the thermodynamic stability was allowed to be included in the fitness model (data not shown).
The influenza virus forecasting system (INFLUCAST)
In the present study, the effectiveness of a potential vaccine strain for a target season of H3N2 human influenza A virus was evaluated retrospectively using the average antigenic distance between the strain and the epidemic viruses sampled in the target season. The most effective vaccine strain was identified mostly in the pre-target season and the effectiveness of actual vaccines appeared to be lower than that of the strains randomly chosen in the pre-target season on average. It was recommended to replace the vaccine strain for every target season with the center-of-mass strain in the pre-target season. Recently, complicated algorithms based on the phylogenetic analysis have been devised to identify the strains that were likely to be more effective than actual vaccines in the pre-target seasons (Luksza and Lassig, 2014, Steinbruck et al., 2014). However, the strains isolated in the pre-target seasons were expected to be more effective than actual vaccines as indicated above, and thus it may be more meaningful to identify the most effective vaccine strains in the pre-target seasons. The procedure described in the present study provides a simple way directed toward this purpose.
The procedure of selecting vaccine strains for future epidemic seasons described in the present study was implemented in the influenza virus forecasting system (INFLUCAST) (http://www.nsc.nagoya-cu.ac.jp/~yossuzuk/influcast.html). The system should be continuously updated with accumulation of HI and sequence data and upgraded by improving the antigenic model (Wilson et al., 1981, Xia et al., 2012), the measure of thermodynamic stability (Wiley et al., 1981, Han and Marasco, 2011, Kryazhimskiy et al., 2011), and the fitness model (Soundararajan et al., 2011, Ekiert et al., 2012, Kobayashi and Suzuki, 2012a, Omori and Sasaki, 2013). It is also important to incorporate missing variations due to incomplete sampling of epidemic viruses and de novo mutations into the system. The last problem arises because inactivated vaccines are administered to induce systemic humoral immunity against variable proteins for human influenza viruses. This problem may be avoided by targeting conserved proteins as in the case of poliovirus (Suzuki, 2004), or by inducing heterologous immunity with live-attenuated vaccines as in the case of rotavirus (Soares-Weiser et al., 2010, Cortese et al., 2013). It should be noted, however, that the evolutionary mechanism of amino acid sequence may be integrated with that of antigenicity (Suzuki, 2013b), suggesting that the system may be extended to predict the antigenic evolution.
The following are the supplementary data related to this article.
HI titers used in the present study.
Antigenic distances used in the present study.
Representatives of epidemic viruses used in the present study.
Acknowledgments
The author thanks two anonymous reviewers for valuable comments. This work was supported by KAKENHI 26650134 to Y.S.
References
- Ahmad S., Gromiha M.M., Fawareh H., Sarai A. ASAView: database and tool for solvent accessibility representation in proteins. BMC Bioinf. 2004;5:51. doi: 10.1186/1471-2105-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y., Bolotov P., Dernovoy D., Kiryutin B., Zaslavsky L., Tatusova T., Ostell J., Lipman D. The Influenza Virus Resource at the National Center for Biotechnology Information. J. Virol. 2008;82:596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford T., Rambaut A., Pascual M. Canalization of the evolutionary trajectory of the human influenza virus. BMC Biol. 2012;10:38. doi: 10.1186/1741-7007-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J.D., Glassman M.J. Inferring stabilizing mutations from protein phylogenies: application to influenza hemagglutinin. PLoS Comput. Biol. 2009;5:e1000349. doi: 10.1371/journal.pcbi.1000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush R.M., Bender C.A., Subbarao K., Cox N.J., Fitch W.M. Predicting the evolution of human influenza A. Science. 1999;286:1921–1925. doi: 10.1126/science.286.5446.1921. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention . Vol. 53. 2004. pp. 284–287. (Update: influenza activity—United States, 2003–2004 season). [Google Scholar]
