Abstract
In many areas of molecular biology there is a need to rapidly extract and analyze genetic information; however, current technologies for DNA sequence analysis are slow and labor intensive. We report here how modern photolithographic techniques can be used to facilitate sequence analysis by generating miniaturized arrays of densely packed oligonucleotide probes. These probe arrays, or DNA chips, can then be applied to parallel DNA hybridization analysis, directly yielding sequence information. In a preliminary experiment, a 1.28 x 1.28 cm array of 256 different octanucleotides was produced in 16 chemical reaction cycles, requiring 4 hr to complete. The hybridization pattern of fluorescently labeled oligonucleotide targets was then detected by epifluorescence microscopy. The fluorescence signals from complementary probes were 5-35 times stronger than those with single or double base-pair hybridization mismatches, demonstrating specificity in the identification of complementary sequences. This method should prove to be a powerful tool for rapid investigations in human genetics and diagnostics, pathogen detection, and DNA molecular recognition.
Full text
PDF
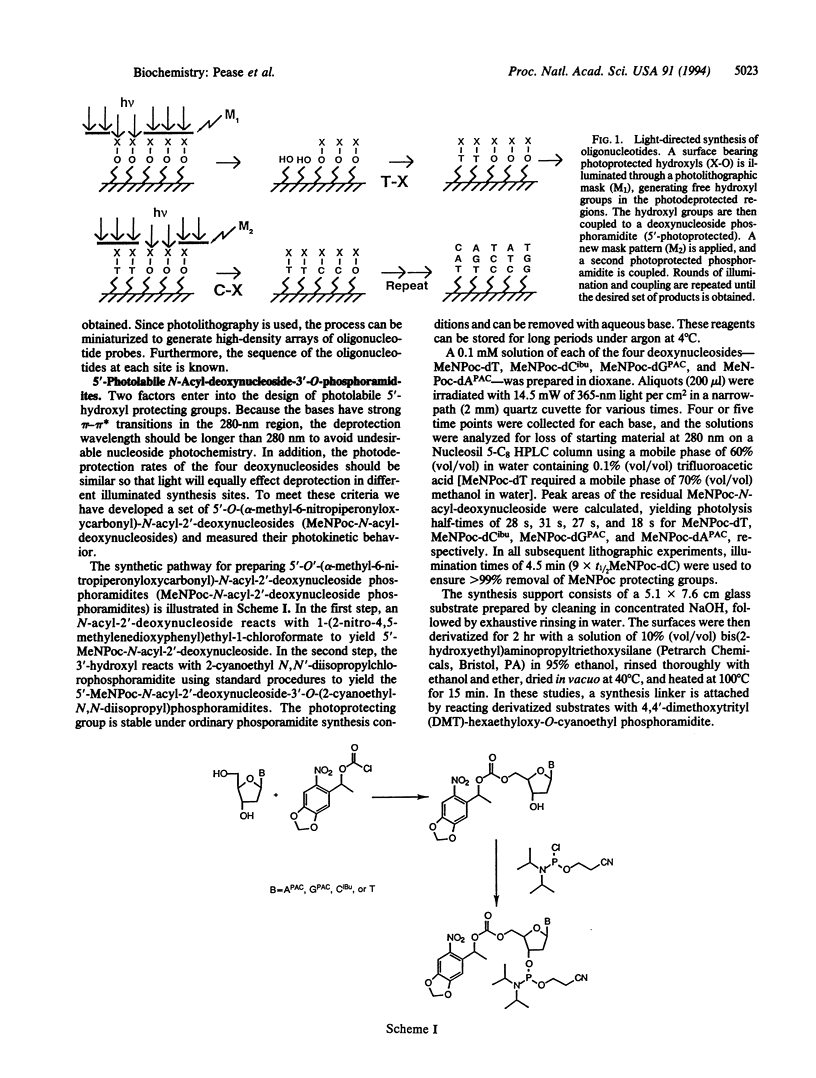

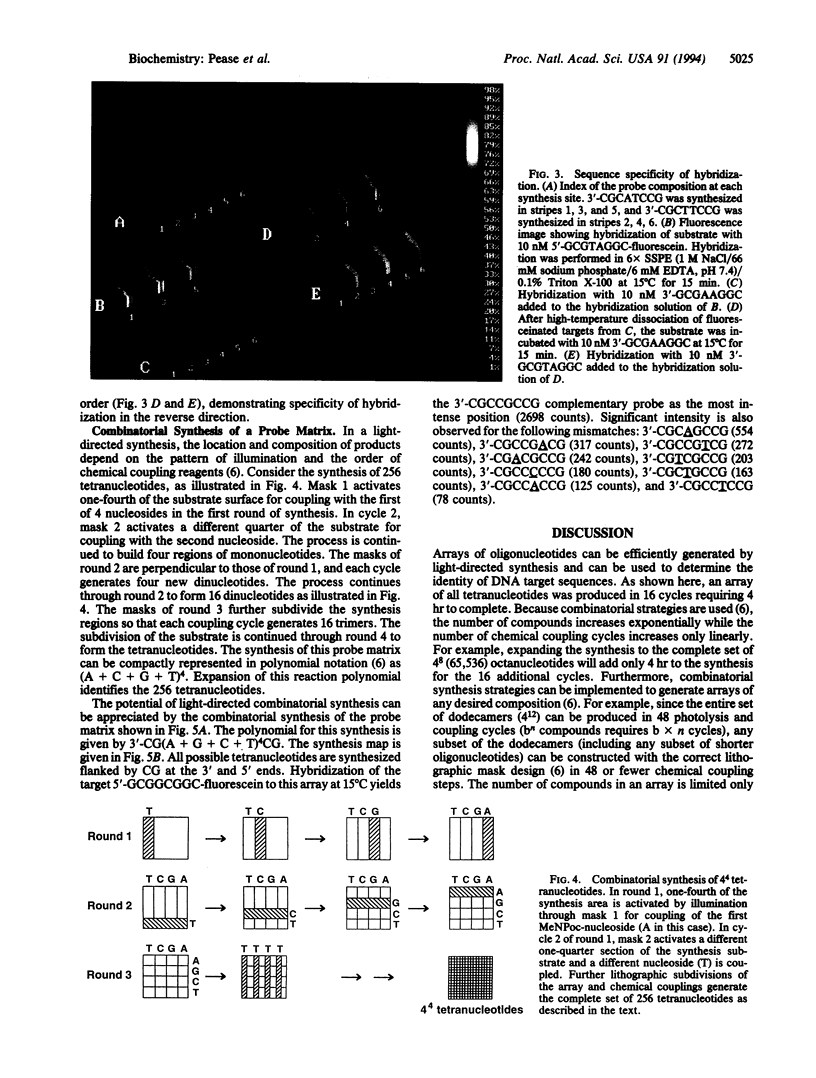
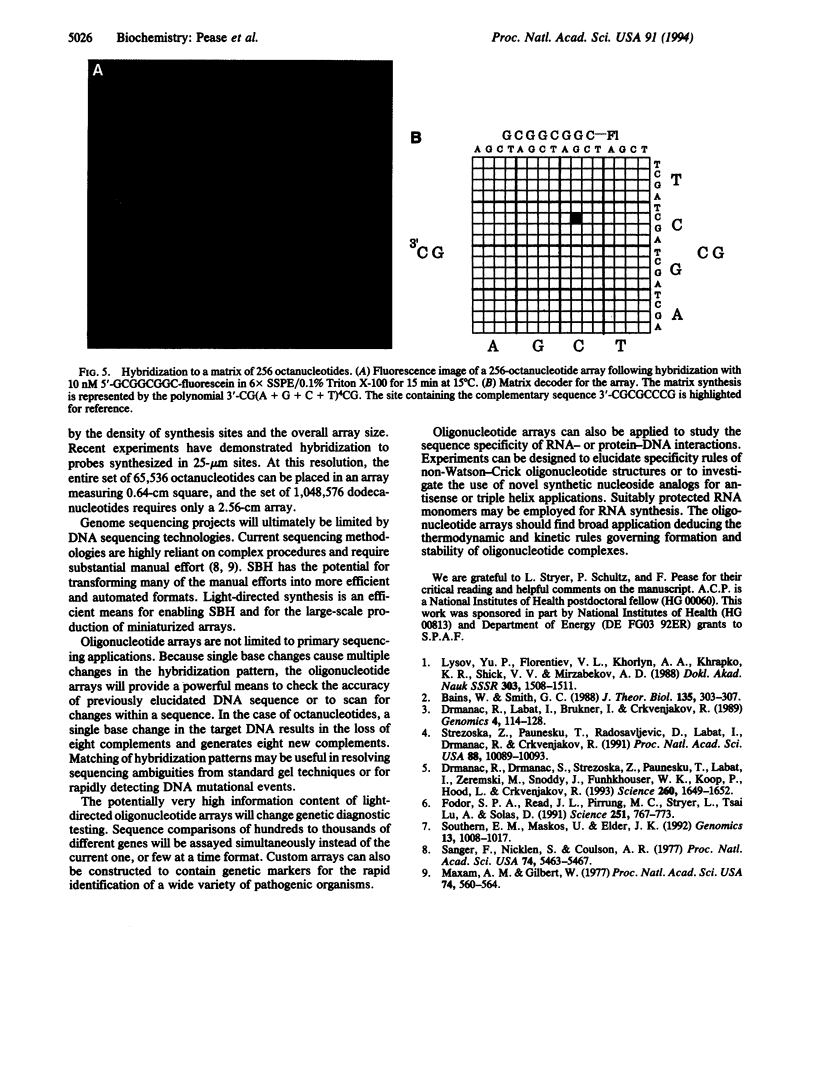
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bains W., Smith G. C. A novel method for nucleic acid sequence determination. J Theor Biol. 1988 Dec 7;135(3):303–307. doi: 10.1016/s0022-5193(88)80246-7. [DOI] [PubMed] [Google Scholar]
- Drmanac R., Drmanac S., Strezoska Z., Paunesku T., Labat I., Zeremski M., Snoddy J., Funkhouser W. K., Koop B., Hood L. DNA sequence determination by hybridization: a strategy for efficient large-scale sequencing. Science. 1993 Jun 11;260(5114):1649–1652. doi: 10.1126/science.8503011. [DOI] [PubMed] [Google Scholar]
- Drmanac R., Labat I., Brukner I., Crkvenjakov R. Sequencing of megabase plus DNA by hybridization: theory of the method. Genomics. 1989 Feb;4(2):114–128. doi: 10.1016/0888-7543(89)90290-5. [DOI] [PubMed] [Google Scholar]
- Fodor S. P., Read J. L., Pirrung M. C., Stryer L., Lu A. T., Solas D. Light-directed, spatially addressable parallel chemical synthesis. Science. 1991 Feb 15;251(4995):767–773. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M., Maskos U., Elder J. K. Analyzing and comparing nucleic acid sequences by hybridization to arrays of oligonucleotides: evaluation using experimental models. Genomics. 1992 Aug;13(4):1008–1017. doi: 10.1016/0888-7543(92)90014-j. [DOI] [PubMed] [Google Scholar]
- Strezoska Z., Paunesku T., Radosavljević D., Labat I., Drmanac R., Crkvenjakov R. DNA sequencing by hybridization: 100 bases read by a non-gel-based method. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10089–10093. doi: 10.1073/pnas.88.22.10089. [DOI] [PMC free article] [PubMed] [Google Scholar]