Abstract
Background
Genome-wide association studies (GWAS) have shown that the common single nucleotide polymorphism (SNP) rs6800541 located in SCN10A, encoding the voltage-gated Nav1.8 sodium channel, is associated with PR–interval prolongation and atrial fibrillation (AF). SNP rs6800541 is in high linkage disequilibrium with the non-synonymous variant in SCN10A, rs6795970 (V1073A, r2=0.933). We therefore sought to determine whether common and rare SCN10A variants are associated with early onset AF.
Methods and Results
SCN10A was sequenced in 225 AF patients in whom there was no evidence of other cardiovascular disease or dysfunction (lone AF). In an association study of the rs6795970 SNP variant, we included 515 AF patients, and two control cohorts of 730 individuals free of AF and 6,161 randomly sampled individuals. Functional characterization of SCN10A variants was performed by whole-cell patch-clamping. In the lone AF cohort, nine rare missense variants and one splice site donor variant were detected. Interestingly, AF patients were found to have higher G allele frequency of rs6795970 which encodes the alanine variant at position 1073 (described from here on as A1073, odds ratio = 1.35 [1.16–1.54]; p=2.3×10−05). Both of the common variants, A1073 andP1092, induced a gain-of-channel function, while the rare missense variants, V94G and R1588Q, resulted in a loss-of-channel function.
Conclusions
The common variant A1073 is associated with increased susceptibility to AF. Both rare and common variants have impact on the function of the channel, indicating that these variants influence susceptibility to AF. Hence, our study suggests that SCN10A variations are involved in the genesis of AF.
Keywords: atrial fibrillation arrhythmia, genetic polymorphism, electrophysiology, genotyping, Genome Wide Association Study, lone atrial fibrillation, SCN10A, voltage gated sodium channel alpha subunit Nav1.8, rs6795970, functional characterization
Introduction
Atrial fibrillation (AF), which is the most commonly sustained cardiac arrhythmia, is a global health problem accounting for increasing morbidity, mortality, and healthcare costs.1–3 Identifying and understanding the genetic basis of AF and/or association of genomic regions with AF will provide valuable insight into the pathogenesis of AF, and potentially improve the risk stratification and therapeutic options.
Genome wide association studies (GWAS) have identified 10 loci in the human genome that are associated with AF.4 Thus, common genetic variants play a role in the development of this multifactorial disease.5 Several studies have shown PR-interval prolongation on an electrocardiogram to be an independent risk factor for developing AF.6–8 Five independent GWAS publications have shown that genetic variants in SCN10A influence the PR-interval duration.9–13 Pfeufer et al. showed that five out of nine PR-associated loci from GWAS increased the risk of AF.10 They found that the single nucleotide polymorphism (SNP) rs6800541, which is located in an intron of SCN10A, had the strongest association with PR-interval duration and one of the strongest associations with AF among nine other GWAS hits. This SNP is in high and moderate linkage disequilibrium with two common nonsynonymous SNPs in SCN10A: rs6795970 (A1073) and rs12632942 (P1092), respectively.10 The substantial arrhythmogenic potential of genetic variants in SCN10A is underscored by the fact that another SNP (rs10428132) in this gene was the top hit in a GWAS on Brugada syndrome; a condition strongly associated with AF.14,15 Moreover, very recently, a phenome-wide study associated SCN10A, through rs6795970, directly with AF.16 This, however, contradicts with the findings by Holm et al. who did not report an association of rs6795970 with AF.11
SCN10A encodes the voltage-gated sodium channel, Nav1.8. This channel is the predominant tetrodotoxin-resistant sodium channel in primary sensory neurons, with particularly high levels of expression in nociceptive neurons, where it plays a key role in peripheral pain processing.17,18 Expression has also been shown in vagal, but not in sympathetic fibers.19,20
Recently, a number of studies have indicated that SCN10A mRNA is present in both human and mouse heart and that this channel is involved in the cardiac INa current.9,21–23 Yang et al. demonstrated higher expression of Nav1.8 transcripts in mouse atria compared to ventricle.21 Facer et al. detected Nav1.8 protein in both atrial myocytes and nerve fibers in the myocardium.24 Using genetic lineage tracing, others have shown that Nav1.8 is expressed in aortic bodies and the nerves around blood vessels of the heart.20 Interestingly, Verkerk et al. found that Nav1.8 is highly expressed in intracardiac neurons.22 In summary, these studies suggest that Nav1.8 is expressed in both cardiac myocytes and intracardiac neurons.
The recent notion that Nav1.8 might be important for cardiac electrophysiological properties raises the possibility that altered function of this gene may be coupled with cardiac arrhythmias.25 Thus, in the present study, we investigated whether the common SCN10A variant rs6795970 is associated with AF and thereby would be the variant carrying the effect of the GWAS hit. In addition, we screened 225 lone AF patients for SCN10A variants, and characterized the two rarest variants together with the two common variants functionally using patch-clamp electrophysiology.
Methods
Study Subjects
Patient records with the ICD-10 diagnose code I48.9 (atrial fibrillation and flutter) were collected and read. Only 225 patients with “lone AF” and onset of disease before age of 40 years were recruited. Lone AF was defined as AF in absence of clinical or echocardiographic findings of cardiovascular disease, hypertension requiring medical therapy, metabolic- or pulmonary diseases. For the genotyping of the rs6795970 SNP, we recruited a cohort of 358 Scandinavian lone AF patients with onset of AF before the age of 50 (83% male gender, median age of AF onset 34.5 years [interquartile range 28–39 years]) and a cohort of 157 unselected AF patients (68% male gender, median age of 66 years [interquartile range 32–86 years]) (Supplemental Material). Blood samples, ECG and clinical data were collected from all participating subjects. The study conforms to the principles outlined in the Declaration of Helsinki and was approved by the local scientific ethics committees, and all patients provided written informed consent.
Control Population
A total of 730 healthy subjects (52% males, median 66 years [interquartile range 52–76 years]) from two control cohorts (control groups I and II) were included in this study (Supplemental Material). Control group I (complete sequencing of SCN10A) consisted of 216 unrelated healthy Danish blood donors with a normal ECG and without any cardiac symptoms. Control group II comprised 514 ethnically matched, middle-aged men and women without a history of AF or other manifestations of cardiovascular disease; however, with a high prevalence of risk factors for AF. Control groups I and II were previously described in detail.26,27 These control groups were used in the genotyping of rs6795970 using a Taqman assay. To increase the statistical power of our association study, we also used a third control cohort (control group III), comprising 6,161 individuals randomly selected from a Danish cohort study (Inter99, LuCamp).28 Although this control cohort could only provide data on rs6795970, due to the exome-chip which was used in the Inter99 study. This control group is assumed to represent the general population.
SCN10A Screening
The method is available in Supplemental Material.
SNP Genotyping
We genotyped rs6795970, encoding A1073, in 515 AF patients of which 358 were lone AF patients. For comparison, in addition to control groups I and II (nTotal=730), we also used the data from a European-American population (n=4,300) from the Exome Sequencing Project (ESP). Genotyping of control groups I and II was performed as previously described.26 Furthermore, we also used the exome-chip data on the rs6795970 SNP from control group III.