- Centers for Disease Control and Prevention . Vol. 59. 2010. pp. 901–908. (Update: influenza activity—United States, 2009–2010 season). [Google Scholar]
- Coleman M.T., Dowdle W.R., Pereira H.G., Schild G.C., Chang W.K. The Hong Kong/68 influenza A2 variant. Lancet. 1968;292:1384–1386. doi: 10.1016/s0140-6736(68)92683-4. [DOI] [PubMed] [Google Scholar]
- Cortese M.M., Immergluck L.C., Held M., Jain S., Chan T., Grizas A.P., Khizer S., Barrett C., Quaye O., Mijatovic-Rustemapasic S., Gautam R., Bowen M.D., Moore J., Tate J.E., Parashar U.D., Vazquez M. Effectiveness of monovalent and pentavalent rotavirus vaccine. Pediatrics. 2013;132:e25–e33. doi: 10.1542/peds.2012-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert D.C., Kashyap A.K., Steel J., Rubrum A., Bhabha G., Khayat R., Lee J.H., Dillon M.A., O’Neil R.E., Faynboym A.M., Horowitz M., Horowitz L., Ward A.B., Palese P., Webby R., Lerner R.A., Bhatt R.R., Ian A., Wilson I.A. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature. 2012;489:526–532. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch W.M., Leiter J.M., Li X.Q., Palese P. Positive Darwinian evolution in human influenza A viruses. Proc. Natl. Acad. Sci. U. S. A. 1991;88:4270–4274. doi: 10.1073/pnas.88.10.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis T., Jr., Salk J., Pearson H.E., Brown P.N. Protective effect of vaccination against induced influenza A. Proc. Soc. Exp. Biol. Med. 1944;55:104–105. [Google Scholar]
- Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- Guerois R., Nielsen J.E., Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J. Mol. Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- Gupta V., Earl D.J., Deem M.W. Quantifying influenza vaccine efficacy and antigenic distance. Vaccine. 2006;24:3881–3888. doi: 10.1016/j.vaccine.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T., Marasco W.A. Structural basis of influenza virus neutralization. Ann. N. Y. Acad. Sci. 2011;1217:178–190. doi: 10.1111/j.1749-6632.2010.05829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Deem M.W. Low-dimensional clustering detects incipient dominant influenza strain clusters. Protein Eng. Des. Sel. 2010;23:935–946. doi: 10.1093/protein/gzq078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henle W., Henle G., Stokes J., Jr. Demonstration of the efficacy of vaccination against influenza type A by experimental infection of human beings. J. Immunol. 1943;46:163–175. [Google Scholar]
- Hensley S.E., Das S.R., Bailey A.L., Schmidt L.M., Hickman H.D., Jayaraman A., Viswanathan K., Raman R., Sasisekharan R., Bennink J.R., Yewdell J.W. Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science. 2009;326:734–736. doi: 10.1126/science.1178258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.-W., King C.-C., Yang J.-M. Co-evolution positions and rules for antigenic variants of human influenza A/H3N2 viruses. BMC Bioinf. 2009;10:S41. doi: 10.1186/1471-2105-10-S1-S41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Igarashi M., Miyazaki Y., Murakami T., Iida S., Kida H., Takada A. Gnarled-trunk evolutionary model of influenza A virus hemagglutinin. PLoS One. 2011;6:e25953. doi: 10.1371/journal.pone.0025953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Suzuki Y. Compensatory evolution of net-charge in influenza A virus hemagglutinin. PLoS One. 2012;7:e40422. doi: 10.1371/journal.pone.0040422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Suzuki Y. Evidence for N-glycan shielding of antigenic sites during evolution of human influenza A virus hemagglutinin. J. Virol. 2012;86:3445–3451. doi: 10.1128/JVI.06147-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryazhimskiy S., Dushoff J., Bazykin G.A., Plotkin J.B. Prevalence of epistasis in the evolution of influenza A surface proteins. PLoS Genet. 2011;7:e1001301. doi: 10.1371/journal.pgen.1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-S., Chen J.S.-E. Predicting antigenic variants of influenza A/H3N2 viruses. Emerg. Infect. Dis. 2004;10:1385–1390. doi: 10.3201/eid1008.040107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-S., Chen M.-C., Liao Y.-C., Hsiung C.A. Identifying potential immunodominant positions and predicting antigenic variants of influenza A/H3N2 viruses. Vaccine. 2007;25:8133–8139. doi: 10.1016/j.vaccine.2007.09.039. [DOI] [PubMed] [Google Scholar]
