Background: cAMP signaling augments radiation-induced apoptosis of lung cancer cells.
Results: cAMP signaling reduces the expression of sirtuin 6 deacetylase in non-small cell lung cancer cells by promoting ubiquitin-proteasomal degradation via inhibition of the Raf-MEK-ERK pathway.
Conclusion: cAMP signaling reduces sirtuin 6, which augments radiation-induced apoptosis in lung cancer cells.
Significance: cAMP signaling augments radiation-induced apoptosis by modulating epigenetic control.
Keywords: Apoptosis, Cyclic AMP (cAMP), Extracellular Signal-regulated Kinase (ERK), Lung Cancer, Protein Kinase A (PKA), Sirtuin
Abstract
The cAMP signaling system regulates various cellular functions, including metabolism, gene expression, and death. Sirtuin 6 (SIRT6) removes acetyl groups from histones and regulates genomic stability and cell viability. We hypothesized that cAMP modulates SIRT6 activity to regulate apoptosis. Therefore, we examined the effects of cAMP signaling on SIRT6 expression and radiation-induced apoptosis in lung cancer cells. cAMP signaling in H1299 and A549 human non-small cell lung cancer cells was activated via the expression of constitutively active Gαs plus treatment with prostaglandin E2 (PGE2), isoproterenol, or forskolin. The expression of sirtuins and signaling molecules were analyzed by Western blotting. Activation of cAMP signaling reduced SIRT6 protein expression in lung cancer cells. cAMP signaling increased the ubiquitination of SIRT6 protein and promoted its degradation. Treatment with MG132 and inhibiting PKA with H89 or with a dominant-negative PKA abolished the cAMP-mediated reduction in SIRT6 levels. Treatment with PGE2 inhibited c-Raf activation by increasing inhibitory phosphorylation at Ser-259 in a PKA-dependent manner, thereby inhibiting downstream MEK-ERK signaling. Inhibiting ERK with inhibitors or with dominant-negative ERKs reduced SIRT6 expression, whereas activation of ERK by constitutively active MEK abolished the SIRT6-depleting effects of PGE2. cAMP signaling also augmented radiation-induced apoptosis in lung cancer cells. This effect was abolished by exogenous expression of SIRT6. It is concluded that cAMP signaling reduces SIRT6 expression by promoting its ubiquitin-proteasome-dependent degradation, a process mediated by the PKA-dependent inhibition of the Raf-MEK-ERK pathway. Reduced SIRT6 expression mediates the augmentation of radiation-induced apoptosis by cAMP signaling in lung cancer cells.
Introduction
Lung cancer is a malignant tumor arising from the respiratory epithelium. It is a leading cause of cancer death in both males and females worldwide and has the second highest incidence in women and developing countries (1). Lung cancer is divided into two morphologic groups: small cell lung cancer (SCLC)2 and non-small cell lung cancer (NSCLC). NSCLC accounts for ∼85% of all lung cancer cases (2). Lung cancer is treated by surgery, irradiation, and/or chemotherapy; however, the prognosis remains dismal, with a 5-year survival rate for NSCLC of around 15% (3). Thus, much research has been devoted to improving the efficiency of chemotherapy and radiotherapy based on a better understanding of the molecular mechanisms underlying cell proliferation and death; such research should bring substantial medical benefits, including longer survival and/or amelioration of symptoms (4).
3′,5′-Cyclic adenosine monophosphate (cAMP) is a second messenger that activates signaling pathways that regulate a variety of cellular functions, including metabolism, gene transcription, cell proliferation, migration, and death. cAMP is synthesized from ATP by membrane adenylate cyclases (ACs), which are activated by stimulatory GTP-binding proteins after stimulation of cells by numerous hormones and neurotransmitters, and soluble adenylate cyclases, which can be activated by divalent cations. Cyclic nucleotide phosphodiesterases degrade cAMP to 5′-AMP. cAMP binds to and activates cAMP-dependent protein kinase (PKA), exchange factor directly activated by cAMP (EPAC), and cyclic-nucleotide-gated ion channels (CNG) (5). Activated PKA phosphorylates enzymes to regulate both metabolism and transcription factors such as cyclic AMP response element binding protein (CREB). This in turn regulates the expression of other genes (6). cAMP signaling also modulates cancer cell death induced by anticancer drugs and γ-rays (7, 8).
Histone acetylation is one of the major mechanisms underlying the epigenetic regulation of gene expression and results in loosening of tightly packed heterochromatin to the more relaxed euchromatin. Histone acetylation status is determined by the balance between acetylation (by histone acetyltransferases) and deacetylation (by histone deacetylases (HDACs)) (9). The sirtuins, one of four families of HDAC, use nicotinamide adenine dinucleotide (NAD+) as a co-substrate. Mammals express seven sirtuin isoforms, each of which has a different substrate specificity and subcellular localization. The sirtuins are involved in a variety of cellular functions, including genomic stability, life span, aging, energy metabolism, and tumorigenesis (10). Sirtuin 6 (SIRT6) removes an acetyl group from histone H3 lysines 9 and 56 and regulates a broad range of biological functions, including glucose homeostasis, genomic stability, and apoptosis, by controlling gene expression via transcription factors such as NF-κB and HIF-1α (11–13). A recent study identified alterations in SIRT6 expression in various cancers, including NSCLC (14), suggesting that SIRT6 may be involved in carcinogenesis and tumor progression.
Both cAMP signaling and SIRT6 are thought to be involved in regulating cancer cell death induced by anticancer drugs and ionizing radiation. Our own preliminary experiments suggested that cAMP signaling reduces the expression of SIRT6 in lung cancer cells. Thus, we hypothesized that cAMP modulates SIRT6 activity and regulates radiation-induced cell death in lung cancer cells. Here, we examined the effect and the underlying mechanism of cAMP signaling on SIRT6 protein expression and γ-radiation-induced apoptosis in lung cancer cells. We found that cAMP signaling induced the ubiquitin-dependent degradation of SIRT6, thereby reducing its levels within the cell. This process was mediated by the PKA-Raf-MEK-ERK pathways. We also showed that cAMP signaling augmented radiation-induced apoptosis in lung cancer cells, at least in part by down-regulating the expression of SIRT6 protein.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
Human NSCLC cell lines H1299 and A549 (Korea Cell line Bank, Seoul, Korea), were cultured in Dulbecco's modified Eagle's medium (DMEM) for H1299 cells or RPMI 1640 for A549 cells containing 10% fetal bovine serum (Welgene, Gyeongsan, Korea) and 100 units/ml penicillin/streptomycin. The cells were incubated in a 5% CO2 incubator at 37 °C. Isoproterenol, SP600125, PD0325901, and DMSO were purchased from Sigma. PGE2, forskolin, MG132, PD98059, and SB600125 were purchased from Cayman Chemical (Ann Arbor, MI). The FITC-annexin V apoptosis detection kit was purchased from BD Biosciences. Phenylmethanesulfonyl fluoride (PMSF), protease inhibitor mixture, sodium orthovanadate, and sodium fluoride were purchased from Roche Applied Science.
Transient Transfection and Expression Plasmids
H1299 and A549 cells were transfected with a pcDNA3.1 vector harboring a constitutively active mutant of long-form Gαs containing a Glu-Glu tag (GαsQL-EE) (Missouri S&T cDNA Resource Center, MO) using the calcium phosphate method. This mutant harbors a leucine residue instead of glutamine at 227 residues (the latter is essential for the intrinsic GTPase activity). A dominant-negative PKA (dnPKA) was a gift from Dr. G. Stanley McKnight (University of Washington) (15). Wild-type and dominant-negative CREBs (S133A, R287L) were gifts from Dr. Sahng-June Kwak (Dankook University, Cheonan, Korea). Constitutively active MEK1 was a gift from Dr. Pann-Ghill Suh (Ulsan National Institute of Science and Technology, Ulsan, Korea) (16). Wild-type and dominant-negative ERK1 and ERK2 (ERK1 K71R, and ERK2 K52R, respectively) were gifts from Dr. Melanie H. Cobb (University of Texas) (17). Wild-type and dominant-negative SIRT6 were gifts from Dr. Kartin F. Chua (Stanford University) (18). The plasmids containing short hairpin RNA sequences targeting Gαs, CHIP (the carboxyl terminus of Hsp70-interacting protein), ITCH, MDM2, Skp2, and scrambled shRNA (negative control) were purchased from Sigma. Iduna shRNA was a gift from Dr. Yun-Il Lee (Samsung Advanced Institute of Technology, Seoul, Korea) (19). Decoy oligonucleotides for the CRE (CRE decoy) were prepared as described previously (20).