Molecular Biology and In vitro Electrophysiology
Introduction of variants, cell culturing and patch-clamping of transiently transfected Neuro2A cells were performed as previously described.29 A detailed description is available in Supplemental Material.
Bioinformatics and Statistical Analysis
All variants were reviewed in publicly available SNP databases (dbSNP, and ESP6500). We used 4 in silico tools to predict whether the variants were disease causing. The MAF in the 2 case cohorts were compared one by one with the 3 control cohorts using the Chi-square test. Similarly, we performed a pooled analysis where MAF in the 2 case groups were compared with a pooled MAF from our largest control population and ESP. Data are presented as mean ± standard error of mean (SEM) unless otherwise noted. Kolmogorov-Smirnov test was applied to confirm Gaussian distribution. Two-tailed Student’s t-test, one-way or two-way ANOVA combined with a Bonferroni post hoc test, or Chi-square tests, were used as appropriate to test for significant differences. A value of P <0.05 was considered statistically significant. Further description is available at the Supplemental Material.
Results
Genetic screening
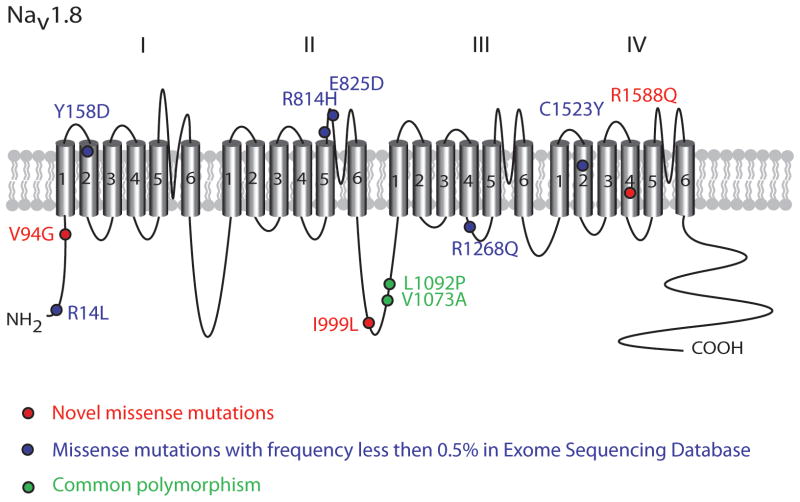
We included 225 unrelated Danish patients with onset of disease before the age of 40 years for full genetic screening (clinical data listed in Supplemental Material). The individual clinical characteristics of the patients with the rare SCN10A variations are listed in Table 1. In Figure 1 the positions of rare and common missense variants found in SCN10A in lone AF are illustrated in the Nav1.8 protein topology. We identified nine rare missense variants (R14L; V94G; Y158D; R814H; E825D; I999L; R1268Q; C1523Y; R1588Q) and one splice site donor variant (rs75991777) (in exon 4 at second position T>C, Table 2). These variants were neither present in our in-house control group (n = 432 alleles), nor have any been previously reported in conjunction with AF. However, except for I999L and rs75991777, all variants were identified in the Exome Sequencing Project database (n=6,503) with minor allele frequency (MAF) less than 0.5% in the European-American population. rs75991777 has been reported in the dbSNP database from the 1000 Genomes Project with a MAF of 0.1%. The amino acid residues altered in the rare variants were found to be highly conserved across eukaryotic species, except for R814H and E825D, which differed in rat and mouse (data not shown). In our co-segregation analysis, we were able to screen the family members of the patients with I999L, C1523Y and rs75991777 variants. None of the family members diagnosed with AF carried the variant identified in the probands. Family members of remaining geno-positive patients were not available. PolyPhen2 prediction software predicted 78% (7 out of 9) of the rare variants to have a functional effect on protein function (Table 2).30 By using the Giudicessi et al. agreement of ≥3 in silico tools on these rare variants, it was predicted that 60% of the variants were damaging (Table 2).31
Table 1.
Clinical characteristics of the lone AF patients with rare variants (MAF < 0.5% in EA in ESP6500)
| Patients | Exon | rs ID | Pos. of variants |
ECG description |
P-wave (ms) |
PR interval (ms) |
QRS interval (ms) |
QTc (ms) |
HR (bpm) |
Type of AF | Onset of AF |
Clinical information | rs6795970 (A1073) |
rs12632942 (P1092) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | E1 | rs141207048 | R14L | SR | 135 | 206 | 98 | 434 | 112 | Paroxysmal | 31 | AVNRT | V/A | L/P |
| 2 | E2 | rs202143516 | V94G | SR | 119 | 144 | 102 | 414 | 57 | Paroxysmal | 28 | Father and sister pacemaker | V/A | L |
| 3 | E2 | rs202143516 | V94G | SR | 94 | 132 | 84 | 396 | 64 | Paroxysmal | 31 | Nonhemorragic stroke after AF diagnosis | V/A | L |
| 4 | E4 E15 |
rs202192818 rs139861061 |
Y158D R814H |
SR, SVPCs with aberration | 131 | 186 | 86 | 418 | 103 | Persistent | 32 | AF RFA times four | V/A | L/P |
| 5 | E4 | rs75991777 | c.599+1 | SR | 135 | 162 | 116 | 421 | 63 | Paroxysmal | 35 | Atrial flutter, AVNRT, inducible VT and ICD. Normal coronary angiography. Family history of SCD and AF. |
A | L |
| 6 | E15 | rs146028829 | E825D | SR | 110 | 172 | 72 | 423 | 84 | Paroxysmal | 18 | AF RFA, AVNRT | V/A | L/P |
| 7 | E16 | unknown | I999L | SR, IRBBB | 118 | 160 | 100 | 412 | 61 | Paroxysmal | 35 | Mother with SVPCs | V/A | L |
| 8 | E21 | rs138832868 | R1268Q | SR, IRBBB | 90 | 152 | 94 | 394 | 59 | Paroxysmal | 30 | Several DC cardioversions | A | L/P |
| 9 | E21 | rs138832868 | R1268Q | AF | - | - | 92 | - | 96 | - | 23 | Atrial flutter type II | A | L/P |
| 10 | E26 | rs142217269 | C1523Y | SR, J-wave in II, III, aVF | 119 | 154 | 96 | 398 | 54 | Paroxysmal | 30 | Family history of AF | A | L |
| 11 | E27 | unknown | R1588Q | SR | 110 | 156 | 96 | 419 | 55 | Paroxysmal | 28 | RFA for AVNRT | A | L/P |
Positions and clinical characteristics of the patients with rare variants with minor allele frequencies (MAF) less than 0.5% in the European-American (EA) in the Exome Sequencing Project (n=6503, ESP6500) server. SR; Sinus rhythm, SVPC; Supraventricular premature complexes, IRBBB; Incomplete right bundle branch block, AF; Atrial fibrillation, AVNRT; AV-nodal reentry tachycardia, RFA; Radiofrequency ablation, ICD; Implantable cardioverter-defibrillator, VT; Ventricular Tachycardia
Figure 1.
Nav1.8 topology. Positions of rare and common variants found in SCN10A in lone AF are indicated on the Nav1.8 protein.
