Background: Pdx1 interacts with the methyltransferase Set7/9 to transactivate β cell genes.
Results: Methylation of Pdx1 residue Lys-131 by Set7/9 augments Pdx1 activity.
Conclusion: The ability of Pdx1 to regulate genes in β cells is partially dependent upon its methylation by Set7/9.
Significance: This study reveals a previously unappreciated role for Lys methylation in the maintenance of Pdx1 activity and β cell function.
Keywords: Diabetes, Gene Knockout, pancreatic Islet, Protein Methylation, Transcription, Pdx1, Set7/9
Abstract
The transcription factor Pdx1 is crucial to islet β cell function and regulates target genes in part through interaction with coregulatory factors. Set7/9 is a Lys methyltransferase that interacts with Pdx1. Here we tested the hypothesis that Lys methylation of Pdx1 by Set7/9 augments Pdx1 transcriptional activity. Using mass spectrometry and mutational analysis of purified proteins, we found that Set7/9 methylates the N-terminal residues Lys-123 and Lys-131 of Pdx1. Methylation of these residues occurred only in the context of intact, full-length Pdx1, suggesting a specific requirement of secondary and/or tertiary structural elements for catalysis by Set7/9. Immunoprecipitation assays and mass spectrometric analysis using β cells verified Lys methylation of endogenous Pdx1. Cell-based luciferase reporter assays using wild-type and mutant transgenes revealed a requirement of Pdx1 residue Lys-131, but not Lys-123, for transcriptional augmentation by Set7/9. Lys-131 was not required for high-affinity interactions with DNA in vitro, suggesting that its methylation likely enhances post-DNA binding events. To define the role of Set7/9 in β cell function, we generated mutant mice in which the gene encoding Set7/9 was conditionally deleted in β cells (SetΔβ). SetΔβ mice exhibited glucose intolerance similar to Pdx1-deficient mice, and their isolated islets showed impaired glucose-stimulated insulin secretion with reductions in expression of Pdx1 target genes. Our results suggest a previously unappreciated role for Set7/9-mediated methylation in the maintenance of Pdx1 activity and β cell function.
Introduction
Deficiency of insulin secretion underlies the transition from normoglycemia to hyperglycemia in both type 1 and type 2 diabetes (1, 2). Circulating insulin arises almost exclusively from islet β cells in the pancreas, but our understanding of the molecular mechanisms governing β cell function in health and disease remains incomplete. A key protein that is essential for β cell function is the homeobox transcription factor Pdx1. During mammalian development, Pdx1 is essential for pancreas organogenesis (3–5). In the adult pancreas, Pdx1 is restricted primarily to β cells and is responsible for the regulation of genes that are essential to β cell function, proliferation, and survival (6, 7). In this context, elucidating the molecular mechanisms of Pdx1 action will be crucial in future attempts to restore β cell function in the setting of diabetes.
The interaction of Pdx1 with other transcription factors and cofactors appears to be important in modulating Pdx1 activity (positively or negatively) at a given target gene. For example, the formation of a transcriptional complex between Pdx1 and the basic helix-loop-helix factor NeuroD1 in β cells results in the formation of a complex with the gene encoding preproinsulin (Ins1/2) that permits synergistic activation of transcription (8, 9). By contrast, interaction of Pdx1 with PCIF1 results in degradation of Pdx1 protein by the proteasome and a consequent reduction in Pdx1 activity (10). Interactions such as these suggest a model in which various aspects of Pdx1 activity, including DNA binding affinity, protein stability, and recruitment of basal transcriptional machinery, are modulated to achieve homeostatic function of the β cell.
Our laboratory has reported previously that the lysine methyltransferase Set7/9 interacts with Pdx1 to promote transactivation of an Ins1/2 minienhancer (11) that contains the classical Pdx1 binding sequence (5′-TAAT-3′) present in elements A3 and A4. Augmentation of transcriptional activity was correlated with an increase in Lys4 methylation of histone 3 (H3) and the conversion of the initiating isoform of RNA polymerase II to its elongating isoform. In recent years, several reports have suggested that the methyltransferase activity of Set7/9 is not only restricted to Lys residues on histones but that it includes Lys residues of other proteins, such as p53, p65, and estrogen receptor α, among others (12–15). These varied methylation events have been shown to alter the activity or half-life of these proteins, emphasizing that Lys methylation (similar to Ser/Thr phosphorylation or Lys acetylation) modulates transcription factor function (16, 17).
In light of these new perspectives on Lys methylation and Set7/9 action, we asked whether the interactions between Set7/9 and Pdx1 might affect Pdx1 activity independently of effects on histones. In this study, our findings reveal a heretofore unappreciated role for Lys-specific methylation of Pdx1 by Set7/9 in the maintenance of normal β cell function.
MATERIALS AND METHODS
Cells, Animals, and Assays
NIH3T3, HEK293, MIN6, and INS-1 cells were cultured as described previously (11, 18–20). All animal studies were reviewed and approved by the Indiana University Institutional Animal Care and Use Committee. Setd7Loxp/+ mice, in which Cre recombinase recognition sequences (Loxp) flank exon 2, were generated by subcontract to Ingenious Targeting Laboratories. The neomycin selection cassette was removed by crossing Setd7Loxp/+ mice to the FLP1 recombinase expressing mouse strain. Mice were backcrossed onto the C57BL/6 background for 10 generations. MIP1-CreERT mice on the C57BL/6 background were provided by Dr. L. Philipson (21). For induction of Cre-mediated recombination, mice were gavaged orally with tamoxifen dissolved in peanut oil (mixed for 1 h at 55 °C) at a dose of 5 mg/day/mouse. Intraperitoneal glucose tolerance tests using 2 g/kg body weight glucose proceeded as described previously (22). Islets were isolated from collagenase-perfused pancreata as described previously (23). Static glucose-stimulated insulin secretion assays using islets were performed as described previously (22), and insulin released into the medium was normalized to total islet insulin content. Insulin was measured using a mouse insulin ELISA kit (Alpco Diagnostics).
Recombinant Plasmids, Mutagenesis, and PCR
Plasmids were generated using standard recombinant techniques. PCR-generated constructs were verified by automated sequencing. Recombinant protein was expressed using the Escherichia coli expression vectors pET21d or pET15b and purified as described previously (24). Recombinant Set7/9 protein was purchased from Prospec. Point mutations were generated using the QuikChange site directed mutagenesis kit (Agilent). The following primers were used to make the respective point mutants: K123R, 5′-CAGCTGCCTTTCCCATGGATGAGGTCGACCAAAGCTCAC-3′ and 5′-GTGAGCTTTGGTCGACCTCATCCATGGGAAAGGCAGCTG-3′; K126R, 5′-TCCCATGGATGAAGTCTACCAGAGCTCACGCGT-3′ and 5′-ACGCGTGAGCTCTGGTAGACTTCATCCATGGGA-3′; and K131R, 5′-CCAAAGCTCACGCGTGGAGAGGCCAGTGG-3′ and 5′-CCACTGGCCTCTCCACGCGTGAGCTTTGG-3′. CMV promoter-driven vectors (pBAT12) were used to express wild-type and mutant Pdx1 in HEK293 and NIH3T3 cells as described previously (9). The CMV promoter-driven vector used to drive Setd7 has been described previously (25). Methods and primers for SYBR Green-based real-time RT-PCR have been described previously (26).
Antibodies and Demethylase Inhibitor
Pdx1 antibody was obtained from Millipore (catalog no. 07-696). Set7/9 antibody was obtained from Cell Signaling Technology (catalog no. 2813). Monoclonal FLAG M2 antibody was obtained from Sigma-Aldrich (catalog no. F1804). Anti-Pdx1(Lys-131-methyl) antibody was generated in rabbits by contract to 21st Century Biochemicals using the following synthetic peptides: methylated peptide 1, C-Ahx-TKAHAW[K-Me1]GQWAG-amide; methylated peptide 2, Ac-AHAW[K-Me1]GQWAGGA-Ahx-KKC-amide. Gapdh antibody was obtained from Ambion (catalog no. AM4300). The fluorophore-labeled secondary antibodies IRDye 700 and IRDye 800 were obtained from Licor Biosciences. The demethylase inhibitor BP-107-7 was synthesized using a synthetic route described previously (27, 28).
