Abstract
Inflammatory bowel disease (IBD) is characterised by an inappropriate chronic immune response against resident gut microbes. This may be on account of distinct changes in the gut microbiota termed as dysbiosis. The role of fungi in this altered luminal environment has been scarcely reported. We studied the fungal microbiome in de-novo paediatric IBD patients utilising next generation sequencing and compared with adult disease and normal controls. We report a distinct difference in fungal species with Ascomycota predominating in control subjects compared to Basidiomycota dominance in children with IBD, which could be as a result of altered tolerance in these patients.
Keywords: Fungal microbiota, Paediatric inflammatory bowel disease, Gut microbiota
1. Introduction
Crohn's disease (CD) and ulcerative colitis (UC) are two types of inflammatory bowel disease (IBD) of unknown aetiology that result in significant morbidity and health expenditure. Recent data suggests that the prevalence of IBD in general practice in the UK has exponentially increased to around 400 per 100,000 [1]. The incidence rate of juvenile onset IBD has risen by nearly 30% in Scotland over the last twenty years with a concerning decline in age at presentation [2]. Paediatric IBD is often considered the ‘purest’ form of disease without many extraneous influences of adult behaviour e.g. smoking or disease comorbidity. Studying the aetiological factors of this form of the disease is perhaps more likely to further our understanding of the initiating events of these debilitating conditions [3].
The current paradigm of IBD pathogenesis suggests an exaggerated immune response against the luminal microbiota in genetically susceptible individuals. The risk-associated genes are critical in the innate immune system that recognises and distinguishes pathogens from commensal microorganisms, are involved in the clearance of pathogens or pertain to the regulation of adaptive immunity [4]. Crucially, a significant proportion of individuals with IBD do not have any evidence of heritable genetic susceptibility indicating that environmental triggers play an important role in the pathogenesis of IBD [5]. The gastrointestinal microbiota has come to the fore in efforts to explain this “pathogenesis gap”. The normal gastrointestinal tract harbours a complex community of commensal microbes that are critical to normal physiological functioning. In fact, the microbial gene set within the gut lumen is 150 times larger than the entire human gene complement [6]. Traditional culture methods have been unsuccessful in characterising the entire microbiota with many microbes going undetected [7]. Newer molecular methods and next generation sequence analysis in particular have vastly enhanced the yield of bacteria and fungi that can be identified within the gut [8–10].
In contrast to luminal bacteria, the exact role of colonising fungi and their pathogenic potential has not been fully explored. From metagenomic studies it has been established that 99.1% of the genetic catalogues from the lumen are of bacterial origin whereas fungal DNA accounts for around 0.02% of the entire mucosa-associated microbiota [6,8,11,12]. It is difficult to ascertain if changes to this component have an impact on the final genesis of inflammation. However, other aspects of the immune response may also explain changes that occur in the fungal microbiome. Anti-Saccharomyces cerevisiae antibodies (ASCA) have been found to have a role in diagnosis, disease phenotype and prognosis, more commonly found in CD patients compared to UC patients and healthy controls [13,14]. These patients have also demonstrated a decreased tolerance to S. cerevisiae [15]. It has now been shown that CD patients with pattern recognition receptor and autophagy gene variants, but not those with genetic variants of IL-23 signalling, were more likely to develop ASCA antibodies [16]. Several questions still remain unanswered in the aetiopathogenesis of IBD, especially the role of intestinal fungi in initiating/driving the abnormal inflammation that is characteristic of IBD. Fungi have received scant attention in the literature of IBD microbiology to date and warrant specific and targeted consideration.
2. Methods
2.1. Patient recruitment, biopsy collection and processing
The 25 children with IBD selected for this study were part of a cohort of children who had been recruited to the Bacteria In Scottish Children Undergoing Investigation before Treatment (BISCUIT) study [17]. The inclusion and exclusion criteria and the modality of assessment of these patients have been reported previously. In short, these children were stringently evaluated, and only those with documented new onset IBD who had not received and IBD treatment at any time or systemic antibiotics in the 3 months prior to their colonoscopy. The comparator groups comprised of 12 control paediatric subjects from the same study, two adult patients with a normal colonoscopy and two adult patients with ulcerative colitis (UC), who were also part of a previously reported study [18].