Western Blot Analysis
Western blotting was performed as previously described (21). Antibodies against SIRT1, -2, and -7, Skp2, ubiquitin, and p-CREB were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), and antibodies against SIRT6, CHIP, Mdm2, p-c-Raf, c-Raf, p-MEK, MEK, p-ERK, ERK, CREB, p-p38, p38, p-JNK, JNK, poly(ADP-ribose) polymerases (PARP), caspase-3, and FLAG were purchased from Cell Signaling Technology (Beverly, MA). An antibody against the Glu-Glu tag was purchased from Covance (Princeton, NJ) and an antibody against β-actin was purchased from Sigma. Proteins were visualized using the Enhanced Chemiluminescence reagent (Menlo Park, CA), and blot images were acquired by an LAS-3000 luminescent image analyzer (Fuji, Tokyo, Japan). Band density was quantified using Multi Gauge v.2.3 software (Fuji) and expressed as a ratio relative to the control band density.
Quantitative Reverse Transcription Polymerase Chain Reaction (qPCR)
qPCR was performed in a C1000 thermal cycler (Bio-Rad) as previously described (22). The sequences of primers used for qPCR were as follows: SIRT6, 5′-CCAAGTTCGACACCACCTTT-3′ and 5′-CGGACGTACTGCGTCTTACA-3′; CHIP, 5′ AGGCCAAGCACGACAAGTACAT-3′ and 5′-CTGATCTTGCCACACAGGTAGT-3′; iduna, 5′-CTACAAACAGGAAAGCGAAC-3′ and 5′-CATGAAGCTCCTTTTACACA-3′; ITCH, 5′-CCTTACGTAGAGGTCACAGTAG-3′ and 5′-CTCCAAGCTGCAAAGTCAC-3′; MDM2, 5′-AATCATCGGACTCAGGTACA-3′ and 5′-GTCAGCTAAGGAAATTTCAGG-3′; Skp2, 5′-CTGTCTCAGTGTTCCAAGTTGCA-3′ and 5′-CAGAACACCCAGAAAGGTTAAGT-3′; GAPDH, 5′-ACCACAGTCCATG-CCATCAC-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′. After 40 cycles of PCR, the average threshold cycle (Ct) values obtained from triplicate qPCR reactions were normalized against the Ct values for GAPDH.
Immunoprecipitation
H1299 cells transfected with GαsQL or empty vector were lysed in lysis buffer containing 20 mm Tris-Cl (pH 7.5), 150 mm NaCl, 0.5% Nonidet P-40, 2 mm EDTA, 2 mm EGTA, 1 mm Na3VO4, 1 mm NaF, 1 mm PMSF, and a protease inhibitor mixture. The cell lysate (800 μg of protein) was precleared by incubation with protein G-agarose (Santa Cruz) for 1 h followed by centrifugation. The supernatant was then incubated with appropriate antibodies (1 μg) overnight at 4 °C on a rotator followed by incubation with 40 μl of protein G-agarose for 2 h. Bound proteins were then precipitated by centrifugation. The precipitate was washed three times before Western blot analysis with an anti-ubiquitin antibody.
Flow Cytometry
Lung cancer cells were exposed to γ-rays (10 gray) and then incubated for 24 h. The cells were then washed twice with phosphate-buffered saline and harvested by trypsinization and centrifugation at 3500 × g for 5 min at 4 °C. The cells were incubated in annexin V buffer containing FITC-annexin V and propidium iodide for 15 min. The fluorescence of 10,000 cells per sample was detected in a FACSCalibur flow cytometer (BD Biosciences).
Data Analysis
All experiments were repeated at least three times, and the data were expressed as the means ± S.E. Data were analyzed using a non-parametric Mann-Whitney U test. A p value < 0.05 was considered statistically significant.
RESULTS
cAMP Signaling Reduces SIRT6 Expression in Lung Cancer Cells
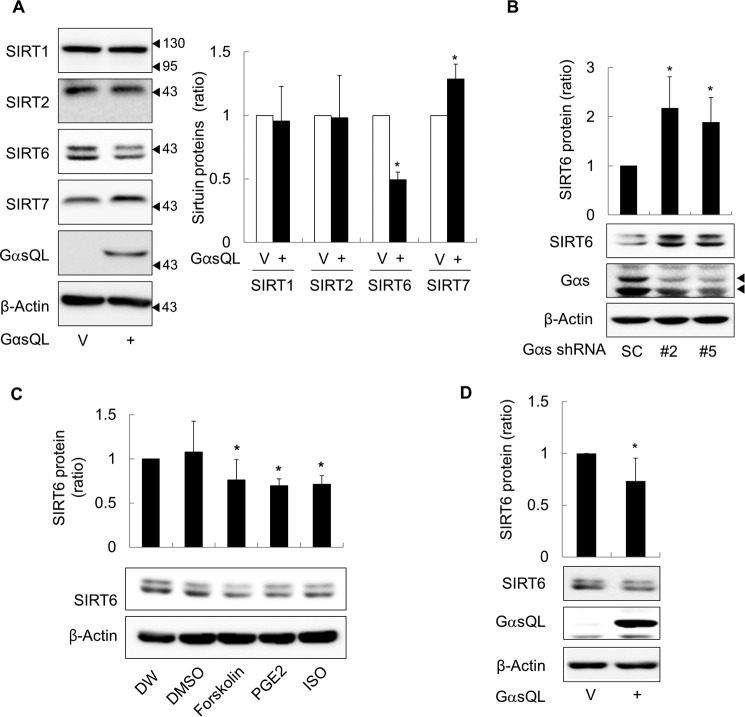
To examine the effect of cAMP signaling on the expression of sirtuins, constitutively active GαsQL was transiently expressed in H1299 NSCLC cells to activate cAMP signaling. The expression of sirtuin isoforms, which are known to localize in nucleus for cytosol for epigenetic control, was then analyzed by Western blotting. Transient expression of GαsQL reduced SIRT6 protein levels in H1299 NSCLC cells (but increased SIRT7 protein levels) compared with those in vector-transfected controls (Fig. 1A). Knocking down Gαs using two different shRNAs increased SIRT6 expression in H1299 cells (Fig. 1B). Activation of cAMP signaling by treatment with Gαs-coupled receptor agonists (PGE2 and isoproterenol) or the adenylyl cyclase activator, forskolin, also reduced SIRT6 expression in cancer cells at 24 h after treatment (Fig. 1C). Expressing GαsQL also reduced SIRT6 protein in A549 lung cancer cells (Fig. 1D). Taken together, these results indicate that cAMP signaling reduces SIRT6 protein expression in lung cancer cells.
FIGURE 1.
