Abstract
Approximately 3-15% of all malignant melanomas (MM) are familial cases. MM is a highly heterogeneous tumour type from a genetic perspective. Pedigrees with disease confined to a single generation of siblings or MM occurring among second- or third-degree relatives suggest multifactorial polygenic inheritance. However, not infrequently, within large families aggregations of MM are consistent with autosomal dominant inheritance, suggesting a hereditary syndrome caused by germline alterations of a single gene. Several different genes are involved in the development of MM. However, even when taken together they are responsible for less than 20% of all MM cases. It is thus necessary to perform association studies focused on genetic markers that could be used in identifying patients with a high risk of MM. Evaluation of aggregations of MM and other malignancies, like breast cancer, could be essential in identifying relatives of MM probands being at high risk of developing malignancies other than MM. The ultimate goal is to apply in these cases prevention recommendations and surveillance protocols to reduce the disease risk.
Keywords: familial melanoma, genetic factors
Epidemiology
Malignant melanoma (MM) is one of the most aggressive human malignancies. The incidence rates differ among Caucasian populations: ~35 per 100 000 in Australia [1], ~10 per 100 000 in the United States [2], ~7-10 per 100 000 in Western Europe [3,4]. Its incidence has increased dramatically over the past years in Caucasian population worldwide up to a 10-fold increase since 1950s [5].
Risk factors
Sun exposure
Well-established environmental risk factor of MM is ultraviolet radiation (UV), sunburn and overexposure to the sun [6]. Armstrong and Kricker estimated that up to 65% of MM could be related to sunburn [7]. Exposure in childhood seems to be an especially important risk factor [8]. In countries with a growing awareness of MM and public health campaigns to avoid UV a downturn in MM incidence has been observed [9].
Genetic factors
It has been long stated in many different reviews that genetic susceptibility is another major MM risk factor. Recent epidemiological studies confirm these suggestions. Evaluation of over eight thousand Swedish probands with primary MM revealed that the familial risk for offspring of affected parents is about 2.57, and even higher (4.22) when a parent has been diagnosed at an age <50 years [10]. Similar results were obtained during the evaluation of 1505 MM probands recorded in Utah (USA) when the familial risk for first-degree relatives (both offspring and siblings) was 2.1, and as high as 6.52 when MM cases were diagnosed under 50 years [11]. Hemminki et al reported that the familial MM risk showed a clear age dependence and higher risk in in situ melanoma than in its invasive counterpart [12].
Familial clustering of MM has also been studied in twins. A population-based study of cancer risk in twins performed by Swerdlow et al indicated an increased risk of the occurrence of MM in opposite-sex co-twins, however studies performed by Hemminki and Milan did not confirm these results [13-15].
Familial aggregations of malignancy may be due to either genetic factors, environmental factors shared by family members, or both. In a recent study Czene et al assessed the genetic and environmental components in the main types of cancer by statistical analyses of family pairs. The importance of genetic effects was indicated by a higher correlation among relatives that were more closely related to each other genetically. Structural equation modelling was used to derive estimates of the importance of environmental and genetic effects. A statistically significant proportion of susceptibility to cancer accounted for by genetic effects was obtained for almost all malignancies, including MM. It was estimated that in the case of MM genetic factors constitute 21% of susceptibility, similarly to other neoplasms, such as cancers of breast, colon or stomach [16].
Familial aggregations of MM
There are two distinct ways of defining familial melanoma: 1) occurrence of melanoma in at least two first-degree relatives; or 2) families with at least two melanomas irrespective of the degree of relationship.
Approximately 3-15% of all MM are familial cases of any type [17]. In Australia, Holman and Armstrong found a positive family history in 15% of 507 cases (9.9% in first-degree relatives) [18]. In the USA MM was present in first-degree relatives in 4.1% of 116 cases [19], in Denmark among 474 patients 4.7% had a relative with MM (3% in first-degree relatives) [20]. In our center among 405 unselected MM patients 12 cases (3%) had at least one first-degree relative affected.
In several families the co-occurrence of melanoma of the skin and the eye is reported [21]. The question whether ocular melanoma is also part of the familial melanoma syndrome remains unanswered.
Familial MM constitutes most probably a heterogeneous group of disorders characterized by occurrence of MM among relatives. The mode of inheritance is controversial and according to Mao is most likely polygenic [22]. Pedigrees with disease confined to a single generation of siblings or MM occurring among second- or third-degree relatives suggest multifactorial polygenic inheritance. However, not infrequently, within large families aggregations of MM are consistent with autosomal dominant inheritance. Anderson et al described MM in at least 15 members of a three-generation kindred. Early age of onset and a tendency for multiple primary lesions were characteristic features [23]. Lynch and Krush (1968) described two families with malignant melanoma in two generations in one family and three generations in the other [24]. Such pedigrees strongly suggest that part of familial MM constitutes hereditary syndrome caused by single-gene germline alterations. In order to identify such families the first definition of familial melanoma described above should be used.
Familial aggregations of MM and malignancies at different sites of origin
Aggregations of MM and other malignancies have been reported by many authors.
Most significantly a high incidence of pancreatic cancer has been reported in melanoma families, either by itself or in association with breast cancer [25-27].
Weston et al described a case with familial aggregation of melanoma, basal cell carcinoma and gastric cancer [28]. Aggregations of MM and cerebral astrocytoma [29], squamous cell carcinoma (SCC) [30] and kidney [31] were described. Recently, an increased risk of breast cancer among first-degree relatives of MM probands from families with strong cancer familial aggregation (showing features of autosomal dominant pattern of inheritance) has been suggested [32].