- Lees W.D., Moss D.S., Shepherd A.J. A computational analysis of the antigenic properties of haemagglutinin in influenza A H3N2. Bioinformatics. 2010;26:1403–1408. doi: 10.1093/bioinformatics/btq160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y.-C., Lee M.-S., Ko C.-Y., Hsiung C.A. Bioinformatics models for predicting antigenic variants of influenza A/H3N2 virus. Bioinformatics. 2008;24:505–512. doi: 10.1093/bioinformatics/btm638. [DOI] [PubMed] [Google Scholar]
- Luksza M., Lassig M. A predictive fitness model for influenza. Nature. 2014;507:57–61. doi: 10.1038/nature13087. [DOI] [PubMed] [Google Scholar]
- Mitnaul L.J., Castrucci M.R., Murti K.G., Kawaoka Y. The cytoplasmic tail of influenza A virus neuraminidase (NA) affects NA incorporation into virions, virion morphology, and virulence in mice but is not essential for virus replication. J. Virol. 1996;70:873–879. doi: 10.1128/jvi.70.2.873-879.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D.L., Cox M.M. 5th edition. W. H. Freeman and Company; New York: 2008. Lehninger Principles of Biochemistry. [Google Scholar]
- Nelson M.I., Holmes E.C. The evolution of epidemic influenza. Nat. Rev. Genet. 2007;8:196–205. doi: 10.1038/nrg2053. [DOI] [PubMed] [Google Scholar]
- Omori R., Sasaki A. Timing of the emergence of new successful viral strains in seasonal influenza. J. Theor. Biol. 2013;329:32–38. doi: 10.1016/j.jtbi.2013.03.027. [DOI] [PubMed] [Google Scholar]
- Osterholm M.T., Kelley M.S., Sommer A., Belongia E.A. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect. Dis. 2012;12:36–44. doi: 10.1016/S1473-3099(11)70295-X. [DOI] [PubMed] [Google Scholar]
- Rambaut A., Pybus O.G., Nelson M.I., Viboud C., Taubenberger J.K., Holmes E.C. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;453:615–619. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell C.A., Jones T.C., Barr I.G., Cox N.J., Garten R.J., Gregory V., Gust I.D., Hampson A.W., Hay A.J., Hurt A.C., de Jong J.C., Kelso A., Klimov A.I., Kageyama T., Komadina N., Lapedes A.S., Lin Y.P., Mosterin A., Obuchi M., Odagiri T., Osterhaus A.D.M.E., Rimmelzwaan G.F., Shaw M.W., Skepner E., Stohr K., Tashiro M., Fouchier R.A.M., Smith D.J. Influenza vaccine strain selection and recent studies on the global migration of seasonal influenza viruses. Vaccine. 2008;26:D31–D34. doi: 10.1016/j.vaccine.2008.07.078. [DOI] [PubMed] [Google Scholar]
- Russell C.A., Jones T.C., Barr I.G., Cox N.J., Garten R.J., Gregory V., Gust I.D., Hampson A.W., Hay A.J., Hurt A.C., de Jong J.C., Kelso A., Klimov A.I., Kageyama T., Komadina N., Lapedes A.S., Lin Y.P., Mosterin A., Obuchi M., Odagiri T., Osterhaus A.D.M.E., Rimmelzwaan G.F., Shaw M.W., Skepner E., Stohr K., Tashiro M., Fouchier R.A.M., Smith D.J. The global circulation of seasonal influenza A (H3N2) viruses. Science. 2008;320:340–346. doi: 10.1126/science.1154137. [DOI] [PubMed] [Google Scholar]
- Schymkowitz J.W.H., Rousseau F., Martins I.C., Ferkinghoff-Borg J., Stricher F., Serrano L. Prediction of water and metal binding sites and their affinities by using the Fold-X force field. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10147–10152. doi: 10.1073/pnas.0501980102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih A.C.-C., Hsiao T.-C., Ho M.-S., Li W.-H. Simultaneous amino acid substitutions at antigenic sites drive influenza A hemagglutinin evolution. Proc. Natl. Acad. Sci. U. S. A. 2007;104:6283–6288. doi: 10.1073/pnas.0701396104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel J.J., Wiley D.C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- Smith D.J., Lapedes A.S., de Jong J.C., Bestebroer T.M., Rimmelzwaan G.F., Osterhaus A.D.M.E., Fouchier R.A.M. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- Soares-Weiser K., Maclehose H., Ben-Aharon I., Goldberg E., Pitan F., Cunliffe N. Vaccines for preventing rotavirus diarrhoea: vaccines in use. Cochrane Database Syst. Rev. 2010:CD008521. doi: 10.1002/14651858.CD008521. [DOI] [PubMed] [Google Scholar]