Gαs inhibits SIRT6 expression in lung cancer cells. A, effects of Gαs on the expression of sirtuin isoforms in H1299 human lung cancer cells. B, effects of Gαs knockdown on the expression of SIRT6. C, effects of cAMP signaling on the expression of SIRT6. D, effects of Gαs on the expression of SIRT6 in A549 human lung cancer cells. Lung cancer cells (H1299 and A549 cells) were transfected with EE-tagged GαsQL (GαsQL) or vector pcDNA3.1 (V) using the calcium phosphate method and then incubated for 24 h. The cells were then harvested, homogenized, and analyzed by Western blotting. Gαs expression was knocked down by transfection of 10 μg of shRNA. Scrambled shRNA was used as a control (SC). The two arrowheads indicate long- and short-forms of Gαs proteins (B). H1299 cells were treated with 40 μm forskolin, 10 μm PGE2, 1 μm isoproterenol (ISO), distilled water (DW), or DMSO for 24 h before Western blotting analysis. An asterisk indicates a statistically significant difference compared with the respective control or vector-transfected cells (*, p < 0.05; Mann-Whitney U test).
cAMP Signaling Promotes Ubiquitin-Proteasome-dependent Degradation of SIRT6 in H1299 Cells
To investigate the mechanism by which cAMP signaling reduces SIRT6 expression, we next used quantitative RT-PCR to examine the effects of GαsQL on the expression of SIRT6 mRNA in H1299 cells. Expressing GαsQL did not significantly alter the levels of SIRT6 mRNA (Fig. 2A). Thus, we next examined the effect of Gαs on SIRT6 degradation in H1299 cells. The results showed that expressing GαsQL in the presence of cycloheximide (an inhibitor of protein synthesis) promoted SIRT6 degradation (Fig. 2B). Thus, to test whether Gαs promoted SIRT6 degradation via the proteasomal pathway, we examined the effect of a proteasomal inhibitor, MG132, on SIRT6 degradation. Treatment with MG132 increased the SIRT6 level in both vector- and GαsQL-transfected cells, completely abolishing the SIRT6-reducing effect of GαsQL (Fig. 2C). Because many proteins are ubiquitinated before proteasomal degradation, we next examined whether Gαs affected SIRT6 ubiquitination. We found that the expression of GαsQL increased the amount of polyubiquitinated SIRT6 that was immunoprecipitated from H1299 cell lysates by an anti-SIRT6 antibody (Fig. 2D). Next, we examined whether MG132 affected the proteasomal degradation of SIRT6 after cAMP signaling activated by either PGE2 or forskolin. The results showed that MG132 completely abolished the PGE2- or forskolin-mediated reduction in SIRT6 expression (Fig. 3, A and B). To identify the ubiquitin E3 ligase involved in SIRT6 degradation, we next analyzed SIRT6 expression after knocking down the E3 ligases CHIP, iduna, ITCH, MDM2, and Skp2 using specific shRNAs. E3 ligase knockdown was confirmed at both the mRNA and protein levels (Fig. 3, C and D). Both basal and the PGE2-induced expression of SIRT6 were increased in the cells in which any of the E3 ligases were knocked down, suggesting that all of the E3 ligases examined play a role in SIRT6 degradation (Fig. 3D). However, PGE2 had a SIRT6-reducing effect after knock down of MDM2 or Skp2. Taken together, these results indicate that cAMP signaling promotes ubiquitin-proteasome-dependent degradation of SIRT6 in H1299 cells and that the process involves several E3 ligases.
FIGURE 2.
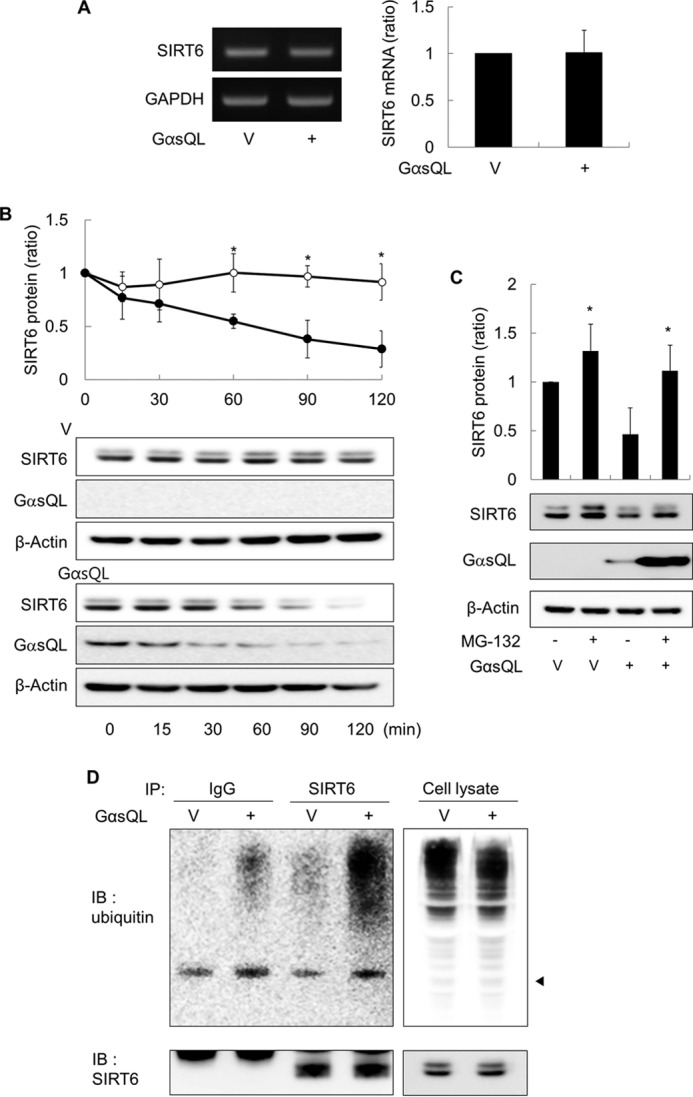
Gαs promotes the proteasome-dependent degradation of SIRT6 in H1299 cells. A, effects of Gαs on the expression of SIRT6 mRNA. H1299 cells were transfected with GαsQL or vector (V), and the expression of SIRT6 mRNA was analyzed by RT-PCR and real-time quantitative PCR 24 h later. B, effects of Gαs on the degradation of SIRT6 protein. H1299 cells were transfected with GαsQL or vector (V) and then treated with 50 μg/ml cycloheximide (CHX) 24 h later. Cells were harvested at the indicated times. C, effect of MG-132 on the SIRT6 expression. H1299 cells were transfected with GαsQL or vector (V) and incubated for 6 h before exposure to 10 μm MG132 for 18 h. Cells were then harvested for analysis. D, effects of Gαs on the ubiquitination of SIRT6. H1299 cells were transfected with GαsQL or vector (V) and incubated 24 h before harvesting. Whole cell lysates (800 μg) were precleared by incubation with protein A-agarose for 1 h followed by centrifugation. IP, immunoprecipitation. The precleared lysate was then incubated with an anti-SIRT6 antibody (1 μg) or with control IgG at 4 °C for 16 h followed by protein A-agarose for 2 h. Finally, the samples were washed three times and analyzed by Western blotting (IB) with antibodies against SIRT6 or ubiquitin. An arrowhead indicates the molecular weight of SIRT6 (D). Asterisks (*) on the histograms indicate a statistically significant difference from the respective control or vector-transfected control cells (p < 0.05, Mann-Whitney U test). One-way analysis of variance analysis was also performed to compare the amount of HDAC6 protein remained following cycloheximide treatment (B).
FIGURE 3.
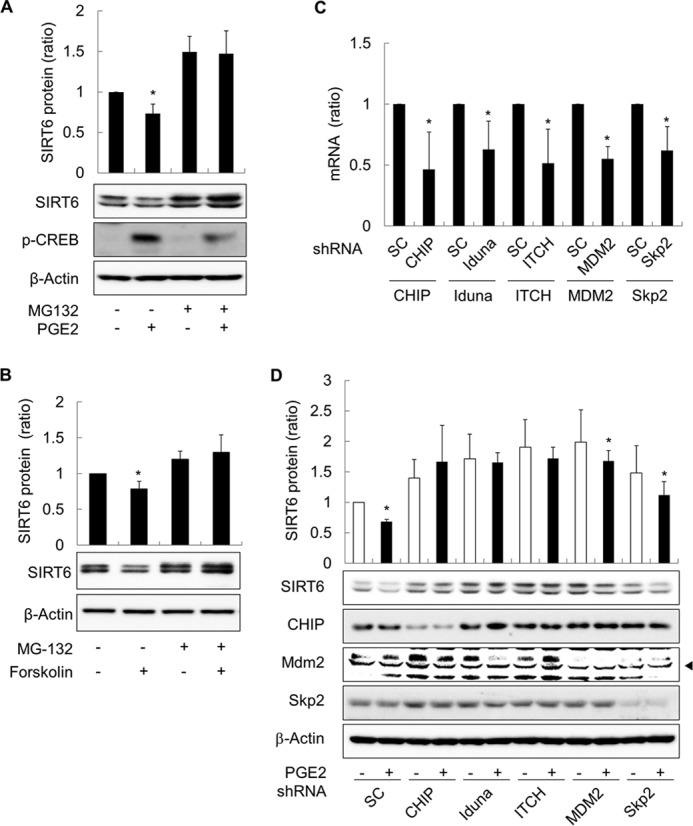
Prostaglandin E2 promotes the proteasome-dependent degradation of SIRT6 in H1299 cells. A and B, effect of PGE2 or forskolin on the degradation of SIRT6 protein. H1299 cells were treated with 10 μm PGE2, 40 μm forskolin, or DMSO in the presence/absence of 10 μm MG132 for 24 h before Western blot analysis. C and D, effects of knocking down E3 ligases on PGE2-promoted degradation of SIRT6 protein. H1299 cells were transfected with shRNA against CHIP, iduna, ITCH, Mdm2, or Skp2 or with scrambled shRNA (SC). To confirm knock down of the target ligases, expression of mRNA for each E3 ligase was analyzed by qPCR at 48 h after transfection (C). At 24 h after transfection, cells were treated with 10 μm PGE2 for 24 h before Western blotting (D). Asterisks (*) on the histograms indicate a statistically significant difference from the respective control cells (p < 0.05, Mann-Whitney U test).