Table 2.
Genetic variations in SCN10A in lone AF patients
| ESP6500
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Exon | rsID | GVS Function |
Amino Acid Pos. |
cDNA Pos. |
EA Allele # | AA Allele # | ALL Allele # | MAF (%) (EA/AA/AII) |
PolyPhen- 2 Prediction |
SIFT | Grantham | Conservation | Agreement ≥3 |
| 1 | rs141207048 | missense | R14L | 41 | A=26/C=8574 | A=2/C=4404 | A=28/C=12978 | 0.30/0.045/0.21 | Probably-damaging | Damaging | 102 | Conserved | D |
| 1 | rs34314583 | coding-synonymous | R15R | 45 | A=92/G=8508 | A=15/G=4391 | A=107/G=12899 | 1.07/0.34/0.82 | - | - | - | Conserved | - |
| 2 | rs202143516 | missense | V94G | 281 | C=1/A=8599 | C=0/A=4406 | C=1/A=13005 | 0.012/0.0/0.0077 | Probably-damaging | Damaging | 109 | Conserved | D |
| 4 | rs202192818 | missense-near-splice | Y158D | 472 | C=5/A=8595 | C=0/A=4406 | C=5/A=13001 | 0.058/0.0/0.038 | Probably-damaging | Damaging | 160 | Conserved | D |
| 4 | rs75991777 | splice-5 | None | c.599+1 | - | - | - | - | - | - | - | Conserved | - |
| 5 | rs74717885 | missense | I206M | 618 | C=137/T=8463 | C=14/T=4392 | C=151/T=12855 | 1.59/0.32/1.16 | Benign | Tolerated | 10 | Conserved | B |
| 9 | rs62244070 | coding-synonymous | E428E | 1284 | T=2158/C=6442 | T=474/C=3932 | T=2632/C=10374 | 25.09/10.76/20.24 | - | - | - | Conserved | - |
| 11 | rs7617919 | coding-synonymous | L492L | 1476 | A=2169/G=6429 | A=471/G=3935 | A=2640/G=10364 | 25.23/10.69/20.30 | - | - | - | Conserved | - |
| 11 | rs7630989 | missense | S509P | 1525 | G=241/A=8359 | G=925/A=3481 | G=1166/A=11840 | 2.80/20.99/8.97 | Benign | Tolerated | 74 | Conserved | B |
| 15 | rs139861061 | missense | R814H | 2441 | T=6/C=8594 | T=1/C=4405 | T=7/C=12999 | 0.07/0.023/0.054 | Possibly-damaging | Damaging | 29 | Not Conserved | B |
| 15 | rs146028829 | missense | E825D | 2475 | G=19/T=8581 | G=2/T=4404 | G=21/T=12985 | 0.22/0.045/0.16 | Benign | Tolerated | 45 | Not Conserved | B |
| 16 | rs7374804 | coding-synonymous | K950K | 2850 | C=557/T=8043 | C=362/T=4044 | C=919/T=12087 | 6.48/8.22/7.07 | - | - | - | Not Conserved | - |
| 16 | rs57326399 | missense | I962V | 2884 | C=2253/T=6347 | C=482/T=3924 | C=2735/T=10271 | 26.20/10.94/21.03 | Benign | Tolerated | 29 | Not Conserved | B |
| 16 | rs59468016 | coding-synonymous | G979G | 2937 | A=2248/G=6352 | A=224/G=4182 | A=2472/G=10534 | 26.14/5.08/19.01 | - | - | - | Not Conserved | - |
| 16 | unknown | missense | I999L | 2995 | - | - | - | - | Benign | Tolerated | 5 | Conserved | B |
| 17 | rs73062575 | missense | P1045T | 3133 | T=273/G=8327 | T=22/G=4384 | T=295/G=12711 | 3.17/0.5/2.27 | Benign | Tolerated | 38 | Not Conserved | B |
| 17 | rs6791171 | coding-synonymous | T1064T | 3192 | T=1216/C=7384 | T=880/C=3526 | T=2096/C=10910 | 14.14/19.97/16.12 | - | - | - | Not Conserved | - |
| 17 | rs6795970 | missense | A1073 | 3218 | G=5176/A=3424 | G=3961/A=445 | G=9137/A=3869 | 39.81/10.01/29.74 | Benign | Tolerated | 64 | Not Conserved | B |
| 18 | rs12632942 | missense | P1092 | 3275 | G=2236/A=6364 | G=625/A=3781 | G=2861/A=10145 | 26.0/14.18/21.99 | Benign | Tolerated | 98 | Not Conserved | B |
| 18 | unknown | coding-synonymous | D1113D | 3339 | - | - | - | - | - | - | - | Not Conserved | - |
| 19 | rs6771157 | coding-synonymous | T1131T | 3393 | C=2238/G=6362 | C=626/G=3780 | C=2864/G=10142 | 26.02/14.21/22.02 | - | - | - | Not Conserved | - |
| 21 | rs138832868 | missense-near-splice | R1268Q | 3803 | T=24/C=8576 | T=3/C=4403 | T=27/C=12979 | 0.279/0.068/0.20 | Probably-damaging | Damaging | 43 | Conserved | D |
| 22 | rs11711062 | missense | S1337T | 4009 | T=56/A=8544 | T=3/A=4403 | T=59/A=12947 | 0.65/0.07/0.45 | Benign | Tolerated | 58 | Conserved | B |
| 25 | rs6790627 | coding-synonymous | K1441K | 4323 | C=1230/T=7370 | C=1500/T=2906 | C=2730/T=10276 | 14.30/34.04/20.99 | - | - | - | Conserved | - |
| 26 | rs142217269 | missense | C1523Y | 4568 | T=23/C=8577 | T=1/C=4405 | T=24/C=12982 | 0.27/0.02/0.18 | Probably-damaging | Damaging | 194 | Conserved | D |
| 27 | rs78425180 | coding-synonymous | T1570T | 4710 | T=122/C=8478 | T=16/C=4390 | T=138/C=12868 | 1.41/0.36/1.06 | - | - | - | Conserved | - |
| 27 | unknown | missense | R1588Q | 4763 | T=0/C=8600 | T=1/C=4405 | T=1/C=13005 | 0.0/0.023/0.008 | Probably-damaging | Damaging | 43 | Conserved | D |
| 27 | rs6599242 | coding-synonymous | S1622S | 4866 | G=7870/A=730 | G=4232/A=174 | G=12102/A=904 | 8.49/3.95/6.95 | - | - | - | Conserved | - |
| 27 | rs77804526 | missense | V1697I | 5089 | T=118/C=8482 | T=13/C=4393 | T=131/C=12875 | 1.37/0.3/1.01 | Benign | Tolerated | 29 | Conserved | B |
| 27 | rs116353929 | coding-synonymous | D1739D | 5217 | A=265/G=8335 | A=36/G=4370 | A=301/G=12705 | 3.08/0.82/2.31 | - | - | - | Conserved | - |
Positions of the variants found in lone AF cohort. The frequency and MAF of the alleles are reported from ESP6500 exome server. PolyPhen-2 prediction reports the possible impact of an amino acid substitution on protein structure and function based on Polymorphism Phenotyping-2 (PolyPhen-2) program. D: Disease Causing; B: Benign; EA: European American; AA: African American; ESP 6500 exomes: Exome Variant Server (chromosomes 1–22, and X); MAF (%) (EA/AA/All): the minor-allele frequency in percent listed in the order of European American (EA), African American (AA) and all populations (All). (delimited by /)
Genotyping of rs6795970 encoding A1073
The result of the SNP genotyping is listed in Table 3. We were able to genotype the SNP rs6795970 in SCN10A in 515 AF patients (358 lone AF and 157 unselected AF patients) with a total call rate of 98.5%. The frequency of the G allele (encoding Alanine (Ala)) was 68.2% in all AF cases compared to 62.2% in 6,161 randomly sampled Danish exomes (Odds Ratio (OR) = 1.28, 95% Confidence Interval (CI) [1.11–1.47]; p=3.9×10−04). A similar result was found in a meta-analysis in which the control group comprised 4,300 European-American exomes from the ESP6500 database along with the 6,161 Danish exomes (OR = 1.35 [1.16–1.54]; p=2.3×10−05, Table 3). These results indicate that rs6795970 increases the risk of developing AF (Table 3).