Methylation Assays in Vitro
Purified Pdx1 protein (150 nm-1.2 μm) and Set7/9 protein (200 nm) were incubated at 30 °C for 3 h in a reaction buffer containing 50 mm Tris (pH 8.5), 4 mm DTT, 5 mm MgCl2, 0.05 mg/ml BSA, 1 μm [3H]AdoMet,3 and 50 μm AdoMet in 20 μl of volume. The reaction was stopped by the addition of 6× SDS gel loading buffer. Analysis by polyacrylamide gel electrophoresis proceeded as described previously (11).
Coimmunoprecipitation Assays
Immunoprecipitations from whole cell lysates using protein A or protein G Dynabeads (Life Technologies) proceeded as described previously (11). Immunoprecipitations involving anti-HA antibody, which was performed using the HA tag immunoprecipitation kit (Pierce). ChIP assays were performed using the Active Motif ChIP-IT® Express enzymatic kit (catalog no. 53009) according to the protocol of the manufacturer.
Immunoblot Analysis
Samples were resolved on 10% polyacrylamide gels and transferred onto a PVDF membrane (Millipore). Membranes were exposed to primary antibody overnight at 4 °C and then processed as described previously (29).
Transient Transfections and Luciferase Reporter Assays
Cells were transfected using Metafectene Pro (Biontex) according to the instructions of the manufacturer in 6-well tissue culture plates. After 48 h, whole cell extracts were used to assess luciferase activity using a commercially available luciferase assay kit (Promega).
Generation and Purification of the Tandem Affinity Purification (TAP) Tag-Pdx1 from MIN6 Cells
The full-length mouse Pdx1 cDNA sequence was inserted in-frame into the pNTAP-C vector (Stratagene), generating TAP-Pdx1 with the TAP tag fused to the N terminus of Pdx1. The TAP-Pdx1 sequence was then subcloned into the pAdTrack-CMV shuttle vector, which was used to generate recombinant adenoviruses expressing N-terminal TAP-tagged, full-length Pdx1 as described previously (30). MIN6 cells were infected with the purified TAP-Pdx1 or control GFP adenovirus as described previously (30, 31) and lysed 48 h later. TAP Pdx1 was purified using the InterPlay TAP purification system (Stratagene) and subjected to mass spectrometry.
EMSAs
The Ins E-A probe 5′-GATCCTTCATCAGGCCATCTGGCCCCTTGTTAATAATCTAATTACCCTAGGTCTAA-3′ was labeled with [γ32P]ATP using T4 polynucleotide kinase and duplexed with an unlabeled complementary strand (5′-GATCTTAGACCTAGGGTAATTAGATTATTAACAAGGGGCCAGATGGCCTGATGAAG-3′). Nuclear extracts from transfected NIH3T3 cells were isolated as described previously (19), and around 2 μg of total nuclear protein was used in each EMSA reaction with the labeled Ins E-A probe. Competition EMSAs were performed using an unlabeled Ins E-A probe. EMSA reactions proceeded at room temperature for 15 min. The EMSA buffer and electrophoresis protocol have been described previously (32). The gel was visualized, and competition EMSAs were quantitated using a PhosphorImager (Molecular Dynamics). Competition EMSAs were modeled on the basis of single-phase exponential dissociation using the non-linear least squares fit algorithm in Prism 5.0 software (GraphPad), and apparent dissociation constants were determined as described previously (24).
Mass Spectrometry Analysis
Samples (proteins from in vitro experiments and from MIN6 cells) were first denatured with 8 m urea, reduced with 10 mm DTT in 10 mm ammonium bicarbonate, alkylated with 55 mm iodoacetamide (prepared in 10 mm ammonium bicarbonate), digested with trypsin, and incubated overnight at 37 °C. The samples in vitro were analyzed using Thermo Fisher Scientific LTQ Orbitrap Velos Pro and Surveyor HPLC. Tryptic peptides were injected onto the C18 column. Peptides were eluted with a linear gradient from 3%-40% acetonitrile (in water with 0.1% trifluoracetic acid) developed over 90 min at room temperature, at a flow rate of 60 μl/min, and the effluent was electrosprayed into the LTQ mass spectrometer. Blanks were run prior to the sample run to make sure that there was no significant signal from solvents or the column. The TSKgel columns (ODS-100 V, 3 μm, 1.0 × 50 mm) were used for the Surveyor HPLC system. The trypsin-digested Pdx1 immunoprecipitants from MIN6 cells were pressure-loaded onto a 12-cm multidimensional protein identification technology column as described previously (33). Peptides were analyzed by MS/MS on an LTQ XL mass spectrometer following elution during a 10-step multidimensional protein identification technology run. Protein database searches were performed using SEQUEST® algorithms within Proteome Discoverer (Thermo) against appropriate FASTA sequence databases obtained from UniProt. Database searches included dynamic modification analysis for mono-, di-, and trimethylated Lys residues and Met residue oxidation.
Statistical Analysis
All data are presented as mean ± S.E. One-way analysis of variance (followed by a Dunnett's post test) was used for comparisons in which two or more conditions were compared, and two-tailed Student's t test was performed when two conditions were compared. Prism 5.0 software (GraphPad) was used for all statistical analyses. Statistical significance was defined as p < 0.05.
RESULTS
Pdx1 Is Methylated by Set7/9 in Vitro at Positions Lys-123 and Lys-131
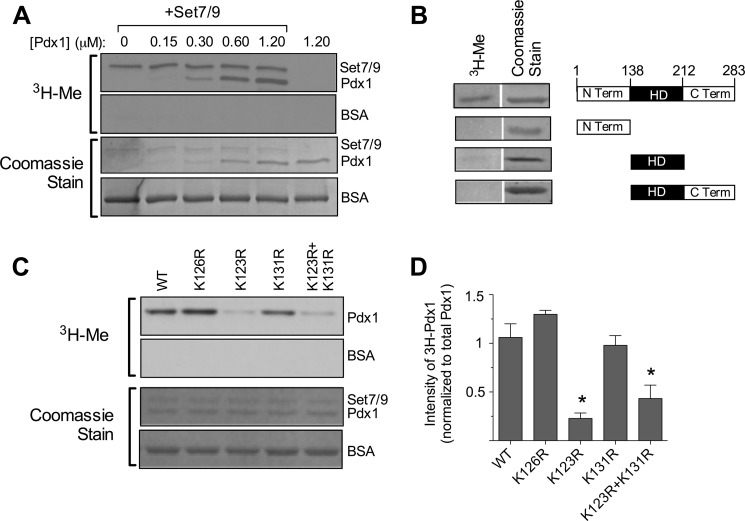
Recent evidence suggesting that Set7/9 methylates non-histone proteins led us to investigate first whether the interaction between Set7/9 and Pdx1 leads to Lys methylation of Pdx1. We performed methylation reactions in vitro using purified Pdx1 and Set7/9 proteins and [3H]AdoMet. Incorporation of the [3H]methyl group into Pdx1 in this reaction is evidence of protein methylation (11). As shown in Fig. 1A, [3H]methyl was incorporated into Pdx1 in a concentration-dependent manner and only in the presence of Set7/9. By contrast, no methylation of bovine serum albumin was observed in the same reaction, suggesting that the methylation is specific for Pdx1 (Fig. 1A).
FIGURE 1.
Pdx1 methylation by Set7/9 in vitro. A, a methylation assay in vitro using recombinant Set7/9, [3H]AdoMet and increasing concentrations of Pdx1 protein was performed, and then reactions were subjected to polyacrylamide gel electrophoresis. The first and second panel show fluorography for 3H, and the third and fourth panels show corresponding Coomassie staining of the same gel. B, methylation assay in vitro using recombinant Set7/9, [3H]AdoMet, and full-length or truncated Pdx1 proteins was performed, followed by polyacrylamide gel electrophoresis. A schematic of truncated mutants of Pdx1 is shown in the right panel, and corresponding 3H fluorography and Coomassie stains are shown in the left panel. Term, terminus; HD, homeodomain. C, methylation assays in vitro using WT and mutated Pdx1 proteins were performed with corresponding quantitation of methylated Pdx1 protein intensities (normalized to total Pdx1 protein by Coomassie staining). All images shown are representative of at least three experiments. *, p < 0.05 compared with wild-type Pdx1.