Molecular background of familial melanoma
Genes conferring high risk of developing MM
CDKN2A
In 1994 the first and so far the most significant high-risk MM susceptibility gene, CDKN2A (9p21, OMIM 600160) was identified [33,34]. One of the four transcripts coded by this gene is the p16 protein. It inhibits the activity of the complex of cyclin D1 with cyclin-dependent kinase 4 (CDK4) or 6 (CDK6), the function of which is to promote cellular proliferation [35]. Thus CDKN2A acts as a tumour suppressor gene by inhibiting cellular proliferation.
Germline mutation frequencies in CDKN2A among members of melanoma families show considerable variation. Soufir et al showed frequent involvement of CDKN2A in French familial melanoma families (46%) whereas Fitzgerald et al identified 18% of US familial melanoma harboured CDKN2A constitutional mutations [36,37]. Among Swedish families only 8% of cases were found to harbour alterations in the CDKN2A gene [38,39]. Germline mutation and large deletion analysis of the CDKN2A/ARF genes in Polish families with multiple melanomas and in families with an aggregation of MM and breast cancer revealed that less than 6% of familial MM can be associated with the CDKN2A/ARF constitutional mutations [40]. In a separate study done by Lamperska et al no CDKN2A mutations have been detected in Polish melanoma-prone families [41].
According to Bressac-de-Paillerets et al the number of CDKN2A alterations correlates with the number of MM cases in the family and a young age at diagnosis (<50) [42]. However, no germline mutations have been reported in childhood melanoma so far [43].
In Dutch melanoma-prone families with a specific recurrent 19 bp deletion of exon 2 of the CDKN2A gene, a high frequency of pancreatic cancer was observed [44]. CDKN2A was the first of the melanoma genes and is associated with an increase in the risk of pancreatic cancer as well [45]. Occurrence of multiple MM, pancreatic cancer and also breast cancer has been reported in families with different recurrent CDKN2A constitutional alterations [46].
In a recent study Bishop et al examined 80 families with documented CDKN2A mutations and multiple cases of cutaneous melanoma to establish the penetrance of the CDKN2A germline mutations. The penetrance was 30% by the age of 50 and 67% by the age of 80. It depended on the geographic origin: at 80 years of age, it was 58% in Europe, 76% in North America and 91% in Australia [47]. These data suggest that UV increases the penetrance of the CDKN2A mutations.
There is no doubt that regardless of the geographical origin, inclusion criteria or study methods, CDKN2A constitutional alterations have been found only in the minority of melanoma-prone families. In families with multigenerational inheritance of MM there remains a possibility that non-coding mutations of CDKN2A or alterations in another tumour suppressor gene predispose to familial melanoma.
ARF
In 1995 two independent groups discovered that CDKN2A shares exons 2 with another gene, ARF [48,49]. The transcript, p14ARF is involved in the regulation of the cell cycle and apoptosis [50]. It has also been implicated in the pathogenesis of MM. The first exon of ARF is unique, whereas its second exon is derived from exon 2 of CDKN2A using a different reading frame of the sequence. Up to now mutations in ARF are reported to be present only in a few families worldwide and are characterized by an aggregation of MM and brain tumours [51]. Hewitt et al described a germline mutation of ARF in patients affected with melanoma or breast cancer from a family with multiple melanomas and breast cancers [52], in an independent study performed in our center no constitutional ARF alterations in cases with familial aggregation of MM and breast cancer were detected [40].
CDK4
In 1996 Zuo et al identified the CDK4 gene (12q13, OMIM 123829) as a third MM susceptibility gene [48]. CDK4 is an oncogene that codes the cyclin-dependent kinase 4 protein. Alterations in this gene are responsible for the occurrence of very small proportions of familial MM. Up to now germline CDK4 mutations have been found in 3 families world-wide [36,53,54].
BRCA2
Mutations in BRCA2 (13q12, OMIM 600185), acting in DNA repair, predispose to a range of cancer types. The Breast Cancer Linkage Consortium estimates the relative risk of melanoma to be 2.58 in BRCA2 carriers [55]. In 2002 Scott et al examined 71 patients with ocular MM and pedigree and clinical data suggestive of genetic background (bilateral cases, positive family history for occurrence of MM, age at diagnosis <50). He estimated the prevalence of possible loss of function changes in BRCA2 in 3% of patients with familial ocular melanoma [56]. No germline BRCA2 mutations have been found in familial skin MM to date. The penetrance of this gene is unknown. Johannsson et al suggested that apart from breast and ovarian cancer, the incidence of other cancer types did not appear to be greatly increased in BRCA2-associated families and did not warrant specific clinical follow-up in mutation carriers [57].
NBS1
In 1998 Varon et al mapped and cloned the gene responsible for Nijmegen breakage syndrome characterized by spontaneous chromosomal instability, immunodeficiency and predisposition to cancer. The NBS1 gene is located on 8q21 (OMIM 602667) [58]. The NBS1 protein product, called nibrin (also referred to as p95), is an integral component of hMRE11/hRAD50/NBS1 nuclease complex, acting in a double-strand break repair of human DNA [59].
Mutations in the NBS1 gene predispose to a range of cancer types. We recently reported a greater than expected prevalence of a founder 657del5 mutation in the NBS1 gene in consecutive MM patients with aggregations of breast cancer but this preliminary finding needs to be confirmed [60]. No germline NBS1 mutations have been found in familial skin MM to date.