Table 3.
Association of rs6795970 frequencies with AF
| Control cohorts G-allele (%) | 553 (37.9)
|
4606 (37.4)
|
8030 (38.4)
|
|||||
|---|---|---|---|---|---|---|---|---|
| Cases vs. Control I and II (n=730) | Cases vs. Control III (n=6161) | Cases vs. Control III and EA ESP (n=10.461) | ||||||
| AF cohorts G allele (%) | OR [95% CI] | p-value | OR [95% CI] | p-value | OR [95% CI] | p-value | ||
|
|
|
|
|
|||||
| All AF Cases n = 508 | 694 (68.2) | 1.30 [1.10–1.56] | 0.002 | 1.28 [1.11–1.47] | 3.9×10−04 | 1.35 [1.16–1.54] | 2.3×10−05 | |
| Lone AF Cases n = 354 | 482 (68.1) | 1.30 [1.10–1.59] | 0.007 | 1.27 [1.10–1.51] | 0.003 | 1.33 [1.12–1.56] | 4.6×10−04 | |
The allele frequencies are presented with Odds ratios (OR) and the p-values for their association with AF. ESP: Exome Sequencing Project, EA: European Americans, AF: Atrial Fibrillation, MAF: Minor Allele Frequency. The Chi-squared tests were used.
Clinical Features
Nine out of 11 of the AF patients harboring an SCN10A variant had paroxysmal AF and several of these patients also had other arrhythmias (Table 1). The R14L variant was identified in a patient with paroxysmal AF with onset of AF at age 31 and AV-nodal re-entry tachycardia (AVNRT). The V94G variant was found in two patients with paroxysmal and persistent AF with an onset of disease at age 28 and 27, respectively. The missense variants Y158D and R814H were identified in a patient with persistent AF with onset at age 31, who had several radiofrequency ablation (RFA) procedures for AF. The paroxysmal AF patient with onset of disease at age 35 had a splice-site donor variant at exon 4. This patient had normal coronary angiography with atrial flutter, AVNRT, inducible ventricular tachycardia and implantable cardioverter-defibrillator. Furthermore, this patient had a family history of SCD and AF. The missense variant E825D was identified in a paroxysmal AF patient with very early onset of disease at age 18. This patient also had AVNRT and several RAF procedures performed. The patient carrying the variant I999L had onset of paroxysmal AF at the age of 35 and also presented incomplete right bundle branch block. This patient also carries the variant L10P in SCN3B, as previously reported.32 The R1268Q variant were identified in two AF patients with onset of disease at age 23 (also had atrial flutter type II) and 31 (also had Incomplete Right Bundle Branch Block (IRBBB) and several DC conversions). The patient with the rare variant C1523Y was diagnosed with paroxysmal AF at age 30, and in another paroxysmal AF patient with onset of disease at age 28, we identified the variant R1588Q. This latter patient had an RFA procedure for AVNRT. Interestingly, four of the rare variant carriers (≈40%) have AVNRT, in addition to AF, suggesting that the AV-node, and perhaps the autonomic nerve system, could play an important role in the genesis of AF in these patients.
Electrophysiology
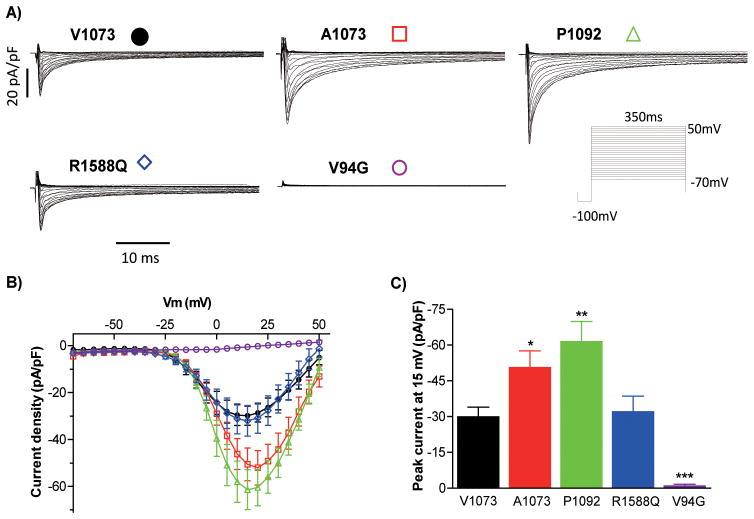
The electrophysiological properties of Nav1.8 V1073, used as reference, and four variants were investigated by whole-cell patch-clamping of Neuro-2A cells (Figure 2, and 3). We chose to analyze the V94G found in two unrelated patients and R1588Q variants based on the lowest variant frequency in the background population (not present in 2,000 non-AF Danish exomes, data not shown), thereby reducing the risk of investigating a random finding. At the time of variant selection for functional studies, the two variants were not found in the Exome Sequencing database (n= 5,400), but later appeared in one exome for each variant when 1,100 additional subjects were included in the database (n= 6,500). We also investigated the two non-synonymous common variants A1073 andP1092. Figure 2A illustrates representative whole-cell currents from the Neuro-2A cells expressing V1073 and variant Nav1.8 channels. Nav1.8 channels are activated by depolarizing potentials more positive than −15 mV, with a fast activating current peaking at +15 mV (Figure 2B). A part of the current is rapidly inactivated, while a long lasting current component of approximately 10 % of the peak current level persists (Figure 2A).
Figure 2.
Current recordings of SCN10A variants. A) Representative whole-cell current traces of Nav1.8, using V1073 as reference construct, and mutant channels. Currents were recorded following a voltage step protocol with 5 mV increments from −70 mV to +50 mV, preceded with a −100 mV step. B) Current-voltage (I-V) relationship. The current is normalized to cell capacitance to provide a measure of Na+ current density. C) Peak current density at 15mV, Two-way ANOVA with Bonferroni post-tests was applied to test for significant differences. Mean ± SEM values are presented in Table 4. B, C) n=13–15 for each group. Asterisks indicate the voltages at which the parameters were statistically different versus V1073 *p<0.05, **p<0.01 and ***p<0.001.
