Background: TNF receptor-associated factor 2 (TRAF2) is a key adaptor molecule in the TNF receptor (TNFR) signaling pathway.
Results: TRAF-interacting protein (TRIP) inhibits Lys63-linked TRAF2 ubiquitination by blocking the binding of the cofactor sphingosine 1-phosphate (S1P) to the TRAF2 RING domain.
Conclusion: TRIP negatively regulates the TRAF2 ubiquitin-dependent pathway by modulating the TRAF2-S1P interaction.
Significance: TRIP is an important cellular regulator of the TNFR-mediated inflammatory response.
Keywords: Inflammation, NF-κB, Sphingosine 1-Phosphate (S1P), TNF Receptor-associated Factor (TRAF), Tumor Necrosis Factor (TNF), Ubiquitylation (Ubiquitination), TNFR, Inflammation, TRAF-interacting Protein
Abstract
The signaling pathway downstream of TNF receptor (TNFR) is involved in the induction of a wide range of cellular processes, including cell proliferation, activation, differentiation, and apoptosis. TNFR-associated factor 2 (TRAF2) is a key adaptor molecule in TNFR signaling complexes that promotes downstream signaling cascades, such as nuclear factor-κB (NF-κB) and mitogen-activated protein kinase activation. TRAF-interacting protein (TRIP) is a known cellular binding partner of TRAF2 and inhibits TNF-induced NF-κB activation. Recent findings that TRIP plays a multifunctional role in antiviral response, cell proliferation, apoptosis, and embryonic development have increased our interest in exploring how TRIP can affect the TNFR-signaling pathway on a molecular level. In our current study, we demonstrated that TRIP is negatively involved in the TNF-induced inflammatory response through the down-regulation of proinflammatory cytokine production. Here, we demonstrated that the TRAF2-TRIP interaction inhibits Lys63-linked TRAF2 ubiquitination by inhibiting TRAF2 E3 ubiquitin (Ub) ligase activity. The TRAF2-TRIP interaction inhibited the binding of sphingosine 1-phosphate, which is a cofactor of TRAF2 E3 Ub ligase, to the TRAF2 RING domain. Finally, we demonstrated that TRIP functions as a negative regulator of proinflammatory cytokine production by inhibiting TNF-induced NF-κB activation. These results indicate that TRIP is an important cellular regulator of the TNF-induced inflammatory response.
Introduction
The downstream pathways of TNF-induced TNF receptor (TNFR)3 signaling are involved in the induction of diverse physiological processes, such as cellular proliferation, activation, differentiation, and apoptosis (1–5). In response to TNF-α stimulation, TNFR1 directly interacts via death domains (DDs) with TNFR1-associated DD protein (TRADD) and receptor-interacting protein 1/2 (RIP1/2) to form a receptor complex (3). The TNFR1-TRADD-RIP1 interaction recruits Fas-associated DD protein, which activates the caspase cascade, leading to the induction of apoptosis. Alternatively, TNFR1 can recruit additional adaptor molecules, including TNFR-associated factor 1/2 (TRAF1/2) and cellular inhibitor of apoptosis protein 1/2 (cIAP1/2) that induce cell survival/antiapoptotic signals through downstream nuclear factor-κB (NF-κB) activation (3, 4). NF-κB activation is crucial for the inducible expression of multiple cellular genes that encode proinflammatory cytokines, such as interleukin-1 (IL-1), IL-6, and TNF-α, in addition to antiapoptotic proteins (4, 5).
TRAF2 ubiquitination is crucial for the regulation of NF-κB activation at multiple levels of the signaling cascade in the TNFR-mediated pathway (1–3). The RING domain of TRAF2 possesses E3 ubiquitin (Ub) ligase activity and is capable of Lys63-linked autoubiquitination, which leads to the activation of downstream adaptors/kinases, including RIP1, cIAP1/2, TAK1 (transforming growth factor-β-activated kinase-1), TAK1-binding protein 2/3 (TAB2/3), NF-κB essential modulator (NEMO), inhibitor of IκB (IκB) kinases (IKKs), and IκB-α (1, 3, 6). TRAF2-mediated signaling is positively or negatively modulated by a number of cellular binding partners (1, 3). Sphingosine kinase 1 (SphK1), which is a lipid kinase that is responsible for the production of sphingosine 1-phosphate (S1P), directly interacts with TRAF2, positively modulating both TRAF2-mediated NF-κB activation and antiapoptotic processes (7). Interestingly, S1P functions as a cofactor for the TRAF2 E3 Ub ligase and is required for the Lys63-linked autoubiquitination of TRAF2 and for the ubiquitination of RIP1 (8). TRAF2-mediated NF-κB activation is negatively modulated by several binding partners, including TRAF-interacting protein (TRIP/TRAIP), TRAF family member-associated NF-κB activator (TANK/I-TRAF), and deubiquitination enzymes (DUBs), such as A20 and the familial cylindromatosis (CYLD) tumor suppressor gene, although some interactions remain controversial (9–15). Of these partners, TRIP is of particular interest because it is a crucial regulator of TRAF2-mediated NF-κB activation.
TRIP is an inhibitor of TRAF2-mediated NF-κB activation via a direct protein-protein interaction with TRAF1/2 in TNFR or CD30 signaling complexes (12). The murine Trip gene encodes a 470-amino acid polypeptide that contains a RING domain, a coiled-coil domain, and a leucine zipper domain in its N-terminal region (12, 16). The coiled-coil and leucine zipper domains of TRIP are important not only for the TRIP-TRAF2 interaction but also for the inhibition of TRAF2-mediated NF-κB activation (12). The RING domain of TRIP exhibits E3 Ub ligase activity, although the cellular targets of this activity and the involvement of this property of TRIP in TNFR signaling have not been studied in detail (17, 18). TRIP is involved in cell proliferation and survival through direct protein-protein interactions with CYLD or with the protein-tyrosine kinase Syk in cancer cells, although the direct interaction between TRIP and CYLD remains controversial (17–20). However, the physiological significance and the precise role of TRIP in TNF-induced inflammatory responses has not yet been clearly identified.
In our current study, we identified the cellular and molecular mechanisms underlying the regulation of TRAF2-mediated NF-κB activation via the TRAF2-TRIP interaction in the TNFR-mediated pathway. We investigated whether the TRAF2-TRIP interaction inhibits TRAF2 ubiquitination by suppressing the binding of S1P to the RING domain of the TRAF2 E3 Ub ligase. Thereby, we demonstrated that TRIP is negatively involved in the down-regulation of proinflammatory cytokine production by inhibiting TNF-induced NF-κB activation. These results indicate that TRIP acts as a negative regulator of the TNF-induced inflammatory response.
EXPERIMENTAL PROCEDURES
Cell Culture, Mice, and Reagents
HeLa, 293T, and PlatE cells were maintained in DMEM (Welgene, Daegue, Korea) supplemented with 10% FBS (Invitrogen) and an antibiotic/antimycotic solution (Welgene) in a 5% CO2 atmosphere at 37 °C. Recombinant human macrophage colony-stimulating factor (M-CSF) was described previously (23). Human TNF-α (hTNF-α) and mouse TNF-α (mTNF-α) were purchased from Peprotech (Rocky Hill, NJ). Glutathione-Sepharose 4B beads (GST beads) and anti-FLAG-M2 affinity gels (FLAG beads) were purchased from Amersham Biosciences and from Sigma-Aldrich, respectively. S1P was purchased from Enzo Life Science (Farmingdale, NY). S1P-coated affinity beads (S1P beads) were purchased from Echelon Biosciences (Salt Lake City, UT). Specific antibodies were purchased from the following commercial sources: anti-FLAG (F), anti-Myc, anti-hemagglutinin (HA), and β-actin from Sigma-Aldrich; Anti-Xpress (Xp) from Invitrogen; anti-GST, anti-B23, anti-α-tubulin, and Ub from Santa Cruz Biotechnology, Inc.; anti-TRAF2, anti-phospho-p65, anti-p65, anti-phospho-IκB-α, anti-IκB-α, anti-phospho-p38, anti-p38, anti-phospho-ERK, anti-ERK, anti-phospho-JNK, and anti-JNK from Cell Signaling Technology (Danvers, MA); anti-RIP1 from BD Biosciences; anti-TRIP from Abcam (Cambridge, MA); and anti-CYLD from Enzo Life Sciences. C57BL6/J mice were purchased from Daehan Biolink (Umsung, Korea). All animal experiments were approved by the Animal Experiment Ethics Committee of Chungnam National University (approval no. CNU-00114).
Plasmids
The epitope-tagged eukaryotic expression plasmids for TRAF2, RIP1, TRADD, TAB2, cIAP1, and TRIP were described previously (12, 16, 24). The expression plasmids for IKK-β, MEKK1, and p65 were kindly provided by Inpyo Choi (Korea Research Institute of Bioscience and Biotechnology, Daejeon, Korea). For the expression of TRIP and its deletion mutants, murine Trip cDNA was amplified via PCR and subcloned into pEBG (12), pcDNA3.1-His (Invitrogen), and pMXs-puro (25). Eukaryotic expression plasmids for SphK1 and Ubc13 were amplified by PCR and subcloned into pCMV2-FLAG (Sigma-Aldrich). For the construction of p65-inducible expression plasmid DNA (pEUI-HA-p65), PCR-amplified p65 tagged with the HA epitope was subcloned into the pEUI vector, which is a tebufenozide-responsive inducible vector that was obtained from Hyunju Ro (Chungnam National University). Eukaryotic expression plasmids for HA-tagged Ub (HA-Ub, wild type (WT)), HA-Ub-Lys48 only (Lys48-only) and HA-Ub-Lys63 only (Lys63-only) were kindly provided by Edward W. Harhaj (University of Miami, Miami, FL); the eukaryotic expression plasmids for HA-Ub-K48R (K48R), HA-Ub-K63R (K63R), and HA-Ub-KO (KO (lysine-deficient mutant)) were described previously (24). For the TRIP-knockdown (KD) plasmid DNA (TRIP-KD), a short hairpin RNA (shRNA) targeting the murine Trip gene was designed as described previously (16) and subcloned into pSuper-retro-puro (Oligoengine, Seattle, WA). The green fluorescent protein (GFP)-KD plasmid DNA (GFP-KD) was described previously (23). For the gene KD assay with siRNA, TRIP siRNA (TRIPi, 5′-UCC ACA UCA AAG GUA GCA UTT-3′), CYLD siRNA (CYLDi, 5′-CAA AGA GAA CUG CAU GAG G-3′), and GFP siRNA (GFPi, 5′-GCA UCA AGG UGA ACU UCA A-3′) were synthesized by Bioneer (Daejeon, Korea).
