Background: Mesp1 and Etv2 are essential transcription factors in the regulation of mesodermal lineage development, but their relationship is unclear.
Results: Mesp1 interacts physically with Creb1 and transcriptionally regulates Etv2 gene expression.
Conclusion: Etv2 is a direct downstream target gene of Mesp1.
Significance: This is the first report to identify Creb1 as a coactivator of Mesp1 to regulate gene expression.
Keywords: Basic Helix-Loop-Helix Transcription Factor (bHLH), ETS Transcription Factor Family, Protein-Protein Interaction, Transcription Regulation, Transcriptional Coactivator
Abstract
Mesoderm posterior 1 (Mesp1) is well recognized for its role in cardiac development, although it is expressed broadly in mesodermal lineages. We have previously demonstrated important roles for Mesp1 and Ets variant 2 (Etv2) during lineage specification, but their relationship has not been defined. This study reveals that Mesp1 binds to the proximal promoter and transactivates Etv2 gene expression via the CRE motif. We also demonstrate the protein-protein interaction between Mesp1 and cAMP-responsive element binding protein 1 (Creb1) in vitro and in vivo. Utilizing transgenesis, lineage tracing, flow cytometry, and immunostaining technologies, we define the lineage relationship between Mesp1- and Etv2-expressing cell populations. We observe that the majority of Etv2-EYFP+ cells are derived from Mesp1-Cre+ cells in both the embryo and yolk sac. Furthermore, we observe that the conditional deletion of Etv2, using a Mesp1-Cre transgenic strategy, results in vascular and hematopoietic defects similar to those observed in the global deletion of Etv2 and that it has embryonic lethality by embryonic day 9.5. In summary, our study supports the hypothesis that Mesp1 is a direct upstream transactivator of Etv2 during embryogenesis and that Creb1 is an important cofactor of Mesp1 in the transcriptional regulation of Etv2 gene expression.
Introduction
During early embryogenesis, mesodermal precursor cells within the yolk sac aggregate to form blood islands and serve as the site for endothelial and blood cell production (1, 2). Within the early yolk sac blood island, centrally located cells differentiate into primitive blood, whereas the more peripheral cells give rise to endothelial cells (3). The differentiation of these progenitor cells to endothelial and hematopoietic lineages is governed via the spatiotemporal regulation of transcription factors (4, 5). Two transcription factors, Mesp1 and Etv2, have been reported to have distinct functions in the specification of blood and endothelial lineages (6–9). Mesp1, a basic helix-loop-helix (bHLH)2 transcription factor, has an essential role in the regulation of cardiac mesoderm as Mesp1 null embryos have perturbed heart development and are nonviable (10–15), and the overexpression of Mesp1 has been shown to induce the cardiac molecular program (10, 11). Recent studies also suggest a broader context-dependent role for Mesp1 in mesodermal lineage determination (9).
Etv2 is a key regulator of endothelial and hematopoietic development (16–19). Previous studies have demonstrated that Etv2 has a narrow window of expression during development and that embryos lacking Etv2 develop severe cardiovascular defects and die by E9.5 (7, 8). We and others have shown that Etv2 is essential for the development of blood and vascular lineages and that Etv2 has a negative effect on the development of the cardiac lineage (20–22). Moreover, studies have demonstrated that Etv2 interacts physically with coexpressed factors (i.e. Foxc2 and/or Gata2) to transactivate downstream targets, including Lmo2, Scl/Tal1, Tie2, CD31/Pecam1, Sox7, Fli1, and others (7, 8, 21, 23–26). Together, these studies support the notion that Mesp1 and Etv2 are important regulators of mesodermal lineages.
cAMP-responsive element-binding protein (Creb1) has been identified as the transcription factor mediating cAMP stimulation (27, 28). Creb1 and its two paralogs, cAMP-responsive element modulator (Crem) and activating transcription factor 1 (ATF1), constitute a subgroup of the bZIP finger proteins on the basis of their conservation of the bZIP domain (29). Genetic studies have revealed the function of these subgroup proteins during murine development. Creb1 null mice have impaired T cell development, a reduction in the size of the corpus callosum, and anterior commissures and die because of respiratory defects (30). Targeted global deletion of CREM results in the absence of spermatogenesis (31, 32). Although the global deletion of ATF1 does not cause any phenotypic abnormalities, the double knockout of ATF1 and Creb1 results in embryonic lethality because of preimplantation defects (33). Therefore, these genes are hypothesized to have redundant roles in a variety of tissues (33, 34). Creb1 has been shown to transactivate gene expression through a specific motif, TGACGTCA, that is referred to as the cAMP-response element (CRE), which is found in the promoter of the genes regulated by cAMP (35). Upon cAMP stimulation, Creb1 is phosphorylated at the Ser-133 residue by PKA and is then able to recruit the Creb-binding protein (CBP) activator complex (36). In addition to cAMP, other signaling pathways may also stimulate Creb1 activation. We have reported previously that the Vegf/Flk1 signaling pathway leads to the activation of the p38-Creb1 cascade, which, in turn, activates Etv2 gene expression during embryogenesis (23). Creb1 binds to the CRE motif as a homodimer and also forms a heterodimer with other bZip factors, such as CCAAT/enhancer-binding protein (C/EBP) and NF-IL6 (27). In addition, Creb1 cooperatively interacts with other transcription factors such as AP1 and Sox9 and other bHLH factors to regulate gene expression (37–40). Interestingly, Myod transactivates its target gene, Rb1, through the proximal CRE motif in the Rb1 promoter by interacting with Creb1 (39).
In this study, our goal is to define the transcriptional and lineage relationship between Mesp1 and Etv2. Using ChIP and transcriptional assays, our study is the first to demonstrate that Mesp1 binds and transactivates Etv2 gene expression. Surprisingly, the E-box motifs are not required for the transactivation of Etv2 by Mesp1. Rather, Mesp1 interacts with Creb1 and regulates Etv2 gene expression through the CRE motif that is harbored in the proximal promoter. Our study demonstrates coexpression of Mesp1-Cre+ cells and Etv2-EYFP+ cells during murine embryonic development, with the majority of the Etv2-EYFP expressing cells at E8.5 and E9.5 arising from Mesp1-Cre+ cells. Finally, we conditionally ablate Etv2 using the Mesp1-Cre mouse model and observe a lethal phenotype similar to the Etv2 global mutants. Collectively, these data indicate that Mesp1 is a direct upstream regulator of Etv2. We further propose that the CRE motif in the proximal Etv2 promoter may serve as an integration site for both Mesp1 and Vegf/Flk1 signaling to coordinately regulate Etv2 gene expression during mesodermal lineage specification.
EXPERIMENTAL PROCEDURES
DNA and RNA Manipulation
DNA subcloning, mutagenesis, RNA extraction, cDNA synthesis, and quantitative PCR were performed as previously described (41). The dominant-negative inhibitor of Creb1 (A-Creb1) was purchased from Addgene (42). TaqMan probes for quantitative PCR were purchased from Applied Biosystems as previously described (43). Creb1 siRNA oligos were purchased from GE Dharmacon.
ChIP
In the ChIP assay, chromatin preparation and immunoprecipitation from EBs were performed as previously described (24). The following primer pairs were used in the ChIP assay: Etv2 proximal promoter, 5′-CTCCCCAAGTTCTTTTCCAAGC-3′ (forward) and 5′-CTGATAGGGGAGGGGGAATTTT-3′ (reverse); Etv2 distal enhancer, 5′-GGGCTAAAGGGCATTTCCTG-3′ (forward) and 5′-CCCCCACACTCTTTACATTACAA-3′ (reverse); and Gapdh, 5′-TGACGTGCCGCCTGGAGAAA-3′(forward) and 5′-AGTGTAGCCCAAGATGCCCTTCAG-3′ (reverse).
Cell Culture and Transcriptional Assays
NIH3T3 cells were maintained in DMEM containing 10% FBS. The day before transfection, 1 × 105 NIH3T3 cells were seeded onto 6-well plates. The Mesp1 expression plasmid, the Etv2 promoter fused to the firefly luciferase reporter, and CMV-Renilla luciferase were transfected into NIH3T3 cells with FuGENE HD (Promega). The siRNA oligo transfection was performed with Lipofectamine 2000 reagent. Cells were lysed 24 h after transfection and examined using the Dual-Luciferase assay (Promega). The firefly luciferase activity was normalized to that of the Renilla luciferase. All of the experiments were repeated at least three times.