cAMP Signaling Reduces SIRT6 Expression in H1299 Cells via PKA and CREB
To identify the signaling pathway involved in the SIRT6-reducing effects of cAMP, we next examined the role of PKA. PKA was inhibited by both a pharmacologic inhibitor (H89) and dnPKA, because H89 can inhibit other protein kinases as well as PKA. Inhibiting PKA with H89 or by expression of dnPKA increased the basal level of SIRT6 expression in H1299 cells and abolished the SIRT6-reducing effects of GαsQL and PGE2 (Fig. 4, A and B). Treatment with an EPAC-selective cAMP analogue (8-(4-chloro-phenylthio)-2′-O-methyladenosine-3′-5′-cyclic monophosphate) did not reduce SIRT6 expression, and knockdown of EPAC did not block SIRT6-reducing effect of PGE2 (data not shown). Because CREB is a well known downstream target of PKA, we next examined whether it plays a role in this process. CREB activation was inhibited by transfecting H1299 cells with dominant-negative CREBs (CREB S133A and CREB R287L), resulting in an increase in both basal and PGE2-induced levels of SIRT6 expression (Fig. 4C). Similarly, blocking the binding of CREB to its target genes by transfecting cells with a CRE decoy oligonucleotide increased SIRT6 expression, which was unaffected by PGE2 (Fig. 4D). The results indicate that cAMP signaling reduces SIRT6 expression in lung cancer cells by activating the PKA and CREB pathways.
FIGURE 4.
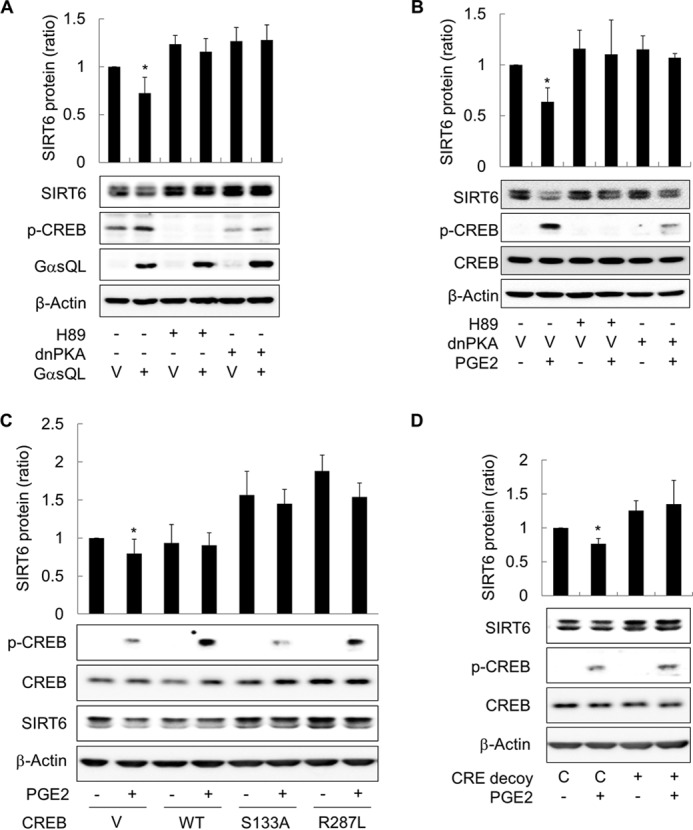
cAMP signaling inhibits SIRT6 expression in H1299 cells via the PKA and CREB pathways. A, inhibiting PKA abolished the effects of GαsQL on SIRT6 expression. B, inhibiting PKA blocked the effects of PGE2 on SIRT6 expression. C, inhibiting CREB activation increased SIRT6 expression. D, inhibiting CREB binding to CRE increased SIRT6 expression. H1299 cells were transfected with GαsQL, a dnPKA, wild-type CREB (WT), dominant-negative CREBs (S133A, R287L), oligonucleotides (CRE decoy, CRE control; C), or respective control vectors (V) and then incubated for 24 h. The cells were then treated with 20 μm H89, 10 μm PGE2, or DMSO for 24 h before Western blot analysis. CREB phosphorylation (pCREB) was analyzed 30 min after exposure to PGE2. Asterisks (*) on the histograms indicate a statistically significant difference from the respective control cells (p < 0.05, Mann-Whitney U test).
cAMP Signaling Reduces SIRT6 Expression in H1299 Cells by Inhibiting the ERK Pathway
To study the signaling pathway that mediates the SIRT6-reducing effect of cAMP signaling, we first examined the time course of SIRT expression in PGE2-treated cells. Treating H1299 cells with PGE2 for 1 h led to a significant reduction in SIRT6 expression after 24 h, and treatment for 2 h reached a maximum reduction in SIRT6 expression (Fig. 5A). This suggests that activation of PGE2 signaling within 2 h after exposure is sufficient to induce a reduction of SIRT6 expression. To examine the signaling pathway that mediates the SIRT6-reducing effect of cAMP in more detail, we next analyzed the effect of mitogen-activated protein kinases (MAPKs) on SIRT6 expression after treatment with specific inhibitors. Treatment with the ERK inhibitor, PD98059, reduced SIRT6 expression in a manner similar to that of PGE2; however, treatment with a JNK inhibitor (SP600125) or a p38 inhibitor (SB203580) had no effect on SIRT6 expression (Fig. 5B). To confirm the effect of ERK signaling on SIRT6 expression, we next inhibited ERK signaling in H1299 cells by expressing dominant-negative ERKs. Expressing dominant-negative ERK1 (dnERK1) or ERK2 (dnERK2) reduced SIRT6 expression (Fig. 5C), whereas expressing constitutively active MEK (caMEK) increased SIRT6 expression and abolished the HDAC6-reducing effect of PGE2 in H1299 cells (Fig. 5D). Treatment with PGE2 reduced activating phosphorylation of c-Raf at Ser-338 but increased the inhibitory phosphorylation of c-Raf at Ser-259 (Fig. 6A). Treatment with PGE2 also reduced activating phosphorylation of downstream MEK and ERK. The temporal pattern of ERK phosphorylation was inversely correlated with CREB phosphorylation after PGE2 treatment (Fig. 6B), suggesting that PKA is involved in the phosphorylation of both ERK and CREB. Expressing dominant-negative PKA abolished the PGE2-mediated inhibitory phosphorylation of c-Raf at Ser-259 (Fig. 6C), resulting in the inhibition of the c-Raf-MEK-ERK signaling pathways. Taken together, these results indicate that cAMP signaling reduces SIRT6 expression in lung cancer cells via PKA-dependent inhibition of the Raf-MEK-ERK signaling pathways.
FIGURE 5.