Luciferase Assays
Analyses of NF-κB reporter assays were performed as described previously (12, 26). Briefly, 293T cells were seeded at 2 × 105 cells/ml into 24-well plates. After 16 h, the 293T cells were cotransfected with F-TRAF2 (0.5 μg) and TRIP (1.0 μg) with or without pEUI-HA-p65 (0.5 μg) in triplicate using TurboFect reagent (Fermentas, Glen Burnie, MD). The plasmids p(κB)3-NF-κB-Luc (0.25 μg) and pcDNA3.0HisLacZ (0.1 μg) (Invitrogen) were used as reporters. At 16 h post-transfection, the expression of HA-tagged NF-κB p65 (HA-p65In) was induced via tebufenozide treatment (1 μm) for 24 h. After 24 h, the cells were harvested and lysed. Luciferase activity was measured using a luciferase assay system (Promega, Madison, WI) and normalized to β-galactosidase activity (Applied Biosystems, Bedford, MA) according to the manufacturers' instructions. The S1P-mediated NF-κB reporter assay was performed as described previously (8). Briefly, HeLa cells were plated at 2 × 105 cells/ml in 24-well plates and transfected with Xp-TRIP (1.0 μg) and reporter plasmids using TurboFect reagent. At 24 h post-transfection, the cells were stimulated using 20 ng/ml hTNF-α in the presence or absence of 10 μm S1P for 24 h. The luciferase assay was performed as described above. For the TNF-induced NF-κB reporter assay, HeLa cells (2 × 105 cells/ml) seeded in 24-well plates were transfected with Xp-TRIP (1–1.5 μg) or TRIP siRNA (TRIPi, 100–200 nm) in triplicate with reporter plasmids as described above. GFP siRNA (GFPi, 200 nm) was used as a control. At 16 h post-transfection, the cells were treated with or without hTNF-α (25 ng/ml) for 7 h, harvested, and lysed. The luciferase assay was performed as described above.
Protein-Protein Interactions
Analyses of protein-protein interactions using immunoprecipitation, a pull-down (PD) assay, and immunoblotting (IB) were performed as described previously (12, 27). Briefly, 293T cells seeded at 3.5 × 105 cells/ml in 6-well plates were cotransfected with GST-TRAF2 (1.5 μg) and Xp-TRIP (1.5 μg) with or without F-RIP1, F-cIAP1, or F-SphK1 (3 μg). At 24 h post-transfection, the cells were harvested and lysed in cell lysis buffer (20 mm Tris-HCl (pH 7.5), 150 mm NaCl, 10% glycerol, 1 mm EDTA, 1% Triton X-100, 2 mm sodium orthovanadate, 2 mm NaF, 1 mm PMSF, and proteinase inhibitor mixture (Roche Applied Science)). The cell lysates were incubated with GST beads at 4 °C for 2–4 h. Then the beads were washed three times with cell lysis buffer and subsequently subjected to immunoblot analysis with antibodies. For the TRAF2-S1P interaction assay, an S1P bead PD assay was performed as described previously (8). Briefly, 293T cells seeded at 3.5 × 105 cells/ml in 6-well plates were cotransfected with F-TRAF2 (1.5 μg) with or without Xp-TRIP (2.5 μg) or TRIP siRNA (TRIPi, 100–200 nm) using TurboFect reagent. At 24 h post-transfection, the cells were lysed in S1P binding buffer (10 mm HEPES (pH 7.4), 150 mm NaCl, 0.5% Nonidet P-40, and proteinase inhibitor mixture). S1P beads (50% slurry) were mixed with the cell lysates, and the mixtures were rocked at 4 °C for 4 h. The beads were washed four times with S1P binding buffer and subsequently subjected to immunoblot analysis with antibodies.
Ubiquitination Assays
Ubiquitination assays were performed as described previously (11, 24). Briefly, the expression plasmids for F-TRAF2 (0.3 μg) or GST-TRAF2 (1.5 μg), HA-Ub (0.5 μg), and Xp-TRIP (1–3 μg) were transfected into 293T cells seeded at 3.5 × 105 cells/ml in 6-well plates. At 24 h post-transfection, the cells were harvested. To detect ubiquitinated TRAF2 under high stringency immunoprecipitation conditions, the cells were lysed with 1% SDS in cell lysis buffer containing 10 mm N-ethylmaleimide. Cell lysates were boiled for 10 min and then diluted to 0.1% SDS with cell lysis buffer. Transfected TRAF2 was pulled down from the cell lysates using FLAG or GST beads and subsequently subjected to immunoblot analysis with antibodies. The endogenous TRAF2 ubiquitination assay was performed as described previously (24, 28). Briefly, HeLa cells were seeded at 2 × 105 cells/ml in 6-well plates and transfected with Xp-TRIP (3 μg) or TRIP siRNA (TRIPi, 100–200 nm) using TurboFect reagent. At 24 h post-transfection, the cells were treated with or without hTNF-α (40 ng/ml) for 45 min. Endogenous TRAF2 in the cell lysates was immunoprecipitated with an anti-TRAF2 antibody and protein G beads (50% slurry) (Amersham Biosciences) overnight at 4 °C and subsequently subjected to immunoblot analysis with antibodies. The TRAF2-mediated endogenous RIP1 ubiquitination assay was performed as described previously (29). Briefly, 293T cells seeded at 3.5 × 105 cells/ml in 6-well plates were cotransfected with HA-Ub (0.5 μg) and F-TRAF2 (1 μg) with or without Xp-TRIP (2.5 μg) using TurboFect reagent. At 24 h post-transfection, the cells were lysed. The cell lysates were rocked at 4 °C for 16 h with a RIP1 antibody and protein G beads (50% slurry), and the mixtures were analyzed as described above.
TNF-induced Signaling Pathway
The activation of MAPKs and NF-κB by TNF-α was analyzed as described previously (23, 24, 26). Briefly, bone marrow (BM) cells were collected from the tibias/femurs of 4–6-week-old C57BL6/J male mice and cultured in α-minimum essential medium (Invitrogen) supplemented with 10% FBS and 150 ng/ml M-CSF for 2 days to differentiate BM-derived macrophages (BMMs). During BMM culture, retroviral supernatant carrying an F-TRIP expression cassette was prepared by transfecting the pMXs-puro-FLAG-TRIP plasmid into PlatE cells as described previously (26, 30). After BMM culture for 2 days, the BMMs were infected with the F-TRIP-expressing retroviral supernatant and further cultured with M-CSF (150 ng/ml) and puromycin (2 μg/ml) for 2 days. The puromycin-resistant BMMs were cultured for 2 h under serum-free conditions and then treated with or without mTNF-α (50 ng/ml) for 5–30 min. Finally, the BMMs were harvested, lysed, and subsequently subjected to immunoblot analysis with antibodies. The relative band intensities were determined by densitometry.
TNF-induced p65 Nuclear Translocation Assay
The TNF-induced p65 nuclear translocation assay was performed as described previously (31). Briefly, F-TRIP-overexpressing BMMs were prepared via retroviral supernatant transduction as described above. The BMMs were stimulated with 50 ng/ml mTNF-α under serum-free conditions for the indicated periods. The cells were washed with buffer A (10 mm HEPES (pH 7.5), 10 mm KCl, 3 mm MgCl2, 1 mm EDTA, 1 mm DTT, 10 mm NaF, 10 mm glycerophosphate, 1 mm Na3VO4, 0.5 mm PMSF, 0.05% Nonidet P-40, and proteinase inhibitor mixture). To separate cytoplasmic fractions, the cells were carefully harvested, lysed with buffer A for 30 min at 4 °C, and separated by centrifugation for 10 min at 3000 × g and 4 °C. The supernatants (i.e. cytoplasmic fractions) were transferred into new tubes. The remaining nuclear pellets were further washed with buffer A containing 0.1% Nonidet P-40 and lysed with radioimmune precipitation assay buffer (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1% Nonidet P-40, 0.1% deoxycholate, and proteinase inhibitor mixture). The nuclear and cytoplasmic fractions were analyzed via IB with antibodies. B23 and α-tubulin were used as nuclear and cytosolic markers, respectively (32). For the subcellular localization of p65, BMMs (7.0 × 104 cells/ml) or HeLa cells (5.0 × 104 cells/ml) were plated on sterile glass coverslips in 24-well plates with 100 ng/ml M-CSF. After 24 h of culture, the cells were incubated with serum-free medium for 16 h. Then the cells were stimulated with 50 ng/ml TNF-α for 30 min, fixed, and stained with anti-p65 (Santa Cruz Biotechnology) and with Cy3-conjugated anti-rabbit IgG secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA). The nuclei were counterstained with DAPI (Sigma-Aldrich) as described previously (27). Finally, the stained cells were analyzed using an Axiovert 135M laser scanning confocal microscope (Carl Zeiss).
Real-time PCR Analysis
Real-time PCR was performed as described previously (26, 30). Briefly, the puromycin-resistant BMMs (3 × 105 cells/ml) transduced with F-TRIP-retroviral supernatant as described above were treated with mTNF-α (10 or 50 ng/ml) under serum-free conditions for 6 h. Then the cells were harvested, and total RNA was isolated using Isol-RNA lysis reagent (5 Prime, Gaithersburg, MD). cDNA was synthesized from 1 μg of total RNA using M-MLV reverse transcriptase (Affymetrix, Santa Clara, CA). Real-time PCR was performed using the IQ SYBR Green Supermix and a Mini-Opticon machine (Bio-Rad). The following primers were used: β-actin, 5′-ATG AAG ATC CTC CTG ACC GAG CG-3′ and 5′-TAC TTG CGC TGA GGA GC-3′; TNF-α, 5′-CAT CTT CTC AAA ATT CGA GTG ACA A-3′ and 5′-TGG GAG TAG ACA AGG TAC AAC CC-3′; IL-1β, 5′-ATG GCA ACT GTT CCT GAA CTC AAC T-3′ and 5′-AGT GAT ACT GCC TGC CTG AAG CTC T-3′; and IL-6, 5′-GAG GAT ACC ACT CCC AAC AGA CC-3′ and 5′-AAG TGC ATC GTT CAT ACA-3′. β-Actin was used as the normalization control. For real-time PCR analysis by TRIP-KD, retroviral supernatants carrying a TRIP-KD shRNA expression cassette (TRIP-KD) were prepared from PlatE cells. BMMs were prepared as described above to optimize the TRIP-KD effect. Then BMMs (3 × 105 cells/ml) transduced with retroviral TRIP-KD supernatant were cultured with M-CSF (150 ng/ml) and puromycin (2 μg/ml) for 2 days. The puromycin-resistant BMMs were stimulated using mTNF-α (10 or 50 ng/ml) under serum-free conditions for 6 h. Finally, the BMMs were harvested, lysed, and subsequently subjected to real-time PCR analysis.
Enzyme-linked Immunosorbent Assay (ELISA)
The puromycin-resistant BMMs (7.5 × 104 cells/ml in 96-well plates) transduced with F-TRIP- or TRIP-KD-retroviral supernatant as described above were treated with or without mTNF-α (10 or 50 ng/ml) under serum-free conditions for 24 h. The culture supernatants were harvested, and inflammatory cytokine levels were measured at 450 nm using an ELISA reader (Bio-Rad) with an IL-6 ELISA kit (BD Bioscience) or an IL-1β ELISA kit (Thermo Scientific, Rockford, IL) according to the manufacturer's instructions.