Prior studies have suggested that Lys residues in a broad range of sequence contexts can serve as substrates for Set7/9 (34). Within Pdx1, Lys residues occur in both the N-terminal transactivation domain (residues Lys-15, Lys-123, Lys-126, and Lys-131) and the DNA binding homeodomain (residues Lys-147, Lys-163, Lys-191, Lys-200, Lys-202, Lys-203, Lys-207, and Lys-208). To narrow the possible Lys residues that may be methylated by Set7/9, we next performed methylation assays using different truncated mutants of Pdx1, as shown in Fig. 1B. Surprisingly, none of the truncated mutants were methylated by Set7/9 in vitro, suggesting that methylation of Pdx1 requires the structural context of the full-length protein.
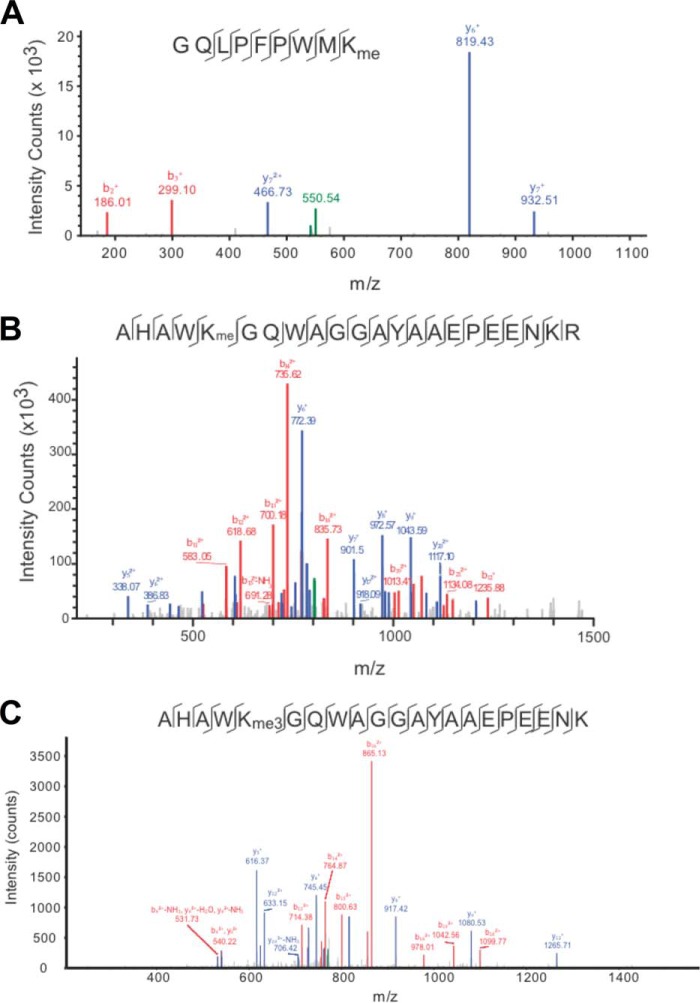
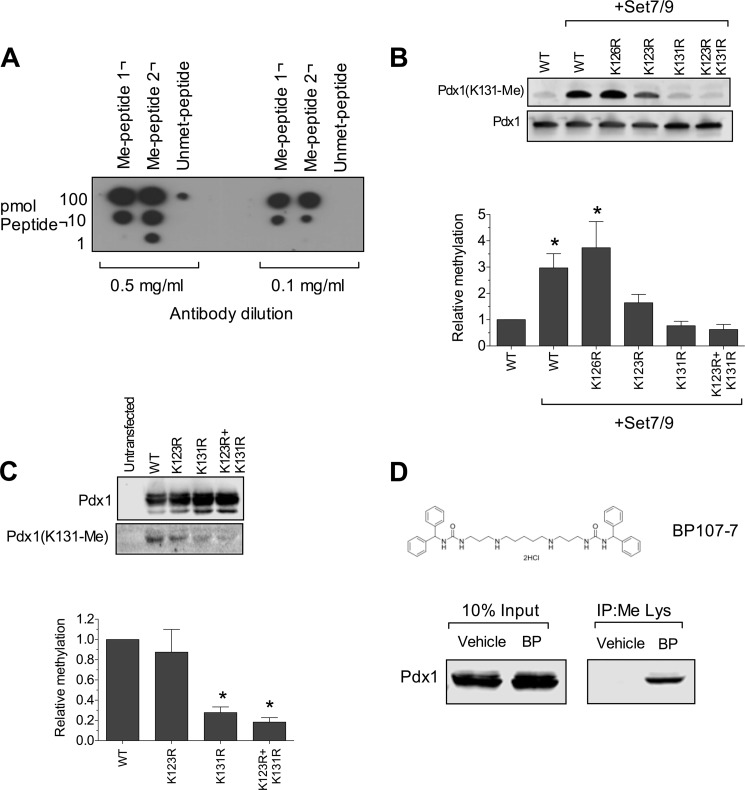
To identify the methylated Lys residue(s) in the full-length protein, we next performed mass spectrometry of methylated full-length Pdx1. Broad coverage of the peptides confirmed the monomethylation of Pdx1 at two Lys residues in the N-terminal region of the protein (residues Lys-123 and Lys-131) (Fig. 2, A and B). Data from the mass spectrometric analysis was verified using mutated Pdx1 proteins in which residues Lys-123, Lys-126, and Lys-131 in the N-terminal domain were mutated to Arg (K123R, K126R, and K131R). As shown in Fig. 1C, reductions in methylation of Pdx1 clearly occurred in the K123R mutant, but no statistically significant reduction was seen in the K131R mutant. Little residual methylation was observed in the K123R/K131R double mutant. Together, these data suggest that residue Lys-123 appears to be the kinetically preferred site for methylation by Set7/9 in vitro, with evidence of lesser methylation occurring at Lys-131.
FIGURE 2.
MS/MS spectra of Pdx1 in vitro and in MIN6 cells. A, tandem mass spectrum of the peptide GQLPFPWMK showing modification of Lys-123 with monomethylation (+14 Da) following treatment with recombinant Set7/9 in vitro. The SEQUEST XCorr for this +2 peptide is 1.83, with a ppm (parts per million) of −1.65. (B) Tandem mass spectrum of the +3 peptide AHAWKGQWAGGAYAAEPEENKR showing monomethylation (+14 Da) of Lys-131 in Pdx1 following treatment with recombinant Set7/9 in vitro. The SEQUEST XCorr for this peptide is 6.19, with a ppm of 0.34. C, representative MS/MS spectrum from an LTQ ion trap for the +2 peptide AHAWKGQWAGGAYAAEPEENKR showing trimethylation at Lys-131 in Pdx1 in transduced MIN6 β cells. The SEQUEST XCorr for this peptide is 2.24, with a ppm of 19.25.