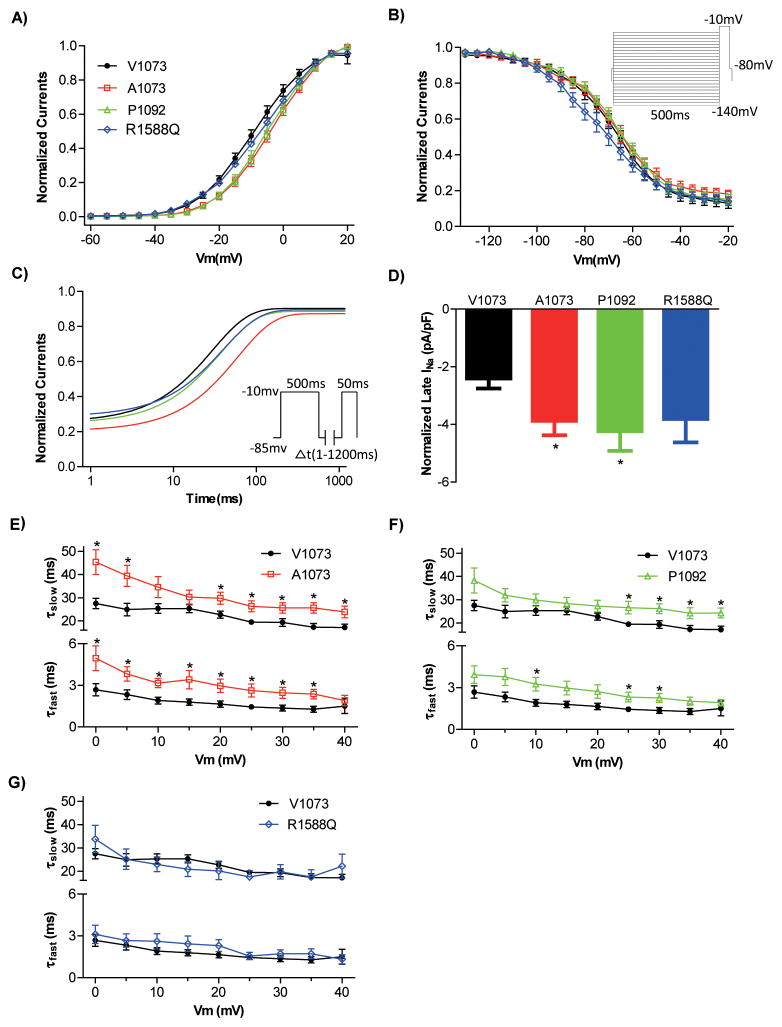
Figure 3.
Activation and inactivation properties of SCN10A variants. A) Steady-state activation curves. Activation properties were determined from I-V relationships by normalizing peak INa to driving force and maximal INa, and plotting normalized conductance vs. Vm. B) Steady-state inactivation curves. Protocol is shown in insert. Boltzmann curves were fitted to both steady-state activation and inactivation data. C) Time course of recovery from inactivation following a pre-potential protocol (insert) was fitted to a one-exponential equation: I/Imax =y0+A x exp(−t/τ), t is the time from the beginning of the test pulse, A and τ=fractional amplitude and time constant, respectively. D) Late (sustained) sodium current was normalized to cell size. E,F,G) Voltage dependence of inactivation time constants. The decaying phase of whole-cell current traces (as in Figure 2A) was fitted with 2 exponential equation: I/Imax =Af x exp(−t/τf) + As x exp(−t/τs). Lower and upper bundles of symbols indicate fast (τf) and slow (τs) time constant values respectively, n=8–11 for each group. A–D) Averaged values and the numbers of cells measured are presented in Table 4. Asterisks indicate the voltages at which the parameters were statistically different versus V1073 (*p<0.05).
The V94G-Nav1.8 channel does not conduct any current. The R1588Q variant showed peak current amplitude similar to reference channels, however, it had a faster time to peak, together with a more than 6 mV negative shift in the steady-state inactivation (V½,V1073 −68.0±1.8 mV, V½,R1588Q −74.4±2.5 mV, Figure 3B). As −74.4 mV is close to the resting membrane potential of both atrial cardiomyocytes and neurons, this shift would be expected to play a major role in the channel availability, reducing the number of available channels as compared to V1073 Nav1.8. The combined electrophysiological characterization of R1588Q would therefore be expected to result in a loss-of-function phenotype.
Compared with V1073 Nav1.8 (−29.9±4.1 pA/pF), the two common variants expressed larger peak currents (A1073, −50.6±7.0 pA/pF; P1092, −61.4±8.5 pA/pF). While the steady-state inactivation properties for A1073 and P1092 were not altered, the steady-state activation properties were shifted to more positive potentials for these two common variants (V½,V1073 1.6±1.3 mV, V½,A1073 7.1±1.3 mV, V½,P1092 6.4±1.5 mV)(Figure 3A). For A1073, the time-dependent recovery from inactivation was decelerated (Figure 3C) and the time-to-peak accelerated (Table 4). Both common variants have a slower current decay at a number of different potentials (Figure 3E and 3F). Interestingly, the absolute sustained current level was increased for both A1073 and P1092 (V1073, 2.4±0.3 pA/pF; A1073, 3.9±0.5 pA/pF; P1092, 4.2±0.7pA/pF) (Figure 3D). For both reference Nav1.8 channels and the two common variants the sustained current level is 7–8 % of the peak current at +15 mV. Hence, the increase in the absolute sustained current level of the variants is probably due to the overall increased activity of these variant channels. Together, we found a depolarized shift in voltage-dependence of steady-state activation on A1073 and P1092 and decelerated recovery from inactivation on the A1073 variant. However, these two common variants also induced dramatically larger peak-current amplitude, a slower current decay phase from inactivation, and a pronounced larger persistent current. Hence, the combined electrophysiological changes of these two common variants would result in gain-of-function phenotypes.
Table 4.
Electrophysiological characterization of SCN10A variants
| V1073 (n=15) | A1073 (n=14) | P1092 (n=13) | R1588Q (n=13) | |
|---|---|---|---|---|
| Peak current at 15 mV (pA/pF) | −29.9±4.1 (n=16) | −50.6±7* (n=13) | −61.4±8.5** (n=14) | −32.0±6.5 (n=12) |
| Steady-state inactivation | ||||
| V1/2 (mV) | −68.0± 1.8 | −67.6± 2.1 | −65.9± 1.4 | −74.4± 2.5* |
| K | 9.8± 1.2 (n=19) | 9.7± 1.6 (n=14) | 9.6±0.5 (n=12) | 11.6±0.8 (n=14) |
| Steady-state activation | ||||
| V1/2 (mV) | 1.6± 1.3 | 7.1±1.3* | 6.4± 1.5* | 5.5±2.4 |
| K | 8.1.± 0.4 (n=12) | 8.0± 0.4 (n=11) | 7.8± 0.4 (n=13) | 9.2± 0.9 (n=11) |
| Time course of recovery from inactivation at −85 mV (ms) | 33.6±5.6 (n=18) | 73.6±14.5* (n=14) | 46.2±5.4 (n=12) | 45.5±8.7 (n=15) |
| Time to Peak Current at 15 mV(ms) | 1.8±0.2 (n=11) | 1.3±0.1* (n=12) | 1.5±0.1 (n=13) | 1.3±0.1* (n=11) |
| Late INa at 15 mV (pA/pF) | −2.4± 0.3 | −3.9± 0.5* | −4.2±0.7* | −3.3±0.8 |
Data are presented as mean ± SEM. Two-way ANOVA combined with Bonferroni post-test was used to test for significant differences of Peak current at 15 mV. The other parameters were tested by Two-tailed Student’s t-test.
p<0.05 and
p<0.01 versus reference (V1073).