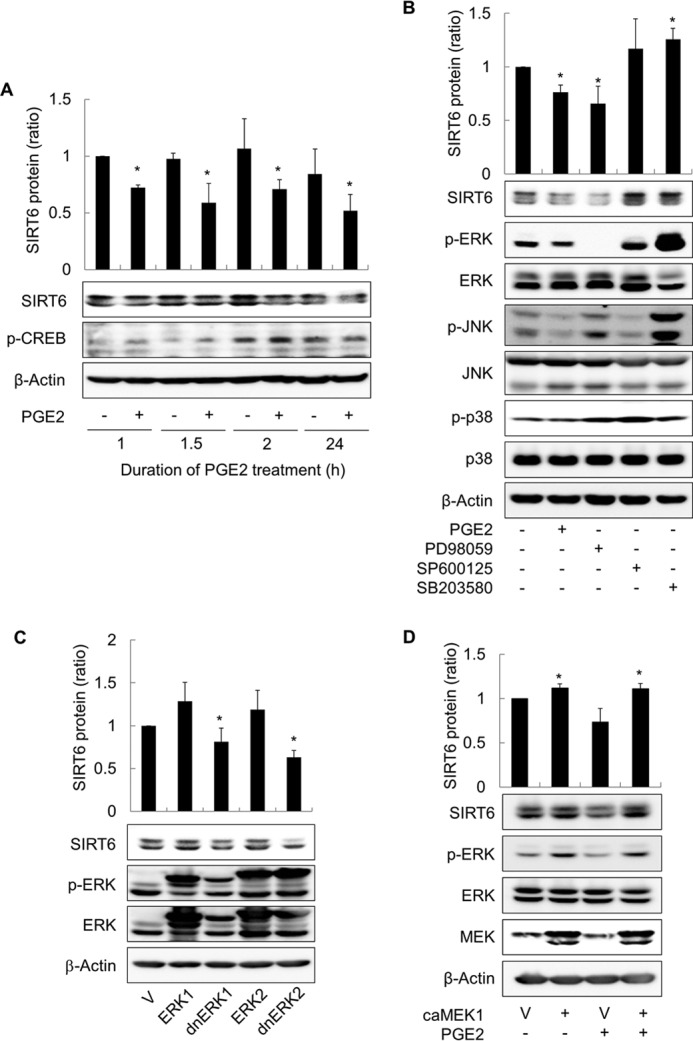
cAMP signaling reduces SIRT6 expression in H1299 cells by inhibiting the ERK pathway. A, effect of PGE2 treatment duration on SIRT6 expression. H1299 cells were treated with 10 μm PGE2 or with DMSO for the indicated times before the culture medium was aspirated. The cells were then washed with DPBS, and fresh medium was added. The cells were harvested 24 h after the start of PGE2 treatment, and SIRT6 expression was analyzed by Western blotting. B, effect of MAPK inhibition on SIRT6 expression. C, effect of ERK inhibition by dominant-negative ERKs (dnERK) on SIRT6 expression. D, effect of ERK activation on PGE2-induced inhibition of SIRT6 expression. H1299 cells were treated with 10 μm PGE2, 40 μm PD98059, 20 μm SP600125, 20 μm SB203580, or DMSO for 2 h and then harvested and analyzed by Western blotting. The expression of SIRT6 was analyzed 24 h after treatment. H1299 cells were also transfected with ERK1, ERK2, dominant-negative ERKs (dnERK1, dnERK2), constitutively active MEK1 (caMEK1), or the respective empty vectors (V) and incubated for 24 h. Transfected cells were treated with 10 μm PGE2 for 30 min (if necessary) before Western blot analysis of signaling molecules. Asterisks (*) on the histograms indicate a statistically significant difference from the respective control cells (p < 0.05, Mann-Whitney U test).
FIGURE 6.
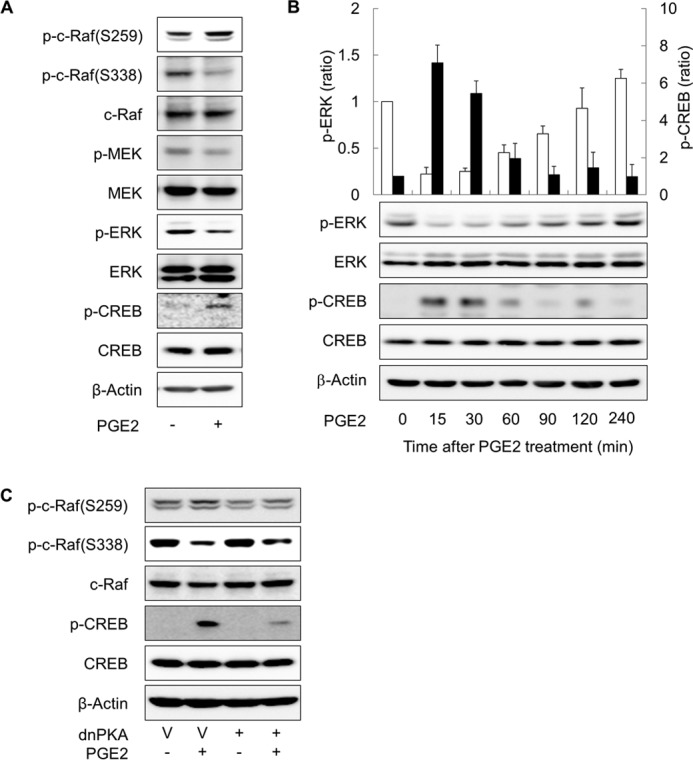
cAMP signaling inhibits the ERK pathway in a PKA-dependent way. A, effect of PGE2 on the activation Raf-MEK-ERK pathway. B, temporal patterns of CREB and ERK phosphorylation after PGE2 treatment. H1299 cells were treated with 10 μm PGE2 or DMSO, and CREB and ERK phosphorylation was examined at the indicated times by Western blotting. The empty bar represents p-ERK and the filled bar p-CREB. C, effect of PKA on PGE2-induced on c-Raf phosphorylation. H1299 cells were treated with 10 μm PGE2 or DMSO for 30 min and then harvested and analyzed by Western blotting. The expression of SIRT6 was analyzed 24 h after treatment. H1299 cells were also transfected with dnPKA or the empty vectors (V) and incubated for 24 h. Transfected cells were treated with 10 μm PGE2 for 30 min (if necessary) before Western blot analysis.
Gαs Augments γ-Ray-induced Apoptosis in Lung Cancer Cells by Reducing SIRT6 Expression
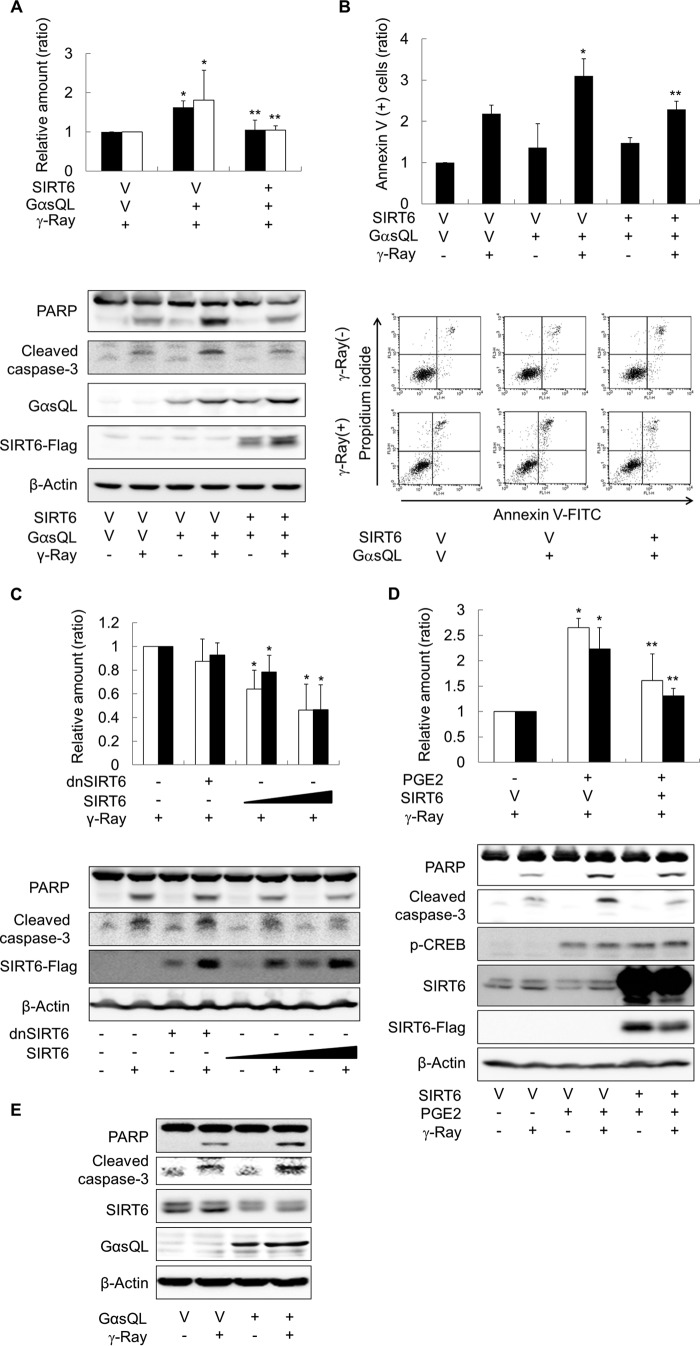
To examine the effects of cAMP-mediated reductions in SIRT6 expression, we next examined γ-ray-induced apoptosis in lung cancer cells. Activation of cAMP signaling via expression of GαsQL increased the cleavage of caspase 3 and PARP, and the percentage of annexin V-positive cells in γ-ray-irradiated H1299 cells, which indicated an increase in apoptosis (Fig. 7, A and B). Co-expression of wild-type SIRT6 abolished these effects. SIRT6 expression alone reduced the amount of γ-ray-induced apoptosis, but expression of dominant-negative SIRT6 failed to reduce the apoptosis significantly (Fig. 7C). PGE2-mediated activation of cAMP signaling also increased the cleavage of caspase 3 and PARP, which was also blocked by expression of wild-type SIRT6 (Fig. 7D). Expressing Gαs in A549 cells also increased the γ-ray-induced cleavage of caspase 3 and PARP (Fig. 7E). These results suggest that cAMP signaling reduces SIRT6 expression in lung cancer cells, which in turn augments γ-ray-induced apoptosis.