Lys-131 of Pdx1 Is Required for Transcriptional Augmentation by Set7/9
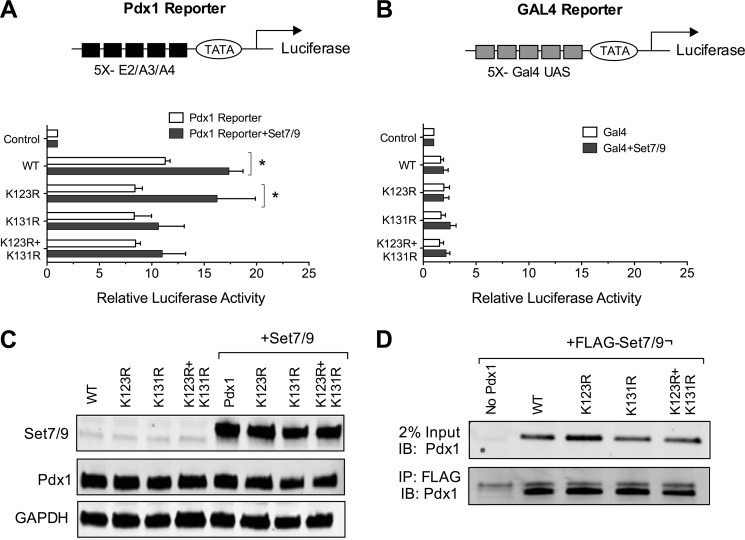
In prior studies, we showed that Set7/9 augments the transcriptional activity of Pdx1 in the context of a Pdx1 reporter (containing five tandem Pdx1 binding sites). This result has been ascribed previously to Set7/9-mediated methylation of H3 (residue Lys-4) at this reporter (11). On the basis of our results in Fig. 1, we considered the possibility that this transcriptional augmentation by Set7/9 might, instead, be a result of Pdx1 Lys methylation. To test this possibility, we performed reporter gene activity assays using wild-type and mutated Pdx1 in the mouse-derived cell line NIH3T3, which is devoid of endogenous Pdx1. As shown in Fig. 3A, when a cDNA encoding wild-type Pdx1 is cotransfected into NIH3T3 cells with a Pdx1 reporter plasmid containing tandem copies of a Pdx1 binding sequence (derived from the rat Ins1 E2/A4/A3 promoter element) driving luciferase (9), ∼12-fold activation of luciferase activity is observed. No activation is observed, however, upon cotransfection of the reporter with a cDNA encoding Set7/9 alone (Fig. 3A). When cDNAs encoding both wild-type Pdx1 and Set7/9 are cotransfected, a more than additive 18-fold activation of the reporter is observed. Although mutation of residue Lys-123 to Arg (K123R) had no effect on reporter gene transactivation, mutation of Lys-131 to Arg (K131R) or the double mutation K123R/K131R resulted in a loss of transcriptional augmentation by Set7/9 (Fig. 3A). These effects were specific for the Pdx1 reporter because no significant effects were observed on the control Gal4 reporter (which contains five tandem copies of the yeast Gal4 upstream activating sequence) (Fig. 3B). Additionally, the effects of the K131R mutation was not caused by changes in Pdx1 levels, because all transfected Pdx1 mutants showed similar intensity on immunoblot analysis (Fig. 3C). These results suggest that, although both Lys-123 and Lys-131 appear to be targets for methylation by Set7/9 in vitro (with the former being preferred), only Lys-131 appears to be required for the transcriptional augmentation of reporter activity in cells.
FIGURE 3.
Transcription augmentation by Set7/9 is dependent upon the Pdx1 residue Lys-131. NIH3T3 cells were transiently cotransfected with Set7/9, WT and mutant Pdx1 proteins, and either the Pdx1 reporter plasmid or the Gal4 reporter plasmid. Cells were harvested 48 h later, and whole cell extracts were used to assay luciferase activity. A, luciferase activities (relative to control transfections without Pdx1) with the Pdx1 reporter plasmid, which contains tandem elements of the Ins E2/A3/A4 element. A schematic of reporter is shown in the top panel. n = 3 transfections in triplicate; *, p < 0.05 for the comparisons shown. B, luciferase activities (relative to control transfections without Pdx1) with the Gal4 reporter plasmid, which contains tandem elements of the yeast Gal4 DNA binding domain. A schematic of the reporter is shown in the top panel). n = 3 transfections in triplicate. UAS, upstream activating sequence. C, immunoblots (for the indicated proteins) from whole cell extract from transfected cells. Results are representative of three transfections. D, NIH3T3 cells were transfected with FLAG-Set7/9 and the indicated Pdx1 proteins, and then nuclear extracts were subjected to immunoprecipitation (IP) using FLAG antibody and immunoblotted (IB) using Pdx1 antibody. Results are representative of two experiments.
Lys methylation has been shown to affect multiple aspects of transcription factor activity, including protein half-life and DNA binding affinity (17, 35). We suspected that the effects of the K131R mutation on Pdx1 half-life were negligible because proteins levels of the transfected proteins were identical for the wild type and mutants (Fig. 3C). To test whether loss of transcriptional augmentation activity between Pdx1 K131R and Set7/9 was due to decreased interaction between the two proteins, a coimmunoprecipitation assay for Set7/9 and Pdx1 using NIH3T3 cells transfected with wild-type or mutant Pdx1 and Set7/9 was performed (Fig. 3D). Neither the K131R mutant nor the K123R/K131R double mutant exhibited any differences in interaction with Set7/9, suggesting that the interaction between Pdx1 and Set7/9 is independent of the target Lys residue being methylated.
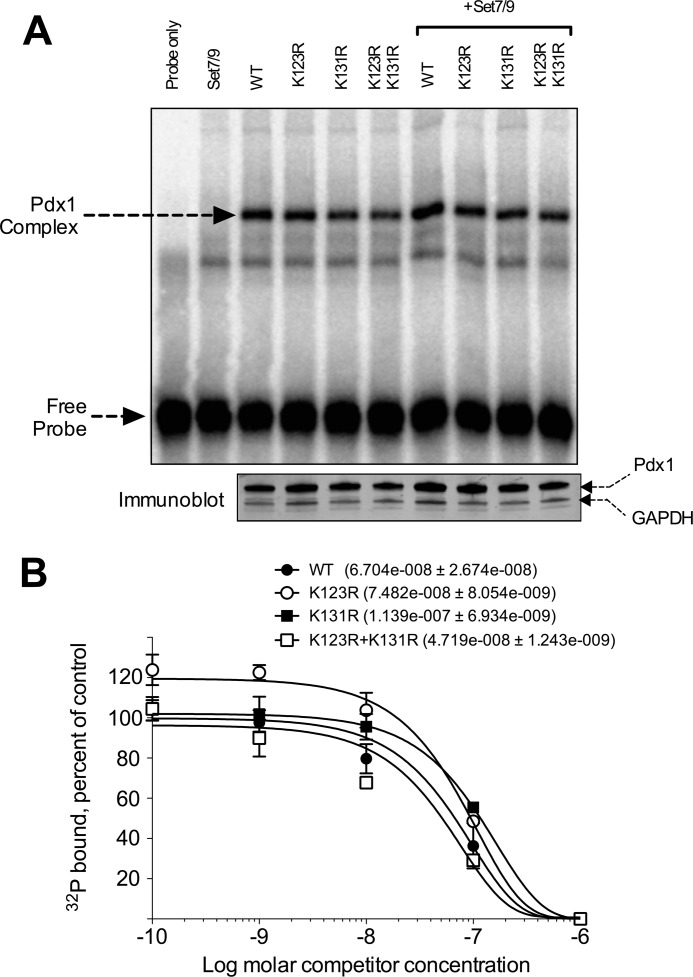
To ascertain the effects of the Pdx1 mutations on DNA binding affinity, we next performed EMSAs. Nuclear extracts from NIH3T3 cells transfected with Pdx1 proteins and Set7/9 were subjected to EMSA with the 32P-labeled rat Ins1 promoter E-A element (9, 36). No apparent differences in the intensity of the Pdx1-specific complex were observed in the EMSAs (Fig. 4A). To quantitate more precisely whether subtle differences in binding affinity exist, we performed competition EMSAs using increasing concentrations of the unlabeled E-A probe. Binding competition curves were subsequently calculated and modeled on the basis of single-phase exponential dissociation (Fig. 4B). As shown in Fig. 4B, none of the mutants exhibited dissociation constants that were different from the wild-type Pdx1 protein. Taken together, the data in Figs. 3 and 4 suggest that the transcriptional augmentation by Set7/9 that is enabled by methylation at Lys-131 likely involves transcriptional events that occur after binding to DNA, such as through the activation of the RNA polymerase II complex at the promoter (11).
FIGURE 4.
Point mutations do not affect DNA binding by Pdx1. A, NIH3T3 cells were cotransfected with the WT or mutant Pdx1 proteins with or without cotransfection of Set7/9. Nuclear extracts from transfected cells were harvested and subjected to EMSA using the 32P-labeled E-A DNA fragment containing a Pdx1 binding site derived from Ins E2/A3/A4. The positions of the Pdx1-containing complex and the unbound, free probe are indicated by arrows. Bottom panel, immunoblots for Pdx1 and GAPDH from the same nuclear extracts as used for the EMSA. B, quantitation and modeling of EMSA reactions (as in A), with increasing concentrations of unlabeled E-A probe. For each Pdx1 mutant, the control condition is the fraction of total probe bound in the absence of competitor. The ordinate represents probe binding (at the given competitor concentration) as a percentage of the control condition. The apparent dissociation constants of the WT and mutant proteins are shown in the inset, with no statistical differences seen. The data shown are from three EMSA experiments.