Western Blot, Coimmunoprecipitation, and GST Pulldown Assays
C2C12 cells were maintained in DMEM with 10% FBS. HA-Creb1 and Myc-Mesp1 overexpression plasmids were transfected into C2C12 cells using Lipofectamine and Plus reagents (Invitrogen). Transfected cells were lysed 24 h later and utilized for Western blot and coimmunoprecipitation assays as described previously (44). For the protein-protein interaction between HA-Mesp1 and endogenous Creb1, EBs were treated with doxycycline from days 3 to 4 and then lysed for immunoprecipitation. Antibodies utilized in the Western blot and coimmunoprecipitation assays included anti-HA serum (3F10) from Roche; anti-HA serum (rabbit), anti-Myc serum (9E10), and anti-Myc serum (rabbit) from Santa Cruz Biotechnology; and anti-Creb1 serum (rabbit) from EMD Millipore. GST pulldown assays were performed following a procedure reported previously (44). Mesp1 deletion constructs were subcloned into the pGEX-4T vector. GST-Mesp1 fusion proteins were purified using B-per buffer (Pierce) and glutathione-Sepharose 4B beads (GE Healthcare). HA-tagged Creb1 deletional constructs were translated in vitro in the presence of [35S]methionine.
Immunohistochemistry
LacZ staining and immunohistochemistry were performed as described previously (45). Primary antibodies used for immunohistochemistry included chicken anti-green fluorescent protein serum (1:500, Abcam, catalog no. ab13970), rabbit anti-Dsred (1:200, Clontech, catalog no. 632496), goat anti-Pecam1 serum (1:500, R&D Systems, catalog no. AF3628), rat anti-endomucin serum (1:500, Abcam, catalog no. ab106100), rat anti-Tie2 serum (1:600, eBiosciences, catalog no. 14-5987-82), and rabbit anti-Flk1 serum (1:500, Cell Signaling Technology, catalog no. 55B11). Secondary antibodies included Alexa Fluor 488-donkey anti-chicken serum, Cy3-donkey anti-goat serum, Cy3-donkey anti-rabbit serum, Cy3-donkey anti-rat serum, and Cy5-donkey anti-rabbit serum, which were diluted 1:800 (all were obtained from Jackson ImmunoResearch Laboratories). Results were imaged on a Zeiss Axio Imager M2 upright microscope. Merged images of color overlay were generated digitally after photographing images in separate channels.
ES Cell Culture, Embryoid Body Differentiation, and FACS Analysis
HA-tagged Mesp1 was subcloned into the p2Lox vector and then electroporated into A2Lox-Cre mES cells to establish an iHA-Mesp1 ES cell line (46). ES cell maintenance and induction of differentiation by EB formation were performed as previously described (47).
Single-cell Quantitative PCR
The 3.9-kb upstream promoter of Etv2 was subcloned into the p2Lox-EYFP vector. This construct was electroporated into A2lox ES cells to obtain the reporter ES cell line Etv2-EYFP clone. EBs were prepared as described above and harvested on day 4. The EBs were treated with trypsin and resuspended in 3% FBS medium. EYFP+ cells were sorted using a BD FACSAria for single-cell quantitative RT-PCR as described previously (25). The sorted cells were stained with LIVE/DEAD reagent and loaded onto a Fluidigm C1 single-cell capture chip. The cells were then lysed for cDNA synthesis and amplification. The lysates from 24 cells were selected for PCR using the Fluidigm BioMark HD chip and TaqMan probes for Creb1, Etv2, and Mesp1. The data were analyzed using Fluidigm real-time PCR analysis software (25).
Animal Husbandry
Etv2-EYFP transgenic mice were engineered in our laboratory as described previously (21). Mesp1-Cre mice were provided by Kenneth Murphy (9). Rosa-TdTomato (stock no. 007914) and Rosa-LacZ (stock no. 003474) reporter lines were purchased from The Jackson Laboratory (48, 49). To generate a conditional Etv2 (Etv2fl/fl) knockout allele, a targeting vector was constructed that included a 6.4-kb-long arm of homology, a 74-bp LoxP cassette, a 2.02-kb short arm of homology, and the 1.8-kb target region, which included exons five through seven, and a neomycin cassette flanked by flippase recognition target (FRT) and LoxP cassettes. The targeting vector was then linearized and electroporated into C57BL/5 × 129/SvEv hybrid ES cells as described previously (50). After antibiotic selection with G418, surviving clones were expanded and screened for correct integration by PCR. Three clones were further confirmed by Southern blot analysis and identified for blastocyst injection. The resulting chimeric animals were bred for germ line transmission. These animals were crossed to Flp mice (The Jackson Laboratory, stock no. 003946) to remove the neomycin cassette (51). Flp was then removed by backcrossing, resulting in Etv2 floxed mice. To confirm the mutation strategy, floxed mice were crossed with EIIA-Cre mice that constitutively express the Cre recombinase (Jackson, stock no. 003724) to generate a germ line mutation (52). Embryos homozygous for this mutation phenocopied the global Etv2 mutant phenotype described previously (data not shown) (8). For conditional knockout studies, Cre transgenic mice were crossed to Etv2fl/fl mice to generate Etv2fl/+; Cre+ mice, which were then mated to Etv2fl/fl mice to obtain tissue-specific knockout animals. All mice were maintained at the University of Minnesota using protocols approved by the institutional animal care and use committee and research animal resources.
Statistics
All data represent the mean ± S.D. of at least three replicates. Statistical significance was tested by Student's t test for two groups and Kruskal-Wallis H test with Dunn's multiple comparison test for more than two groups using Prism5 software (GraphPad).
RESULTS
Etv2 Is a Direct Downstream Target Gene of Mesp1
As outlined in our previous report, Mesp1 can induce Etv2 gene expression in the ES/EB system (9). In this study, utilizing the inducible Mesp1 ES cell model (iMesp1 cells), we further examined the regulation of Etv2 gene expression by Mesp1. We hypothesized that a limited induction of Mesp1 (6 h) should enrich the differentially expressed genes for direct targets. Our results demonstrate that Mesp1 induces Etv2 mRNA levels by 1.5-fold at both 6 and 24 h (Fig. 1A). However, we did not observe any effect on Lmo2 expression, a documented Etv2 direct downstream target, at either time (Fig. 1A) (24). This may be due to the modest induction of Etv2 by Mesp1 (1.5-fold), which is relatively low compared with the overexpressed Etv2 (10-fold) expression. The 3.9-kb Etv2 promoter has been defined and contains regulatory elements that direct reporter expression in a similar spatiotemporal pattern as endogenous Etv2 (21). Moreover, the 3.9-kb Etv2 promoter harbors two conserved regions, CRI and CRII (21). Bioinformatics analysis reveals three E-box motifs within these conserved regions (Fig. 1B). The ChIP assay revealed that Mesp1 binds to the CRII region but not to the CRI region (Fig. 1C).
FIGURE 1.
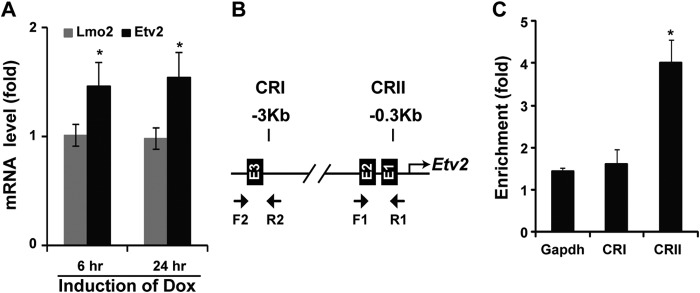
Etv2 is a direct downstream target gene of Mesp1. A, Etv2 is induced by Mesp1 using the iMesp1 ES/EB system treated with doxycycline (Dox) for 6 h. The induction is persistent following 24-h treatment. In contrast, Lmo2 is not induced by Mesp1 following 6- or 24-h treatments (*, p < 0.05). B, bioinformatics analysis reveals three E-box motifs in CRI and CRII of the Etv2 upstream 3.9-kb promoter. Specific primers for the ChIP assay are indicated for CRI and CRII. C, Mesp1 binds to the CRI region but not to the CRII region, as revealed using ChIP assays. Gapdh was a negative control for the ChIP assay (*, p < 0.05).