FIGURE 7.
cAMP signaling increases γ-ray-induced apoptosis by reducing SIRT6 expression in lung cancer cells. A, effects of Gαs and SIRT6 on γ-ray-induced cleavage of caspase 3 and PARP in H1299 cells. B, effects of Gαs and SIRT6 on γ-ray-induced annexin V-staining of H1299 cells. C, effects of SIRT6 on γ-ray-induced apoptosis in H1299 cells. D, effect of PGE2 on γ-ray induced apoptosis in H1299 cells. E, effect of Gαs on γ-ray-induced cleavage of caspase 3 and PARP in A549 cells. H1299 or A549 cells were transfected with GαsQL, wild-type SIRT6 (SIRT6), dominant-negative SIRT6 (dnSIRT6), or empty vector (V) followed by incubation for 24 h. The cells were pretreated (or not) with PGE2 for 30 min and then irradiated with γ-rays (10 gray). Cells were incubated for 24 h before apoptosis was analyzed by Western blotting or flow cytometry after staining with annexin V and propidium iodide. The empty bar represents cleaved caspase 3, and the filled bar represents PARP (A, C, and D). The bar graph shows the proportion of annexin V-positive cells within the whole cell population (B). Asterisks (*) on the histograms indicate a statistically significant difference from the respective control or vector-transfected control cells; the double asterisks (**) represent a statistically significant difference from the GαsQL-transfected or PGE2-treated control cells (p < 0.05, Mann-Whitney U test).
DISCUSSION
Here we examined the effect of cAMP signaling on SIRT6 expression in lung cancer cells. We also examined the underlying molecular mechanisms and their functional significance. We found that 1) cAMP signaling reduced SIRT6 expression by promoting its degradation via the ubiquitin-proteasome pathway, 2) SIRT6 degradation is mediated by the PKA-dependent inhibition of the Raf-MEK-ERK pathways, and 3) reduced SIRT6 expression augments γ-ray-induced apoptosis of NSCLC cells (Fig. 8).
FIGURE 8.
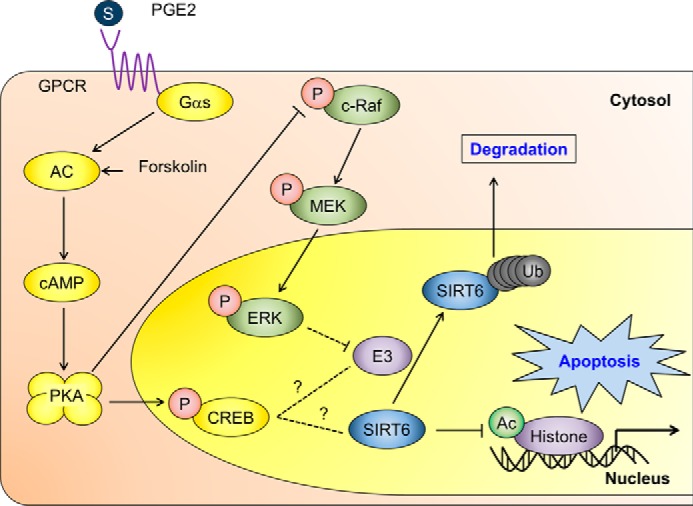
A suggested mechanism by which cAMP signaling reduces SIRT6 expression in lung cancer cells, resulting in augmented apoptosis. The solid lines indicate proven signaling pathways, and the dotted lines indicate potential signaling pathways. GPCR, G-protein-coupled receptor; Ub, ubiquitin.
Our finding that cAMP signaling reduces SIRT6 expression in lung cancer cells was supported by experiments showing that SIRT6 expression was reduced after activation of cAMP signaling via expression of constitutively active Gαs (which stimulates ACs) by treatment with Gαs-coupled receptor agonists (PGE2 and isoproterenol) or by treatment with an adenylate cyclase activator, forskolin, in H1299 and A549 NSCLC. cAMP signaling also increased SIRT7 expression but did not affect the expression of other SIRT isoforms, suggesting that cAMP has an isoform-specific effect. Several molecules, such as microRNAs and peroxisome proliferator-activated receptor γ, were reported to regulate SIRT6 expression (23, 24), and cAMP signaling also stimulates HDAC4 activity (25). However, the effect of cAMP signaling on SIRT6 expression is unclear. Here we show for the first time that cAMP signaling regulates SIRT6 expression in lung cancer cells. Thus, cAMP signaling appears to regulate gene expression by modulating deacetylation via HDACs as well as by activating transcription factors such as CREB.
We also showed that cAMP signaling reduces SIRT6 expression by promoting the degradation of SIRT6 via the ubiquitin-proteasome pathway. Although cAMP signaling did not decrease SIRT6 mRNA levels, it increased the ubiquitination of SIRT6. By contrast, inhibiting the proteasome abolished the cAMP-mediated proteasomal degradation of SIRT6. Several studies show that SIRT6 expression is regulated by ubiquitin-proteasome-dependent degradation (26–28). cAMP regulates the ubiquitin-proteasomal degradation of several proteins by controlling ubiquitin E3 ligases, including atrogin-1 (29), NEDD4L (30), and SCF-type ubiquitin E3 ligase (31). We also found that the ubiquitin E3 ligases MDM2, CHIP, iduna, ITCH, and Skp2 were involved in SIRT6 degradation in lung cancer cells. However, the specific E3 ligase that mediates the cAMP-mediated degradation of SIRT6 remains to be identified.