- Soundararajan V., Zheng S., Patel N., Warnock K., Raman R., Wilson I.A., Raguram S., Sasisekharan V., Sasisekharan R. Networks link antigenic and receptor-binding sites of influenza hemagglutinin: mechanistic insight into fitter strain propagation. Sci. Rep. 2011;1:200. doi: 10.1038/srep00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbruck L., Klingen T.R., McHardy A.C. Computational prediction of vaccine strains for human influenza A (H3N2) viruses. J. Virol. 2014;88:12123–12132. doi: 10.1128/JVI.01861-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y. Negative selection on neutralization epitopes of poliovirus surface proteins: implications for prediction of candidate epitopes for immunization. Gene. 2004;328:127–133. doi: 10.1016/j.gene.2003.11.020. [DOI] [PubMed] [Google Scholar]
- Suzuki Y. Natural selection on the influenza virus genome. Mol. Biol. Evol. 2006;23:1902–1911. doi: 10.1093/molbev/msl050. [DOI] [PubMed] [Google Scholar]
- Suzuki Y. Positive selection operates continuously on hemagglutinin during evolution H3N2 human influenza A virus. Gene. 2008;427:111–116. doi: 10.1016/j.gene.2008.09.012. [DOI] [PubMed] [Google Scholar]
- Suzuki Y. Detection of positive selection eliminating effects of structural constraints in hemagglutinin of H3N2 human influenza A virus. Infect. Genet. Evol. 2013;16:93–98. doi: 10.1016/j.meegid.2013.01.017. [DOI] [PubMed] [Google Scholar]
- Suzuki Y. Predictability of antigenic evolution for H3N2 human influenza A virus. Genes Genet. Syst. 2013;88:225–232. doi: 10.1266/ggs.88.225. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Nei M. Origin and evolution of influenza virus hemagglutinin genes. Mol. Biol. Evol. 2002;19:501–509. doi: 10.1093/oxfordjournals.molbev.a004105. [DOI] [PubMed] [Google Scholar]
- Tomita M., Hashimoto K., Takahashi K., Matsuzaki Y., Matsushima R., Saito K., Yugi K., Miyoshi F., Nakano H., Tanida S., Saito Y., Kawase A., Watanabe N., Shimizu T., Nakayama Y. The E-CELL project: towards integrative simulation of cellular processes. N. Gener. Comput. 2000;18:1–12. [Google Scholar]
- Treanor J. Weathering the influenza vaccine crisis. N. Engl. J. Med. 2004;351:2037–2040. doi: 10.1056/NEJMp048290. [DOI] [PubMed] [Google Scholar]
- Whittle J.R.R., Zhang R., Khurana S., King L.R., Manischewitz J., Golding H., Dormitzer P.R., Haynes B.F., Walter E.B., Moody M.A., Kepler T.B., Liao H.-X., Harrison S.C. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc. Natl. Acad. Sci. U. S. A. 2011;108:14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley D.C., Wilson I.A., Skehel J.J. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- Wilson I.A., Skehel J.J., Wiley D.C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- World Health Organization Influenza (Seasonal) 2009. http://www.who.int/mediacentre/factsheets/fs211/en/index.html
- World Health Organization Recommended composition of influenza virus vaccines for use in the 2014–2015 northern hemisphere influenza season. Wkly. Epidemiol. Rec. 2014;89:93–104. [PubMed] [Google Scholar]
- World Health Organization Recommended composition of influenza virus vaccines for use in the 2015 southern hemisphere influenza season. Wkly. Epidemiol. Rec. 2014;89:441–452. [PubMed] [Google Scholar]
- Xia Z., Jin G., Zhu J., Zhou R. Using a mutual information-based site transition network to map the genetic evolution of influenza A/H3N2 virus. Bioinformatics. 2009;25:2309–2317. doi: 10.1093/bioinformatics/btp423. [DOI] [PubMed] [Google Scholar]
- Xia Z., Huynh T., Kang S.-g., Zhou R. Free-energy simulations reveal that both hydrophobic and polar interactions are important for influenza hemagglutinin antibody binding. Biophys. J. 2012;102:1453–1461. doi: 10.1016/j.bpj.2012.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HI titers used in the present study.
Antigenic distances used in the present study.
Representatives of epidemic viruses used in the present study.