Cytokine Expression Array
The analysis of the cytokine expression array was performed using a mouse cytokine array kit (R&D Systems, Minneapolis, MN) according to the manufacturer's instructions. Briefly, the puromycin-resistant BMMs that were transduced with the retroviral F-TRIP supernatant as described above were cultured on 100-mm dishes (2 × 105 cells/ml) with M-CSF (50 ng/ml) for 1 day and subsequently treated with mTNF-α (50 ng/ml) under serum-free conditions for 1 day. After 1 day, the culture supernatants were harvested, and the cytokine levels were measured using an antibody-based luminescence system and densitometry.
Statistical Analysis
All experiments were performed at least three times. The data represent the mean ± S.D. (n = 3/group). Student's t test was used to determine the significance of the differences observed between the experimental samples (*, p < 0.05; **, p < 0.01).
RESULTS
TRIP Inhibits Lys63-linked TRAF2 Ubiquitination via the TRAF2-TRIP Interaction
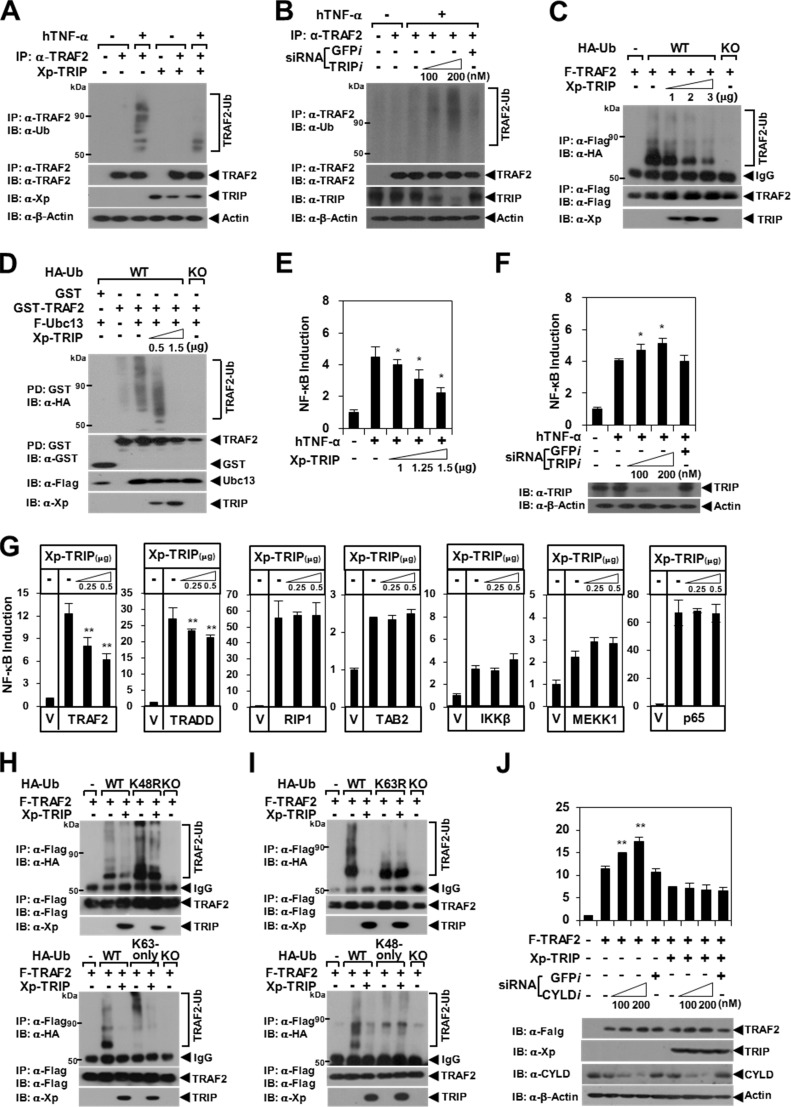
To determine the precise role of the TRAF2-TRIP interaction in TNF-induced NF-κB signaling, we conducted a TRAF2 ubiquitination assay with TRIP overexpression. After TNF-α stimulation, the ubiquitination of endogenous TRAF2 was significantly inhibited by the forced expression of TRIP in HeLa cells (Fig. 1A). In contrast, TNF-induced endogenous TRAF2 ubiquitination was enhanced by TRIP-KD (Fig. 1B). TRAF2 autoubiquitination induced by TRAF2 overexpression was also inhibited by TRIP in a dose-dependent manner (Fig. 1C). Lys63-linked TRAF2 ubiquitination has been reported to be mediated by the interaction of the E2 Ub-conjugating enzyme Ubc13/Uev1A with TRAF2 (33). Thus, we performed a Ubc13-dependent TRAF2 ubiquitination assay. We observed that Ubc13-dependent TRAF2 ubiquitination was inhibited by TRIP in a dose-dependent manner (Fig. 1D). Consistent with these results, TNF-induced, TRADD-mediated (i.e. an upstream signal of TRAF2), and TRAF2-mediated NF-κB activation was reduced by TRIP in a dose-dependent manner, but TNF-induced NF-κB activation was enhanced by TRIP-KD (Fig. 1, E–G).
FIGURE 1.
Inhibition of Lys63-linked TRAF2 ubiquitination by TRIP. A, inhibition of endogenous TRAF2 ubiquitination by TRIP. HeLa cells transfected with vector alone or with Xp-TRIP (3 μg) were stimulated for 45 min using hTNF-α (40 ng/ml). Endogenous TRAF2 was immunoprecipitated (IP) with anti-(α)-TRAF2 antibody. Ubiquitinated TRAF2 (TRAF2-Ub) was visualized via anti-Ub IB. β-Actin was used as a loading control. B, the effects of TRIP-KD on TNF-induced endogenous TRAF2 ubiquitination. TRIP expression was knocked down using siRNA corresponding to mRNA sequences of the Trip gene (TRIPi (100–200 nm)). GFPi was used as a control. Endogenous TRIP was visualized via anti-TRIP IB. C, inhibition of TRAF2 autoubiquitination by TRIP. 293T cells were transfected with F-TRAF2 (0.3 μg) and HA-Ub (0.3 μg, WT) with or without Xp-TRIP (1–3 μg). F-TRAF2 was immunoprecipitated using FLAG beads, and TRAF2-Ub was visualized via anti-HA IB. HA-Ub-KO (KO; lysine-deficient mutant) was used as a negative control. IgG, heavy chain of the anti-FLAG antibody. D, inhibition of Ubc13-dependent TRAF2 ubiquitination by TRIP. 293T cells were transfected with GST-TRAF2 (1.5 μg), F-Ubc13 (0.5 μg), and HA-Ub (0.3 μg) with or without Xp-TRIP (0.5–1.5 μg). GST-TRAF2 was pulled down (PD) using GST beads, and TRAF2-Ub was visualized via anti-HA IB. E, effects of TRIP on TNF-induced NF-κB activation. HeLa cells were transfected with Xp-TRIP (1–1.5 μg), p(κB)3-NF-κB-Luc (0.2 μg), and pcDNA3.0HisLacZ (0.1 μg) as reporter plasmids. The cells were treated with or without hTNF-α (25 ng/ml) for 7 h. Each data point was obtained in triplicate, and transfection was normalized relative to β-galactosidase activity. *, p < 0.05. F, the effects of TRIP-KD on TNF-induced NF-κB activation. G, effect of TRIP on NF-κB activation via the self-stimulation of TNFR-signaling molecules. 293T cells were transfected with a signaling molecule expression plasmid (0.5 μg) with or without Xp-TRIP (0.25–0.5 μg). At 24 h post-transfection, the cells were analyzed using luciferase assays. V, vector alone. **, p < 0.01. H, inhibition of Lys63-linked TRAF2 ubiquitination by TRIP. 293T cells were transfected with F-TRAF2 (0.3 μg) and HA-Ub (0.3 μg) with or without Xp-TRIP (3 μg). The effects of TRIP on TRAF2 ubiquitination using HA-Ub-K48R (K48R) and HA-Ub-Lys63 only (Lys63-only) are shown in the top and bottom panels, respectively. I, TRIP does not affect Lys48-linked TRAF2 ubiquitination. 293T cells were transfected with F-TRAF2 (0.3 μg) and HA-Ub (0.3 μg) with or without Xp-TRIP (3 μg). The effects of TRIP on TRAF2 ubiquitination using HA-Ub-K63R (K63R) and HA-Ub-Lys48 only (Lys48-only) are shown in the top and bottom panels, respectively. J, the effects of CYLD-KD on TRAF2-mediated NF-κB activation. 293T cells were transfected with F-TRAF2 (0.5 μg) with or without Xp-TRIP (0.5 μg). CYLD expression was knocked down via CYLDi (100–200 nm) transfection. GFPi was used as a control. *, p < 0.05. Error bars, S.D.
Lys63-linked TRAF2 ubiquitination is crucial for the activation of downstream signaling kinases (1, 2). In contrast, Lys48-linked TRAF2 ubiquitination is targeted to the protein degradation process via a proteasome-dependent mechanism; TRAF2 degradation is required for macrophage differentiation (1, 2). To determine the type of Ub-linked chain on TRAF2 that is inhibited by TRIP, we examined the inhibitory effect of TRIP on TRAF2 ubiquitination using HA-Ub. Lys63-linked TRAF2 ubiquitination was significantly inhibited by TRIP (Fig. 1H); however, Lys48-linked ubiquitination was not altered by TRIP (Fig. 1I). Consistent with these results, we observed no effects of TRIP on TRAF2 stability, self-oligomerization, and macrophage differentiation (data not shown). Because CYLD has been reported to interact directly with TRIP (19), we examined whether the DUB activity of CYLD is involved in the inhibitory effect of TRIP on TRAF2-mediated NF-κB activation. However, we observed that the inhibition of TRAF2-mediated NF-κB activation by TRIP was not affected by CYLD-KD (Fig. 1J). Hence, these results indicate that TRIP inhibits TRAF2-mediated NF-κB activation by suppressing Lys63-linked TRAF2 ubiquitination in the TNFR-mediated pathway.