CHK2
Malignant melanoma has been suggested to belong to the clinical spectrum of Li-Fraumeni syndrome (LFS, OMIM151623). In 1999 Blasina et al and Chaturvedi et al independently identified cell-cycle-checkpoint kinase 2 (CHK2, MIM 604373), one of the key mediators of cellular responses to DNA damage [61,62]. The germline recurrent mutation in the CHK2 gene (1100delC in exon 10) has been found among two families with a phenotype suggestive of Li-Fraumeni syndrome [63]. The tumour spectrum of one of these families included MM. In a recent study germline 1100delC mutation in the CHK2 gene was reported in a patient with MM of the skin and three metachronous malignancies belonging to LFS tumour spectrum [64]. Thus, it seems that 1100delC of the CHK2 gene may be responsible for occurrence of very small proportions of MM cases, especially in families with Li-Fraumeni syndrome or Li-Fraumeni-like syndrome. No germline CHK2 mutations have been found in familial skin MM to date.
MLH1, MSH2
Germline mutations in the MLH1 (3p21, OMIM 120436) and MSH2 (2p22, OMIM 120435) genes can lead to Muire-Torre syndrome, a variant of hereditary non-polyposis colorectal cancer syndrome with an increased frequency of occurrence of skin malignancies, including MM. Both genes participate in mismatch repair system of DNA. There is a report of biallelic somatic inactivation of the MLH1 gene in a primary skin melanoma [65]. It seems that a small proportion of MLH1/MSH2 carriers could be at risk of developing MM. No germline MLH1/MSH2 mutations have been found in familial skin MM to date.
XP-genes
MM, as well as a large number of basal cell carcinoma and squamous cell cancer of the skin, is a characteristic feature of xeroderma pigmentosum syndrome (XP). XP is a group of disorders characterized by high sensitivity to sunlight with the development of skin malignancies at an early age. There are several variants of XP caused by different XP susceptibility genes, all of these genes are involved in UV-damaged DNA repair. Three most common forms of XP variants are: XPA (OMIM 278700), XPC (OMIM 278720) and XPD (OMIM 278730). Recently an increased risk of cutaneous melanoma in carriers of XPD polymorphisms has been reported [66,67].
In the literature we found no data about germline mutations of XPA, XPC and XPD genes in familial melanoma to date.
Genes conferring a low risk of developing MM
MC1R
The MC1R gene (16q24, OMIM 155555) codes a protein that acts as the receptor for melanocyte-stimulating hormone (MSH). It has been reported that some germline allelic variants of the MC1R gene (Arg151Cys, Arg160Trp, Asp294His) conferred to fair skin/red hair phenotype associated with an increased risk of MM [68-70]. The same allelic variants independently of skin type are associated with an increased MM risk [69]. They also act as modifiers of the melanoma risk in carriers of the CDKN2A mutations by increasing the penetrance from 50% to 84% in Australia and from 18% to 55% in the Netherlands [71,72].
Other factors
Inherited traits explaining part of the familial risk of MM by modulating the response to UV radiation also include:
1) nevi, the risk of developing a melanoma increases with the number of nevi and their clinically atypical aspects (increase in size, red hue and/or ABCDE criteria: (A) asymmetry, (B) vague border, (C) variegated pigmentation, (D) diameter exceeding 5 mm, (E) elevation) [73,74]. This risk is especially high in dysplastic nevus syndrome [75]. Recent twin studies indicate the contribution of both genetic factors and sun exposure to the number of nevi [76]. Analysis of familial transmission of nevi suggests a more complex mechanism than a single-gene effect [77]. Atypical nevi, present either in the familial or sporadic setting are regarded by dermatologists as strong indicators of an increased melanoma risk and should be either monitored or excised.
It should be mentioned that MM, especially a nodular type, can arise in apparently healthy skin;
2) skin type (liability to tan), the risk of MM is increased in persons with fair skin (limited capabilities of tanning) and red hair [78].
Conclusions
MM is a highly heterogeneous tumour type from a genetic perspective. Pedigree and clinical data of a proportion of familial MM suggest a hereditary syndrome caused by germline alterations of a single gene. Several different genes are involved in the development of MM. However, even when taken together they are responsible for less than 20% of all MM cases. It is thus necessary to perform association studies focused on identifying genetic markers that could be used in identifying patients with a high risk of MM. Evaluation of aggregations of MM and other malignancies, like breast cancer, could be essential in identifying relatives of MM probands being at high risk of developing malignancies other than MM. The ultimate goal is to apply in these cases prevention recommendations and surveillance protocols to reduce the disease risk.
Prevention recommendations
The use of higher sun protection factor sunscreens, avoidance of physical trauma of nevi.
Surveillance protocols
In the case of familial MM: melanoma patient, his first- and second degree relatives: half-yearly careful skin examination by the dermatologist. In cases with atypical nevi: skin examination should be performed every three months. Random removal of a large number of atypical nevi is not recommended. The removal of lesions in susceptible individuals should be indicated by clinical signs of malignant transformation (increase of size, red hue, bleeding, pruritus, etc.).
Additionally the melanoma patient and his relatives should be screened for pancreatic cancer with an annual ultrasonography and computer tomography every three years.
In cases with familial aggregations with MM and malignancies (at least two cases) at different sites of origin, especially with MM diagnosed under the age of 55: breast cancer surveillance is suggested (yearly physical examination, ultrasonography and finally mammography starting from the age of 25).
Figure 1.