Biopsies were taken from a single site, from the distal colon (rectum/sigmoid), and in the case of IBD subjects macroscopic inflammation had to be present at the site. 2-3 Biopsies were collected using standard endoscopic forceps into a sterile 1.5 ml Eppendorf container and placed immediately onto ice before transfer to −80 °C storage.
DNA extraction of mucosal biopsies was performed using the commercially available Qiagen QIAamp Mini kit (Qiagen, Crawley, UK) with minor modifications [18]. Ethical approval was granted by North of Scotland Research Ethics Service on behalf of all participating centres and written informed consent was obtained from the adult subjects and from the parents of the paediatric patients. Informed assent was also obtained from older children who were deemed capable of understanding the nature of the study. The BISCUIT study is publically registered on the United Kingdom Clinical Research Network Portfolio (9633).
2.2. Preparation of samples for pyrosequencing
Biopsy DNA was quantified by Nanodrop mass spectrophotometry before dilution to 25 ng/μl. Initial PCR amplification was undertaken with FastStart High Fidelity PCR reagents (Roche, Penzberg, Germany) utilising a per-reaction mix containing 50 ng DNA template. The 18S rDNA primers were taken from Ott et al., [7]. No identifier was added to the reverse primer. Hence the 540 bp PCR product was flanked by a 40 bp fusion primer/multiplex identifier sequence at the forward end and a 30 bp fusion primer at the reverse end (Table 1). After confirmation of successful PCR amplification, products were purified as per the recommended Agencourt AMPure (Beckman Coulter, Beverly, MA, USA) purification method for 454 sequencing and sequenced on Roche 454 Titanium (454 life sciences, Branford, CT, USA) by NewGene (Newcastle, UK).
Table 1.
List of fungal 18S rRNA specific Next Generation sequencing primers.

2.3. Bioinformatic and statistical analysis
Data analysis of the 454 sequence data was performed using QIIME version 1.3.0 workflow for 18S data using the Qiime compatible version of the silva-104 release (downloaded from http://www.arb-silva.de/download/archive/qiime/) for template based alignment and taxonomic assignment: Sequences were binned according to sample-specific barcode, denoised (fast denoiser) and clustered with uclust into de-novo operational taxonomic units (OTUs) at 97% sequence similarity. Representative sequences were picked for each OTU and aligned with Pynast using the Silva template alignment core_Silva_aligned.fasta and chimera check was performed with ChimeraSlayer for removal of potential chimeric sequences. Taxonomy assignment of each OTU was performed by blasting against the taxonomic mapping file Silva_taxa_mapping_104set_97_otus.txt followed by construction of OTU tables at different taxonomic levels. OTU tables were rarefied at 3000 and results were plotted at phylum and genus level. All novel sequence data was deposited at NCBI's Sequence Read Archive under accession number PRJEB7438.
2.4. Quantitative real time PCR
Quantitative PCR was done to estimate the amount of fungal rDNA present in all seven paediatric biopsy samples (6 IBD and 1 control) and three adult biopsy control samples. The 18S region was amplified from 50 ng of sample DNA using methods described previously [8]. Standard curve was generated from 10-fold serial dilutions of amplified fungal 18S rRNA genes from Candida albicans strain.
3. Results
A total of 37 paediatric patient colonic biopsies were included within the study. This included 12 control patients, 13 with CD and 12 with UC. Fungal DNA was amplifiable from 8 patient samples, 6 children with a diagnosis of IBD – 4 with Crohn's disease, 2 children with ulcerative colitis and 2 children without IBD.