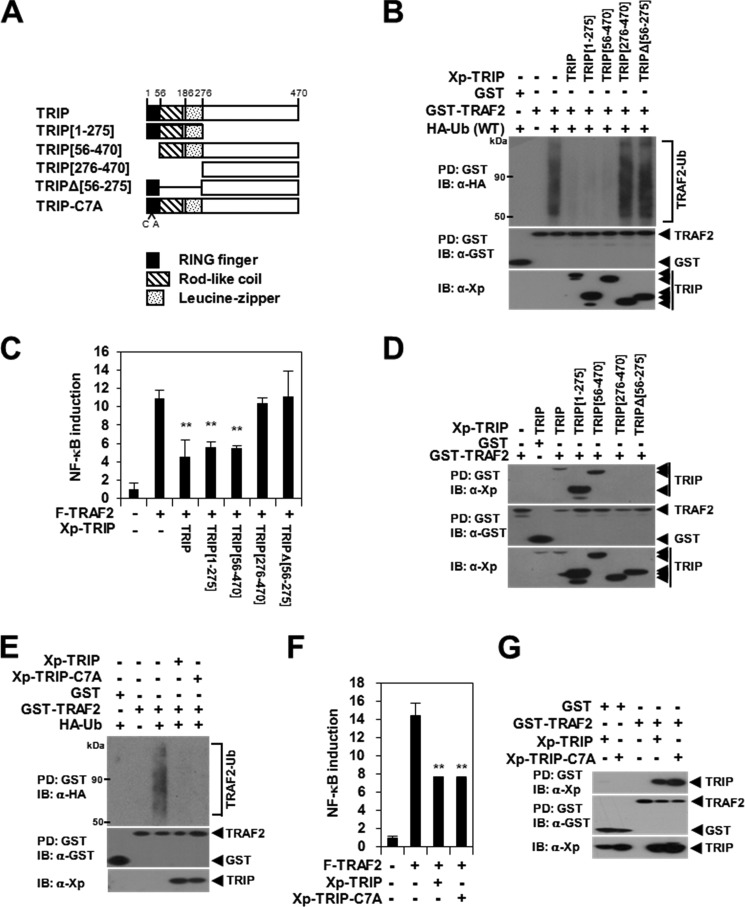
To determine the precise domain of TRIP that is involved in the inhibition of TRAF2 ubiquitination, we constructed expression plasmids for truncated TRIP proteins (Fig. 2A). In a TRAF2 ubiquitination assay, we observed that TRAF2 ubiquitination was significantly inhibited by the N-terminal TRIP(1–275) mutant but not by the C-terminal TRIP(276–470) mutant (Fig. 2B). Furthermore, TRAF2 ubiquitination was not affected by the non-TRAF2-binding mutant TRIPΔ(56–275) (Fig. 2B), indicating that the coiled-coil and leucine zipper domains of TRIP are required for the inhibition of TRAF2 ubiquitination. We also observed that TRAF2 ubiquitination, TRAF2-mediated NF-κB activation, and the TRAF2-TRIP interaction were not altered by the deletion of the RING domain (TRIP(56–470)) or by a C7A mutation (i.e. a mutation of the Cys residue in position 7 in the RING domain to Ala) in the N-terminal region of TRIP (Fig. 2, B–G). Taken together, these data demonstrate that the inhibition of TNF-induced NF-κB activation by TRIP is mediated by the suppression of Lys63-linked TRAF2 ubiquitination via the TRAF2-TRIP interaction.
FIGURE 2.
TRAF2 ubiquitination is inhibited by TRAF2-TRIP interaction through the rodlike coiled-coil and leucine zipper domain. A, schematic representation of TRIP mutants. The RING finger (black box), coiled-coil structure (hatched box), and leucine zipper domain (punctate box) are indicated. The number of amino acid residues and the single point mutation (C7A) are indicated below the diagram. The internal deletion is shown using lines. B, inhibition of TRAF2 ubiquitination by TRIP deletion mutants. 293T cells were transfected with GST-TRAF2 (1.5 μg) and HA-Ub (0.3 μg) with or without Xp-TRIP mutants (1.5 μg). GST-TRAF2 was pulled down (PD) using GST beads, and the ubiquitinated TRAF2 (TRAF2-Ub) was visualized via anti-HA IB. C, the effects of TRIP deletion mutants on TRAF2-mediated NF-κB activation. 293T cells were cotransfected with F-TRAF2 (0.5 μg) with or without Xp-TRIP or its mutants (0.5 μg). Each data point was performed in triplicate, and transfection was normalized relative to β-galactosidase activity. **, p < 0.01. D, mapping the interaction domain of TRIP with TRAF2 by deletion mutation. 293T cells were cotransfected with GST-TRAF2 (0.5 μg) with or without Xp-TRIP or its mutant (1.5 μg). GST alone or GST-TRAF2 was pulled down using GST beads, and the pulled down GST-TRAF2 was visualized via anti-GST IB. The protein bound to the TRAF2 was visualized via anti-Xp IB. E, the effect of the TRIP-C7A mutation on TRAF2 ubiquitination. 293T cells were transfected with GST-TRAF2 (1.5 μg) and HA-Ub (0.3 μg) with or without Xp-TRIP or Xp-TRIP-C7A (1.5 μg). F, the effect of TRIP-C7A mutant on TRAF2-mediated NF-κB activation. 293T cells were cotransfected with F-TRAF2 (0.5 μg) with or without Xp-TRIP or Xp-TRIP-C7A (1.5 μg) and reporter plasmids. G, effect of the TRIP C7A mutation on the TRAF2-TRIP interaction. 293T cells were cotransfected with GST-TRAF2 (0.5 μg) with or without Xp-TRIP or Xp-TRIP-C7A (1.5 μg). Error bars, S.D.
TRIP Negatively Regulates TRAF2-mediated RIP1 Ubiquitination
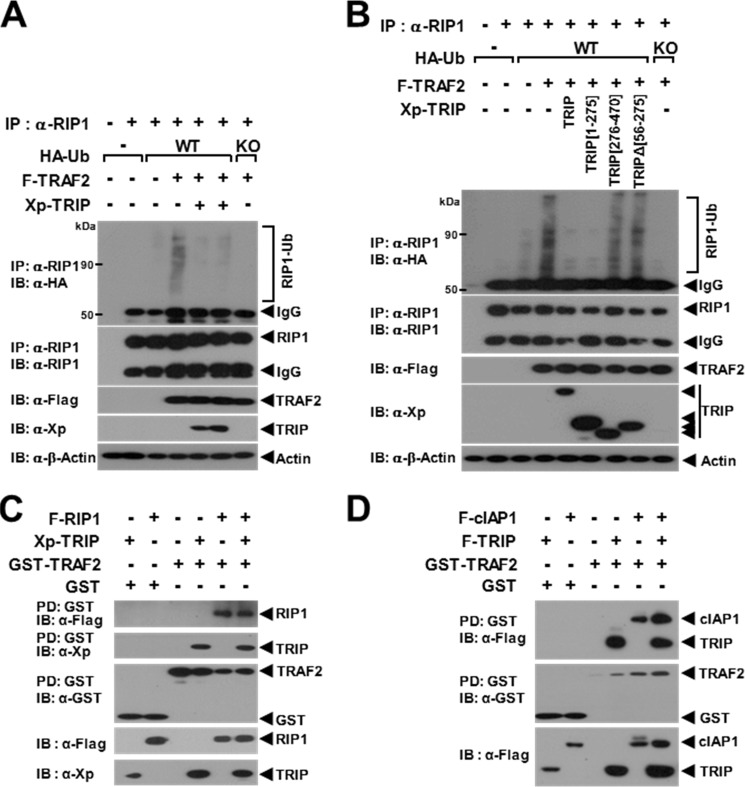
Because the TRAF2 E3 Ub ligase promotes Lys63-linked RIP1 ubiquitination (29), we conducted a TRAF2-mediated RIP1 ubiquitination assay with TRIP overexpression. When endogenous RIP1 ubiquitination was enhanced by TRAF2, TRAF2-mediated endogenous RIP1 ubiquitination was significantly inhibited by TRIP (Fig. 3A). In the deletion mutant assay, TRAF2-mediated endogenous RIP1 ubiquitination was inhibited by TRIP(1–275) but not by the non-TRAF2-binding mutants TRIP(276–470) and TRIPΔ(56–275) (Fig. 3B). Because RIP1 binds directly to the TRAF domain of TRAF2 (34), we examined whether TRIP inhibits the TRAF2-RIP1 interaction. The interaction of TRAF2 and RIP1 was not altered by TRIP (Fig. 3C). In the TNFR-signaling pathway, cIAP1/2 is also involved in both Lys48- and Lys63-linked RIP1 ubiquitination through a direct interaction with TRAF2 (1, 35). Therefore, we next examined whether TRIP blocks the interaction between TRAF2 and cIAP1. The interaction between TRAF2 and cIAP1 was not inhibited by TRIP (Fig. 3D). Thus, these data indicate that TRAF2-mediated RIP1 ubiquitination is inhibited by the TRAF2-TRIP interaction but that TRIP does not block the recruitment of RIP1 or cIAP1 to TRAF2.
FIGURE 3.
Inhibition of TRAF2-mediated endogenous RIP1 ubiquitination by TRIP. A, the effect of TRIP on TRAF2-mediated RIP1 ubiquitination. 293T cells were transfected with F-TRAF2 (1 μg) and HA-Ub (0.5 μg) with or without Xp-TRIP (2.5 μg). Endogenous RIP1 was immunoprecipitated (IP) with an anti-RIP1 antibody. Ubiquitinated RIP1 (RIP1-Ub) was visualized via anti-HA IB. B, inhibition of TRAF2-mediated RIP1 ubiquitination by TRIP deletion mutants. 293T cells were transfected with F-TRAF2 (1 μg) and HA-Ub (0.3 μg) with or without the Xp-TRIP deletion mutant (2.5 μg). C, TRAF2-RIP1 interaction is not inhibited by TRIP. 293T cells were transfected with GST-TRAF2 (0.5 μg) and F-RIP1 (1 μg) with or without Xp-TRIP (1.5 μg). GST-TRAF2 was pulled down (PD) using GST beads, and the pulled down GST-TRAF2 was visualized via anti-GST IB. The protein bound to TRAF2 was visualized via anti-FLAG (RIP1) or anti-Xp (TRIP) IB. D, TRAF2-cIAP1 interaction is not inhibited by TRIP. 293T cells were transfected with GST-TRAF2 (0.5 μg) and F-cIAP1 (1 μg) with or without F-TRIP (1.5 μg). The protein bound to TRAF2 was visualized via anti-FLAG (cIAP1 or TRIP) IB.
Binding of S1P to TRAF2 Is Inhibited by TRIP
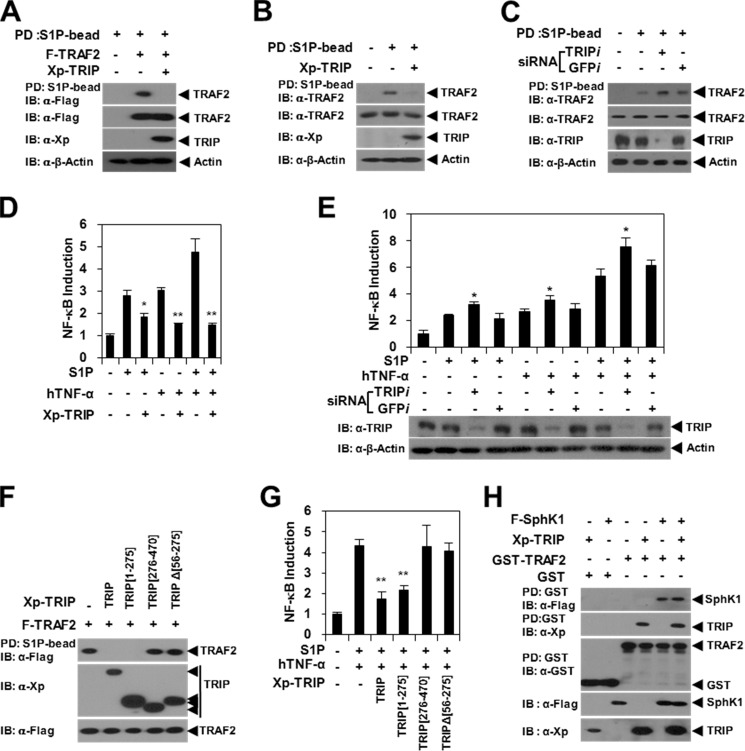
S1P functions as a cofactor for the TRAF2 E3 Ub ligase via specific binding to the TRAF2 RING domain, which induces efficient Lys63-linked TRAF2 autoubiquitination and TRAF2-mediated RIP1 ubiquitination (8). To determine whether TRIP blocks the TRAF2-S1P interaction, we conducted an S1P bead PD assay. We observed that the pull-down of TRAF2 using S1P beads was significantly inhibited by TRIP (Fig. 4A). The endogenous TRAF2-S1P interaction was also reduced by TRIP (Fig. 4B), but the endogenous TRAF2-S1P interaction was enhanced by TRIP-KD (Fig. 4C). Consistent with these results, we observed that S1P- or TNF-S1P-induced NF-κB activation was modulated by TRIP overexpression or TRIP-KD (Fig. 4, D and E).