cAMP signaling reduced SIRT6 expression via PKA-dependent inhibition of the Raf-MEK-ERK pathways. We found that inhibiting PKA by either treatment with a PKA inhibitor or by expression of dominant-negative PKA abolished the cAMP-mediated reduction in SIRT6 expression in lung cancer cells and that EPAC was not involved in the reduction of SIRT6. This finding indicates that cAMP signaling reduced SIRT6 expression via PKA (32). The results suggest that PKA phosphorylates specific target proteins to promote the ubiquitination of SIRT6; thus we tried to identify the signaling molecules that mediate this effect. We found that the cAMP-mediated reduction in SIRT6 expression is mediated via the Raf-MEK-ERK signaling pathways. This was confirmed by our findings that inhibiting ERK reduced SIRT6 expression, that activating ERK abolished the SIRT6-reducing effect of PGE2, and that PGE2 inhibited Raf-MEK-ERK signaling in a PKA-dependent manner. PGE2 inhibited c-Raf activation by increasing the inhibitory phosphorylation of c-Raf at Ser-259 and by reducing activating phosphorylation at Ser-338. This, in turn, inhibited downstream MEK-ERK signaling, thereby reducing SIRT6 expression. These results agree with those published in previous reports showing that cAMP increases inhibitory Raf-1 phosphorylation at Ser-259 and reduces activating Raf-1 phosphorylation at Ser-338 in a PKA-dependent manner, thereby inducing ERK deactivation (33, 34). ERK activates SIRT1 in chondrocytes (35) and induces SIRT1 expression in human endothelial cells (36); however, the effect of ERK on SIRT6 remains unknown. The present study showed that inhibiting the Raf-MEK-ERK pathway also reduces SIRT6 expression in lung cancer cells. The Ras-ERK cascade inhibits the ubiquitination and degradation of GATA3 protein during T cell differentiation (37); thus it is plausible that ERK also inhibits the ubiquitination and degradation of SIRT6 in lung cancer cells. In addition to the ERK-dependent reduction of SIRT6 expression, we found that PKA activates CREB and reduces SIRT6 expression by regulating the expression of CRE-containing genes. The expression of dominant-negative CREB or CRE decoy oligonucleotides increased SIRT6 expression both before and after PGE2 treatment. Thus, cAMP signaling seems to reduce SIRT6 expression by promoting its proteasomal degradation and by regulating its gene expression via CREB.
Finally, we examined the effect of reduced SIRT6 expression on apoptosis. Reduced SIRT6 expression mediated augmentation of radiation-induced apoptosis in lung cancer cells by cAMP signaling. Experiments showed that 1) cAMP signaling augmented radiation-induced apoptosis in lung cancer cells, 2) that cAMP signaling reduced SIRT6 expression, 3) that exogenous expression of SIRT6 abolished the cAMP signaling-mediated effects on apoptosis, and 4) that exogenous expression of SIRT6 reduced the level of radiation-induced apoptosis. We previously showed that cAMP signaling regulates apoptosis in various cancer cells, including lung cancer cells, by modulating the expression of Bcl-2 family proteins (8), the expression of X-linked inhibitor of apoptosis protein (22), and the activity of ataxia-telangiectasia mutated (ATM) protein kinase (21). Here, we describe a novel mechanism through which cAMP signaling augments apoptosis, reducing SIRT6 expression. The anti-apoptotic effect of SIRT6 suggests that it may have potentially oncogenic roles in the tumorigenesis and progression of lung cancer. Our findings support those of a recent report showing that SIRT6 functions as an oncogene in the murine epidermis (38) prostate cancer (39).
Novel approaches to obtain antiproliferative effects or apoptosis-inducing effects of various cancer cells by controlling cAMP signaling are vigorously investigated (40), and many HDAC inhibitors are explored to develop anti-cancer drugs (41). This study might contribute to the development of new strategies to improve cancer radiotherapy by modulating the activity of cAMP signaling and HDACs.
In conclusion, the present study showed that 1) cAMP signaling reduces SIRT6 expression by promoting its degradation via the ubiquitin-proteasome pathway, 2) that this process is mediated by PKA-dependent inhibition of the ERK pathway, and 3) that reduced SIRT6 expression mediates the augmentation of radiation-induced apoptosis by cAMP signaling in lung cancer cells. Thus, we have identified a novel mechanism by which cAMP signaling regulates SIRT6 expression and apoptosis in lung cancer cells, suggesting a regulatory role for cAMP signaling in epigenetic control of gene expression.
This work was supported by the Basic Science Research Program through the National Research Foundation funded by the Ministry of Education, Science, and Technology (2012R1A1A2044374), a grant from the National R&D Program for Cancer Control, Ministry of Health and Welfare, Republic of Korea (0720540), and by the Education and Research Supporting Fund from Seoul National University Hospital.
- SCLC
- small cell lung cancer
- NSCLC
- non-small cell lung cancer
- AC
- adenylate cyclase
- CREB
- cAMP response element (CRE)-binding protein
- Gαs
- stimulatory α subunit of G protein
- GαsQL
- constitutively active mutant long form of the α subunit of stimulatory heterotrimeric GTP-binding protein
- PKA
- cAMP-dependent protein kinase
- PARP
- poly(ADP-ribose) polymerase
- qPCR
- quantitative PCR
- siRNA
- small interfering RNA
- SIRT6
- sirtuin 6
- HDAC
- histone deacetylase
- PGE2
- prostaglandin E2
- dn
- dominant negative.
REFERENCES
- 1. Jemal A., Bray F., Center M. M., Ferlay J., Ward E., Forman D. (2011) Global cancer statistics. CA Cancer J. Clin. 61, 69–90 [DOI] [PubMed] [Google Scholar]
- 2. Zochbauer-Muller S., Gazdar A. F., Minna J. D. (2002) Molecular pathogenesis of lung cancer. Annu. Rev. Physiol. 64, 681–708 [DOI] [PubMed] [Google Scholar]
- 3. Govindan R., Page N., Morgensztern D., Read W., Tierney R., Vlahiotis A., Spitznagel E. L., Piccirillo J. (2006) Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the surveillance, epidemiologic, and end results database. J. Clin. Oncol. 24, 4539–4544 [DOI] [PubMed] [Google Scholar]
- 4. Johnson D. H., Schiller J. H., Bunn P. A., Jr. (2014) Recent clinical advances in lung cancer management. J. Clin. Oncol. 32, 973–982 [DOI] [PubMed] [Google Scholar]
- 5. Lefkimmiatis K., Zaccolo M. (2014) cAMP signaling in subcellular compartments. Pharmacol. Ther. 143, 295–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Taylor S. S., Zhang P., Steichen J. M., Keshwani M. M., Kornev A. P. (2013) PKA: lessons learned after twenty years. Biochim. Biophys. Acta 1834, 1271–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Choi Y. J., Kim S. Y., Oh J. M., Juhnn Y. S. (2009) Stimulatory heterotrimeric G protein augments gamma ray-induced apoptosis by up-regulation of Bak expression via CREB and AP-1 in H1299 human lung cancer cells. Exp. Mol. Med. 41, 592–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Choi Y. J., Oh J. M., Kim S. Y., Seo M., Juhnn Y. S. (2009) Stimulatory heterotrimeric GTP-binding protein augments cisplatin-induced apoptosis by upregulating Bak expression in human lung cancer cells. Cancer Sci. 100, 1069–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shahbazian M. D., Grunstein M. (2007) Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76, 75–100 [DOI] [PubMed] [Google Scholar]
- 10. Finkel T., Deng C. X., Mostoslavsky R. (2009) Recent progress in the biology and physiology of sirtuins. Nature 460, 587–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kawahara T. L., Michishita E., Adler A. S., Damian M., Berber E., Lin M., McCord R. A., Ongaigui K. C., Boxer L. D., Chang H. Y., Chua K. F. (2009) SIRT6 links histone H3 lysine 9 deacetylation to NF-κB-dependent gene expression and organismal life span. Cell 136, 62–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Michishita E., McCord R. A., Berber E., Kioi M., Padilla-Nash H., Damian M., Cheung P., Kusumoto R., Kawahara T. L., Barrett J. C., Chang H. Y., Bohr V. A., Ried T., Gozani O., Chua K. F. (2008) SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452, 492–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhong L., D'Urso A., Toiber D., Sebastian C., Henry R. E., Vadysirisack D. D., Guimaraes A., Marinelli B., Wikstrom J. D., Nir T., Clish C. B., Vaitheesvaran B., Iliopoulos O., Kurland I., Dor Y., Weissleder R., Shirihai O. S., Ellisen L. W., Espinosa J. M., Mostoslavsky R. (2010) The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1α. Cell 140, 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu T. P., Tsai M. H., Lee J. M., Hsu C. P., Chen P. C., Lin C. W., Shih J. Y., Yang P. C., Hsiao C. K., Lai L. C., Chuang E. Y. (2010) Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer Epidemiol. Biomarkers Prev. 19, 2590–2597 [DOI] [PubMed] [Google Scholar]