Transactivation of Etv2 by Mesp1 is Mediated by the CRE Motif
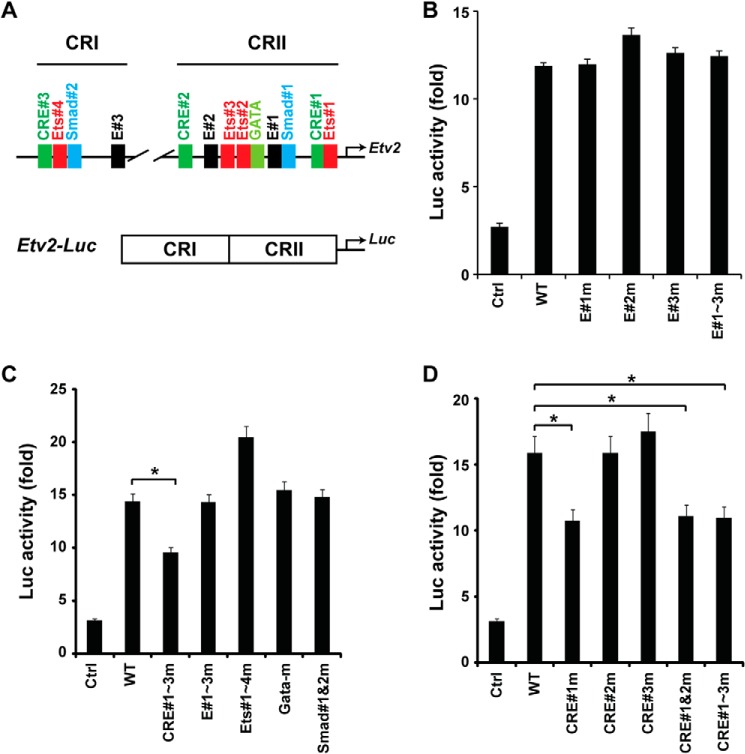
Mesp1 has been shown to be a potent transactivator of gene expression in multiple cell lines (9). As shown above, our studies demonstrated the protein-DNA interaction between Mesp1 and E-box motifs in vivo. To further examine the transcriptional regulation of Etv2 by Mesp1, we utilized the Etv2 promoter, which harbors a number of conserved motifs in the CRI and CRII regions, including E-box, CRE, Ets, Smad, and Gata motifs (Fig. 2A, top panel). CRI and CRII were constructed adjacently of the luciferase gene as the reporter Etv2-luc (Fig. 2A, bottom panel). As shown in Fig. 2B, Mesp1 transactivated the Etv2-luc reporter ∼12-fold. However, mutation of the E-box motifs, individually or in combination, did not attenuate the transactivation by Mesp1 (Fig. 2B). Furthermore, transcriptional assays with the mutation of these conserved motifs demonstrated that only mutation of the CRE motifs, but not Ets, Gata, or Smad motifs, resulted in attenuation of the reporter gene activation by Mesp1 (Fig. 2C). To identify the specificity of the CRE motif, each CRE motif was mutated. As shown in Fig. 2D, the transactivation was down-regulated only upon CRE#1 mutation but not CRE#2 or CRE#3 mutations. Importantly, the combinatorial mutations of CRE#2 or CRE#3 did not add to the effect of the CRE#1 mutation, indicating that CRE#1 is the primary element necessary for Mesp1-mediated transactivation of the Etv2 promoter.
FIGURE 2.
Mesp1 transactivates Etv2 gene expression via the CRE motif. A, top panel, the conserved motifs present in the CRI and CRII regions of the Etv2 3.9-kb promoter. ETS, Ets factor-binding motif; Smad, Smad-binding motif; E, E-box motif; GATA, Gata factor-binding motif. Bottom panel, the Etv2-luc reporter is constructed with CRI and CRII in front of the luciferase gene. B, using transcriptional assays, Mesp1 can transactivate the Etv2-luc reporter up to 12-fold. However, mutation of the E-box motifs, individually or in combination, does not attenuate the transactivation. Ctrl, control. C, CRE motifs are required for Mesp1 transactivation of the Etv2 promoter using mutagenesis screening. Note that mutagenesis of the Ets, Gata, or Smad motifs does not reduce the transactivation by Mesp1 (*, p < 0.05). D, mutation of CRE#1, instead of CRE#2 or CRE#3, attenuates the transactivation by Mesp1. Additional mutations of CRE#2 or CRE#3 motifs do not further enhance the effect of CRE#1 on the transcriptional activation (*, p < 0.05).
Mesp1 Interacts with Creb1
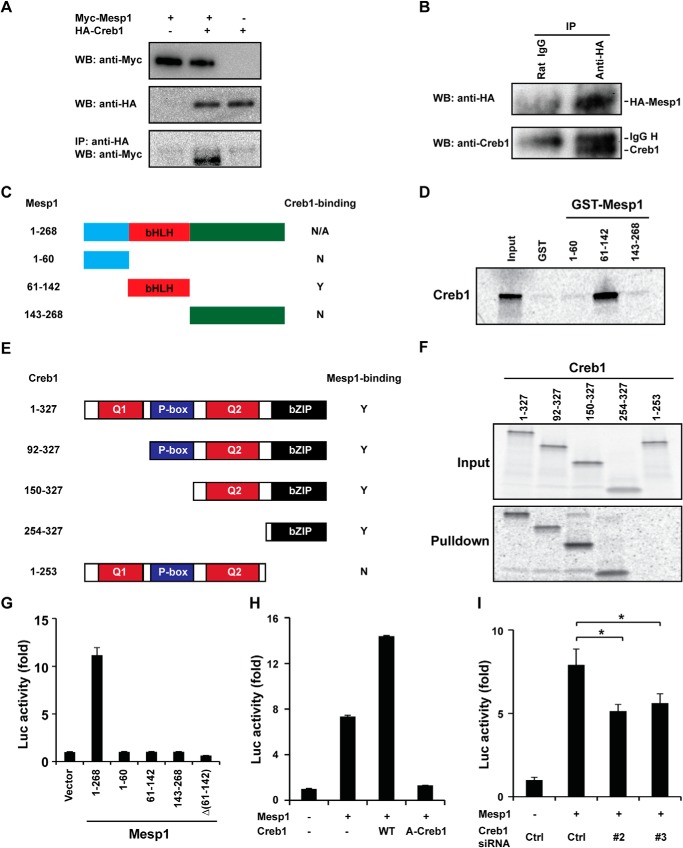
We have reported previously that Vegf/Flk1 signaling induces Etv2 gene expression through the p38-Creb1 cascade via the CRE motif (23). Our transcriptional assays described above suggested that Mesp1 interacts with Creb1, thereby regulating Etv2 gene expression. To test our hypothesis, we overexpressed Myc-tagged Mesp1 and HA-tagged Creb1 in cultured cells. As shown in Fig. 3A, Myc-Mesp1 was coimmunoprecipitated with HA-Creb1 using a HA antibody. Next we examined whether Mesp1 interacts with endogenous Creb1 in vivo. Because no antibody is available to immunoprecipitate Mesp1, we generated an inducible HA-tagged Mesp1 ES cell line using the p2Lox system. We confirmed that HA-Mesp1 is induced to a similar level as endogenous Mesp1 upon doxycycline treatment (data not shown). The immunoprecipitation was successful, as shown in Fig. 3B. We detected endogenous Creb1 in the immunoprecipitated complex using Western blot analysis (Fig. 3B). We further mapped the interacting domains of Mesp1 and Creb1 by performing GST pulldown assays. Purified GST-Mesp1 middle region (harboring the bHLH domain), but not the N-terminal or C-terminal domains (Fig. 3C), was able to pull down Creb1 (Fig. 3D), and the results are summarized in Fig. 3C. Creb1 deletional constructs are shown in Fig. 3E. These data support the notion that the Creb1 C-terminal region (bZIP domain) is essential for the interaction with Mesp1 (Fig. 3, E and F). The deletion of the bZIP domain results in a complete loss of the pulldown (Fig. 3F), as summarized in Fig. 3E. To define the region required for the transactivation by Mesp1, a series of Mesp1 constructs was generated, as shown in Fig. 3G. Only full-length Mesp1 has transactivation activity, and deletion of any of the domains results in complete loss of function (Fig. 3G). Our studies suggest that Mesp1 transactivates the Etv2 reporter through its interaction with Creb1. A-Creb1 has been reported as the dominant negative inhibitor of wild-type Creb1 (42). As shown in Fig. 3H, wild-type Creb1 enhances the transactivation by Mesp1 from 7-fold to 14-fold. However, A-Creb1 represses the transactivation by Mesp1 to baseline levels. In addition, we identified two siRNA oligos (#2 and #3) that down-regulate endogenous Creb1 efficiently (data not shown). Cotransfection of these siRNA oligos (Creb1 siRNA #2 and #3) attenuate the transactivation by Mesp1 from 8-fold to ∼5-fold (Fig. 3I).
FIGURE 3.