FIGURE 4.
TRAF2-S1P binding and S1P-induced NF-κB activation are inhibited by the TRAF2-TRIP interaction. A, TRAF2-S1P binding is inhibited by TRIP. 239T cells were transfected with F-TRAF2 (1.5 μg) with or without Xp-TRIP (2.5 μg). S1P-binding protein was pulled down (PD) using S1P beads, and the pulled down TRAF2 was visualized via anti-FLAG IB. B, TRIP inhibits S1P binding to endogenous TRAF2. Xp-TRIP (2.5 μg) or its vector control was transfected into 293T cells. The endogenous TRAF2 derived from the PD assay using S1P beads was visualized via anti-TRAF2 IB. C, S1P binding to endogenous TRAF2 was enhanced by TRIP-KD. TRIP expression was knocked down using siRNA corresponding to mRNA sequences of the Trip gene (TRIPi (200 nm)). GFPi was used as a control. Endogenous TRIP was visualized via anti-TRIP IB. D, TRIP inhibits S1P-induced NF-κB activation. HeLa cells were transfected with or without Xp-TRIP (1 μg) in the presence or absence of S1P (10 μm). A luciferase assay was performed with hTNF-α (20 ng/ml) stimulation for 24 h. *, p < 0.05; **, p < 0.01. E, effects of TRIP-KD on S1P-induced NF-κB activation. TRIP expression was knocked down using siRNA (TRIPi (200 nm)). GFPi was used as a control. F, TRAF2-TRIP interaction inhibits S1P binding to TRAF2. 293T cells were transfected with F-TRAF2 (1.5 μg) with or without Xp-TRIP (2.5 μg). TRAF2 derived from the PD assay using S1P beads was visualized via anti-FLAG IB. G, TRAF2-TRIP interaction inhibits S1P-induced NF-κB activation. HeLa cells were transfected with or without Xp-TRIP (1 μg) in the presence or absence of S1P (10 μm). A luciferase assay was performed with hTNF-α (20 ng/ml) stimulation for 24 h. H, TRAF2-SphK1 interaction is not affected by TRIP. 293T cells were transfected with GST-TRAF2 (1.5 μg) and SphK1 (1.5 μg) with or without Xp-TRIP (2.5 μg). GST-TRAF2 was pulled down using GST beads, and TRAF2-binding proteins were visualized via anti-FLAG (SphK1) or anti-Xp (TRIP) IB. Error bars, S.D.
Next, we conducted S1P bead PD assays with TRIP deletion mutants. The pull-down of TRAF2 by S1P beads was significantly inhibited by the TRIP(1–275) mutant, but the TRAF2-S1P interaction was not affected by the non-TRAF2-binding mutants TRIP(276–470) and TRIPΔ(56–275) (Fig. 4F). We obtained similar results in the TNF-S1P-induced NF-κB activation assay (Fig. 4G). Because TRAF2 binds directly to SphK1, which is a lipid kinase that is responsible for the production of S1P (7), we next examined whether TRIP inhibits the TRAF2-SphK1 interaction. However, the interaction of TRAF2 and SphK1 was not altered by TRIP (Fig. 4H). These results indicate that TRIP is negatively involved in TNF-induced NF-κB activation through the inhibition of S1P cofactor binding to the RING domain of the TRAF2 E3 Ub ligase.
TRIP Inhibits the Phosphorylation and Nuclear Translocation of p65
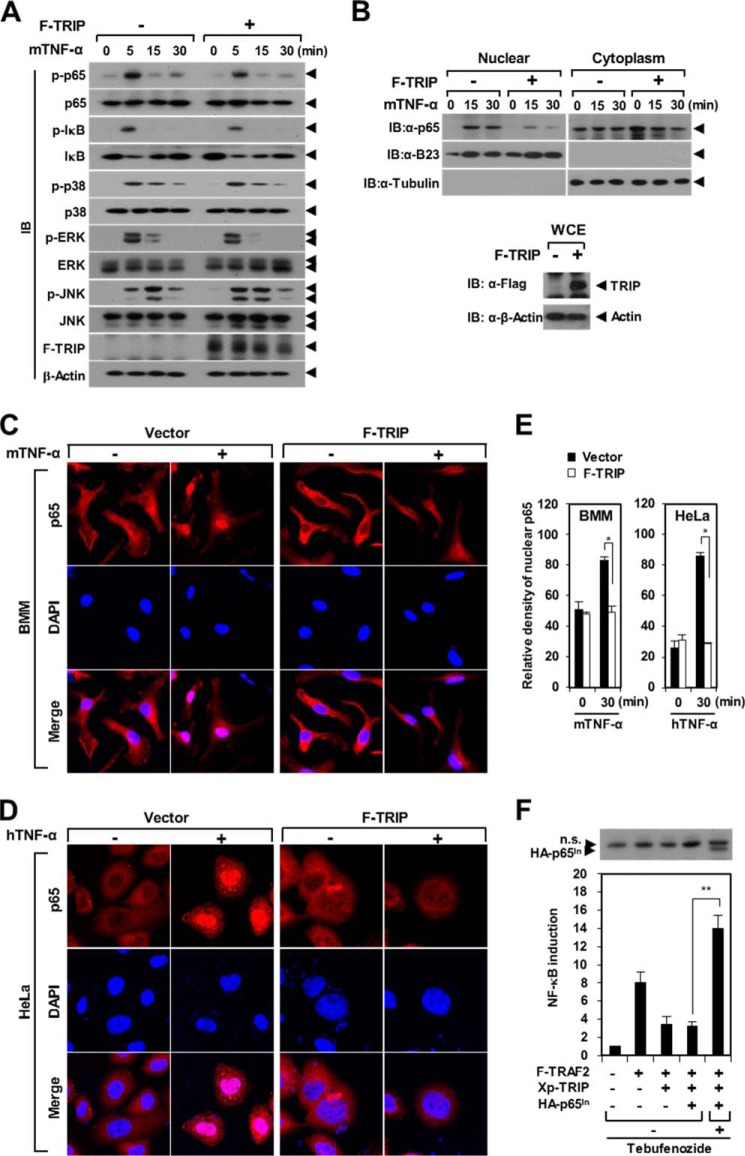
TNF ligation to the TNFR activates NF-κB and MAPK signaling (1, 3). Thus, we examined the effect of TRIP on the TNF-induced signaling pathway. After TNF-α stimulation, the phosphorylation of NF-κB p65, p38, JNK, and ERK was significantly enhanced in a time-dependent manner in BMMs (Fig. 5A). Upon TNF-α stimulation for 5 min, the phosphorylation of NF-κB p65 and IκB-α was reduced (∼53 and ∼47%, respectively) by TRIP; however, the phosphorylation of other targets was unchanged or slightly changed (Fig. 5A). To further examine the effect of TRIP on TNF-induced NF-κB p65 activation, we performed a nuclear fractionation assay. The extent of nuclear NF-κB p65 was significantly reduced by TRIP expression, whereas the level of cytoplasmic NF-κB p65 was not altered by TRIP (Fig. 5B). Consistent with these results, the nuclear translocation of NF-κB p65 was reduced by TRIP in BMMs stimulated with TNF-α (Fig. 5, C and E). We observed similar results in HeLa cells after hTNF-α stimulation (Fig. 5, D and E). Next, we generated tebufenozide-responsive NF-κB p65-inducible expression plasmid DNA based on an ecdysone-inducible system and performed a rescue experiment for the inhibitory effect of TRIP on TRAF2-mediated NF-κB activation. The inhibition of TRAF2-mediated NF-κB activation by TRIP was rescued by the inducible expression of downstream NF-κB p65 derived from tebufenozide induction (Fig. 5F). These results indicate that TRIP is involved in the process of NF-κB p65 activation via the down-regulation of p65 phosphorylation and nuclear import.
FIGURE 5.
TRIP inhibits TNF-induced NF-κB signaling. A, inhibition of NF-κB p65 and IκB-α phosphorylation by TRIP. Puromycin-selected BMMs that were transduced with TRIP retroviral supernatants were cultured for 2 h under serum-free conditions. BMMs were stimulated with mTNF-α (50 ng/ml) and subsequently analyzed via IB with antibodies recognizing phosphorylated and total p65, IκB-α, p38, ERK, and JNK. B, reduction of nuclear NF-κB p65 by TRIP in BMMs stimulated with TNF-α. BMMs transduced with F-TRIP retroviral supernatants were stimulated with mTNF-α (50 ng/ml). Cytosolic and nuclear extracts from BMMs were analyzed via IB with anti-p65, anti-B23, and anti-α-tubulin antibodies (top). B23 and α-tubulin served as positive controls for the nuclear and cytoplasmic fractions, respectively. The expression of F-TRIP in a whole cell extract (WCE) was analyzed via anti-FLAG IB (bottom). C, inhibition of the nuclear translocation of NF-κB p65 by TRIP in BMMs stimulated with mTNF-α. BMMs cultured in serum-free medium were subjected to mTNF-α (50 ng/ml) stimulation for 30 min. The cells were stained with anti-p65 antibody and a Cy3-conjugated anti-rabbit IgG secondary antibody. The nuclei were counterstained with DAPI. D, inhibition of the nuclear translocation of NF-κB p65 by TRIP in HeLa cells stimulated with hTNF-α. HeLa cells cultured in serum-free medium were subjected to hTNF-α (20 ng/ml) stimulation for 30 min. E, summary of NF-κB p65 nuclear translocation by TRIP. *, p < 0.05. F, ability of NF-κB p65-inducible expression to rescue the inhibition of TRAF2-mediated NF-κB activation by TRIP. 293T cells were transfected with F-TRAF2 (0.5 μg) and Xp-TRIP (1.0 μg) with or without pEUI-HA-p65 (0.5 μg). At 16 h post-transfection, the inducible expression of HA-tagged NF-κB p65 (HA-p65In) was stimulated via tebufenozide treatment (1 μm) for 24 h. The inducible expression of NF-κB p65 (HA-p65In) was analyzed via IB with anti-HA antibodies (top). n.s., nonspecific bands. **, p < 0.01. Error bars, S.D.