Fungal diversity was assessed in all paediatric samples alongside four adult samples to act as a further comparison. The adult samples comprised 2 patients with ulcerative colitis and 2 patients with negative colonoscopy findings. Fungal DNA could be amplified in one of the adult patients with de-novo UC and both the adult controls.
Pyrosequencing generated ∼90,000 individual sequencing reads in total with a mean yield of 5245 reads per sample after bioinformatic processing but before rarefaction. The minimum read score was BISCUIT 27 with 3385 reads therefore rarefaction analysis was performed at a threshold of 3000 reads to allow subject-to-subject comparison. Two patient samples (BISCUIT 64 and 1UC15) were discarded from further analysis as sequence data was confirmed to be Eukaryotic but could not subsequently be matched to fungi. Almost all sequences from both patients were identical and matched only to uncultured ‘Banisveld eukaryote’ (∼72%), identified from Banisveld water and thought to relate to a distinct phylum [19].
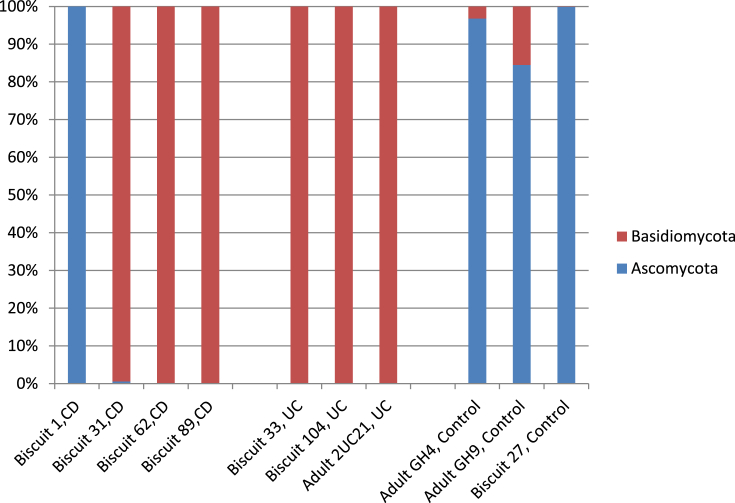
Comparisons were made at both phylum and genus levels (Figs. 1 and 2). Phylum level analysis indicated that fungal sequences almost exclusively belonged to the Ascomycota and Basidiomycota phyla (Fig. 1). The most abundant phylum was Basidiomycota which was responsible for 100% of detectable fungal sequences in 5 of the 6 paediatric IBD patient samples (3/4 CD and 2/2 UC). In comparison the phylum data from the three adult samples showed that the two healthy control samples comprised >80% Ascomycota sequences whilst the UC patient contained exclusively Basidiomycota sequences.
Fig. 1.
Individual patient phylum-level diversity assessment as stacked bars.
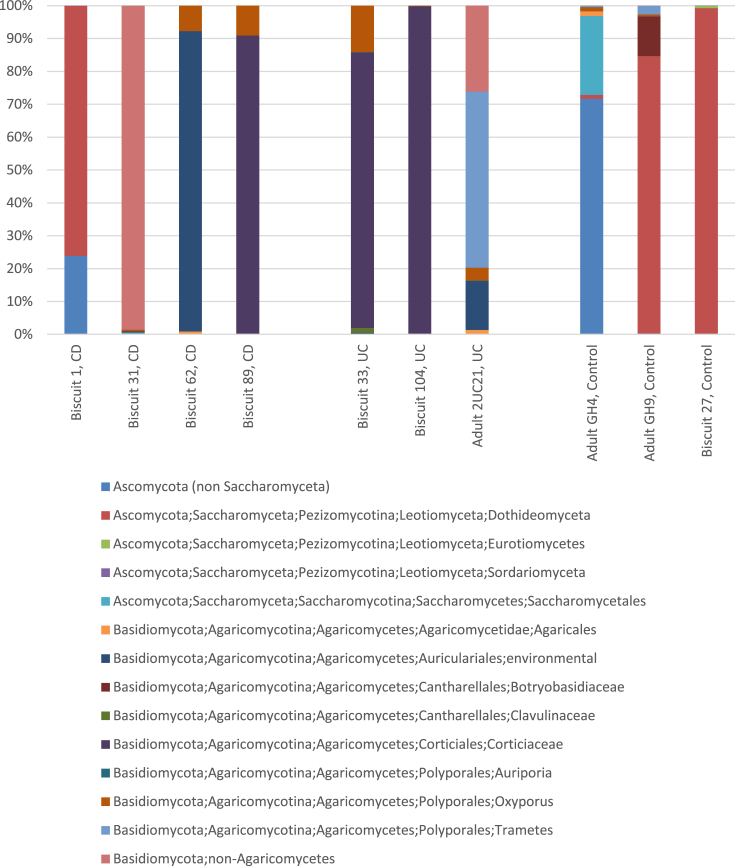
Fig. 2.