N: the numbers of cells measured; T: time constant; V1/2: midpoint potential; k: slope factor.
Discussion
In the present study, we found 10 rare missense SCN10A variants in 225 lone AF patients in which the minor allele frequencies were less than 0.5% in the ESP. Furthermore, we showed that the A1073 variant (rs6795970) increases the risk of AF. Functional characterization of the two rarest variants of rs6795970 (with the lowest MAF) found in lone AF revealed reduced activity of Nav1.8 while, conversely, the A1073 variant was found to increase activity of the channel.
The variant rs6800541, identified in previous GWAS studies, has been associated with PR-interval duration and is in close proximity to SCN10A.10 This variant also decreases the odds ratio for AF (OR=0.92, p=1.4×10−03).10 Others have tested the association of rs6795970, which is in high linkage disequilibrium with rs6800541, with AF and showed the opposite trend, however, the association was not statistically significant.10,11 In the present study, we found that the G allele of rs6795970 increases the risk of AF in both lone AF (OR=1.33, p=4.6×10−04) and general AF (OR=1.35, p=2.3×10−05, Table 3). Functional studies of A1073 revealed a number of altered biophysical parameters, with the greatest one being increased peak and sustained current as well as a slowing of fast inactivation. We therefore suggest that this variant has a gain-of-function phenotype, which seems to increase the risk of AF.
In lone AF patients, we identified 9 rare variants in SCN10A with MAF less than 0.5% in the ESP and one splice site donor variant in exon 4 (Table 1). We performed electrophysiological patch-clamp studies on the two rarest variants, V94G and R1588Q, which initially were found to be novel. Later, both variants were reported in ESP6500, but it should be noted that this is a nonselective database, where AF patients are not excluded. Since voltage-gated sodium channels are inactivated at voltage potentials close to the resting membrane potential, a negative shift in the steady-state inactivation, as seen for R1588Q, will result in a decreased availability of the channels. The V94G variant did not conduct any current. Hence, both tested variants found in lone AF patients have a loss-of-function phenotype. These data on the rare variants combined with the data on the A1073 common variant suggest that both gain and loss of functionality of the Nav1.8 current may be involved in the development of AF. This has previously also been reported for the Nav1.5 current 29,33–35.
The I999L variant is a novel variant; however, since the patient also has a SCNB3 variant suspected to be disease causing, we did not include functional data of this variant.32
Currently, the role of Nav1.8 in cardiac electrophysiology remains unclear. If the primary role of Nav1.8 channels is in the cardiomyocytes, both loss- and gain-of-function phenotypes could give rise to AF. This is analogous to what has been observed for Nav1.5 channels, where both decreased NaV1.5 current has been associated with AF via augmented propensity for conduction delay with a subsequent increased risk of re-entrant arrhythmias, and augmented current have been suggested to increase triggered activity.29,35,36 Another possibility is that the Nav1.8 peak current does not have a major impact, as it is quantitatively much smaller than the Nav1.5 peak current. But, since the sustained (late) Nav1.8 current is 20–50 fold higher than the corresponding Nav1.5 late current, this depolarizing current could have a significant impact on the action potential duration and refractory period, and thereby protect against AF.37
Given the sparse expression of Nav1.8 channels in cardiac myocytes, the association of SCN10A variants to AF could be mediated through neuronal input. One study has suggested that rs6795970 in SCN10A may modulate the ventricular heart rate response during AF through a modulation of the AV-node.38 The AV-node is highly innervated by parasympathetic nerve fibers where Nav1.8 is expressed.39–41 Recognition of disease mechanisms overlap between the AF and AV-nodal re-entry tachycardia and the fact that AVNRT may be the triggering factor for AF has been reported in several studies.42–45 Consistent with this notion, several of our paroxysmal AF patients with the rare variants have AVNRT (Table 1).
In AF patients, there has been substantial evidence of sympathetic tone-dependent AF.46 Changes in autonomic tone, also known as sympathovagal imbalance, are important triggers in some forms of paroxysmal AF and also in the generation and maintenance of persistent AF.41–43 With the expression of Nav1.8 channels in vagal fibers and their absence in sympathetic fibers, it is possible that the observed effects on Nav1.8 function of the different variants alters the sympathovagal balance.19,20,22
We were able to examine 3 families for co-segregation of SCN10A variants identified in the respective probands (Table 1), but none of the family members diagnosed with AF carried the variant of the proband. It is, however, not surprising that a monogenetic segregation pattern is absent since only a few reports have shown familial co-segregation of rare variants in AF.47 In line with our study, it has been suggested that variation in the SCN5A, is not the main cause of familial AF.34 Whether a person with a rare variant develops AF probably depends on both the genetic background and environmental factors. Thus, the rare loss-of-function variants found in our study should most likely be regarded as important modifiers for the genesis of AF.
Perspective
In summary, this study reveals a correlation between variations in SCN10A and AF. The results thereby support the notion of SCN10A being important in cardiac physiology as genetic variations now have been found to be implicated in cardiac conduction, Brugada syndrome, and AF. The fact that SCN10A variations could play a promoting role in lone AF, as well as other types of AF, highlights the importance of further studies on the cellular and electrophysiological factors involved in the development of AF. Hence, our results further contribute to understanding the complexity of cardiac electrophysiology and suggest that SCN10A genotyping in the future could improve risk prediction.
Supplementary Material
Acknowledgments
We wish to thank Lasse Skibsbye, Mina Ghasemilee and Anne Katrine Kastberg for technical assistance. We also wish to thank LuCamp, The Lundbeck Foundation Centre for Applied Medical Genomics in Personalized Disease Prediction, Prevention and Care for providing Danish population controls, as well as Christian Theil Have for bioinformatics support. For the LuCamp partner list, see the supplementary materials.
Funding Sources: This work was supported by the Research Committee of Rigshospitalet (University of Copenhagen), The Danish Heart Foundation, The John and Birthe Meyer foundation, The Arvid Nilsson Foundation, The Novo Nordisk Foundation, Fondsbørsvekselerer Henry Hansen’s Legat, Director Ib Henriksens Foundation, NIH grants R01HL092577, R01HL104156, K24HL105780, and an AHA Established Investigator Award 13EIA14220013.
Footnotes
Conflict of Interest Disclosures: Javad Jabbari is employed at LEO Pharma A/S. Anders G. Holst and Bo Liang are employed at Novo Nordisk A/S, Morten Grunnet at Lundbeck A/S.