TRIP Inhibits the Expression of Proinflammatory Cytokines
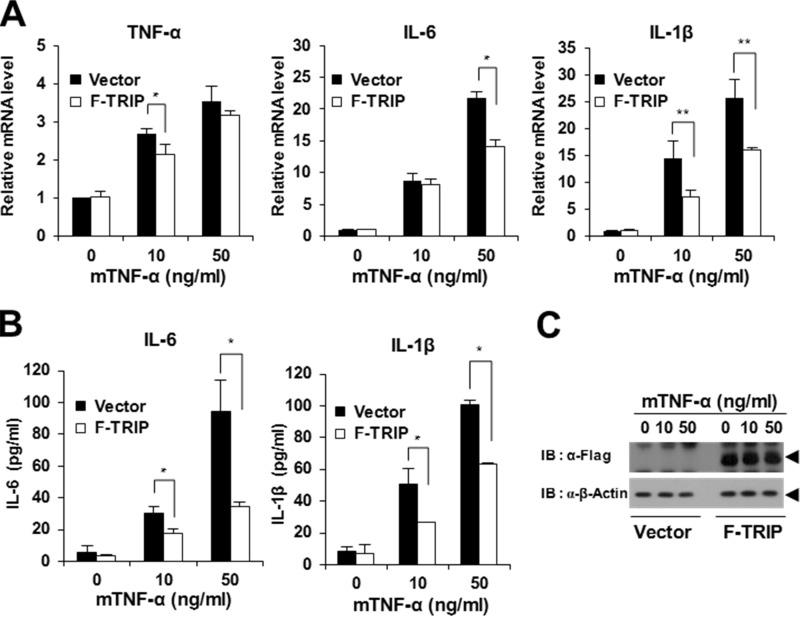
TNFR members play important roles in the production of immunomodulating cytokines that control diverse aspects of immune function (1, 3). Thus, we examined whether TRIP is involved in the production of immunomodulating cytokines. As a preliminary experiment, we prepared conditioned medium from TRIP-overexpressing BMMs after TNF-α stimulation and analyzed the production of 40 different cytokines using a commercial multiplex cytokine array kit. After TNF-α stimulation, we observed that the production of 27 cytokines derived from the conditioned medium of TRIP-overexpressing BMMs was decreased relative to the control; however, no cytokines were found to be significantly up-regulated by TRIP (data not shown). In the quantification analysis performed using densitometry, we further narrowed this list to 18 candidates that exhibited expression levels less than 15% of those of the control samples (Table 1). In particular, we found that the production of proinflammatory cytokines, such as IL-1α, IL-1β, IL-6, and IL-17, were down-regulated by TRIP. Because NF-κB activation is considered a crucial modulator of inflammation via its ability to induce the transcription of proinflammatory cytokine genes (4, 5), we further examined the effect of TRIP on the expression of proinflammatory cytokines. During real-time PCR analysis, we observed that TNF-α, IL-6, and IL-1β mRNA expression levels significantly increased in BMMs after TNF-α stimulation (Fig. 6A). However, the increased expression of these proinflammatory cytokines after TNF-α stimulation was inhibited by TRIP (Fig. 6A). Furthermore, we observed that the secretion of the cytokines IL-6 and IL-1β into the culture supernatant of BMMs after TNF-α stimulation was significantly reduced by TRIP (Fig. 6B). Under these experimental conditions, we examined TRIP expression using immunoblot analysis (Fig. 6C).
TABLE 1.
Cytokines down-regulated by TRIP overexpression in BMMs after TNF-α stimulation
The cytokines down-regulated (i.e. expression ∼15% lower than that of the vector control) by TRIP overexpression were selected (boldface type). ND, not detected. Blc, B lymphocyte attractant; C5a, complement component 5a; G-csf, granulocyte colony-stimulating factor; I-309, inflammatory cytokine 309; sICAM-1, soluble intracellular adhesion molecule-1; Il-1ra, interleukin-1 receptor antagonist; Ip-10, interferon γ-induced protein 10; I-tac, interferon-inducible T cell α chemoattractant; Kc, keratinocyte cell-derived chemokine; Je, junctional epithelium chemokine; Mcp-5, monocyte chemoattractant protein-5; Mig, monocyte interferon γ-inducing factor; Mip, macrophage inflammatory protein; Rantes, regulated on activation, normal T cell expressed and secreted; Sdf-1, stromal cell-derived factor 1; Tarc, thymus- and activation-regulated chemokine; Timp-1, tissue inhibitor of metalloproteinase-1; Trem-1, triggering receptor expressed on myeloid cells.
| Gene | -Fold change | Gene | -Fold change |
|---|---|---|---|
| Blc | 0.63 ± 0.03 | Il-16 | 0.75 ± 0.13 |
| C5a | 0.47 ± 0.11 | Il-17 | 0.77 ± 0.07 |
| G-csf | 0.61 ± 0.08 | Il-23 | 0.67 ± 0.02 |
| Gm-csf | 1.05 ± 0.01 | Il-27 | 0.24 ± 0.01 |
| I-309 | 0.78 ± 0.02 | Ip-10 | 0.96 ± 0.01 |
| Eotaxin | 0.69 ± 0.07 | I-tac | 0.83 ± 0.45 |
| sICAM-1 | 0.87 ± 0.05 | Kc | 0.99 ± 0.01 |
| Ifn-γ | 0.42 ± 0.15 | M-csf | 0.74 ± 0.26 |
| Il-1α | 0.56 ± 0.04 | Je | 1.02 ± 0.0 |
| Il-1β | 0.38 ± 0.02 | Mcp-5 | 0.97 ± 0.08 |
| Il-1ra | 0.86 ± 0.02 | Mig | 0.64 ± 0.13 |
| Il-2 | 0.68 ± 0.17 | Mip-1α | 0.92 ± 0.0 |
| Il-3 | 0.60 ± 0.14 | Mip-1β | 0.94 ± 0.0 |
| Il-4 | 0.59 ± 0.15 | Mip-2 | 0.92 ± 0.0 |
| Il-5 | ND | Rantes | 0.91 ± 0.0 |
| Il-6 | 0.82 ± 0.01 | Sdf-1 | ND |
| Il-7 | 0.98 ± 0.01 | Tarc | ND |
| Il-10 | 0.57 ± 0.20 | Timp-1 | 0.91 ± 0.02 |
| Il-13 | 0.45 ± 0.05 | Tnf-α | 1.02 ± 0.01 |
| Il-12 p70 | N.D. | Trem-1 | 0.78 ± 0.58 |
FIGURE 6.
TRIP inhibits proinflammatory cytokine production in response to TNF stimulation. A, inhibition of proinflammatory cytokine expression by TRIP. Puromycin-selected BMMs transduced with TRIP retroviral supernatants or vector alone were cultured under serum-free conditions for 6 h with or without mTNF-α (10 or 50 ng/ml) treatment. Proinflammatory cytokine expression was then analyzed by real-time PCR analysis. Open box, F-TRIP-transduced BMMs. Black box, retroviral vector-transduced BMMs. *, p < 0.05; **, p < 0.01. B, inhibition of IL-6 and IL-1β secretion by TRIP expression. Proinflammatory cytokine secretion was analyzed using an ELISA kit. C, analysis of TRIP expression in retrovirally transduced BMMs via immunoblot analysis. Error bars, S.D.
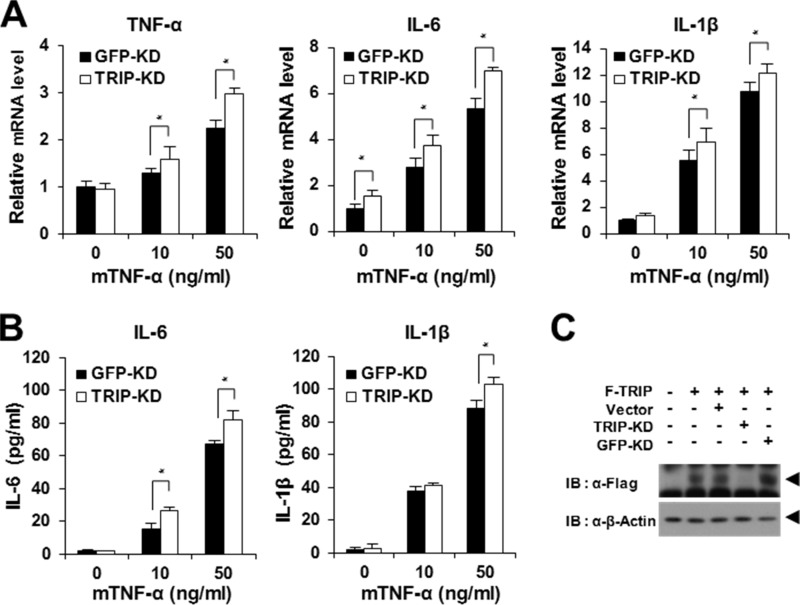
Next, we investigated whether TRIP-KD is involved in the expression of proinflammatory cytokines in BMMs stimulated with TNF-α. To examine the effects of TRIP-KD, BMMs transduced with TRIP-KD retroviral supernatant were selected using puromycin, and the selected cells were further tested for the expression of proinflammatory cytokines using real-time PCR and ELISA analysis. We observed a significant increase in the gene expression of proinflammatory cytokines in TRIP-KD BMMs after TNF-α stimulation (Fig. 7A). Furthermore, the secretion of IL-6 and IL-1β in the culture supernatant of BMMs was significantly enhanced by TRIP-KD (Fig. 7B). Finally, we examined the reduced level of TRIP expression after TRIP-KD using immunoblot analysis (Fig. 7C). Taken together, these data demonstrate that TRIP is a negative regulator of the expression of TNF-induced proinflammatory cytokine genes.
FIGURE 7.
Enhancement of proinflammatory cytokine production by TRIP-KD in response to TNF stimulation. A, promotion of proinflammatory cytokine expression by TRIP-KD. Puromycin-selected BMMs transduced with retroviral supernatants from TRIP-KD were cultured under serum-free conditions for 6 h with or without mTNF-α (10 or 50 ng/ml) treatment. Then proinflammatory cytokine expression was analyzed by real-time PCR analysis. Open box, TRIP-KD-transduced BMMs. Black box, GFP-KD (control)-transduced BMMs. *, p < 0.05. B, enhancement of IL-6 and IL-1β secretion by TRIP-KD. Proinflammatory cytokine secretion was analyzed using an ELISA kit. C, analysis of TRIP-KD effects via immunoblot analysis. TRIP-KD was visualized via anti-FLAG IB. GFP-KD was used as a control. Error bars, S.D.
DISCUSSION
Studies performed over the past decade have demonstrated convincingly that Ub-dependent signaling is a centrally important mechanism that governs the NF-κB pathway during inflammatory responses (1, 3, 6). Thus far, a number of E2 Ub-conjugating enzymes (e.g. Ubc13 and Ubc5), E3 Ub ligases (e.g. TRAFs and cIAPs), DUBs (e.g. A20 and CYLD), and Ub-binding adaptors (e.g. TABs and NEMO) have been demonstrated to be involved in diverse NF-κB pathways (1, 3, 6). In particular, E3 Ub ligases are responsible for dictating substrate specificity and are the key control points for Ub-dependent NF-κB pathways (36).