Individual patient genus-level diversity assessment as stacked bars.
Genus level analysis was undertaken to compare the Basidiomycota sequences between the paediatric IBD patient samples (BISCUIT 31, 62, 89, 33, 104; Fig. 2). The number of genera detected in paediatric samples varied from 3 to 8 genera and showed that both UC patients and one of the CD patient samples were predominated by Corticiales sequences (Fig. 2). Adult samples contained between 7 and 9 genera. The Corticiales are one of the most ecologically diverse groups of Basidiomycota containing saprobes, plant and fungal pathogens, and lichens. The other 2 CD patients were predominated by an uncultured Basidiomycota sequence (BISCUIT 31) and an Auriculariales sequence (BISCUIT 62). The remaining two paediatric patient samples (BISCUIT 1 (CD) and BISCUIT 27 (control)) were heavily represented by Dothideomyceta species, although the CD patient also contained some unclassified Ascomycota sequences (∼24%). When genus level analysis was explored within the adult samples, there was representation from more genera than in the paediatric samples. There was no similarity between individual diversity profiles; the control patients overlapped in containing Dothideomyceta species with adult GH4 also containing unidentified Ascomyceta, Saccharomycetales, Agaricales and Oxyporus. Adult GH9 also contained Botryobasidiaceae and Trametes. Patient 2UC21 (adult de-novo UC) contained Agaricales, Auriculariales, Oxyporus, Trametes and uncultured environmental Basidiomycota species.
Quantitative PCR to document relative fungal load was performed on all fungal positive biopsy samples and the results are summarised in Supplementary Table 2. Five out of the seven paediatric samples showed much lower fungal load as compared to the adult samples. The two samples which showed comparable results were both from patients with CD (BISCUIT1 and BISCUIT89).
4. Discussion
This study elucidates the fungal microbiome or mycobiome in de-novo paediatric IBD patients. It is quite interesting to note that an amplified fungal PCR product was available for only 8 of the 37 paediatric subjects. This could potentially be due to the DNA extraction method, which was optimised for bacterial DNA extraction. The extraction protocol did not subject biopsy samples to mechanical disruption, which is recommended for some fungal species. Nevertheless, a variety of fungal species were identified. This cohort has been subject to an extensive (and successful) bacterial diversity analysis by next-generation sequencing which has been reported elsewhere, and the relative paucity of fungi in this cohort is indeed a novel finding [20]. It is possible that aspects of fungal diversity have therefore been under-reported within this study due to these limitations, however biopsy samples from both the paediatric and adult cohorts were processed in the same way therefore comparisons made between the various study groups reflects genuine differences. It is also possible that the low fungal diversity reported in this study is a confounder of bowel preparation prior to colonoscopy, which all recruits were subjected to. Whilst this may well be the case, it has not impacted to a similar degree on reported bacterial diversity in the same cohort [20]. It would also be difficult to ethically justify accessing the colonic mucosa for research sampling in children without adequate bowel preparation, hence these children were undergoing colonoscopy for diagnostic purposes and bowel preparation was therefore essential.