Residue Lys-131 of Pdx1 Is Methylated in Cells
We next asked whether Pdx1 residue Lys-131 is methylated by Set7/9 in cells. To assess methylation in cells, we generated a peptide-based polyclonal antibody against methylated Lys-131 of Pdx1. Three different peptides (two methylated and one unmethylated at Lys residues) corresponding to the Pdx1 sequence containing Lys-131 were subjected to dot blots using varying dilutions of the antibody. As shown in Fig. 5A, the antibody exhibited specificity for the Lys-methylated peptides (Fig. 5A). A corresponding specificity of the antibody for methylated full-length Pdx1 was observed using recombinant Pdx1 proteins methylated by Set7/9 in vitro (Fig. 5B). As expected, an almost complete loss of immunoreactivity was observed with the K131R mutation. However, some cross-reactivity of the antibody was observed with methylated Lys-123 because the K123R mutation showed a slight reduction in immunoreactivity (Fig. 5B). These data confirm that Lys-131 is a site for methylation by Set7/9 in vitro. This antibody was then used to assess the methylation of Pdx1 proteins immunoprecipitated from cells. Wild-type and mutant Pdx1 constructs containing an N-terminal HA) tag were transfected along with Set7/9 into HEK293 cells, which contain no endogenous Pdx1. Pdx1 proteins were immunoprecipitated using an anti-HA antibody and then subjected to immunoblotting using the anti-Pdx1(Lys-131-Me) antibody. As shown in Fig. 5C, second lane, wild-type Pdx1 was methylated in cells, and the K131R and K123R/K131R double mutants showed a clear reduction in methylation. A slight reduction in the K123R mutant is consistent with cross-reactivity of the antibody for methylated Lys-123. Taken together, these data suggest that Lys-131 (and possibly Lys-123) are methylated in cell lines.
FIGURE 5.
Residue Lys-131 of Pdx1 is methylated in cells. A, dot blot analysis using two dilutions of anti-Pdx1(Lys-131-Me) antibodies and methylated and unmethylated peptides corresponding to the N terminus of Pdx1 containing residue Lys-131. B, representative immunoblots (top panel) using anti-Pdx1(Lys-131-Me) antibody or anti-Pdx1 antibody on recombinant Pdx1 proteins methylated by Set7/9 in vitro. The quantitation of immunoblots (n = 3) is shown in the bottom panel. *, p < 0.05 compared with Pdx1 alone. C, HEK293 cells were transfected with Set7/9 and WT or mutant HA-tagged Pdx1 proteins, and then whole cell extracts were immunoprecipitated using anti-HA antibody, followed by immunoblot analysis using anti-Pdx1 and anti-Pdx1(Lys-131-Me) antibodies (top panel). The quantitation of methylated Pdx1 (relative to total immunoprecipitated Pdx1, n = 3) is shown in the bottom panel. *, p < 0.05 compared with WT Pdx1. D, top panel, chemical structure of the Lys demethylase inhibitor BP-107-7 (BP). Bottom panel, INS-1 832/13 β cells were treated with vehicle or BP-107-7 (10 μm) overnight, and then nuclear extracts were immunoprecipitated (IP) using anti-methyl Lys antibody followed by immunoblot using anti-Pdx1 antibody.
To determine whether Lys-131 is endogenously methylated in β cells, we immunoprecipitated methylated Lys proteins from INS-1(832/13) insulinoma cells and then subjected the immunoprecipitate to immunoblotting using an anti-Pdx1 antibody. Initial experiments from INS-1 cells revealed no recovery of methylated Pdx1, raising the concern that β cells may contain demethylases that diminish the ability to detect Pdx1 methylation. To circumvent this possibility, we pretreated INS-1 cells with the Lys demethylase inhibitor BP-107-7 (Fig. 5D, top panel). INS-1 cells pretreated with 10 μm BP-107-7 overnight were subjected to immunoprecipitation using anti-Pdx1 antibody, and the resulting immunoprecipitate was immunoblotted with anti-methylated Lys antibody. As shown in Fig. 5D (bottom panel), although INS-1 cells treated with vehicle showed no recovery of methylated Pdx1, cells treated with BP-107-7 showed a clear signal for Pdx1. These data suggest that Pdx1 is endogenously methylated and demethylated in β cells. To verify methylation at residue Lys-131 in β cell lines, TAP-tagged Pdx1 was overexpressed in MIN6 β cells using adenoviral gene transfer. The tagged Pdx1 protein was then purified from MIN6 β cells and analyzed by LC/MS/MS. Pdx1 was found to be both di- or trimethylated at residue Lys-131 (Fig. 2C shows data for the trimethylated protein). Collectively, these data suggest that Lys-131 is a target of methylation in cells.
Set7/9 Is Required for the Maintenance of Normal β Cell Function and Glucose Homeostasis in Vivo
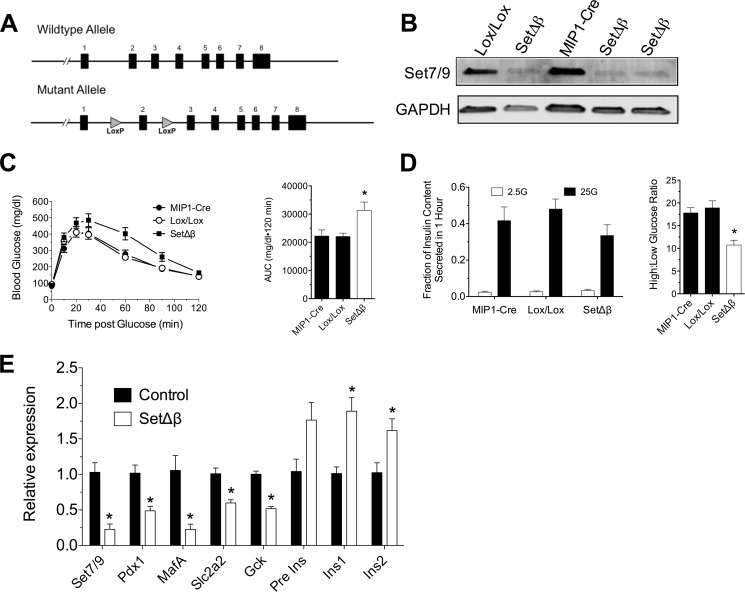
In prior studies, our group showed that transient knockdown of Set7/9 in islet β cells results in a reduction in several gene targets of Pdx1 (26), but the link between Set7/9 and β cell function in vivo has not been explored. To address this link more directly, we generated mice in which Cre recombinase recognition (Loxp) sites flanked exon 2 of the gene encoding Set7/9 (Setd7) so that conditional excision of the exon by Cre recombinase would result in a frameshift and premature termination of the mRNA (Fig. 6A). This strategy ensures the deletion of both the putative Pdx-1 interaction domain (at the N terminus) and the methyltransferase domain (the SET domain) at the C terminus. Setd7Loxp/+ mice were crossed to mice that express the Cre transgene exclusively in islet β cells when treated with tamoxifen (MIP1-CreERT (21). Setd7Loxp/Loxp;MIP1-CreERT mice and littermate controls (Setd7+/+;MIP1-CreERT and Setd7Loxp/Loxp) were treated with five daily doses of tamoxifen at 8 weeks of age to generate β cell-specific Set7/9 knockout mice (SetΔβ) and controls, respectively. At 12 weeks of age (∼3 weeks after conclusion of tamoxifen treatment), islets isolated from SetΔβ mice showed a clear reduction in Set7/9 levels by immunoblot analysis (Fig. 6B).
FIGURE 6.
β Cell-specific deletion of Set7/9 in mice. A, schematic of WT and mutant alleles present in control and mutant mice, respectively, showing positions of the exons (numbered) and the Loxp sites. B, Immunoblot analysis of whole cell lysates of islets isolated from Setd7+/+;MIP1-CreERT (MIP1-Cre), Setd7Loxp/Loxp (Lox/Lox), and MIP1-CreERT;Setd7Loxp/Loxp (SetΔβ) mice for Set7/9 (top panel) and GAPDH (bottom panel). C, results of intraperitoneal glucose tolerance test on MIP1-Cre (n = 12), Lox/Lox (n = 9), and SetΔβ (n = 12) mice (left panel) with a corresponding area under the curve (AUC) analysis (right panel). *, p < 0.05 compared with either MIP1-Cre or Lox/Lox. D, results of glucose-stimulated insulin release assays from islets isolated from MIP1-Cre, Lox/Lox, and SetΔβ mice (left panel), and the corresponding ratio of insulin secreted at high glucose relative to low glucose is shown in the right panel. n = 3, *, p < 0.05 compared with either MIP1-Cre or Lox/Lox. E, results of real-time RT-PCR for the indicated genes from islets isolated from control (MIP1-Cre and Lox/Lox) versus SetΔβ mice. Data are normalized to Actb mRNA levels; n = 4 independent islet isolations; *, p < 0.05 compared with control.