- 15. Clegg C. H., Correll L. A., Cadd G. G., McKnight G. S. (1987) Inhibition of intracellular cAMP-dependent protein kinase using mutant genes of the regulatory type I subunit. J. Biol. Chem. 262, 13111–13119 [PubMed] [Google Scholar]
- 16. Kim Y. K., Kim H. J., Kwon C. H., Kim J. H., Woo J. S., Jung J. S., Kim J. M. (2005) Role of ERK activation in cisplatin-induced apoptosis in OK renal epithelial cells. J. Appl. Toxicol. 25, 374–382 [DOI] [PubMed] [Google Scholar]
- 17. Robbins D. J., Zhen E., Owaki H., Vanderbilt C. A., Ebert D., Geppert T. D., Cobb M. H. (1993) Regulation and properties of extracellular signal-regulated protein kinases 1 and 2 in vitro. J. Biol. Chem. 268, 5097–5106 [PubMed] [Google Scholar]
- 18. Michishita E., McCord R. A., Boxer L. D., Barber M. F., Hong T., Gozani O., Chua K. F. (2009) Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle 8, 2664–2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kang H. C., Lee Y. I., Shin J. H., Andrabi S. A., Chi Z., Gagné J. P., Lee Y., Ko H. S., Lee B. D., Poirier G. G., Dawson V. L., Dawson T. M. (2011) Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc. Natl. Acad. Sci. U.S.A. 108, 14103–14108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim S. Y., Seo M., Kim Y., Lee Y. I., Oh J. M., Cho E. A., Kang J. S., Juhnn Y. S. (2008) Stimulatory heterotrimeric GTP-binding protein inhibits hydrogen peroxide-induced apoptosis by repressing BAK induction in SH-SY5Y human neuroblastoma cells. J. Biol. Chem. 283, 1350–1361 [DOI] [PubMed] [Google Scholar]
- 21. Cho E. A., Kim E. J., Kwak S. J., Juhnn Y. S. (2014) cAMP signaling inhibits radiation-induced ATM phosphorylation leading to the augmentation of apoptosis in human lung cancer cells. Mol. Cancer 13, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cho E. A., Oh J. M., Kim S. Y., Kim Y., Juhnn Y. S. (2011) Heterotrimeric stimulatory GTP-binding proteins inhibit cisplatin-induced apoptosis by increasing X-linked inhibitor of apoptosis protein expression in cervical cancer cells. Cancer Sci. 102, 837–844 [DOI] [PubMed] [Google Scholar]
- 23. Dávalos A., Goedeke L., Smibert P., Ramírez C. M., Warrier N. P., Andreo U., Cirera-Salinas D., Rayner K., Suresh U., Pastor-Pareja J. C., Esplugues E., Fisher E. A., Penalva L. O., Moore K. J., Suárez Y., Lai E. C., Fernández-Hernando C. (2011) miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc. Natl. Acad. Sci. U.S.A. 108, 9232–9237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang S. J., Choi J. M., Chae S. W., Kim W. J., Park S. E., Rhee E. J., Lee W. Y., Oh K. W., Park S. W., Kim S. W., Park C. Y. (2011) Activation of peroxisome proliferator-activated receptor γ by rosiglitazone increases sirt6 expression and ameliorates hepatic steatosis in rats. PLoS ONE 6, e17057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luan B., Goodarzi M. O., Phillips N. G., Guo X., Chen Y. D., Yao J., Allison M., Rotter J. I., Shaw R., Montminy M. (2014) Leptin-mediated increases in catecholamine signaling reduce adipose tissue inflammation via activation of macrophage HDAC4. Cell Metab. 19, 1058–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lin Z., Yang H., Tan C., Li J., Liu Z., Quan Q., Kong S., Ye J., Gao B., Fang D. (2013) USP10 antagonizes c-Myc transcriptional activation through SIRT6 stabilization to suppress tumor formation. Cell Rep. 5, 1639–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ronnebaum S. M., Wu Y., McDonough H., Patterson C. (2013) The ubiquitin ligase CHIP prevents SirT6 degradation through noncanonical ubiquitination. Mol. Cell. Biol. 33, 4461–4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thirumurthi U., Shen J., Xia W., LaBaff A. M., Wei Y., Li C. W., Chang W. C., Chen C. H., Lin H. K., Yu D., Hung M. C. (2014) MDM2-mediated degradation of SIRT6 phosphorylated by AKT1 promotes tumorigenesis and trastuzumab resistance in breast cancer. Sci. Signal 7, ra71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gonçalves D. A., Lira E. C., Baviera A. M., Cao P., Zanon N. M., Arany Z., Bedard N., Tanksale P., Wing S. S., Lecker S. H., Kettelhut I. C., Navegantes L. C. (2009) Mechanisms involved in 3′,5′-cyclic adenosine monophosphate-mediated inhibition of the ubiquitin-proteasome system in skeletal muscle. Endocrinology 150, 5395–5404 [DOI] [PubMed] [Google Scholar]
- 30. Fu J., Akhmedov D., Berdeaux R. (2013) The short isoform of the ubiquitin ligase NEDD4L is a CREB target gene in hepatocytes. PLoS ONE 8, e78522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wiseman S. L., Shimizu Y., Palfrey C., Nairn A. C. (2013) Proteasomal degradation of eukaryotic elongation factor-2 kinase (EF2K) is regulated by cAMP-PKA signaling and the SCFβTRCP ubiquitin E3 ligase. J. Biol. Chem. 288, 17803–17811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Skalhegg B. S., Tasken K. (2000) Specificity in the cAMP/PKA signaling pathway. Differential expression, regulation, and subcellular localization of subunits of PKA. Front Biosci. 5, D678–D693 [DOI] [PubMed] [Google Scholar]
- 33. Dhillon A. S., Pollock C., Steen H., Shaw P. E., Mischak H., Kolch W. (2002) Cyclic AMP-dependent kinase regulates Raf-1 kinase mainly by phosphorylation of serine 259. Mol. Cell. Biol. 22, 3237–3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li Y., Takahashi M., Stork P. J. (2013) Ras-mutant cancer cells display B-Raf binding to Ras that activates extracellular signal-regulated kinase and is inhibited by protein kinase A phosphorylation. J. Biol. Chem. 288, 27646–27657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hong E. H., Yun H. S., Kim J., Um H. D., Lee K. H., Kang C. M., Lee S. J., Chun J. S., Hwang S. G. (2011) Nicotinamide phosphoribosyltransferase is essential for interleukin-1β-mediated dedifferentiation of articular chondrocytes via SIRT1 and extracellular signal-regulated kinase (ERK) complex signaling. J. Biol. Chem. 286, 28619–28631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Polidoro L., Properzi G., Marampon F., Gravina G. L., Festuccia C., Di Cesare E., Scarsella L., Ciccarelli C., Zani B. M., Ferri C. (2013) Vitamin D protects human endothelial cells from H2O2 oxidant injury through the Mek/Erk-Sirt1 axis activation. J. Cardiovasc. Transl. Res. 6, 221–231 [DOI] [PubMed] [Google Scholar]
- 37. Yamashita M., Shinnakasu R., Asou H., Kimura M., Hasegawa A., Hashimoto K., Hatano N., Ogata M., Nakayama T. (2005) Ras-ERK MAPK cascade regulates GATA3 stability and Th2 differentiation through ubiquitin-proteasome pathway. J. Biol. Chem. 280, 29409–29419 [DOI] [PubMed] [Google Scholar]
- 38. Ming M., Han W., Zhao B., Sundaresan N. R., Deng C. X., Gupta M. P., He Y. Y. (2014) SIRT6 promotes COX-2 expression and acts as an oncogene in skin cancer. Cancer Res. 74, 5925–5933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu Y., Xie Q. R., Wang B., Shao J., Zhang T., Liu T., Huang G., Xia W. (2013) Inhibition of SIRT6 in prostate cancer reduces cell viability and increases sensitivity to chemotherapeutics. Protein Cell 9, 702–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Spina A., Di Maiolo F., Esposito A., D'Auria R., Di Gesto D., Chiosi E., Sorvillo L., Naviglio S. (2013) Integrating leptin and cAMP signalling pathways in triple-negative breast cancer cells. Front Biosci. (Landmark Ed.) 18, 133–144 [DOI] [PubMed] [Google Scholar]
- 41. Venugopal B., Evans T. R. (2011) Developing histone deacetylase inhibitors as anti-cancer therapeutics. Curr. Med. Chem. 18, 1658–1671 [DOI] [PubMed] [Google Scholar]