The evolution of the fungal microbiome from childhood to adulthood is not known, but it can be surmised that with increased and varied dietary exposure, adults might have a greater quantity and diversity of fungi in their lumen. This is clearly demonstrated with all adults in this study showing higher fungal loads than the majority of children. An increased diversity was demonstrated in the two adult controls that were studied. Clearly a larger adult study is required to support this finding.
The single paediatric CD recruit with an Ascomycota predominance was clinically reviewed in relation to the other IBD recruits in this study with respect to any critical differences that might explain this finding. All the CD biopsies amplified in this study were taken from macroscopically and microscopically inflamed sites and from recruits with granulomatous changes somewhere in their gastrointestinal tract, though not necessarily at the same site as sampled. Perhaps intriguingly, BISCUITs 31, 62 and 89 (Basidiomycota-predominant) had endoscopic evidence of aphthous ulceration at the site sampled for microbial analysis whereas BISCUIT 1 had inflamed mucosa with erythema and mucosal breaks but no ulcers, though granulomata and chronic inflammation were evident on histology. Bacterial diversity assessment from BISCUIT 1 was not dissimilar at phylum-level to the other CD recruits [20]. Clearly the small number of cases represented in this study, and single incidence of an absence of aphthous ulceration and coincidental Ascomycota predominance prevent any firm conclusion being drawn, but the association of microbial/fungal changes with endoscopic/microscopic disease should be a direction for further research.
The current paradigm in the pathogenesis of IBD suggests a perturbation in the relationship of the host innate immunity and the resident luminal gut microbiota. This study has shown a distinct dichotomy in the fungal microbiota between control patients and patients with IBD with a predominance of Ascomycota sequences (>80% of sequences in all patients) in the former group whilst a majority of IBD patients (6/7) contained exclusively members from the Basidiomycota phylum. Nevertheless, it is acknowledged that this is a small study and further studies are needed to validate these findings. The biopsy samples from these patients were assiduously collected prior to the institution of immunosuppressive treatment and without the co-administration of antibiotics and probably therefore represents the native condition as accurately as ethically permissible in both cases and controls. This stringency in case selection is critical in the assessment of the fungal microbiome. One of the key members of Ascomycetes, Candida is a normal commensal in the gut, which has been documented to overgrow in patients treated with antibiotics [21]. Therefore this shift from the Ascomycota-predominant microbiota in normal subjects to a distinctly different fungal spectrum with predominance of Basidomycetes in patients with de-novo IBD without the conflicting influence of immunosuppression or antibiotics might have pathogenic relevance.
Fungal DNA accounts for around 0.02% of the entire mucosa-associated microbiota as assessed from quantitative analyses from mucosal biopsies [8]. The fact that only 8 out of the paediatric cohort of 37 patients that were included in the previously published bacterial diversity study were positive for fungal PCR and could therefore be included in this study, supports the challenge of assessing fungal diversity within mucosal samples. The finding of distinctly lower mucosal fungal load in the paediatric cohort could partly explain this difficulty. A recent fungal mycobiome analysis of healthy subject faecal samples demonstrated positivity in all subject samples tested (around 100 subjects) finding 66 genera, with generally mutually exclusive presence of either the phyla Ascomycota or Basiodiomycota [22]. The difference in methodology of this study, which targeted the ITS region and our study and that by Ott et al that utilised amplification of the 18S rRNA needs to be acknowledged. Despite this, the findings reflects the data from our study in terms of phylum diversity on an individual basis. It also potentially highlights the fact that most fungal species within the gut are not associated with the mucosa, remaining predominantly within the luminal contents and most likely having limited interaction with the host. We maintain that in terms of clinical relevance, assessing diversity within fungal species that are associated with mucosal tissues, especially when assessing diversity in relation to IBD, a mucosal disease, is the most appropriate sample choice.