SetΔβ mice displayed impaired glucose tolerance following an intraperitoneal glucose challenge (Fig. 6C). To test the possibility that impaired glucose tolerance was caused by β cell dysfunction, we isolated islets from SetΔβ mice and controls and performed glucose stimulated insulin secretion studies in vitro. As shown in Fig. 6D, islets from SetΔβ mice displayed diminished glucose-stimulated insulin secretion compared with islets from controls, even when corrected for islet insulin content. These data suggest a defect in insulin secretory mechanisms and are reminiscent of defects seen in Pdx1-deficient islets (37, 38). Gene expression analysis of isolated islets revealed reductions in the mRNA levels of key Pdx1-regulated genes, including MafA, Slc2a2, Gck, and Pdx1 itself in SetΔβ islets compared with controls (Fig. 6E). The pre-mRNA and mRNAs encoding preproinsulin showed no change and an ∼2-fold increase, respectively, in SetΔβ islets compared with controls, suggesting that the response of this gene may either be delayed compared with the other Pdx1 targets or that it exhibits a more complex regulation in the absence of Set7/9. Taken together, the data from SetΔβ mice suggest a role for Set7/9 in the maintenance of key Pdx1 target genes and overall glucose homeostasis in vivo and parallel the phenotype seen with reductions in Pdx1 itself.
DISCUSSION
Pdx1 is required for both the development and function of islet β cells, and, as such, the mechanism underlying its regulation of transcription has been the subject of numerous studies. Nevertheless, the full extent of the mechanisms governing Pdx1 action remains to be elucidated. Set7/9 is a Lys methyltransferase that has been shown to interact with Pdx1 and augment Pdx1 action at several target genes (26). In this study, we provide data suggesting that Set7/9-mediated methylation of Pdx1 augments Pdx1 transcriptional activity. Although other studies have shown that Pdx1 can be regulated by phosphorylation (39–43), O-GlcNAcylation (44), and sumoylation (45), ours is the first study to show that Pdx1 can be methylated. Moreover, we show a close linkage between Pdx1 and Set7/9 in vivo, where the loss of Set7/9 in β cells closely parallels the phenotype seen with the reduction in Pdx1 levels.
In recent years, the number of identified non-histone targets of Set7/9 methylation has been increasing. Depending upon the protein, Lys methylation appears to affect a variety of functions at the transcriptional level, including protein-protein interactions, protein stability, and DNA binding affinity (17, 35). Notable proteins that are substrates of Set7/9 Lys methylation include p53 (12), estrogen receptor α (14), nuclear factor κB (13, 15, 46), and TATA box binding protein-associated factor 10 (47), among others. In this study, we show evidence that Pdx1 is methylated at Lys residues in vitro and in β cells. An important finding in vitro was that recombinant Pdx1 was methylated by Set7/9 but occurred only in the context of the full-length protein. This requirement for context-dependent methylation seemingly differs from those reported in the literature, where Set7/9 has been shown to methylate Lys residues of specific small peptides whose sequences could be subsequently used to predict the corresponding full-length protein targets (34, 48). To our knowledge, none of the small peptide sequences presented in those studies correspond to sequences in Pdx1. As such, our studies emphasize that targets of Set7/9 and possibly other SET domain-containing methyltransferases may not be predictable on the basis of peptide sequence specificity. More detailed structural studies will be necessary to identify how specific secondary and tertiary structural folds of Pdx1 allow for its methylation by Set7/9.
Our mass spectrometric and mutational analyses in vitro demonstrate that Pdx1 is methylated at two Lys residues in the N-terminal transactivation domain, Lys-123 and Lys-131, with Lys-123 being the apparent preferential target for Set7/9 in vitro. Interestingly, Lys-131 seems to be the more relevant target of methylation when Pdx1 is expressed in cells, and this suggests that Lys-131 methylation has greater functional consequences in the context of the cellular milieu. Our finding that Pdx1 methylation becomes more apparent when cells are treated with Lys demethylase inhibitors also suggests that methylation may be transient in cells or perhaps regulated depending upon the target gene or functions of Pdx1. In this regard, there are several reports that show that proteins methylated by Set7/9 are reciprocally regulated by the demethylase LSD-1 (49–52).
Although methylation of Pdx1 by Set7/9 in vitro resulted in monomethylation of Lys-131 (and Lys-123), our overexpression studies in β cells showed that Lys-131 was di- and trimethylated. This observation raises an important issue that has been discussed in the literature on the stoichiometry of Lys methylation by Set7/9. Some studies have suggested that Set7/9 functions as a dimethyltransferase (53), whereas others suggest that it is exclusively a monomethyltransferase (54). These seemingly contradictory observations could be explained by at least three possibilities. 1) mono- versus dimethylation could be sequence-dependent. In this respect, studies have shown that some peptide sequences are dimethylated by Set7/9 in vitro, whereas other peptide sequences are monomethylated (34). 2) In β cells, interacting proteins may alter the nature of Lys methylation by Set7/9. For example, the methyltransferase G9a exhibits differing stoichiometries of methylation in vivo depending on the presence of interacting proteins (55). 3) Set7/9 may be the first in a cascade of different methyltransferases that achieves di- and trimethylation of Pdx1 residue Lys-131 in cells.
Set7/9 has been shown previously to augment Pdx1 transcriptional activity in reporter gene assays (11). Here we observed that the Pdx1 residue Lys-131 is necessary for this augmentation because mutation of this residue to Arg (K131R) abrogates transcriptional augmentation. Although the transcriptional augmentation by Set7/9 is only about 40%, the effects of such reductions may have biologically significant consequences in the longer term. We next probed possible reasons for how the K131R mutation abrogates transcriptional augmentation by Set7/9. We found that mutation did not hamper Pdx1-Set7/9 interactions (as determined by coimmunoprecipitation experiments) or DNA binding affinity (on the basis of EMSA analysis). However, It remains possible that binding to target genes in the nucleus may be subtly altered by the mutations. Nevertheless, on the basis of our data, we believe that Pdx1 methylation at Lys-131 may affect the interactions of the Pdx1 transcriptional complex with basal transcriptional machinery. In support of this possibility, we showed previously that the Pdx1-Set7/9 interaction was associated with activation of RNA polymerase II (11, 56).
Finally, to interrogate the relationship between Set7/9 and Pdx1 target genes in β cells in vivo, we generated mice with a tamoxifen-inducible β cell-specific deletion of Set7/9 (SetΔβ). Similar to mice with haploinsufficiency of Pdx1, SetΔβ mice exhibited glucose intolerance and β cell dysfunction (37). Several Pdx1 target genes were reduced in SetΔβ mice, consistent with the close functional linkage between Set7/9 and Pdx1. Our results are similar to those we published previously using RNA interference to deplete Set7/9 acutely in islets (26). Although these studies do not definitively prove a link between Set7/9 and methylated Pdx1 (because the loss of Set7/9 may have consequences on broader gene expression), these results do suggest that Pdx1 may be dependent upon the action of Set7/9 for full activity in vivo.
Taken together, our data suggest a previously unappreciated posttranslational modification of Pdx1 that appears to affect its transcriptional activity. Our studies show the methylation of Pdx1 residue Lys-131, the requirement of Lys-131 for the augmentation of gene transcription in reporter assays, the occurrence of Lys-131 methylation in β cell lines, and the requirement for Set7/9 in the maintenance of at least a subset of Pdx1 target genes in vivo. Some limitations and alternative interpretations of our study must be acknowledged. Most notably, it remains possible that the effects observed in our reporter assays or in SetΔβ mice are not directly related to methylation of Pdx1 per se but, rather, to a separate requirement for residue Lys-131 or Set7/9, respectively, on gene transcription. Gene knockin studies in mice (using the K131R mutation of Pdx1) may be more appropriate to address this issue in the future. Also, our studies do not exclude the possibility in vivo that Set7/9 is directly necessary for the maintenance of chromatin structure (histone methylation) at several of the Pdx1 target genes examined or that it acts in concert with β cell transcription factors other than Pdx1. Nevertheless, our studies provide a framework to better understand the nuances of transcriptional regulation by Pdx1. We propose a model in which Pdx1 methylation at residue Lys-131 via Set7/9 is required for the maintenance of transcription at selected target genes. Although this model does not exclude the possibility (as purported in prior studies) that histone methylation by Set7/9 also contributes to transcriptional activity, it emphasizes that transcription factor-cofactor interactions can achieve functional transcriptional complexes in many ways.