Mesp1 interacts directly with Creb1. A, Myc-Mesp1 and HA-Creb1 are overexpressed in C2C12 cells. Overexpression of Myc-Mesp1 and HA-Creb1 is detected using Western blot analysis and anti-Myc or anti-HA sera, respectively. Myc-Mesp1 is coimmunoprecipitated (IP) with HA-Creb1 using a HA antibody (Western blot (WB), anti-Myc). B, HA-Mesp1 is overexpressed to a similar level of endogenous Mesp1 in EBs on day 4. HA-Mesp1 is immunoprecipitated successfully by anti-HA (top panel). Endogenous Creb1 is detected using an anti-Creb serum (bottom panel). C, the deletional constructs of Mesp1. N, no; Y, yes; N/A, not available. D, 35S-labeled Creb1 is pulled down by GST-Mesp1 (61–142) but not Mesp1 (1–60) or Mesp1 (143–268), as summarized in C. E, the Creb1 deletional construct. Q1 and Q2, Glu-rich domains 1 and 2; P-box, phosphorylation domain. F, deletional Creb1 constructs were translated in vitro in the presence of [35S]methione (Input) and then utilized for a pulldown assay with GST-Mesp1 (61–142). All of the deletions harboring the bZIP domain can be pulled down, whereas the constructs lacking the bZIP domain cannot be pulled down, as summarized in E. G, transcriptional activity of Mesp1 deletions. Full-length Mesp1 transactivates the Etv2 reporter, whereas each domain of Mesp1 or deletion of bHLH domain Δ(61–142) does not transactivate the Etv2 reporter. H, wild-type Creb1 augments the activity of Mesp1. The dominant negative inhibitor A-Creb1 represses the activity of Mesp1 to the baseline level. WT, wild-type Creb1. I, knockdown of Creb1 by siRNA #2 and #3 reduces the activity of Mesp1. Ctrl, control, referring to the RNA-induced silencing complex-free siRNA (*, p < 0.05).
Mesp1 and Etv2 Are Coexpressed during Embryogenesis
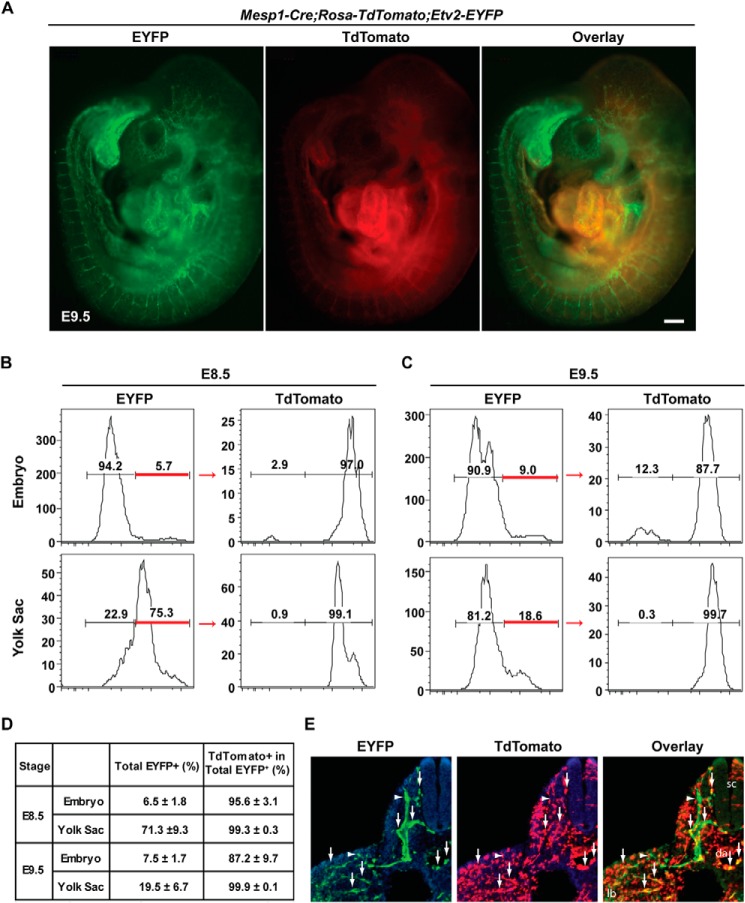
To examine the lineage relationship between the Etv2-EYFP+ cells and Mesp1-Cre+ cells (the Mesp1-Cre lineage), we generated compound embryos, Mesp1-Cre+/−; Rosa-TdTomato+; Etv2-EYFP+, and analyzed the expression of TdTomato and EYFP. In these embryos, EYFP expression was driven by the Etv2 promoter, and TdTomato marked the lineage of Mesp1-expressing cells (Fig. 4A). Whole-mount epifluorescence images reveal a robust TdTomato signal in the heart region and a weak signal in the intersomitic vessels (Fig. 4A, center panel). EYFP fluorescence appeared to largely overlap with the TdTomato signal in both embryonic heart and intersomitic vessels (Fig. 4A, compare the left and right panels). To quantify the overlap, we performed flow cytometric analysis of these embryos at E8.5 and E9.5. On average, 6.5% of cells in the embryo and 71.3% of cells in the yolk sac were EYFP+ at E8.5, and 7.5% of cells in the embryo and 19.5% of cells in the yolk sac were EYFP+ at E9.5 (Fig. 4, B and C). Of these EYFP+ cells, 95.6% in the embryo and 99.3% in the yolk sac (n = 4) were TdTomato+ cells (Mesp1-Cre lineage) at E8.5. At E9.5, the percentage dropped slightly to 87.2% in the embryo, whereas the percentage remained constant at 99.9% in the yolk sac (Fig. 4D). Further immunostaining revealed the colocalization of EYFP and TdTomato in the endothelial cells (Fig. 4E). Here we demonstrate that the majority of the Etv2-EYFP+ cells arises from the Mesp1-Cre+ lineage in both the embryo and yolk sac during embryogenesis. To further define the endogenous gene expression of Etv2 and Mesp1, we utilized the 3.9-kb Etv2-EYFP reporter ES cell line (Fig. 5A). EYFP+ cells were sorted from 3.9-kb Etv2EYFP EBs at day 4. Single-cell quantitative PCR was performed to define the expression of Creb1, Etv2, and Mesp1. As shown in Fig. 5B, Creb1 was detected in all 24 cells, whereas Etv2 was expressed in 11 cells and Mesp1 in 17 cells. The expression of Etv2 in 11 of the total 24 cells may be due to the extended half-life of EYFP protein compared with Etv2 mRNA expression. Overall, we observed coexpression of Creb1, Etv2, and Mesp1 in 9 of 24 cells (37.5%) (Fig. 5B). In summary, our studies demonstrate that Creb1 and Mesp1 are coexpressed with Etv2 during embryogenesis.
FIGURE 4.
The majority of Etv2-EYFP-expressing cells at E8.5-E9.5 arise from Mesp1-Cre+ cells. A, whole-mount images of Mesp1-Cre; Rosa-TdTomato; Etv2-EYFP compound embryos at E9.5. Scale bar = 200 μm. B, FACS analysis of lineage-traced Mesp1-Cre+ cells and Etv2-EYFP+ cells at E8.5 and (C) E9.5 in both the yolk sac and embryo. D, total EYFP+ cells (percent) and TdTomato+ in the total Etv2-EYFP+ population (percent) at E8.5 and E9.5. Note that 95.6% of Etv2-EYFP+ cells at E8.5 and 87.2% of Etv2-EYFP+ cells at E9.5 are derived from cells that have expressed Mesp1-Cre. Note that, in the yolk sac, 98% of Etv2-EYFP+ cells at E8.5 and E9.5 are descendants from Mesp1-Cre+ cells (n = 4). E, immunostaining of EYFP (green, left panel) and TdTomato (red, center panel) in the Mesp1-Cre;Rosa-TdTomato;Etv2-EYFP embryo. The coexpression of EYFP and TdTomato is shown as the overlay (yellow, right panel). The arrows indicate double-positive cells, and the arrowheads point to GFP single positives. sc, spinal cord; da, dorsal aorta; lb, limb bud. The section is at the forelimb level.
FIGURE 5.
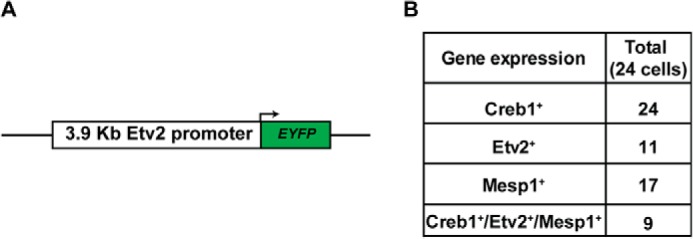
Coexpression of Etv2 and Mesp1. A, the 3.9-kb Etv2 promoter-EYFP construct utilized in the A2lox ES construction. B, gene expression of Creb1, Etv2, and Mesp1 in the 24 cells sorted from the reporter cell line at EB day 4. Creb1 was detected in all 24 cells, Etv2 in 11 cells, and Mesp1 in 17 cells. These three genes were detected in 9 of 24 cells (37.5%).