It is difficult to ascertain if changes to the fungal component have an impact on the final genesis of inflammation. Other aspects of the immune response may however explain changes that occur in the fungal microbiome. There has been plenty of interest in the role of one of the key members of the phylum Ascomycota, namely S. cerevisiae. The role of Anti-S. cerevisiae antibodies (ASCA) has been found to be an important phenotypic determinant, especially in CD patients [13]. These antibodies may be as a result of loss of tolerance against this fungus or another member of the phylum Ascomycota, perhaps C. albicans [23]. If the loss of tolerance hypothesis can be extended to the Ascomycota phylum it may explain why there is less representation in IBD patients and a relative shift towards Basidomycetes. Our study does not have the strength of numbers to definitely prove this hypothesis but it indicates that a radical shift occurs in the fungal microbiota of patients with IBD as opposed to controls, however with the caveat that fungal DNA amplification and diversity assessment appears more limited than conventional bacterial approaches. The initiating event in IBD is still a matter of debate and it is tantalising to suggest that this may in some cases be an inappropriate response to a fungus.
Conflict of interest
None.
Acknowledgements
We are grateful for the expertise of our sequencing provider NewGene. We appreciate the generosity of the families who freely gave their time and samples to make this study possible and the theatre staff of all centres who allowed time for sample collection during busy endoscopy lists. This work was funded by a Clinical Academic Training Fellowship from the Chief Scientist Office in Scotland (CAF/08/01) which also funded the salary of RH, the Broad Medical Research programme and a project grant from NHS Grampian Endowments. The Yorkhill IBD team is generously supported by the Catherine McEwan Foundation and the Yorkhill IBD fund. RKR is supported by an NHS research Scotland fellowship. RKR has received support from a Medical Research Council (MRC) patient research cohorts initiative grant (G0800675) for PICTS.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Stone M.A., Mayberry J.F., Baker R. Prevalence and management of inflammatory bowel disease: a cross-sectional study from central England. Eur J Gastroenterol Hepatol. 2003;15:1275–1280. doi: 10.1097/00042737-200312000-00004. [DOI] [PubMed] [Google Scholar]
- 2.Armitage E., Drummond H.E., Wilson D.C., Ghosh S. Increasing incidence of both juvenile-onset Crohn's disease and ulcerative colitis in Scotland. Eur J Gastroenterol Hepatol. 2001;13:1439–1447. doi: 10.1097/00042737-200112000-00007. [DOI] [PubMed] [Google Scholar]
- 3.Hansen R., Thomson J.M., El-Omar E.M., Hold G.L. The role of infection in the aetiology of inflammatory bowel disease. J Gastroenterol. 2010;45:266–276. doi: 10.1007/s00535-009-0191-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jostins L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henderson P., Satsangi J. Genes in inflammatory bowel disease: lessons from complex diseases. Clin Med. 2011;11:8–10. doi: 10.7861/clinmedicine.11-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckburg P.B., Bik E.M., Bernstein C.N., Purdom E., Dethlefsen L., Sargent M. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ott S.J., Kuhbacher T., Musfeldt M., Rosenstiel P., Hellmig S., Rehman A. Fungi and inflammatory bowel diseases: alterations of composition and diversity. Scand J Gastroenterol. 2008;43:831–841. doi: 10.1080/00365520801935434. [DOI] [PubMed] [Google Scholar]