Acknowledgments
We thank N. Stull, K. Benninger, and M. Robertson of the Islet Core of the Indiana Diabetes Research Center at Indiana University for isolation of mouse pancreatic islets. We also thank Dr. M. Wang of the Indiana University Mass Spectrometry core for assistance with mass spectrometry studies. We also thank Dr. L. Philipson (University of Chicago) for the MIP1-CreERT mice.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DK083583 and R01 DK060581 (to R. G. M.) and R01 CA149095 (to P. M. W.). This work was also supported by an American Diabetes Association junior faculty award (to S. A. T.).
- AdoMet
- S-adenosylmethionine.
REFERENCES
- 1. Ferrannini E., Mari A., Nofrate V., Sosenko J. M., Skyler J. S., and DPT-1 Study Group (2010) Progression to diabetes in relatives of type 1 diabetic patients: mechanisms and mode of onset. Diabetes 59, 679–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ferrannini E., Gastaldelli A., Miyazaki Y., Matsuda M., Mari A., DeFronzo R. A. (2005) β-Cell function in subjects spanning the range from normal glucose tolerance to overt diabetes: a new analysis. J. Clin. Endocrinol. Metab. 90, 493–500 [DOI] [PubMed] [Google Scholar]
- 3. Jonsson J., Carlsson L., Edlund T., Edlund H. (1994) Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 371, 606–609 [DOI] [PubMed] [Google Scholar]
- 4. Offield M. F., Jetton T. L., Labosky P. A., Ray M., Stein R. W., Magnuson M. A., Hogan B. L., Wright C. V. (1996) PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development 122, 983–995 [DOI] [PubMed] [Google Scholar]
- 5. Stoffers D. A., Zinkin N. T., Stanojevic V., Clarke W. L., Habener J. F. (1997) Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat. Genet. 15, 106–110 [DOI] [PubMed] [Google Scholar]
- 6. Babu D. A., Deering T. G., Mirmira R. G. (2007) A feat of metabolic proportions: Pdx1 orchestrates islet development and function in the maintenance of glucose homeostasis. Mol. Genet. Metab. 92, 43–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Khoo C., Yang J., Weinrott S. A., Kaestner K. H., Naji A., Schug J., Stoffers D. A. (2012) Research resource: the pdx1 cistrome of pancreatic islets. Mol. Endocrinol. 26, 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Babu D. A., Chakrabarti S. K., Garmey J. C., Mirmira R. G. (2008) Pdx1 and BETA2/neuroD1 participate in a transcriptional complex that mediates short-range DNA looping at the insulin gene. J. Biol. Chem. 283, 8164–8172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ohneda K., Mirmira R. G., Wang J., Johnson J. D., German M. S. (2000) The homeodomain of PDX-1 mediates multiple protein-protein interactions in the formation of a transcriptional activation complex on the insulin promoter. Mol. Cell. Biol. 20, 900–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Claiborn K. C., Sachdeva M. M., Cannon C. E., Groff D. N., Singer J. D., Stoffers D. A. (2010) Pcif1 modulates Pdx1 protein stability and pancreatic β cell function and survival in mice. J. Clin. Invest. 120, 3713–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Francis J., Chakrabarti S. K., Garmey J. C., Mirmira R. G. (2005) Pdx-1 links histone H3-Lys-4 methylation to RNA polymerase II elongation during activation of insulin transcription. J. Biol. Chem. 280, 36244–36253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chuikov S., Kurash J. K., Wilson J. R., Xiao B., Justin N., Ivanov G. S., McKinney K., Tempst P., Prives C., Gamblin S. J., Barlev N. A., Reinberg D. (2004) Regulation of p53 activity through lysine methylation. Nature 432, 353–360 [DOI] [PubMed] [Google Scholar]
- 13. Ea C.-K., Baltimore D. (2009) Regulation of NF-κB activity through lysine monomethylation of p65. Proc. Natl. Acad. Sci. U.S.A. 106, 18972–18977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Subramanian K., Jia D., Kapoor-Vazirani P., Powell D. R., Collins R. E., Sharma D., Peng J., Cheng X., Vertino P. M. (2008) Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol. Cell 30, 336–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang X.-D., Huang B., Li M., Lamb A., Kelleher N. L., Chen L.-F. (2009) Negative regulation of NF-κB action by Set9-mediated lysine methylation of the RelA subunit. EMBO J. 28, 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Keating S. T., Ziemann M., Okabe J., Khan A. W., Balcerczyk A., El-Osta A. (2014) Deep sequencing reveals novel Set7 networks. Cell Mol. Life Sci. 71, 4471–4486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee D. Y., Teyssier C., Strahl B. D., Stallcup M. R. (2005) Role of protein methylation in regulation of transcription. Endocr. Rev. 26, 147–170 [DOI] [PubMed] [Google Scholar]
- 18. Maier B., Ogihara T., Trace A. P., Tersey S. A., Robbins R. D., Chakrabarti S. K., Nunemaker C. S., Stull N. D., Taylor C. A., Thompson J. E., Dondero R. S., Lewis E. C., Dinarello C. A., Nadler J. L., Mirmira R. G. (2010) The unique hypusine modification of eIF5A promotes islet β cell inflammation and dysfunction in mice. J. Clin. Invest. 120, 2156–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mirmira R. G., Watada H., German M. S. (2000) β-Cell differentiation factor Nkx6.1 contains distinct DNA Binding interference and transcriptional repression domains. J. Biol. Chem. 275, 14743–14751 [DOI] [PubMed] [Google Scholar]
- 20. Chakrabarti S. K., Francis J., Ziesmann S. M., Garmey J. C., Mirmira R. G. (2003) Covalent histone modifications underlie the developmental regulation of insulin gene transcription in pancreatic β cells. J. Biol. Chem. 278, 23617–23623 [DOI] [PubMed] [Google Scholar]
- 21. Tamarina N. A., Roe M. W., Philipson L. (2014) Characterization of mice expressing Ins1 gene promoter driven CreERT recombinase for conditional gene deletion in pancreatic β-cells. Islets 6, e27685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Evans-Molina C., Robbins R. D., Kono T., Tersey S. A., Vestermark G. L., Nunemaker C. S., Garmey J. C., Deering T. G., Keller S. R., Maier B., Mirmira R. G. (2009) PPAR-γ activation restores islet function in diabetic mice through reduction of ER stress and maintenance of euchromatin structure. Mol. Cell. Biol. 29, 2053–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stull N. D., Breite A., McCarthy R. C., Tersey S. A., Mirmira R. G. (2012) Mouse islet of Langerhans isolation using a combination of purified collagenase and neutral protease. J. Vis. Exp. 67, e4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Taylor D. G., Babu D., Mirmira R. G. (2005) The C-terminal domain of the β cell homeodomain factor Nkx6.1 enhances sequence-selective DNA binding at the insulin promoter. Biochemistry 44, 11269–11278 [DOI] [PubMed] [Google Scholar]
- 25. Nishioka K., Chuikov S., Sarma K., Erdjument-Bromage H., Allis C. D., Tempst P., Reinberg D. (2002) Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 16, 479–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Deering T. G., Ogihara T., Trace A. P., Maier B., Mirmira R. G. (2009) Methyltransferase Set7/9 maintains transcription and euchromatin structure at islet-enriched genes. Diabetes 58, 185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sharma S. K., Wu Y., Steinbergs N., Crowley M. L., Hanson A. S., Casero R. A., Woster P. M. (2010) (Bis)urea and (bis)thiourea inhibitors of lysine-specific demethylase 1 as epigenetic modulators. J. Med. Chem. 53, 5197–5212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Verlinden B. K., Niemand J., Snyman J., Sharma S. K., Beattie R. J., Woster P. M., Birkholtz L.-M. (2011) Discovery of novel alkylated (bis)urea and (bis)thiourea polyamine analogues with potent antimalarial activities. J. Med. Chem. 54, 6624–6633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Robbins R. D., Tersey S. A., Ogihara T., Gupta D., Farb T. B., Ficorilli J., Bokvist K., Maier B., Mirmira R. G. (2010) Inhibition of deoxyhypusine synthase enhances islet β cell function and survival in the setting of endoplasmic reticulum stress and type 2 diabetes. J. Biol. Chem. 285, 39943–39952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mosley A. L., Ozcan S. (2003) Adenoviral gene transfer into β-cell lines. Methods Mol. Med. 83, 73–79 [DOI] [PubMed] [Google Scholar]