Etv2fl/fl;Mesp1-Cre+ Embryos Are Nonviable, and Most of Them Die by E9.5
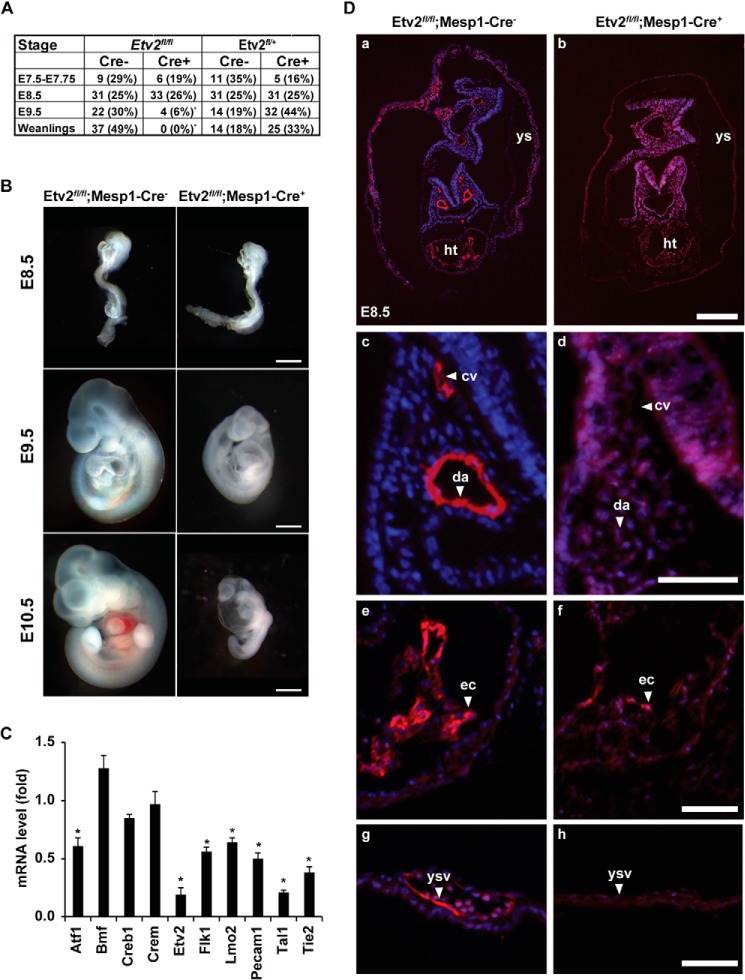
Colocalization of the Mesp1 lineage labeled by TdTomato and Etv2-EYFP prompted us to hypothesize a lineage relationship between Mesp1- and Etv2-expressing cells. To test our hypothesis and assay for gene function in vivo, we utilized the Cre/loxP recombination strategy and conditionally ablated Etv2 with the Mesp1-Cre driver (Fig. 6). We crossed Etv2fl/+;Mesp1-Cre+ mice with Etv2fl/fl mice and analyzed their genetic offspring and embryos at various stages during development. From 76 weanlings, we obtained no offspring of the Etv2fl/fl; Mesp1-Cre+ genotype (Fig. 7A). To further characterize this lethal phenotype, we analyzed E7.5, E8.5, E9.5, and E10.5 embryos during embryogenesis. We observed viable embryos at E7.5 and E8.5, whereas, by E9.5, there was a significant reduction in Etv2fl/fl; Mesp1-Cre+ genotypes (Fig. 7A). Furthermore, morphological assessment of the E9.5 embryos revealed that Etv2fl/fl; Mesp1-Cre+ embryos were significantly smaller than their control littermates (Fig. 7B). The embryos appeared pale and reduced in size and displayed pericardial edema. This phenotype was further manifested at E10.5 (Fig. 7B). To examine the gene expression profile related to this phenotype, we performed quantitative RT-PCR of RNA isolated from E8.5 embryos. As shown in Fig. 7C, Etv2 was significantly down-regulated in the Etv2fl/fl;Mesp1-Cre+ embryos, which confirmed the efficient deletion of Etv2 by the Mesp1-Cre driver. Both endothelial genes (Flk1 and Tie2) and hematopoietic genes (Lmo2, Pecam1, and Tal1) were down-regulated, which photocopied the gene expression profile in the Etv2 null embryo (8). We did not observe any change in Creb1 and Crem expression.
FIGURE 6.
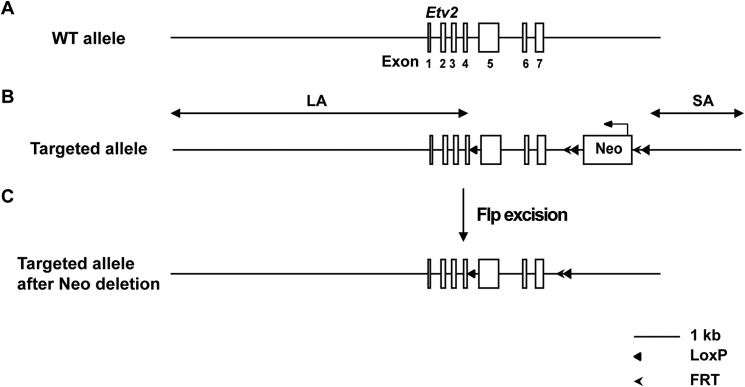
Conditional knock-out of Etv2. A, the seven exons of the Etv2 gene are schematized. B, the conditional targeted allele of Etv2. One LoxP site was placed between exons 4 and 5. The neomycin selection cassette (Neo) is flanked with the FRT-LoxP sequences and placed downstream of exon 7. The schematic of Etv2 conditional knockout reveals a 6.4-kb long arm of homology (LA), a 74-bp LoxP cassette, a 1.85-kb target region (including exons 5–7 and a neomycin cassette flanked with FRT and LoxP), and a 2.02-kb short arm of homology (SA).
FIGURE 7.
Etv2fl/fl;Mesp1-Cre+ embryos are nonviable, and most of them die by E9.5 because of vascular and hematopoietic defects. A, genotypes at E7.5, E8.5, and E9.5 and weanlings from the respective breeding. Note the significant reduction of the Etv2fl/fl; Mesp1-Cre+ embryos at E9.5. *, p < 0.001. B, whole-mount analysis of E8.5-E10.5 embryos. Note the reduced size of the Etv2fl/fl;Mesp1-Cre+ embryos and the pericardial edema and anemia at E10.5 compared with the littermate controls. Scale bars = 500 μm. C, gene expressions in the Etv2fl/fl; Mesp1-Cre+ mutant embryos at E8.5. Note that both endothelial and hematopoietic genes are down-regulated, whereas the expression of Creb1 and Crem is unaffected. Bmf serves as a control. *, p < 0.05. D, histological analysis of Etv2fl/fl; Mesp1-Cre− and Etv2fl/fl; Mesp1-Cretg/+ Etv2fl/fl; Mesp1-Cre+ mouse embryos at E8.5 stained with the endomucin antibody (a–h) reveals the presence of endothelial lineages in control embryos that are largely absent in the Etv2fl/fl; Mesp1-Cre+ mutant embryos. Shown are representative sections of dorsal aortae and cardinal veins (c and d), endocardium (e and f), and yolk sac vasculature (g and h). cv, cardinal vein; da, dorsal aorta; ec, endocardium; ht, heart; ys, yolk sac; ysv, yolk sac vasculature. Scale bars = 200 μm (a and b) and 50 μm (c–h).