The TRAF2 E3 Ub ligase is a key player in cell survival or apoptosis in the TNF-induced signaling pathway (3, 37). Upon TNF-α stimulation, TRAF2 is recruited directly to TNFR2 or via TRADD to TNFR1, which results in the activation of downstream NF-κB and MAPKs (1, 3, 6). Genetic evidence indicates that the activity of the TRAF2 E3 Ub ligase is required for Lys63-linked TRAF2 autoubiquitination and TRAF2-mediated RIP1 ubiquitination, which lead to the activation of downstream NF-κB signaling (6). Moreover, TRAF2 promotes cell survival in response to TNF by recruiting cIAP1/2 to the TNFR1 complex (38, 39). The signaling complexes of TRAF2 and cIAP1/2 are required for RIP1 ubiquitination; however, the Ub chains synthesized by cIAP1/2 are not restricted to the Lys63 linkage (40, 41). Interestingly, TNFR2-mediated signaling enhances TNFR1-induced apoptosis by reducing TRAF2 protein levels (42). The degradation of TRAF2 by the Lys48-Ub-dependent proteasome machinery downstream of cIAP1 E3 Ub ligase activity terminates antiapoptotic signals to activate TNF-induced apoptosis (42, 43). Therefore, in the TNFR-mediated pathway, an available pool of TRAF2 in cells may regulate the balance between cell death and survival signals (3). Considering that the functional role of TRAF2 has emerged as a crucial control point in the TNFR-mediated pathway, investigations of the modulation of TRAF2 function by cellular proteins in response to TNF-α stimulation are of interest.
Because TRIP was initially cloned from a T cell hybridoma after a yeast two-hybrid assay with TRAF2 (12), recent progress in understanding the functional role of TRIP has revealed that TRIP plays multiple roles in cell signaling, proliferation, differentiation, apoptosis, and embryonic development (18). The RING domain of TRIP is highly conserved among its homologs and possesses E3 Ub ligase activity for autoubiquitination in vitro (17). However, in the TNF-induced signaling pathway, validated in vivo substrates of the TRIP E3 Ub ligase have not been well studied (18). Interestingly, recent research demonstrated that TBK1 (TANK-binding kinase 1) is the molecular target for the TRIP E3 Ub ligase (44). TRIP physically interacts with TBK1, and this interaction promotes Lys48-linked TBK1 ubiquitination (44). Thus, the E3 Ub ligase activity of TRIP is negatively involved in antiviral responses because it down-regulates IFN-β production by promoting Lys48-Ub-linked proteasomal degradation of TBK1 (44). Interestingly, the possible involvement of TRIP E3 Ub ligase activity in Drosophila embryonic development has been suggested (45). A genetic study in Drosophila demonstrated that the E3 ligase function of NOPO, which is the Drosophila ortholog of TRIP, is required for the preservation of genomic integrity during early embryogenesis through the formation of the BEN-UEV1A (i.e. an E2 heterodimer) and NOPO (i.e. an E3 ligase) complex (45). We reported previously that the targeted disruption of the Trip gene caused excessive cell death and proliferation defects in mouse embryos, indicating that TRIP is required for mouse embryonic development (16). Interestingly, according to a recent genetic study in Drosophila, the E3 ligase function of NOPO is required for Egr (i.e. Eiger, which is the Drosophila ortholog of TNF)-induced apoptosis (46). TRIP has also been reported to enhance TNF-induced apoptosis in mammalian cells or tissue (21). However, the precise role of TRIP E3 Ub ligase activity in mammalian cells remains unclear. TNF-induced apoptosis in L929 mouse fibroblast cells is not altered by the C7A mutation in the RING domain of the TRIP E3 Ub ligase (17, 18). Similarly, in our current study, we observed that the inhibitory function of TRIP for TRAF2 ubiquitination and TRAF2-mediated NF-κB activation was not altered by the RING deletion or the C7A mutation (Fig. 2). Thus, whether TRIP E3 Ub ligase activity is directly involved in the modulation of TNF-induced apoptosis in mammalian cells is of interest. Further studies are required to determine the role of the RING domain of the TRIP E3 Ub ligase in mammalian cells.
The role of TRIP as a modulator of the NF-κB pathway has been documented by many groups (18). In particular, TRIP has been reported to block the TNF-induced NF-κB pathway in mouse brain tissue and cancer cell lines, such as MCF7, HeLa, N42, and 293T cells (12, 19, 20, 22). However, the molecular mechanisms and the functional consequences of negative modulation by TRIP for the TNF-induced NF-κB pathway remain unknown. Thus, our current question is how TRIP is involved in modulation of the TNF-induced NF-κB pathway. Interestingly, the tumor suppressor CYLD or Syk has been reported to be a novel cellular binding partner of TRIP in cancer cells, although the precise roles of these interactions remain unclear (19, 20). In particular, CYLD possessing a DUB domain directly interacts with TRAF2/6, thereby negatively regulating NF-κB activation of the TNFR superfamily via deubiquitinating activity toward the Lys63-Ub linkages of TRAF2/6 or NEMO (9, 11, 15). Thus, we postulated that the negative regulation of TRAF2 ubiquitination by TRIP is mediated by the recruitment of CYLD to the TRAF2-TRIP complex to form a ternary complex that leads to TRAF2 deubiquitination. However, in our current study, we demonstrated that CYLD knockdown has a negligible effect on the inhibitory function of TRIP for TRAF2-mediated NF-κB activation, indicating that the negative regulation of TRAF2-mediated NF-κB activation by TRIP is not mediated by the recruitment of CYLD to the TRAF2-TRIP complex. S1P was recently demonstrated to be a missing cofactor for TRAF2 E3 Ub ligase activity (8). S1P generated intracellularly by SphK1 is required for optimal TRAF2 autoubiquitination and TRAF2-mediated RIP1 ubiquitination through the Lys63-Ub linkage after TNF-α stimulation (7, 8). The target site of S1P binding to TRAF2 is the RING domain (8). In our current study, we demonstrated that the access of S1P to the RING domain of TRAF2 is blocked by TRAF2-TRIP physical interaction. Based on these findings, we postulate that TRIP is a cellular regulator that controls the binding balance between TRAF2 and its cofactor S1P. However, the manner in which the TRAF2-TRIP physical interaction is involved in the inhibition of TRAF2-S1P binding remains unknown. The direct interaction domain of TRAF2 for binding to TRIP is the TRAF domain rather than the RING domain of TRAF2. Thus, studies exploring whether the TRAF2-TRIP physical interaction can structurally modify the S1P-binding motif in the RING domain of TRAF2 to block S1P access are of interest.
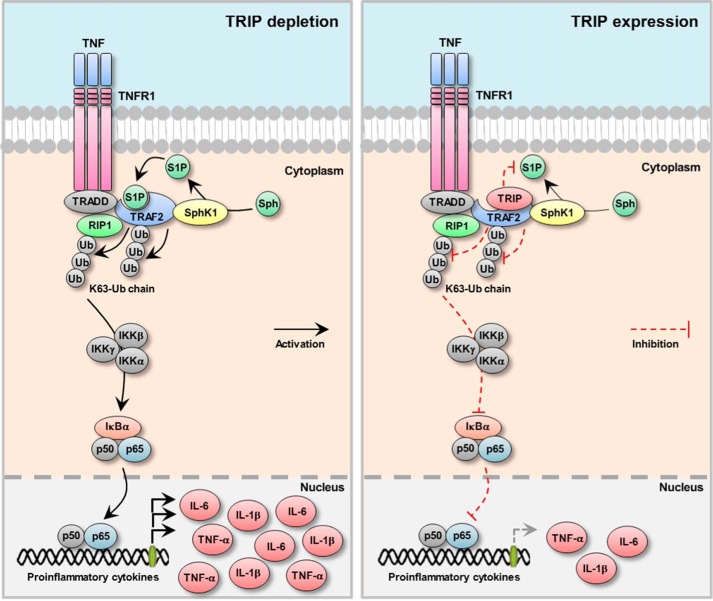
In summary, in our current study, we elucidated the precise inhibitory role of TRIP for TRAF2-mediated NF-κB activation using the TRAF2-TRIP interaction. Our current data suggest that TRIP is negatively involved in Lys63-linked TRAF2 ubiquitination by blocking the binding of the cofactor S1P to the TRAF2 RING domain via the TRAF2-TRIP physical interaction (illustrated in Fig. 8). Finally, we demonstrated that TRIP functions as a negative regulator of the production of proinflammatory cytokines by inhibiting TNF-induced NF-κB activation. Therefore, we conclude that TRIP is an important cellular regulator of the TNFR-mediated inflammatory response.
FIGURE 8.
Proposed model for the involvement of TRIP in the TNF-induced inflammatory response. Black arrows, activation signals; red dashed bars, inhibition signals.
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) and was funded by Ministry of Education Grant NRF-2011-0021771, Korea Basic Science Institute Grant D33400 (to J. S. C.), the research fund of Chungnam National University, and Ministry of National Defense Foundation Grant ADD: 14-01-06-06.
- TNFR
- tumor necrosis factor receptor
- TRAF
- TNFR-associated factor
- TRIP
- TRAF-interacting protein
- NF-κB
- nuclear factor-κB
- RIP1
- receptor-interacting protein 1
- SphK1
- sphingosine kinase 1
- S1P
- sphingosine-1-phosphate
- Ub
- ubiquitin
- CYLD
- cylindromatosis
- DUB
- deubiquitination enzyme
- cIAP
- cellular inhibitor of apoptosis protein
- KD
- knockdown
- DD
- death domain
- TRADD
- TNFR1-associated DD protein
- TAB
- TAK1-binding protein
- NEMO
- NF-κB essential modulator
- IκB
- inhibitor of IκB
- IKK
- IκB kinase
- TANK
- TRAF family member-associated NF-κB activator
- hTNF-α
- human TNF-α
- mTNF-α
- mouse TNF-α
- IB
- immunoblot
- PD
- pull-down
- BM
- bone marrow
- BMM
- BM-derived macrophage.