- 31. Mosley A. L., Corbett J. A., Ozcan S. (2004) Glucose regulation of insulin gene expression requires the recruitment of p300 by the β-cell-specific transcription factor Pdx-1. Mol. Endocrinol. 18, 2279–2290 [DOI] [PubMed] [Google Scholar]
- 32. German M. S., Moss L. G., Wang J., Rutter W. J. (1992) The insulin and islet amyloid polypeptide genes contain similar cell-specific promoter elements that bind identical β-cell nuclear complexes. Mol. Cell. Biol. 12, 1777–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mosley A. L., Sardiu M. E., Pattenden S. G., Workman J. L., Florens L., Washburn M. P. (2011) Highly reproducible label free quantitative proteomic analysis of RNA polymerase complexes. Mol. Cell. Proteomics 10, M110.000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dhayalan A., Kudithipudi S., Rathert P., Jeltsch A. (2011) Specificity analysis-based identification of new methylation targets of the SET7/9 protein lysine methyltransferase. Chem. Biol. 18, 111–120 [DOI] [PubMed] [Google Scholar]
- 35. Herz H.-M., Garruss A., Shilatifard A. (2013) SET for life: biochemical activities and biological functions of SET domain-containing proteins. Trends Biochem. Sci. 38, 621–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chakrabarti S. K., James J. C., Mirmira R. G. (2002) Quantitative assessment of gene targeting in vitro and in vivo by the pancreatic transcription factor, Pdx1: importance of chromatin structure in directing promoter binding. J. Biol. Chem. 277, 13286–13293 [DOI] [PubMed] [Google Scholar]
- 37. Brissova M., Shiota M., Nicholson W. E., Gannon M., Knobel S. M., Piston D. W., Wright C. V., Powers A. C. (2002) Reduction in pancreatic transcription factor PDX-1 impairs glucose-stimulated insulin secretion. J. Biol. Chem. 277, 11225–11232 [DOI] [PubMed] [Google Scholar]
- 38. Johnson J. D., Ahmed N. T., Luciani D. S., Han Z., Tran H., Fujita J., Misler S., Edlund H., Polonsky K. S. (2003) Increased islet apoptosis in Pdx1+/− mice. J. Clin. Invest. 111, 1147–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. An R., da Silva Xavier G., Semplici F., Vakhshouri S., Hao H.-X., Rutter J., Pagano M. A., Meggio F., Pinna L. A., Rutter G. A. (2010) Pancreatic and duodenal homeobox 1 (PDX1) phosphorylation at serine-269 is HIPK2-dependent and affects PDX1 subnuclear localization. Biochem. Biophys. Res. Commun. 399, 155–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Boucher M.-J., Selander L., Carlsson L., Edlund H. (2006) Phosphorylation marks IPF1/PDX1 protein for degradation by glycogen synthase kinase 3-dependent mechanisms. J. Biol. Chem. 281, 6395–6403 [DOI] [PubMed] [Google Scholar]
- 41. Frogne T., Sylvestersen K. B., Kubicek S., Nielsen M. L., Hecksher-Sørensen J. (2012) Pdx1 is post-translationally modified in vivo and serine 61 is the principal site of phosphorylation. PLoS ONE 7, e35233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Humphrey R. K., Yu S.-M., Flores L. E., Jhala U. S. (2010) Glucose regulates steady-state levels of PDX1 via the reciprocal actions of GSK3 and AKT kinases. J. Biol. Chem. 285, 3406–3416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Semache M., Zarrouki B., Fontés G., Fogarty S., Kikani C., Chawki M. B., Rutter J., Poitout V. (2013) Per-Arnt-Sim kinase regulates pancreatic duodenal homeobox-1 protein stability via phosphorylation of glycogen synthase kinase 3β in pancreatic β-cells. J. Biol. Chem. 288, 24825–24833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kebede M., Ferdaoussi M., Mancini A., Alquier T., Kulkarni R. N., Walker M. D., Poitout V. (2012) Glucose activates free fatty acid receptor 1 gene transcription via phosphatidylinositol-3-kinase-dependent O-GlcNAcylation of pancreas-duodenum homeobox-1. Proc. Natl. Acad. Sci. U.S.A. 109, 2376–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kishi A., Nakamura T., Nishio Y., Maegawa H., Kashiwagi A. (2003) Sumoylation of Pdx1 is associated with its nuclear localization and insulin gene activation. Am. J. Physiol. Endocrinol. Metab. 284, E830-E840 [DOI] [PubMed] [Google Scholar]
- 46. Li Y., Reddy M. A., Miao F., Shanmugam N., Yee J. K., Hawkins D., Ren B., Natarajan R. (2008) Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-κB-dependent inflammatory genes: relevance to diabetes and inflammation. J. Biol. Chem. 283, 26771–26781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kouskouti A., Scheer E., Staub A., Tora L., Talianidis I. (2004) Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol. Cell 14, 175–182 [DOI] [PubMed] [Google Scholar]
- 48. Couture J. F., Collazo E., Hauk G., Trievel R. C. (2006) Structural basis for the methylation site specificity of SET7/9. Nat. Struct. Mol. Biol. 13, 140–146 [DOI] [PubMed] [Google Scholar]
- 49. Huang J., Sengupta R., Espejo A. B., Lee M. G., Dorsey J. A., Richter M., Opravil S., Shiekhattar R., Bedford M. T., Jenuwein T., Berger S. L. (2007) p53 is regulated by the lysine demethylase LSD1. Nature 449, 105–108 [DOI] [PubMed] [Google Scholar]
- 50. Kontaki H., Talianidis I. (2010) Lysine methylation regulates E2F1-induced cell death. Mol. Cell 39, 152–160 [DOI] [PubMed] [Google Scholar]
- 51. Sakane N., Kwon H.-S., Pagans S., Kaehlcke K., Mizusawa Y., Kamada M., Lassen K. G., Chan J., Greene W. C., Schnoelzer M., Ott M. (2011) Activation of HIV transcription by the viral Tat protein requires a demethylation step mediated by lysine-specific demethylase 1 (LSD1/KDM1). PLoS Pathog. 7, e1002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang J., Hevi S., Kurash J. K., Lei H., Gay F., Bajko J., Su H., Sun W., Chang H., Xu G., Gaudet F., Li E., Chen T. (2009) The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 41, 125–129 [DOI] [PubMed] [Google Scholar]
- 53. Kwon T., Chang J. H., Kwak E., Lee C. W., Joachimiak A., Kim Y. C., Lee J., Cho Y. (2003) Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet. EMBO J. 22, 292–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xiao B., Jing C., Wilson J. R., Walker P. A., Vasisht N., Kelly G., Howell S., Taylor I. A., Blackburn G. M., Gamblin S. J. (2003) Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature 421, 652–656 [DOI] [PubMed] [Google Scholar]
- 55. Sampath S. C., Marazzi I., Yap K. L., Sampath S. C., Krutchinsky A. N., Mecklenbräuker I., Viale A., Rudensky E., Zhou M. M., Chait B. T., Tarakhovsky A. (2007) Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol. Cell 27, 596–608 [DOI] [PubMed] [Google Scholar]
- 56. Iype T., Francis J., Garmey J. C., Schisler J. C., Nesher R., Weir G. C., Becker T. C., Newgard C. B., Griffen S. C., Mirmira R. G. (2005) Mechanism of insulin gene regulation by the pancreatic transcription factor Pdx-1: application of pre-mRNA analysis and chromatin immunoprecipitation to assess formation of functional transcriptional complexes. J. Biol. Chem. 280, 16798–16807 [DOI] [PubMed] [Google Scholar]