References
- 1.Kim MH, Johnston SS, Chu B-C, Dalal MR, Schulman KL. Estimation of total incremental health care costs in patients with atrial fibrillation in the United States. Circ Cardiovasc Qual Outcomes. 2011;4:313–320. doi: 10.1161/CIRCOUTCOMES.110.958165. [DOI] [PubMed] [Google Scholar]
- 2.Magnani JW, Rienstra M, Lin H, Sinner MF, Lubitz SA, McManus DD, et al. Atrial Fibrillation Current Knowledge and Future Directions in Epidemiology and Genomics. Circulation. 2011;124:1982–1993. doi: 10.1161/CIRCULATIONAHA.111.039677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lloyd-Jones D, Adams RJ, Brown TM, Carnethon M, Dai S, Simone GD, et al. Executive Summary: Heart Disease and Stroke Statistics—2010 Update A Report From the American Heart Association. Circulation. 2010;121:948–954. doi: 10.1161/CIRCULATIONAHA.109.192666. [DOI] [PubMed] [Google Scholar]
- 4.Tucker NR, Ellinor PT. Emerging directions in the genetics of atrial fibrillation. Circ Res. 2014;114:1469–1482. doi: 10.1161/CIRCRESAHA.114.302225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fatkin D, Otway R, Vandenberg JI. Genes and Atrial Fibrillation: A New Look at an Old Problem. Circulation. 2007;116:782–792. doi: 10.1161/CIRCULATIONAHA.106.688889. [DOI] [PubMed] [Google Scholar]
- 6.Cheng S, Keyes MJ, Larson MG, McCabe EL, Newton-Cheh C, Levy D, et al. Long-term Outcomes in Individuals With Prolonged PR Interval or First-Degree Atrioventricular Block. JAMA. 2009;301:2571–2577. doi: 10.1001/jama.2009.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nielsen JB, Pietersen A, Graff C, Lind B, Struijk JJ, Olesen MS, et al. Risk of atrial fibrillation as a function of the electrocardiographic PR interval: results from the Copenhagen ECG Study. Heart Rhythm. 2013;10:1249–1256. doi: 10.1016/j.hrthm.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Oberti C, Wang L, Li L, Dong J, Rao S, Du W, et al. Genome-Wide Linkage Scan Identifies a Novel Genetic Locus on Chromosome 5p13 for Neonatal Atrial Fibrillation Associated With Sudden Death and Variable Cardiomyopathy. Circulation. 2004;110:3753–3759. doi: 10.1161/01.CIR.0000150333.87176.C7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambers JC, Zhao J, Terracciano CMN, Bezzina CR, Zhang W, Kaba R, et al. Genetic variation in SCN10A influences cardiac conduction. Nat Genet. 2010;42:149–152. doi: 10.1038/ng.516. [DOI] [PubMed] [Google Scholar]
- 10.Pfeufer A, van Noord C, Marciante KD, Arking DE, Larson MG, Smith AV, et al. Genome-wide association study of PR interval. Nat Genet. 2010;42:153–159. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holm H, Gudbjartsson DF, Arnar DO, Thorleifsson G, Thorgeirsson G, Stefansdottir H, et al. Several common variants modulate heart rate, PR interval and QRS duration. Nat Genet. 2010;42:117–122. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 12.Denny JC, Ritchie MD, Crawford DC, Schildcrout JS, Ramirez AH, Pulley JM, et al. Identification of Genomic Predictors of Atrioventricular Conduction Using Electronic Medical Records as a Tool for Genome Science. Circulation. 2010;122:2016–2021. doi: 10.1161/CIRCULATIONAHA.110.948828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butler AM, Yin X, Evans DS, Nalls MA, Smith EN, Tanaka T, et al. Novel Loci Associated With PR Interval in a Genome-Wide Association Study of 10 African American Cohorts. Circ Cardiovasc Genet. 2012;5:639–646. doi: 10.1161/CIRCGENETICS.112.963991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bezzina CR, Barc J, Mizusawa Y, Remme CA, Gourraud J-B, Simonet F, et al. Common variants at SCN5A-SCN10A and HEY2 are associated with Brugada syndrome, a rare disease with high risk of sudden cardiac death. Nat Genet. 2013;45:1044–1049. doi: 10.1038/ng.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francis J, Antzelevitch C. Atrial Fibrillation and Brugada Syndrome. J Am Coll Cardiol. 2008;51:1149–1153. doi: 10.1016/j.jacc.2007.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ritchie MD, Denny JC, Zuvich RL, Crawford DC, Schildcrout JS, Bastarache L, et al. Genome- and phenome-wide analyses of cardiac conduction identifies markers of arrhythmia risk. Circulation. 2013;127:1377–1385. doi: 10.1161/CIRCULATIONAHA.112.000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukuoka T, Kobayashi K, Yamanaka H, Obata K, Dai Y, Noguchi K. Comparative study of the distribution of the alpha-subunits of voltage-gated sodium channels in normal and axotomized rat dorsal root ganglion neurons. J Comp Neurol. 2008;510:188–206. doi: 10.1002/cne.21786. [DOI] [PubMed] [Google Scholar]
- 18.Renganathan M, Cummins TR, Waxman SG. Contribution of Na(v)1.8 sodium channels to action potential electrogenesis in DRG neurons. J Neurophysiol. 2001;86:629–640. doi: 10.1152/jn.2001.86.2.629. [DOI] [PubMed] [Google Scholar]
- 19.Ringkamp M, Johanek LM, Borzan J, Hartke TV, Wu G, Pogatzki-Zahn EM, et al. Conduction properties distinguish unmyelinated sympathetic efferent fibers and unmyelinated primary afferent fibers in the monkey. PLoS ONE. 2010;5:e9076. doi: 10.1371/journal.pone.0009076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gautron L, Sakata I, Udit S, Zigman JM, Wood JN, Elmquist JK. Genetic tracing of Nav1.8-expressing vagal afferents in the mouse. J Comp Neurol. 2011;519:3085–3101. doi: 10.1002/cne.22667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang T, Atack TC, Stroud DM, Zhang W, Hall L, Roden DM. Blocking Scn10a channels in heart reduces late sodium current and is antiarrhythmic. Circ Res. 2012;111:322–332. doi: 10.1161/CIRCRESAHA.112.265173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verkerk AO, Remme CA, Schumacher CA, Scicluna BP, Wolswinkel R, de Jonge B, et al. Functional NaV1.8 Channels in Intracardiac Neurons: The Link Between SCN10A and Cardiac Electrophysiology. Circ Res. 2012;111:333–343. doi: 10.1161/CIRCRESAHA.112.274035. [DOI] [PubMed] [Google Scholar]
- 23.Sotoodehnia N, Isaacs A, de Bakker PIW, Dörr M, Newton-Cheh C, Nolte IM, et al. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat Genet. 2010;42:1068–1076. doi: 10.1038/ng.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Facer P, Punjabi PP, Abrari A, Kaba RA, Severs NJ, Chambers J, et al. Localisation of SCN10A gene product Na(v)1.8 and novel pain-related ion channels in human heart. Int Heart J. 2011;52:146–152. doi: 10.1536/ihj.52.146. [DOI] [PubMed] [Google Scholar]