REFERENCES
- 1. Karin M., Gallagher E. (2009) TNFR signaling: ubiquitin-conjugated TRAFfic signals control stop-and-go for MAPK signaling complexes. Immunol. Rev. 228, 225–240 [DOI] [PubMed] [Google Scholar]
- 2. Chen Z. J. (2005) Ubiquitin signalling in the NF-κB pathway. Nat. Cell Biol. 7, 758–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ha H., Han D., Choi Y. (2009) TRAF-mediated TNFR-family signaling. Curr. Protoc. Immunol. 10.1002/0471142735.im1109ds87 [DOI] [PubMed] [Google Scholar]
- 4. Wajant H., Scheurich P. (2011) TNFR1-induced activation of the classical NF-κB pathway. FEBS J. 278, 862–876 [DOI] [PubMed] [Google Scholar]
- 5. Skaug B., Jiang X., Chen Z. J. (2009) The role of ubiquitin in NF-κB regulatory pathways. Annu. Rev. Biochem. 78, 769–796 [DOI] [PubMed] [Google Scholar]
- 6. Chen Z. J. (2012) Ubiquitination in signaling to and activation of IKK. Immunol. Rev. 246, 95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xia P., Wang L., Moretti P. A., Albanese N., Chai F., Pitson S. M., D'Andrea R. J., Gamble J. R., Vadas M. A. (2002) Sphingosine kinase interacts with TRAF2 and dissects tumor necrosis factor-α signaling. J. Biol. Chem. 277, 7996–8003 [DOI] [PubMed] [Google Scholar]
- 8. Alvarez S. E., Harikumar K. B., Hait N. C., Allegood J., Strub G. M., Kim E. Y., Maceyka M., Jiang H., Luo C., Kordula T., Milstien S., Spiegel S. (2010) Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature 465, 1084–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brummelkamp T. R., Nijman S. M., Dirac A. M., Bernards R. (2003) Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-κB. Nature 424, 797–801 [DOI] [PubMed] [Google Scholar]
- 10. Cheng G., Baltimore D. (1996) TANK, a co-inducer with TRAF2 of TNF- and CD 40L-mediated NF-κB activation. Genes Dev. 10, 963–973 [DOI] [PubMed] [Google Scholar]
- 11. Kovalenko A., Chable-Bessia C., Cantarella G., Israël A., Wallach D., Courtois G. (2003) The tumour suppressor CYLD negatively regulates NF-κB signalling by deubiquitination. Nature 424, 801–805 [DOI] [PubMed] [Google Scholar]
- 12. Lee S. Y., Lee S. Y., Choi Y. (1997) TRAF-interacting protein (TRIP): a novel component of the tumor necrosis factor receptor (TNFR)- and CD30-TRAF signaling complexes that inhibits TRAF2-mediated NF-κB activation. J. Exp. Med. 185, 1275–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rothe M., Xiong J., Shu H. B., Williamson K., Goddard A., Goeddel D. V. (1996) I-TRAF is a novel TRAF-interacting protein that regulates TRAF-mediated signal transduction. Proc. Natl. Acad. Sci. U.S.A. 93, 8241–8246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Song H. Y., Rothe M., Goeddel D. V. (1996) The tumor necrosis factor-inducible zinc finger protein A20 interacts with TRAF1/TRAF2 and inhibits NF-κB activation. Proc. Natl. Acad. Sci. U.S.A. 93, 6721–6725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Trompouki E., Hatzivassiliou E., Tsichritzis T., Farmer H., Ashworth A., Mosialos G. (2003) CYLD is a deubiquitinating enzyme that negatively regulates NF-κB activation by TNFR family members. Nature 424, 793–796 [DOI] [PubMed] [Google Scholar]
- 16. Park E. S., Choi S., Kim J. M., Jeong Y., Choe J., Park C. S., Choi Y., Rho J. (2007) Early embryonic lethality caused by targeted disruption of the TRAF-interacting protein (TRIP) gene. Biochem. Biophys. Res. Commun. 363, 971–977 [DOI] [PubMed] [Google Scholar]
- 17. Besse A., Campos A. D., Webster W. K., Darnay B. G. (2007) TRAF-interacting protein (TRIP) is a RING-dependent ubiquitin ligase. Biochem. Biophys. Res. Commun. 359, 660–664 [DOI] [PubMed] [Google Scholar]
- 18. Chapard C., Hohl D., Huber M. (2012) The role of the TRAF-interacting protein in proliferation and differentiation. Exp. Dermatol. 21, 321–326 [DOI] [PubMed] [Google Scholar]
- 19. Regamey A., Hohl D., Liu J. W., Roger T., Kogerman P., Toftgard R., Huber M. (2003) The tumor suppressor CYLD interacts with TRIP and regulates negatively nuclear factor κB activation by tumor necrosis factor. J. Exp. Med. 198, 1959–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou Q., Geahlen R. L. (2009) The protein-tyrosine kinase Syk interacts with TRAF-interacting protein TRIP in breast epithelial cells. Oncogene 28, 1348–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Almeida S., Ryser S., Obarzanek-Fojt M., Hohl D., Huber M. (2011) The TRAF-interacting protein (TRIP) is a regulator of keratinocyte proliferation. J. Invest. Dermatol. 131, 349–357 [DOI] [PubMed] [Google Scholar]
- 22. Krishnan S., Intlekofer K. A., Aggison L. K., Petersen S. L. (2009) Central role of TRAF-interacting protein in a new model of brain sexual differentiation. Proc. Natl. Acad. Sci. U.S.A. 106, 16692–16697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee S. H., Kim T., Park E. S., Yang S., Jeong D., Choi Y., Rho J. (2008) NHE10, an osteoclast-specific member of the Na+/H+ exchanger family, regulates osteoclast differentiation and survival [corrected]. Biochem. Biophys. Res. Commun. 369, 320–326 [DOI] [PubMed] [Google Scholar]
- 24. Walsh M. C., Kim G. K., Maurizio P. L., Molnar E. E., Choi Y. (2008) TRAF6 autoubiquitination-independent activation of the NFκB and MAPK pathways in response to IL-1 and RANKL. PLoS One 3, e4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kitamura T., Koshino Y., Shibata F., Oki T., Nakajima H., Nosaka T., Kumagai H. (2003) Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp. Hematol. 31, 1007–1014 [PubMed] [Google Scholar]
- 26. Yu J., Choi S., Park E. S., Shin B., Yu J., Lee S. H., Takami M., Kang J. S., Meong H., Rho J. (2012) d-chiro-Inositol negatively regulates the formation of multinucleated osteoclasts by down-regulating NFATc1. J. Clin. Immunol. 32, 1360–1371 [DOI] [PubMed] [Google Scholar]
- 27. Yu J., Shin B., Park E. S., Yang S., Choi S., Kang M., Rho J. (2010) Protein arginine methyltransferase 1 regulates herpes simplex virus replication through ICP27 RGG-box methylation. Biochem. Biophys. Res. Commun. 391, 322–328 [DOI] [PubMed] [Google Scholar]
- 28. Shembade N., Harhaj N. S., Yamamoto M., Akira S., Harhaj E. W. (2007) The human T-cell leukemia virus type 1 Tax oncoprotein requires the ubiquitin-conjugating enzyme Ubc13 for NF-κB activation. J. Virol. 81, 13735–13742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wertz I. E., O'Rourke K. M., Zhou H., Eby M., Aravind L., Seshagiri S., Wu P., Wiesmann C., Baker R., Boone D. L., Ma A., Koonin E. V., Dixit V. M. (2004) De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature 430, 694–699 [DOI] [PubMed] [Google Scholar]
- 30. Lee S. H., Rho J., Jeong D., Sul J. Y., Kim T., Kim N., Kang J. S., Miyamoto T., Suda T., Lee S. K., Pignolo R. J., Koczon-Jaremko B., Lorenzo J., Choi Y. (2006) v-ATPase V0 subunit d2-deficient mice exhibit impaired osteoclast fusion and increased bone formation. Nat. Med. 12, 1403–1409 [DOI] [PubMed] [Google Scholar]
- 31. Tse A. K., Wan C. K., Shen X. L., Yang M., Fong W. F. (2005) Honokiol inhibits TNF-α-stimulated NF-κB activation and NF-κB-regulated gene expression through suppression of IKK activation. Biochem. Pharmacol. 70, 1443–1457 [DOI] [PubMed] [Google Scholar]
- 32. Murano K., Okuwaki M., Hisaoka M., Nagata K. (2008) Transcription regulation of the rRNA gene by a multifunctional nucleolar protein, B23/nucleophosmin, through its histone chaperone activity. Mol. Cell. Biol. 28, 3114–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shi C. S., Kehrl J. H. (2003) Tumor necrosis factor (TNF)-induced germinal center kinase-related (GCKR) and stress-activated protein kinase (SAPK) activation depends upon the E2/E3 complex Ubc13-Uev1A/TNF receptor-associated factor 2 (TRAF2). J. Biol. Chem. 278, 15429–15434 [DOI] [PubMed] [Google Scholar]
- 34. Takeuchi M., Rothe M., Goeddel D. V. (1996) Anatomy of TRAF2: distinct domains for nuclear factor-κB activation and association with tumor necrosis factor signaling proteins. J. Biol. Chem. 271, 19935–19942 [DOI] [PubMed] [Google Scholar]
- 35. Varfolomeev E., Goncharov T., Fedorova A. V., Dynek J. N., Zobel K., Deshayes K., Fairbrother W. J., Vucic D. (2008) c-IAP1 and c-IAP2 are critical mediators of tumor necrosis factor alpha (TNFα)-induced NF-κB activation. J. Biol. Chem. 283, 24295–24299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wertz I. E., Dixit V. M. (2008) Ubiquitin-mediated regulation of TNFR1 signaling. Cytokine Growth Factor Rev. 19, 313–324 [DOI] [PubMed] [Google Scholar]
- 37. Xia Z. P., Chen Z. J. (2005) TRAF2: a double-edged sword? Sci. STKE 2005, pe7. [DOI] [PubMed] [Google Scholar]
- 38. Zhang L., Blackwell K., Shi Z., Habelhah H. (2010) The RING domain of TRAF2 plays an essential role in the inhibition of TNFα-induced cell death but not in the activation of NF-κB. J. Mol. Biol. 396, 528–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bertrand M. J., Milutinovic S., Dickson K. M., Ho W. C., Boudreault A., Durkin J., Gillard J. W., Jaquith J. B., Morris S. J., Barker P. A. (2008) cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol. Cell 30, 689–700 [DOI] [PubMed] [Google Scholar]
- 40. Bertrand M. J., Lippens S., Staes A., Gilbert B., Roelandt R., De Medts J., Gevaert K., Declercq W., Vandenabeele P. (2011) cIAP1/2 are direct E3 ligases conjugating diverse types of ubiquitin chains to receptor interacting proteins kinases 1 to 4 (RIP1–4). PLoS One 6, e22356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xu M., Skaug B., Zeng W., Chen Z. J. (2009) A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFα and IL-1β. Mol. Cell 36, 302–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li X., Yang Y., Ashwell J. D. (2002) TNF-RII and c-IAP1 mediate ubiquitination and degradation of TRAF2. Nature 416, 345–347 [DOI] [PubMed] [Google Scholar]
- 43. Yang Y. L., Li X. M. (2000) The IAP family: endogenous caspase inhibitors with multiple biological activities. Cell Res. 10, 169–177 [DOI] [PubMed] [Google Scholar]
- 44. Zhang M., Wang L., Zhao X., Zhao K., Meng H., Zhao W., Gao C. (2012) TRAF-interacting protein (TRIP) negatively regulates IFN-β production and antiviral response by promoting proteasomal degradation of TANK-binding kinase 1. J. Exp. Med. 209, 1703–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Merkle J. A., Rickmyre J. L., Garg A., Loggins E. B., Jodoin J. N., Lee E., Wu L. P., Lee L. A. (2009) no poles encodes a predicted E3 ubiquitin ligase required for early embryonic development of Drosophila. Development 136, 449–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ma X., Huang J., Yang L., Yang Y., Li W., Xue L. (2012) NOPO modulates Egr-induced JNK-independent cell death in Drosophila. Cell Res. 22, 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]