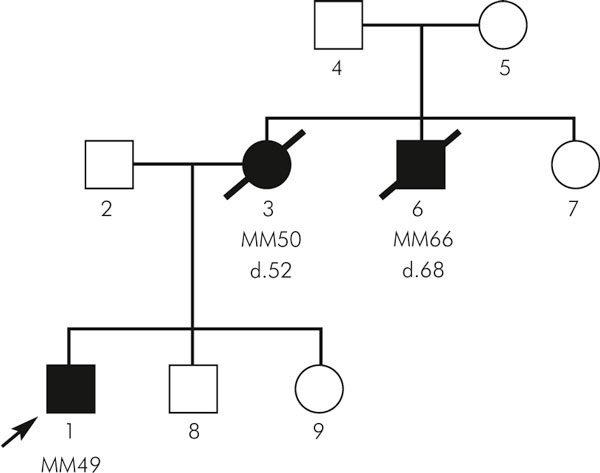
Pedigree of a family with melanoma diagnosed among first-degree relatives registered in our center.
Figure 2.
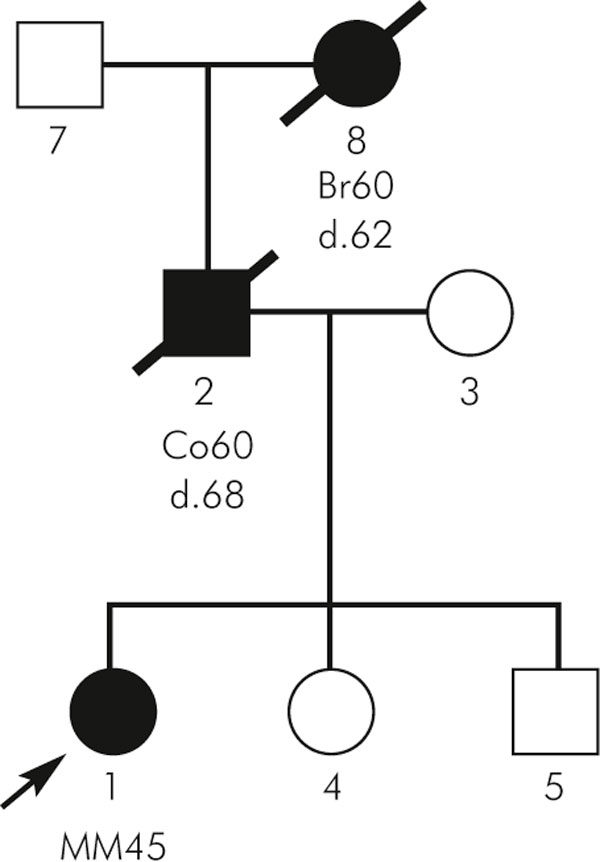
Pedigree of a family with familial aggregations with MM and malignancies of different site of origin.
References
- Marret LD, Nguyen HL, Armstrong BK. Trends in the incidence of cutaneous malignant melanoma in New South Wales, 1983-1996. Int J Cancer. 2001;92(3):457–462. doi: 10.1002/ijc.1203. [DOI] [PubMed] [Google Scholar]
- Jemal A, Devesa SS, Hartge P, Tucker MA. Recent trends in cutaneous melanoma incidence among whites in the United States. J Natl Cancer Inst. 2001;93(9):678–683. doi: 10.1093/jnci/93.9.678. [DOI] [PubMed] [Google Scholar]
- Micheli A, Mugno E, Krogh V, Quinn MJ, Coleman M, Hakulinen T, Gatta G, Berrino F, Capocaccia R. EUROPREVAL Working Group. Cancer prevalence in European registry areas. Ann Oncol. 2002;13(6):840–845. doi: 10.1093/annonc/mdf127. [DOI] [PubMed] [Google Scholar]
- Armstrong BK, Kricker A. Cutaneous melanoma. Cancer Survey. 1994;19:219–240. [PubMed] [Google Scholar]
- Weinstock MA. Issues in the epidemiology of melanoma. Hematol Oncol Clin North Am. 1998;12:681–698. doi: 10.1016/S0889-8588(05)70018-6. [DOI] [PubMed] [Google Scholar]
- English DR, Armstrong BK, Kricker A, Fleming C. Sunlight and cancer. Review Cancer Causes Control. 1997;8(3):271–283. doi: 10.1023/A:1018440801577. [DOI] [PubMed] [Google Scholar]
- Armstrong BK, Kricker A. How much melanoma is caused by sun exposure? Melanoma Res. 1993;3:395–401. doi: 10.1097/00008390-199311000-00002. [DOI] [PubMed] [Google Scholar]
- Whiteman DC, Whiteman AC, Green AC. Childhood sun exposure as a risk factor for melanoma: a systematic review of epidemiological studies. Cancer Causes Control. 2001;12:69–82. doi: 10.1023/A:1008980919928. [DOI] [PubMed] [Google Scholar]
- Bataille V. Genetic epidemiology of melanoma. Eur J Cancer. 2003;39:1341–1347. doi: 10.1016/S0959-8049(03)00313-7. [DOI] [PubMed] [Google Scholar]
- Hemminki K, Li X, Plna K, Granstrom C, Vaittinen P. The nation-wide Swedish family-cancer database-updated structure and familial rates. Acta Oncol. 2001;40(6):772–777. doi: 10.1080/02841860152619214. [DOI] [PubMed] [Google Scholar]
- Goldgar DE, Easton DF, Cannon-Albright LA, Skolnick MH. Systematic population-based assessment of cancer risk in first-degree relatives of cancer probands. J Natl Cancer Inst. 1994;86(21):1600–1608. doi: 10.1093/jnci/86.21.1600. [DOI] [PubMed] [Google Scholar]
- Hemminki K, Zhang H, Czene K. Familial and attributable risks in cutaneous melanoma: effects of proband and age. J Invest Dermat. 2002;120:217–223. doi: 10.1046/j.1523-1747.2003.12041.x. [DOI] [PubMed] [Google Scholar]
- Swerdlow AJ, De Stavola B, Maconochie N, Siskind V. A population-based study of cancer risk in twins: relationships to birth order and sexes of the twin pair. Int J Cancer. 1996;67(4):472–478. doi: 10.1002/(SICI)1097-0215(19960807)67:4<472::AID-IJC2>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Hemminki K, Li X. Cancer risks in twins: results from the Swedish family-cancer database. Int J Cancer. 2002;99(6):873–878. doi: 10.1002/ijc.10441. [DOI] [PubMed] [Google Scholar]
- Milan T, Verkasalo PK, Kaprio J, Koskenvuo M, Pukkala E. Malignant skin cancers in the Finnish Twin Cohort: a population-based study, 1976-97. Br J Dermatol. 2002;147(3):509–512. doi: 10.1046/j.1365-2133.2002.04870.x. [DOI] [PubMed] [Google Scholar]
- Czene K, Lichtenstein P, Hemminki K. Environmental and heritable causes of cancer among 9.6 million individuals in the Swedish Family-Cancer Database. Int J Cancer. 2002;99(2):260–266. doi: 10.1002/ijc.10332. [DOI] [PubMed] [Google Scholar]
- Greene MH, Fraumeni JF. In: The hereditary variant of familial melanoma. Clarh WH, Goldman LI, Mastrangelo MJ, editor. Human Malignant Melanoma, Grune and Stratton, New York; [Google Scholar]
- Holman CD, Armstrong BK. Pigmentary traits, ethnic origin, benign nevi, and family history as risk factors for cutaneous malignant melanoma. J Natl Cancer Inst. 1984;72(2):257–266. [PubMed] [Google Scholar]