- 25.London B. Whither Art Thou, SCN10A, and What Art Thou Doing? Circ Res. 2012;111:268–270. doi: 10.1161/CIRCRESAHA.112.275032. [DOI] [PubMed] [Google Scholar]
- 26.Jabbari J, Olesen MS, Holst AG, Nielsen JB, Haunso S, Svendsen JH. Common polymorphisms in KCNJ5 [corrected] are associated with early-onset lone atrial fibrillation in Caucasians. Cardiology. 2011;118:116–120. doi: 10.1159/000323840. [DOI] [PubMed] [Google Scholar]
- 27.Olesen MS, Jabbari J, Holst AG, Nielsen JB, Steinbrüchel DA, Jespersen T, et al. Screening of KCNN3 in patients with early-onset lone atrial fibrillation. Europace. 2011;13:963–967. doi: 10.1093/europace/eur007. [DOI] [PubMed] [Google Scholar]
- 28.Glümer C, Jørgensen T, Borch-Johnsen K Inter99 study. Prevalences of diabetes and impaired glucose regulation in a Danish population: the Inter99 study. Diabetes Care. 2003;26:2335–2340. doi: 10.2337/diacare.26.8.2335. [DOI] [PubMed] [Google Scholar]
- 29.Olesen MS, Yuan L, Liang B, Holst AG, Nielsen N, Nielsen JB, et al. High Prevalence of Long QT Syndrome–Associated SCN5A Variants in Patients With Early-Onset Lone Atrial Fibrillation. Circ Cardiovasc Genet. 2012;5:450–459. doi: 10.1161/CIRCGENETICS.111.962597. [DOI] [PubMed] [Google Scholar]
- 30.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giudicessi JR, Kapplinger JD, Tester DJ, Alders M, Salisbury BA, Wilde AAM, et al. Phylogenetic and physicochemical analyses enhance the classification of rare nonsynonymous single nucleotide variants in type 1 and 2 long-QT syndrome. Circ Cardiovasc Genet. 2012;5:519–528. doi: 10.1161/CIRCGENETICS.112.963785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olesen MS, Jespersen T, Nielsen JB, Liang B, Møller DV, Hedley P, et al. Mutations in sodium channel β-subunit SCN3B are associated with early-onset lone atrial fibrillation. Cardiovasc Res. 2011;89:786–793. doi: 10.1093/cvr/cvq348. [DOI] [PubMed] [Google Scholar]
- 33.Darbar D, Kannankeril PJ, Donahue BS, Kucera G, Stubblefield T, Haines JL, et al. Cardiac sodium channel (SCN5A) variants associated with atrial fibrillation. Circulation. 2008;117:1927–1935. doi: 10.1161/CIRCULATIONAHA.107.757955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ellinor PT, Nam EG, Shea MA, Milan DJ, Ruskin JN, MacRae CA. Cardiac sodium channel mutation in atrial fibrillation. Heart Rhythm. 2008;5:99–105. doi: 10.1016/j.hrthm.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 35.Olesen MS, Nielsen MW, Haunsø S, Svendsen JH. Atrial fibrillation: the role of common and rare genetic variants. Eur J Hum Genet. 2014;22:297–306. doi: 10.1038/ejhg.2013.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lemoine MD, Duverger JE, Naud P, Chartier D, Qi XY, Comtois P, et al. Arrhythmogenic left atrial cellular electrophysiology in a murine genetic long QT syndrome model. Cardiovasc Res. 2011;92:67–74. doi: 10.1093/cvr/cvr166. [DOI] [PubMed] [Google Scholar]
- 37.Allessie MA, Bonke FI, Schopman FJ. Circus movement in rabbit atrial muscle as a mechanism of tachycardia. III. The “leading circle” concept: a new model of circus movement in cardiac tissue without the involvement of an anatomical obstacle. Circ Res. 1977;41:9–18. doi: 10.1161/01.res.41.1.9. [DOI] [PubMed] [Google Scholar]
- 38.Delaney JT, Muhammad R, Shi Y, Schildcrout JS, Blair M, Short L, et al. Common SCN10A variants modulate PR interval and heart rate response during atrial fibrillation. Europace. 2014;16:485–490. doi: 10.1093/europace/eut278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiou C-W, Eble JN, Zipes DP. Efferent Vagal Innervation of the Canine Atria and Sinus and Atrioventricular Nodes The Third Fat Pad. Circulation. 1997;95:2573–2584. doi: 10.1161/01.cir.95.11.2573. [DOI] [PubMed] [Google Scholar]
- 40.Martin P. The influence of the parasympathetic nervous system on atrioventricular conduction. Circ Res. 1977;41:593–599. doi: 10.1161/01.res.41.5.593. [DOI] [PubMed] [Google Scholar]
- 41.Quan KJ, Lee JH, Van Hare GF, Biblo LA, Mackall JA, Carlson MD. Identification and characterization of atrioventricular parasympathetic innervation in humans. J Cardiovasc Electrophysiol. 2002;13:735–739. doi: 10.1046/j.1540-8167.2002.00735.x. [DOI] [PubMed] [Google Scholar]
- 42.Palma EC, Ferrick KJ, Gross JN, Kim SG, Fisher JD. Transition From Atrioventricular Node Reentry Tachycardia to Atrial Fibrillation Begins in the Pulmonary Veins. Circulation. 2000;102:937–937. doi: 10.1161/01.cir.102.8.937. [DOI] [PubMed] [Google Scholar]
- 43.Hurwitz JL, German LD, Packer DL, Wharton JM, McCarthy EA, Wilkinson WE, et al. Occurrence of atrial fibrillation in patients with paroxysmal supraventricular tachycardia due to atrioventricular nodal reentry. Pacing Clin Electrophysiol. 1990;13:705–710. doi: 10.1111/j.1540-8159.1990.tb02094.x. [DOI] [PubMed] [Google Scholar]
- 44.Brugada J, Mont L, Matas M, Navarro-López F. Atrial fibrillation induced by atrioventricular nodal reentrant tachycardia. Am J Cardiol. 1997;79:681–682. doi: 10.1016/s0002-9149(96)00842-9. [DOI] [PubMed] [Google Scholar]
- 45.Sauer WH, Alonso C, Zado E, Cooper JM, Lin D, Dixit S, et al. Atrioventricular Nodal Reentrant Tachycardia in Patients Referred for Atrial Fibrillation Ablation Response to Ablation That Incorporates Slow-Pathway Modification. Circulation. 2006;114:191–195. doi: 10.1161/CIRCULATIONAHA.106.621896. [DOI] [PubMed] [Google Scholar]
- 46.Arora R. Recent insights into the role of the autonomic nervous system in the creation of substrate for atrial fibrillation: implications for therapies targeting the atrial autonomic nervous system. Circ Arrhythm Electrophysiol. 2012;5:850–859. doi: 10.1161/CIRCEP.112.972273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Darbar D, Herron KJ, Ballew JD, Jahangir A, Gersh BJ, Shen W-K, et al. Familial atrial fibrillation is a genetically heterogeneous disorder. J Am Coll Cardiol. 2003;41:2185–2192. doi: 10.1016/s0735-1097(03)00465-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.