Etv2fl/fl;Mesp1-Cre+ Embryos Have Impaired Vascular Development
To determine whether the conditional deletion of Etv2 by the Mesp1-Cre driver results in vascular defects similar to the Etv2 global mutants, we examined the Etv2fl/fl;Mesp1-Cre conditional knockout (Fig. 7D, a, c, e, g) and control (Fig. 7D, b, d, f, and h) embryos using immunohistochemistry. Transverse heart level sections stained with the endomucin antibody (Fig. 7D, a) revealed a positive signal in the endothelial cells lining the dorsal aortae and cardinal veins (Fig. 7D, c, arrowheads), endocardium (Fig. 7D, e, arrowheads) and yolk sac vessels (Fig. 7D, g, arrowheads) of control (Cre−) embryos. In contrast, these cells appeared to be absent or reduced significantly, and the vascular structures failed to develop properly in the Etv2fl/fl; Mesp1-Cre+ embryos (Fig. 7D, b, d, f, and h). Additional immunohistochemical analysis of the embryos stained with antibodies against Tie2, Flk1, and CD31/Pecam1 revealed similar results (data not shown). In summary, the analysis of Etv2fl/fl;Mesp1-Cre+ conditional knockout embryos demonstrated nonviable embryos by E9.5, and these embryos displayed vascular and hematopoietic defects similar to the Etv2 global mutants. This phenotype was consistent with the hypothesis that a majority of the Etv2+ cells arise from the Mesp1-Cre lineage. To substantiate our findings, we stained sections of a conditional mutant embryo of Etv2 (deleted using the Mesp1-Cre mouse model) as well as Cre-negative controls at E8.5 for three additional endothelial markers, Flk1, Tie2, and Pecam (Fig. 8). The Flk1 antibody revealed strong expression in the endothelial lineages, including the cardinal veins, dorsal aortae, and endocardium, and yolk sac vessels (Fig. 8a). It also stained cells in the head mesenchyme (mes) and the outer layer of the looping heart (ht) (Fig. 8a). All of these cells, except for the mesenchymal cells, were missing or did not express Flk1 in the conditional mutant (Fig. 8b). Similarly, Tie2 and Pecam1 were expressed in the endothelial cells of the control embryos and were missing in the conditional mutant (Fig. 8, compare c and d to e and f), except for a few Pecam-positive cells in the endocardium and the yolk sac (Fig. 8f, ec and asterisk). Consistent with Fig. 8d, we did not observe vessel-like structures in the conditional mutant embryo, indicating that the absence of immunoreactivity is not due to down-regulation of antigens but, rather, an absence of the endothelial cells.
FIGURE 8.
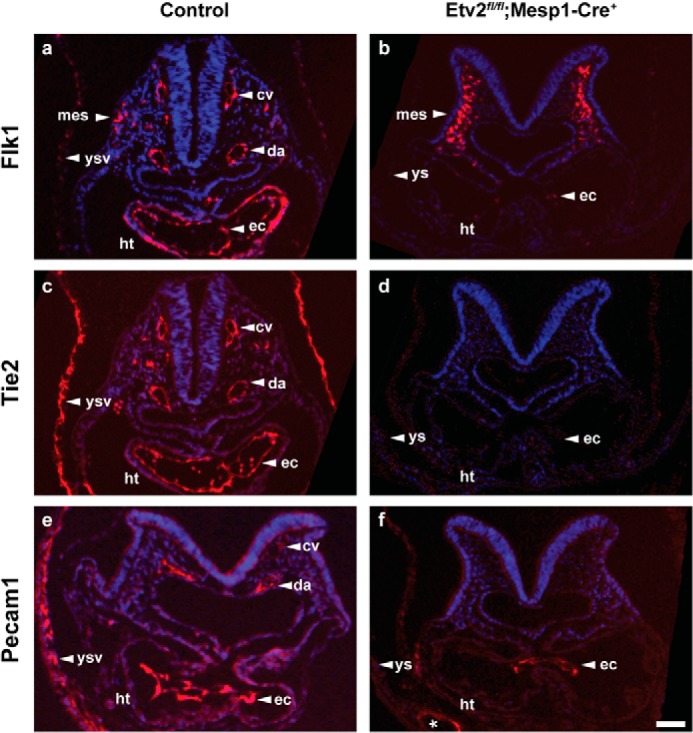
Control embryos (a and c, Etv2fl/+; Mesp1-Cre−; e, Etv2fl/fl; Mesp1-Cre−) and an Etv2fl/fl;Mesp1-Cre+ conditional mutant embryo (b, d, and f) at E8.5 were sectioned transversely at the heart level and immunostained for antibodies against Flk1 (a and b), Tie2 (c and d), and Pecam1 (e and f). The asterisk (f) indicates occasional vessel-like structures in the yolk sac that are Pecam1-positive. Note that the majority of endothelial lineages are missing in the Etv2fl/fl;Mesp1-Cre+ conditional mutants. cv, cardinal vein; da, dorsal aorta; ec, endocardium; ht, heart; mes, mesenchyme, ys, yolk sac; ysv, yolk sac vasculature. Scale bar = 50 μm.
DISCUSSION
Etv2 was initially identified as a direct downstream target gene of Nkx2–5 (8). More recent studies have demonstrated that the functional role for Etv2 is not limited to the endocardium but, rather, that it functions as an essential regulator of hematopoietic and endothelial progenitors (7, 8). Foxc2 has been shown to physically interact with Etv2 and promote the endothelial program. Our laboratory also identified Gata2 as a key binding partner that amplifies transcriptional activity and regulates the hematoendothelial lineages (53). A number of downstream targets of Etv2 have been identified, including Lmo2, Tal1, Tie2, and Pecam1 (18). However, the transcriptional regulation of Etv2 gene expression has not been well characterized. Studies in zebrafish have demonstrated that Foxc1a/b is an upstream regulator of Etsrp (54). However, there is no equivalent Forkhead-binding motif in the 3.9-kb promoter of the murine Etv2 gene (data not shown). In the murine model, the PKA and Vegf/Flk1 signaling cascades have been shown to transactivate Etv2 gene expression via Creb1 during embryogenesis (23, 55).
Previous studies support the notion that Mesp1 is a master regulator of the cardiac lineage during embryogenesis (12). Recent studies have shown that Mesp1 regulates multiple lineages through a context-dependent manner (9). Mesp1 belongs to the bHLH family of proteins, members of which form heterodimers with E12 or E47 proteins and bind to the E-box motifs (CANNTG) (6). Mesp1 has been reported to interact with the E-box motif in the promoters of a number of genes, including Hand2, Myocardin, Gata4, and Dkk1 (10, 11, 13), although a rigorous biochemical assessment for this transcription factor has not been undertaken. In this study, we demonstrated that, although Mesp1 is capable of binding to the E-box of the proximal promoter of Etv2, transcriptional and mutagenesis analyses revealed that these E-box motifs were dispensable for Mesp1 transcription activity. Rather, mutation of the CRE motifs resulted in the attenuation of Mesp1 transactivation but not the mutation of Ets, Gata, or Smad motifs. Moreover, the CRE#1 motif is the only CRE motif required for Mesp1 transcriptional activity. Previous reports suggested that the CRE motif is only functional if it is located near the proximal region of the transcription start site (56). These data are also consistent with our previous report, in which we demonstrated that CRE#1 is the only motif that is essential for the VEGF/Flk1-p38 signaling cascade and Etv2 transcriptional activity (23). Our studies further support the hypothesis that Mesp1 is recruited by the Creb1/CRE#1 activation complex as a coactivator and the E-box motif serves as the initial docking site for Mesp1. In this model, the E-box motif may enhance or stabilize the protein-protein interaction between Mesp1 and Creb1. In the future, it will be interesting to examine the coexistence of the E-box and CRE motifs in the promoters of additional target genes of Mesp1 to further examine this model.
Our transcriptional assays have led us to the hypothesis that Mesp1 interacts with Creb1. Further studies have defined that the protein-protein interaction between Mesp1 and Creb1 is mediated through the bHLH domain of Mesp1 and the bZIP domain of Creb1. Both the bHLH domain and the bZIP domain can form homodimers and heterodimers with bHLH or bZIP factors, respectively. It should be noted that the bHLH and bZIP heterodimer may represent a novel type of heterodimer. It has been reported that the dimer formed between bHLH and bZIP factors may augment or inhibit the function of bZIP or bHLH factors (40). For example, Creb1 serves as a coactivator of Myod, which forms a heterodimer with Creb1 to transactivate Rb gene expression (39). ATF4 blocks the activity of Paraxis or Scleraxis in Sertoli cells by preventing the formation of the Paraxis or Scleraxis-E12 heterodimer complex (40). In this study, we show that Mesp1 regulates the gene expression of Etv2 through the CRE motif by interacting with the coactivator Creb1. It has been reported previously that the VEGF/Flk1-p38 cascade regulates the activity of Creb1 through protein phosphorylation, and phosphorylated Creb1 then recruits the CBP complex (23). Collectively, we propose that Creb1 integrates the signaling from both Mesp1 and the VEGF/Flk1-p38 cascade through two distinct mechanisms, protein-protein interaction and protein modification, thereby transactivating Etv2 gene expression to specify mesodermal lineages. Our studies have also shown that mutation of CRE motifs results in partial attenuation of Mesp1 activity, which indicates that additional motifs are also important in the transcriptional activity of Mesp1 by recruiting other factors. In this study, we reported Creb1 as the first coactivator of Mesp1. Our data warrant further identification of additional Mesp1 cofactors to explore its functional role in the specification of mesodermal lineages.