- Cristofolini M, Franceschi S, Tasin L, Zumiani G, Piscioli F, Talamini R, La Vecchia C. Risk factors for cutaneous malignant melanoma in a northern Italian population. Int J Cancer. 1987;39(2):150–154. doi: 10.1002/ijc.2910390205. [DOI] [PubMed] [Google Scholar]
- Osterlind A, Tucker MA, Hou-Jensen K, Stone BJ, Engholm G, Jensen OM. The Danish case-control study of cutaneous malignant melanoma. I. Importance of host factors. Int J Cancer. 1988;42(2):200–206. doi: 10.1002/ijc.2910420210. [DOI] [PubMed] [Google Scholar]
- de Snoo FA, Bergman W, Gruis NA. Familial melanoma: a complex disorder leading to controversy on DNA testing. Fam Cancer. 2003;2:109–116. doi: 10.1023/A:1025758527675. [DOI] [PubMed] [Google Scholar]
- Tsao H. Update on familial cancer syndromes and the skin. J Am Acad Dermatol. 2000;42:939–971. doi: 10.1016/S0190-9622(00)90285-8. [DOI] [PubMed] [Google Scholar]
- Anderson DE, Smith JL Jr, McBride CM. Hereditary aspects of malignant melanoma. JAMA. 1967;200(9):741–746. doi: 10.1001/jama.200.9.741. [DOI] [PubMed] [Google Scholar]
- Lynch HT, Krush AJ. Heredity and malignant melanoma: implications for early cancer detection. Canad Med Assoc J. 1968;99(1):17–21. [PMC free article] [PubMed] [Google Scholar]
- Whelan AJ, Bartsch D, Goodfellow PJ. Brief report: a familial syndrome of pancreatic cancer and melanoma with a mutation in the CDKN2 tumor-suppressor gene. New Eng J Med. 1995;33:975–977. doi: 10.1056/NEJM199510123331505. [DOI] [PubMed] [Google Scholar]
- Parker JF, Florell SR, Alexander A, DiSario JA, Shami PJ, Leachman SA. Pancreatic carcinoma surveillance in patients with familial melanoma. Arch Dermatol. 2003;139(8):1019–1025. doi: 10.1001/archderm.139.8.1019. [DOI] [PubMed] [Google Scholar]
- Borg A, Sandberg T, Nilsson K, Johannsson O, Klinker M, Masback A, Westerdahl J, Olsson H, Ingvar C. High frequency of multiple melanomas and breast and pancreas carcinomas in CDKN2A mutation-positive melanoma families. J Natl Cancer Inst. 2000;92(15):1260–1266. doi: 10.1093/jnci/92.15.1260. [DOI] [PubMed] [Google Scholar]
- Weston B, Grufferman S, Kostvu D, Burton CS, Grant J. Familial aggregation of melanoma, basal cell carcinoma and gastric adenocarcinoma. Cancer. 1986;57:2230–2234. doi: 10.1002/1097-0142(19860601)57:11<2230::AID-CNCR2820571126>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Kaufman DK, Kimmel DW, Parisi JE, Michels VV. A familial syndrome with cutaneous malignant melanoma and cerebral astrocytoma. Neurology. 1993;43:1728–1731. doi: 10.1212/wnl.43.9.1728. [DOI] [PubMed] [Google Scholar]
- Wassberg C, Thorn M, Yuen J, Hakulinen T, Ringborg U. Cancer risk in patients with earlier diagnosis of cutaneous melanoma in situ. Int J Cancer. 1999;83:314–317. doi: 10.1002/(SICI)1097-0215(19991029)83:3<314::AID-IJC5>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Schmid-Wendter MH, Baumert J, Wendterb CM, Plewig G, Volkenandt M. Risk of second primary malignancies in patients with cutaneous melanoma. Br J Dermatol. 2001;145:981–985. doi: 10.1046/j.1365-2133.2001.04507.x. [DOI] [PubMed] [Google Scholar]
- Debniak T, Gorski B, Cybulski C, Jakubowska A, Kurzawski G, Kladny J, Zaluga E, Fiedorowicz J, Debniak B, Lubinski J. Increased risk of breast cancer in relatives of malignant melanoma patients from families with strong cancer familial aggregation. Eur J Cancer Prev. 2003;12:241–245. doi: 10.1097/00008469-200306000-00013. [DOI] [PubMed] [Google Scholar]
- Hussussian CJ, Struewing JP, Goldstein AM, Higgins PA, Ally DS, Sheahan MD, Clark WH Jr, Tucker MA, Dracopoli NC. Germline p16 mutations in familial melanoma. Nat Genet. 1994;8:15–21. doi: 10.1038/ng0994-15. [DOI] [PubMed] [Google Scholar]
- Kamb A, Shattuck-Eidens D, Eeles R, Liu Q, Gruis NA, Ding W, Hussey C, Tran T, Miki Y, Weaver-Feldhaus J. et al. Analysis of the p16 gene (CDKN2) as a candidate for the chromosome 9p melanoma susceptibility locus. Nat Genet. 1994;8:23–26. doi: 10.1038/ng0994-22. [DOI] [PubMed] [Google Scholar]
- Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- Soufir N, Avril MF, Chompret A, Demenais F, Bombled J, Spatz A, Stoppa-Lyonnet D, Benard J, Bressac-de Paillerets B. Prevalence of p16 and CDK4 germline mutations in 48 melanoma-prone families in France. The French Familial Melanoma Study Group. Hum Mol Genet. 1998;7:941. doi: 10.1093/hmg/7.2.209. [DOI] [PubMed] [Google Scholar]
- FitzGerald MG, Harkin DP, Silva-Arrieta S, MacDonald DJ, Lucchina LC, Unsal H, O'Neill E, Koh J, Finkelstein DM, Isselbacher KJ, Sober AJ, Haber DA. Prevalence of germ-line mutations in p16, p19ARF, and CDK4 in familial melanoma: analysis of a clinic-based population. Proc Natl Acad Sci USA. 1996;93:8541–8545. doi: 10.1073/pnas.93.16.8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platz A, Hansson J, Ringborg U. Screening of germline mutations in the CDK4, CDKN2C and TP53 genes in familial melanoma: a clinic-based population study. Int J Cancer. 1998;25:13–15. doi: 10.1002/(SICI)1097-0215(19980925)78:1<13::AID-IJC3>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Platz A, Hansson J, Mansson-Brahme E, Lagerlof B, Linder S, Lundqvist E, Sevigny P, Inganas M, Ringborg U. Screening of germline mutations in the CDKN2A and CDKN2B genes in Swedish families with hereditary cutaneous melanoma. J Natl Cancer Inst. 1997;89:697–702. doi: 10.1093/jnci/89.10.697. [DOI] [PubMed] [Google Scholar]
- Debniak T, Gorski B, Scott RJ, Cybulski C, Medrek K, Zowocka E, Kurzawski G, Debniak B, Kadny J, Bielecka-Grzela S, Maleszka R, Lubinski J. Germline mutation and large deletion analysis of the CDKN2A and ARF genes in families with multiple melanoma or an aggregation of malignant melanoma and breast cancer. Int J Cancer. 2004;110:558–562. doi: 10.1002/ijc.20163. [DOI] [PubMed] [Google Scholar]
- Lamperska K, Karezewska A, Kwiatkowska E, Mackiewicz A. Analysis of mutations in the p16/CDKN2A gene in sporadic and familial melanoma in the Polish population. Acta Biochim Pol. 2002;49:369–376. [PubMed] [Google Scholar]
- Bressac-de-Paillerets B, Avril MF, Chompret A, Demenais F. Genetic and environmental factors in cutaneous malignant melanoma. Biochemie. 2002;84:67–74. doi: 10.1016/S0300-9084(01)01360-8. [DOI] [PubMed] [Google Scholar]
- Whiteman DC, Miligan A, Welch J, Green AC, Hayward NK. Germline CDKN2A mutations in childhood melanoma. J Natl Cancer Inst. 1997;89:1460–1465. doi: 10.1093/jnci/89.19.1460. [DOI] [PubMed] [Google Scholar]
- Vasen HF, Gruis NA, Frants RR, Velden PA van der, Hille ET, Bergman W. Risk of developing pancreatic cancer in families with familial atypical multiple mole melanoma associated with a specific 19 deletion of p16 (p16-Leiden) Int J Cancer. 2000;87:809–811. doi: 10.1002/1097-0215(20000915)87:6<809::AID-IJC8>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Goldstein AM, Fraser MC, Struewing JP, Hussussian CJ, Ranade K, Zametkin DP, Fontaine LS, Organic SM, Dracopoli NC, Clark WH Jr. et al. Increased risk of pancreatic cancer in melanoma-prone families with p16INK4 mutations. New Engl J Med. 1995;333:970–974. doi: 10.1056/NEJM199510123331504. [DOI] [PubMed] [Google Scholar]
- Borg A, Sandberg T, Nilsson K, Johannsson O, Klinker M, Masback A, Westerdahl J, Olsson H, Ingvar C. High frequency of multiple melanomas and breast and pancreas carcinomas in CDKN2A mutation-positive melanoma families. J Natl Cancer Inst. 2000;92:1260–1266. doi: 10.1093/jnci/92.15.1260. [DOI] [PubMed] [Google Scholar]
- Bishop DT, Demenais F, Goldstein AM, Bergman W, Bishop JN, Bressac-de Paillerets B, Chompret A, Ghiorzo P, Gruis N, Hansson J, Harland M, Hayward N, Holland EA, Mann GJ, Mantelli M, Nancarrow D, Platz A, Tucker MA. Melanoma Genetics Consortium. Geographical variation in the penetrance of CDKN2A mutations for melanoma. J Natl Cancer Inst. 2002;94:894–903. doi: 10.1093/jnci/94.12.894. [DOI] [PubMed] [Google Scholar]
- Duro D, Bernard O, Della Valle V, Berger R, Larsen CJ. A new type of p16INK4/MTS1 gene transcript expressed in B-cell malignancies. Oncogene. 1995;11:21–29. [PubMed] [Google Scholar]