- 9.Zoetendal E.G., von Wright A., Vilpponen-Salmela T., Ben-Amor K., Akkermans A.D., de Vos W.M. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol. 2002;68:3401–3407. doi: 10.1128/AEM.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayashi H., Sakamoto M., Benno Y. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol Immunol. 2002;46:535–548. doi: 10.1111/j.1348-0421.2002.tb02731.x. [DOI] [PubMed] [Google Scholar]
- 11.Bernhardt H., Knoke M. Mycological aspects of gastrointestinal microflora. Scand J Gastroenterol Suppl. 1997;222:102–106. doi: 10.1080/00365521.1997.11720731. [DOI] [PubMed] [Google Scholar]
- 12.Simon G.L., Gorbach S.L. Intestinal flora in health and disease. Gastroenterology. 1984;86:174–193. [PubMed] [Google Scholar]
- 13.Kaul A., Hutfless S., Liu L., Bayless T.M., Marohn M.R., Li X. Serum anti-glycan antibody biomarkers for inflammatory bowel disease diagnosis and progression: a systematic review and meta-analysis. Inflamm Bowel Dis. 2012;18:1872–1884. doi: 10.1002/ibd.22862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russell R.K., Ip B., Aldhous M.C., MacDougall M., Drummond H.E., Arnott I.D. Anti-Saccharomyces cerevisiae antibodies status is associated with oral involvement and disease severity in Crohn disease. J Pediatr Gastroenterol Nutr. 2009;48:161–167. doi: 10.1097/MPG.0b013e318183e112. [DOI] [PubMed] [Google Scholar]
- 15.Landers C.J., Cohavy O., Misra R., Yang H., Lin Y.C., Braun J. Selected loss of tolerance evidenced by Crohn's disease-associated immune responses to auto- and microbial antigens. Gastroenterology. 2002;123:689–699. doi: 10.1053/gast.2002.35379. [DOI] [PubMed] [Google Scholar]
- 16.Murdoch T.B., Xu W., Stempak J.M., Landers C., Targan S.R., Rotter J.I. Pattern recognition receptor and autophagy gene variants are associated with development of antimicrobial antibodies in Crohn's disease. Inflamm Bowel Dis. 2012;18:1743–1748. doi: 10.1002/ibd.22884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen R., Berry S.H., Mukhopadhya I., Thomson J.M., Saunders K.A., Nicholl C.E. The microaerophilic microbiota of de-novo paediatric inflammatory bowel disease: the BISCUIT study. PLoS One. 2013;8:e58825. doi: 10.1371/journal.pone.0058825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomson J.M., Hansen R., Berry S.H., Hope M.E., Murray G.I., Mukhopadhya I. Enterohepatic helicobacter in ulcerative colitis: potential pathogenic entities? PLoS One. 2011;6:e17184. doi: 10.1371/journal.pone.0017184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brad T., van Breukelen B.M., Braster M., van Straalen N.M., Roling W.F. Spatial heterogeneity in sediment-associated bacterial and eukaryotic communities in a landfill leachate-contaminated aquifer. FEMS Microbiol Ecol. 2008;65:534–543. doi: 10.1111/j.1574-6941.2008.00533.x. [DOI] [PubMed] [Google Scholar]
- 20.Hansen R., Russell R.K., Reiff C., Louis P., McIntosh F., Berry S.H. Microbiota of de-novo pediatric IBD: increased faecalibacterium prausnitzii and reduced bacterial diversity in Crohn's but not in ulcerative colitis. Am J Gastroenterol. 2012;107:1913–1922. doi: 10.1038/ajg.2012.335. [DOI] [PubMed] [Google Scholar]
- 21.Mavromanolakis E., Maraki S., Cranidis A., Tselentis Y., Kontoyiannis D.P., Samonis G. The impact of norfloxacin, ciprofloxacin and ofloxacin on human gut colonization by Candida albicans. Scand J Infect Dis. 2001;33:477–478. doi: 10.1080/00365540152030006. [DOI] [PubMed] [Google Scholar]
- 22.Hoffmann C., Dollive S., Grunberg S., Chen J., Li H., Wu G.D. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One. 2013;8:e66019. doi: 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Standaert-Vitse A., Jouault T., Vandewalle P., Mille C., Seddik M., Sendid B. Candida albicans is an immunogen for anti-Saccharomyces cerevisiae antibody markers of Crohn's disease. Gastroenterology. 2006;130:1764–1775. doi: 10.1053/j.gastro.2006.02.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.