Utilizing an array of assays, we demonstrated that Mesp1 is an upstream activator of Etv2 gene expression during embryogenesis. In support of this hypothesis, the conditional knockout of Etv2 using Mesp1-Cre model demonstrated a similar phenotype as the global Etv2 null mice. Mesp1 is an essential transcription factor for cardiac lineage specification, and it transactivates a number of cardiac genes, including Hand2, Myocardin, and Gata4 (10, 11, 13). In contrast, when Mesp1 activates Etv2 gene expression, these Mesp1+/Etv2+ progenitors become restricted to the hematopoietic/endothelial lineages and do not contribute to the cardiac lineage. Therefore, in these Mesp1+/Etv2+ progenitors, the cardiac molecular program will not be induced. Future studies will examine whether Etv2 negatively regulates the function of Mesp1 to further subdivide Mesp1+ mesodermal lineages.
Our studies also indicate that the Mesp1-Cre+ lineage gives rise to more than 95% of the Etv2-EYFP+ cells during embryogenesis at E8.5. It is important to emphasize that this number drops to 87% in E9.5 embryos compared with 99% at the E8.5 or E9.5 yolk sac. Presumably, these EYFP+/TdTomato− cells in the E9.5 embryo are not descendants of the Mesp1-Cre+ lineage. One possibility is that, because Mesp1 lineages are more dominant in the anterior half of the embryo, the posterior vascular cells may originate from a Mesp1-independent source at E9.5 (Fig. 4A) (6). Another explanation that is not mutually exclusive would be that there is another wave of angioblasts during embryogenesis and that these late-born angioblasts are derived from the Mesp1− lineage. In either case, a number of questions will need to be addressed to further define the biology, specification, and transcriptional regulation of the second source of Etv2+ cells. In summary, our study is the first to report Creb1 as the coactivator of Mesp1 and to define a mechanism whereby Mesp1 has an important functional role in the transcriptional regulation of Etv2 gene expression.
Acknowledgments
We thank Michael Przybilla and Rachel Gohla for animal husbandry. We also thank Breyen Coffin for the histology analysis and Jerry Daniel and the BMGC facility for single-cell quantitative RT-PCR analysis and Kenneth Murphy for Mesp1-Cre mice.
This work was supported, in whole or in part, by National Institutes of Health Grants U01 HL100407 and R01 HL122576 (to D. J. G.). This work was also supported by the American Heart Association (Jon Holden DeHaan Foundation Grant 0970499N) (to D. J. G.).
- bHLH
- basic helix-loop-helix
- bZIP
- basic leucine zipper
- CRE
- cAMP response element
- E
- embryonic day
- EB
- embryoid body
- CRI
- conserved region I
- CRII
- conserved region II
- EYFP
- enhanced yellow fluorescent protein.
REFERENCES
- 1. Keller G., Lacaud G., Robertson S. (1999) Development of the hematopoietic system in the mouse. Exp. Hematol. 27, 777–787 [DOI] [PubMed] [Google Scholar]
- 2. Ferkowicz M. J., Yoder M. C. (2005) Blood island formation: longstanding observations and modern interpretations. Exp. Hematol. 33, 1041–1047 [DOI] [PubMed] [Google Scholar]
- 3. Li W., Ferkowicz M. J., Johnson S. A., Shelley W. C., Yoder M. C. (2005) Endothelial cells in the early murine yolk sac give rise to CD41-expressing hematopoietic cells. Stem. Cells Dev. 14, 44–54 [DOI] [PubMed] [Google Scholar]
- 4. Wilson N. K., Foster S. D., Wang X., Knezevic K., Schütte J., Kaimakis P., Chilarska P. M., Kinston S., Ouwehand W. H., Dzierzak E., Pimanda J. E., de Bruijn M. F., Göttgens B. (2010) Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell 7, 532–544 [DOI] [PubMed] [Google Scholar]
- 5. Moignard V., Woodhouse S., Fisher J., Göttgens B. (2013) Transcriptional hierarchies regulating early blood cell development. Blood Cells Mol. Dis. 51, 239–247 [DOI] [PubMed] [Google Scholar]
- 6. Saga Y., Hata N., Kobayashi S., Magnuson T., Seldin M. F., Taketo M. M. (1996) MesP1: a novel basic helix-loop-helix protein expressed in the nascent mesodermal cells during mouse gastrulation. Development 122, 2769–2778 [DOI] [PubMed] [Google Scholar]
- 7. Lee D., Park C., Lee H., Lugus J. J., Kim S. H., Arentson E., Chung Y. S., Gomez G., Kyba M., Lin S., Janknecht R., Lim D. S., Choi K. (2008) ER71 acts downstream of BMP, Notch, and Wnt signaling in blood and vessel progenitor specification. Cell Stem Cell 2, 497–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ferdous A., Caprioli A., Iacovino M., Martin C. M., Morris J., Richardson J. A., Latif S., Hammer R. E., Harvey R. P., Olson E. N., Kyba M., Garry D. J. (2009) Nkx2–5 transactivates the Ets-related protein 71 gene and specifies an endothelial/endocardial fate in the developing embryo. Proc. Natl. Acad. Sci. U.S.A. 106, 814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chan S. S., Shi X., Toyama A., Arpke R. W., Dandapat A., Iacovino M., Kang J., Le G., Hagen H. R., Garry D. J., Kyba M. (2013) Mesp1 patterns mesoderm into cardiac, hematopoietic, or skeletal myogenic progenitors in a context-dependent manner. Cell Stem Cell 12, 587–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bondue A., Lapouge G., Paulissen C., Semeraro C., Iacovino M., Kyba M., Blanpain C. (2008) Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell 3, 69–84 [DOI] [PubMed] [Google Scholar]
- 11. Lindsley R. C., Gill J. G., Murphy T. L., Langer E. M., Cai M., Mashayekhi M., Wang W., Niwa N., Nerbonne J. M., Kyba M., Murphy K. M. (2008) Mesp1 coordinately regulates cardiovascular fate restriction and epithelial-mesenchymal transition in differentiating ESCs. Cell Stem Cell 3, 55–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bondue A., Blanpain C. (2010) Mesp1: a key regulator of cardiovascular lineage commitment. Circ. Res. 107, 1414–1427 [DOI] [PubMed] [Google Scholar]
- 13. David R., Brenner C., Stieber J., Schwarz F., Brunner S., Vollmer M., Mentele E., Müller-Höcker J., Kitajima S., Lickert H., Rupp R., Franz W. M. (2008) MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling. Nat. Cell Biol. 10, 338–345 [DOI] [PubMed] [Google Scholar]
- 14. Saga Y., Miyagawa-Tomita S., Takagi A., Kitajima S., Miyazaki J., Inoue T. (1999) MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development 126, 3437–3447 [DOI] [PubMed] [Google Scholar]
- 15. Kitajima S., Takagi A., Inoue T., Saga Y. (2000) MesP1 and MesP2 are essential for the development of cardiac mesoderm. Development 127, 3215–3226 [DOI] [PubMed] [Google Scholar]
- 16. Kataoka H., Hayashi M., Nakagawa R., Tanaka Y., Izumi N., Nishikawa S., Jakt M. L., Tarui H. (2011) Etv2/ER71 induces vascular mesoderm from Flk1+PDGFRα+ primitive mesoderm. Blood 118, 6975–6986 [DOI] [PubMed] [Google Scholar]
- 17. Palencia-Desai S., Kohli V., Kang J., Chi N. C., Black B. L., Sumanas S. (2011) Vascular endothelial and endocardial progenitors differentiate as cardiomyocytes in the absence of Etsrp/Etv2 function. Development 138, 4721–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lammerts van Bueren K., Black B. L. (2012) Regulation of endothelial and hematopoietic development by the ETS transcription factor Etv2. Curr. Opin. Hematol. 19, 199–205 [DOI] [PubMed] [Google Scholar]
- 19. Wareing S., Eliades A., Lacaud G., Kouskoff V. (2012) ETV2 expression marks blood and endothelium precursors, including hemogenic endothelium, at the onset of blood development. Dev. Dyn. 241, 1454–1464 [DOI] [PubMed] [Google Scholar]