- Stone S, Jiang P, Dayananth P, Tavtigian SV, Katcher H, Parry D, Peters G, Kamb A. Complex structure and regulation of the P16 (MTS1) locus. Cancer Res. 1995;55:2988–2994. [PubMed] [Google Scholar]
- Eymin B, Karayan L, Seite P, Brambilla C, Brambilla E, Larsen CJ, Gazzeri S. Human ARF binds E2F1 and inhibits its transcriptional activity. Oncogene. 2001;20:1033–1041. doi: 10.1038/sj.onc.1204220. [DOI] [PubMed] [Google Scholar]
- Randerson-Moor JA, Harland M, Williams S, Cuthbert-Heavens D, Sheridan E, Aveyard J, Sibley K, Whitaker L, Knowles M, Bishop JN, Bishop DT. A germline deletion of p14 (ARF) but not CDKN2A in a melanoma-neural system tumour syndrome family. Hum Mol Genet. 2001;10:55–62. doi: 10.1093/hmg/10.1.55. [DOI] [PubMed] [Google Scholar]
- Hewitt C, Lee Wu C, Evans G, Howell A, Elles RG, Jordan R, Sloan P, Read AP, Thakker N. Germline mutation of ARF in a melanoma kindred. Hum Mol Genet. 2002;11:1273–1279. doi: 10.1093/hmg/11.11.1273. [DOI] [PubMed] [Google Scholar]
- Zuo L, Weger J, Yang Q, Goldstein AM, Tucker MA, Walker GJ, Hayward N, Dracopoli NC. Germline mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nat Genet. 1996;12:97–9. doi: 10.1038/ng0196-97. [DOI] [PubMed] [Google Scholar]
- Goldstein AM, Chidambaram A, Halpern A, Holly EA, Guerry D IV, Sagebiel R, Elder DE, Tucker MA. Rarity of CDK4 germline mutations in familial melanoma. Melanoma Res. 2002;12:51–55. doi: 10.1097/00008390-200202000-00008. [DOI] [PubMed] [Google Scholar]
- The Breast Cancer Linkage Consortium. Cancer risks in BRCA2 mutation carriers. The Breast Cancer Linkage Consortium. J Natl Cancer Inst. 1999;91:1310–1316. doi: 10.1093/jnci/91.15.1310. [DOI] [PubMed] [Google Scholar]
- Scott RJ, Vajdic CM, Armstrong BK, Ainsworth CJ, Meldrum CJ, Aitken JF, Kricker A. BRCA2 mutations in a population-based series of patients with ocular melanoma. Int J Cancer. 2002;102:188–191. doi: 10.1002/ijc.10693. [DOI] [PubMed] [Google Scholar]
- Johannsson O, Loman N, Moller T, Kristoffersson U, Borg A, Olsson H. Incidence of malignant tumours in relatives of BRCA1 and BRCA2 germline mutation carriers. Eur J Cancer. 1999;35:1248–1257. doi: 10.1016/S0959-8049(99)00135-5. [DOI] [PubMed] [Google Scholar]
- Varon R, Vissinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, Saar K, Beckmann G, Seemanova E, Cooper PR, Nowak NJ, Stumm M, Weemaes CM, Gatti RA, Wilson RK, Digweed M, Rosenthal A, Sperling K, Concannon P, Reis A. Nibrin a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/S0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, Hays L, Morgan WF, Petrini JH. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/S0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- Debniak T, Gorski B, Cybulski C, Jakubowska A, Kurzawski G, Lener M, Mierzejewski M, Masojc B, Medrek K, Kladny J, Zaluga E, Maleszka R, Chosia M, Lubinski J. Germline 657del5 mutation in the NBS1 gene in patients with malignant melanoma of the skin. Melanoma Res. 2003;13:365–370. doi: 10.1097/00008390-200308000-00005. [DOI] [PubMed] [Google Scholar]
- Blasina A, de Weyer IV, Laus MC, Luyten WH, Parker AE, McGowan CH. A human homologue of the checkpoint kinase Cds1 directly inhibits Cdc25 phosphatase. Curr Biol. 1999;14:1–10. doi: 10.1016/S0960-9822(99)80041-4. [DOI] [PubMed] [Google Scholar]
- Chaturvedi P, Eng WK, Zhu Y, Mattern MR, Mishra R, Hurle MR, Zhang X, Annan RS, Lu Q, Faucette LF, Scott GF, Li X, Carr SA, Johnson RK, Winkler JD, Zhou BB. Mammalian Chk2 is a downstream effector of the ATM-dependent DNA damage checkpoint pathway. Oncogene. 1999;18:4047–4054. doi: 10.1038/sj.onc.1202925. [DOI] [PubMed] [Google Scholar]
- Vahteristo P, Tamminen A, Karvinen P, Eerola H, Eklund C, Aaltonen LA, Blomqvist C, Aittomaki K, Nevanlinna H. p53, CHK2, and CHK1 genes in Finnish families with Li-Fraumeni syndrome: further evidence of CHK2 in inherited cancer predisposition. Cancer Res. 2001;61:5718–5722. [PubMed] [Google Scholar]
- Debniak T, Gorski B, Cybulski C, Kurzawski G, Zlowocka E, Kladny J, Chosia M, Lubinski J. Rarity of germline 1100delC mutation in CHK2 in patients with malignant melanoma of the skin. Melanoma Res. 2004;14:121–124. doi: 10.1097/00008390-200404000-00007. [DOI] [PubMed] [Google Scholar]
- Castiglia D, Pagani E, Alvino E, Vernole P, Marra G, Cannavo E, Jiricny J, Zambruno G, D'Atri S. Biallelic somatic inactivation of the mismatch repair gene MLH1 in a primary skin melanoma. Genes Chromosomes Cancer. 2003;37:165–175. doi: 10.1002/gcc.10193. [DOI] [PubMed] [Google Scholar]
- Baccarelli A, Calista D, Minghetti P, Marinelli B, Albetti B, Tseng T, Hedayati M, Grossman L, Landi G, Struewing JP, Landi MT. XPD gene polymorphism and host characteristics in the association with cutaneous malignant melanoma risk. Br J Cancer. 2004;90:497–502. doi: 10.1038/sj.bjc.6601385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomescu D, Kavanagh G, Ha T, Campbell H, Melton DW. Nucleotide excision repair gene XPD polymorphisms and genetic predisposition to melanoma. Carcinogenesis. 2001;22:403–408. doi: 10.1093/carcin/22.3.403. [DOI] [PubMed] [Google Scholar]
- Valverde P, Healy E, Jackson I, Rees JL, Thody AJ. Variants of the melanocyte-stimulating hormone receptor gene are associated with red hair and fair skin in humans. Nat Genet. 1995;11:328–330. doi: 10.1038/ng1195-328. [DOI] [PubMed] [Google Scholar]
- Palmer JS, Duffy DL, Box NF, Aitken JF, O'Gorman LE, Green AC, Hayward NK, Martin NG, Sturm RA. Melanocortin-1 receptor polymorphisms and risk of melanoma: is the association explained solely by pigmentation phenotype? Am J Hum Genet. 2000;66:176–186. doi: 10.1086/302711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box NF, Wyeth JR, O'Gorman LE, Martin NG, Sturm RA. Characterization of melanocyte stimulating hormone receptor variant alleles in twins with red hair. Hum Mol Genet. 1997;6:1891–1897. doi: 10.1093/hmg/6.11.1891. [DOI] [PubMed] [Google Scholar]
- Box NF, Duffy DL, Chen W, Stark M, Martin NG, Sturm RA, Hayward NK. MC1R genotype modifies risk of melanoma in families segregating CDKN2A mutations. Am J Hum Genet. 2001;69:765–773. doi: 10.1086/323412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velden PA van der, Sandkuijl LA, Bergman W, Pavel S, van Mourik L, Frants RR, Gruis NA. Melanocortin-1 receptor variant R151C modifies melanoma risk in Dutch families with melanoma. Am J Hum Genet. 2001;69:774–779. doi: 10.1086/323411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataille V, Bishop JA, Sasieni P, Swerdlow AJ, Pinney E, Griffiths K, Cuzick J. Risk of cutaneous melanoma in relation to the numbers, types and sites of naevi: a case-control study. Br J Cancer. 1996;73(12):1605–1611. doi: 10.1038/bjc.1996.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briollais L, Chompret A, Guilloud-Bataille M, Feingold N, Avril MF, Demenais F. Genetic and epidemiological risk factors for a malignant melanoma-predisposing phenotype: the great number of nevi. Genet Epidemiol. 1996;13(4):385–402. doi: 10.1002/(SICI)1098-2272(1996)13:4<385::AID-GEPI7>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Wachsmuth RC, Harland M, Bishop JA. The atypical-mole syndrome and predisposition to melanoma. N Engl J Med. 1998;339(5):348–349. doi: 10.1056/NEJM199807303390515. [DOI] [PubMed] [Google Scholar]
- Bataille V, Snieder H, MacGregor AJ, Sasieni P, Spector TD. Genetics of risk factors for melanoma: an adult twin study of nevi and freckles. J Natl Cancer Inst. 2000;92(6):457–463. doi: 10.1093/jnci/92.6.457. [DOI] [PubMed] [Google Scholar]
- Goldgar DE, Cannon-Albright LA, Meyer LJ, Piepkorn MW, Zone JJ, Skolnick MH. Inheritance of nevus number and size in melanoma and dysplastic nevus syndrome kindreds. J Natl Cancer Inst. 1991;83(23):1726–1733. doi: 10.1093/jnci/83.23.1726. [DOI] [PubMed] [Google Scholar]
- Grange F, Chompret A, Guilloud-Bataille M, Guillaume JC, Margulis A, Prade M, Demenais F, Avril MF. Comparison between familial and nonfamilial melanoma in France. Arch Dermatol. 1995;131(10):1154–1159. doi: 10.1001/archderm.131.10.1154. [DOI] [PubMed] [Google Scholar]