- 20. Liu F., Kang I., Park C., Chang L. W., Wang W., Lee D., Lim D. S., Vittet D., Nerbonne J. M., Choi K. (2012) ER71 specifies Flk-1+ hemangiogenic mesoderm by inhibiting cardiac mesoderm and Wnt signaling. Blood 119, 3295–3305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rasmussen T. L., Kweon J., Diekmann M. A., Belema-Bedada F., Song Q., Bowlin K., Shi X., Ferdous A., Li T., Kyba M., Metzger J. M., Koyano-Nakagawa N., Garry D. J. (2011) ER71 directs mesodermal fate decisions during embryogenesis. Development 138, 4801–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schupp M. O., Waas M., Chun C. Z., Ramchandran R. (2014) Transcriptional inhibition of etv2 expression is essential for embryonic cardiac development. Dev. Biol. 393, 71–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rasmussen T. L., Shi X., Wallis A., Kweon J., Zirbes K. M., Koyano-Nakagawa N., Garry D. J. (2012) VEGF/Flk1 signaling cascade transactivates Etv2 gene expression. PloS ONE 7, e50103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Koyano-Nakagawa N., Kweon J., Iacovino M., Shi X., Rasmussen T. L., Borges L., Zirbes K. M., Li T., Perlingeiro R. C., Kyba M., Garry D. J. (2012) Etv2 is expressed in the yolk sac hematopoietic and endothelial progenitors and regulates Lmo2 gene expression. Stem Cells 30, 1611–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Behrens A. N., Zierold C., Shi X., Ren Y., Koyano-Nakagawa N., Garry D. J., Martin C. M. (2014) Sox7 is regulated by ETV2 during cardiovascular development. Stem Cells Dev. 23, 2004–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abedin M. J., Nguyen A., Jiang N., Perry C. E., Shelton J. M., Watson D. K., Ferdous A. (2014) Fli1 acts downstream of Etv2 to govern cell survival and vascular homeostasis via positive autoregulation. Circ. Res. 114, 1690–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shaywitz A. J., Greenberg M. E. (1999) CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu. Rev. Biochem. 68, 821–861 [DOI] [PubMed] [Google Scholar]
- 28. Sands W. A., Palmer T. M. (2008) Regulating gene transcription in response to cyclic AMP elevation. Cell. Signal. 20, 460–466 [DOI] [PubMed] [Google Scholar]
- 29. Fujii Y., Shimizu T., Toda T., Yanagida M., Hakoshima T. (2000) Structural basis for the diversity of DNA recognition by bZIP transcription factors. Nat. Struct. Biol. 7, 889–893 [DOI] [PubMed] [Google Scholar]
- 30. Rudolph D., Tafuri A., Gass P., Hämmerling G. J., Arnold B., Schütz G. (1998) Impaired fetal T cell development and perinatal lethality in mice lacking the cAMP response element binding protein. Proc. Natl. Acad. Sci. U.S.A. 95, 4481–4486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Blendy J. A., Kaestner K. H., Weinbauer G. F., Nieschlag E., Schütz G. (1996) Severe impairment of spermatogenesis in mice lacking the CREM gene. Nature 380, 162–165 [DOI] [PubMed] [Google Scholar]
- 32. Nantel F., Monaco L., Foulkes N. S., Masquilier D., LeMeur M., Henriksén K., Dierich A., Parvinen M., Sassone-Corsi P. (1996) Spermiogenesis deficiency and germ-cell apoptosis in CREM-mutant mice. Nature 380, 159–162 [DOI] [PubMed] [Google Scholar]
- 33. Bleckmann S. C., Blendy J. A., Rudolph D., Monaghan A. P., Schmid W., Schütz G. (2002) Activating transcription factor 1 and CREB are important for cell survival during early mouse development. Mol. Cell. Biol. 22, 1919–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mantamadiotis T., Lemberger T., Bleckmann S. C., Kern H., Kretz O., Martin Villalba A., Tronche F., Kellendonk C., Gau D., Kapfhammer J., Otto C., Schmid W., Schütz G. (2002) Disruption of CREB function in brain leads to neurodegeneration. Nat. Genet. 31, 47–54 [DOI] [PubMed] [Google Scholar]
- 35. Benbrook D. M., Jones N. C. (1994) Different binding specificities and transactivation of variant CRE's by CREB complexes. Nucleic Acids Res. 22, 1463–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mayr B., Montminy M. (2001) Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2, 599–609 [DOI] [PubMed] [Google Scholar]
- 37. Matsuo N., Tanaka S., Gordon M. K., Koch M., Yoshioka H., Ramirez F. (2006) CREB-AP1 protein complexes regulate transcription of the collagen XXIV gene (Col24a1) in osteoblasts. J. Biol. Chem. 281, 5445–5452 [DOI] [PubMed] [Google Scholar]
- 38. Zhao L., Li G., Zhou G. Q. (2009) SOX9 directly binds CREB as a novel synergism with the PKA pathway in BMP-2-induced osteochondrogenic differentiation. J. Bone Miner. Res. 24, 826–836 [DOI] [PubMed] [Google Scholar]
- 39. Magenta A., Cenciarelli C., De Santa F., Fuschi P., Martelli F., Caruso M., Felsani A. (2003) MyoD stimulates RB promoter activity via the CREB/p300 nuclear transduction pathway. Mol. Cell. Biol. 23, 2893–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Muir T., Wilson-Rawls J., Stevens J. D., Rawls A., Schweitzer R., Kang C., Skinner M. K. (2008) Integration of CREB and bHLH transcriptional signaling pathways through direct heterodimerization of the proteins: role in muscle and testis development. Mol. Reprod. Dev. 75, 1637–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bowlin K. M., Embree L. J., Garry M. G., Garry D. J., Shi X. (2013) Kbtbd5 is regulated by MyoD and restricted to the myogenic lineage. Differentiation 86, 184–191 [DOI] [PubMed] [Google Scholar]
- 42. Ahn S., Olive M., Aggarwal S., Krylov D., Ginty D. D., Vinson C. (1998) A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol. Cell. Biol. 18, 967–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rasmussen T. L., Martin C. M., Walter C. A., Shi X., Perlingeiro R., Koyano-Nakagawa N., Garry D. J. (2013) Etv2 rescues Flk1 mutant embryoid bodies. Genesis 51, 471–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shi X., Seldin D. C., Garry D. J. (2012) Foxk1 recruits the Sds3 complex and represses gene expression in myogenic progenitors. Biochem. J. 446, 349–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shi X., Garry D. J. (2010) Myogenic regulatory factors transactivate the Tceal7 gene and modulate muscle differentiation. Biochem. J. 428, 213–221 [DOI] [PubMed] [Google Scholar]
- 46. Iacovino M., Bosnakovski D., Fey H., Rux D., Bajwa G., Mahen E., Mitanoska A., Xu Z., Kyba M. (2011) Inducible cassette exchange: a rapid and efficient system enabling conditional gene expression in embryonic stem and primary cells. Stem Cells 29, 1580–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Iacovino M., Roth M. E., Kyba M. (2014) Rapid genetic modification of mouse embryonic stem cells by inducible cassette exchange recombination. Methods Mol. Biol. 1101, 339–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Madisen L., Zwingman T. A., Sunkin S. M., Oh S. W., Zariwala H. A., Gu H., Ng L. L., Palmiter R. D., Hawrylycz M. J., Jones A. R., Lein E. S., Zeng H. (2010) A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Soriano P. (1999) Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70–71 [DOI] [PubMed] [Google Scholar]
- 50. Garry D. J., Meeson A., Elterman J., Zhao Y., Yang P., Bassel-Duby R., Williams R. S. (2000) Myogenic stem cell function is impaired in mice lacking the forkhead/winged helix protein MNF. Proc. Natl. Acad. Sci. U.S.A. 97, 5416–5421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Farley F. W., Soriano P., Steffen L. S., Dymecki S. M. (2000) Widespread recombinase expression using FLPeR (flipper) mice. Genesis 28, 106–110 [PubMed] [Google Scholar]
- 52. Lakso M., Pichel J. G., Gorman J. R., Sauer B., Okamoto Y., Lee E., Alt F. W., Westphal H. (1996) Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. U.S.A. 93, 5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shi X., Richard J., Zirbes K. M., Gong W., Lin G., Kyba M., Thomson J. A., Koyano-Nakagawa N., Garry D. J. (2014) Cooperative interaction of Etv2 and Gata2 regulates the development of endothelial and hematopoietic lineages. Dev. Biol. 389, 208–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Veldman M. B., Lin S. (2012) Etsrp/Etv2 is directly regulated by Foxc1a/b in the zebrafish angioblast. Circ. Res. 110, 220–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yamamizu K., Matsunaga T., Katayama S., Kataoka H., Takayama N., Eto K., Nishikawa S., Yamashita J. K. (2012) PKA/CREB signaling triggers initiation of endothelial and hematopoietic cell differentiation via Etv2 induction. Stem Cells 30, 687–696 [DOI] [PubMed] [Google Scholar]
- 56. Zhang X., Odom D. T., Koo S. H., Conkright M. D., Canettieri G., Best J., Chen H., Jenner R., Herbolsheimer E., Jacobsen E., Kadam S., Ecker J. R., Emerson B., Hogenesch J. B., Unterman T., Young R. A., Montminy M. (2005) Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. U.S.A. 102, 4459–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]