Abstract
Single nucleotide polymorphisms (SNPs) are the benchmark molecular markers for modern genomics. Until recently, relatively few SNPs were known in the zebrafish genome. The use of next-generation sequencing for the positional cloning of zebrafish mutations has increased the number of known SNP positions dramatically. Still, the identified SNP variants remain under-utilized, owing to scant annotation of strain specificity and allele frequency. To address these limitations, we surveyed SNP variation in three common laboratory zebrafish strains using whole-genome sequencing. This survey identified an average of 5.04 million SNPs per strain compared with the Zv9 reference genome sequence. By comparing the three strains, 2.7 million variants were found to be strain specific, whereas the remaining variants were shared among all (2.3 million) or some of the strains. We also demonstrate the broad usefulness of our identified variants by validating most in independent populations of the same laboratory strains. We have made all of the identified SNPs accessible through ‘SNPfisher’, a searchable online database (snpfisher.nichd.nih.gov). The SNPfisher website includes the SNPfisher Variant Reporter tool, which provides the genomic position, alternate allele read frequency, strain specificity, restriction enzyme recognition site changes and flanking primers for all SNPs and Indels in a user-defined gene or region of the zebrafish genome. The SNPfisher site also contains links to display our SNP data in the UCSC genome browser. The SNPfisher tools will facilitate the use of SNP variation in zebrafish research as well as vertebrate genome evolution.
Keywords: Danio rerio, Zebrafish, Genome, SNP, Variation, Next-generation sequencing, Whole-genome sequencing
SUMMARY: SNP variation in three common laboratory zebrafish strains is analyzed using whole-genome sequencing and is organized into a user-friendly and searchable online database.
INTRODUCTION
Next-generation sequencing (NGS) has revolutionized the acquisition of large amounts of genomic data (Turner et al., 2009). The ability to sequence entire genomes has spawned projects like the ‘Thousand Genomes Project’, which aims to identify variation within human populations to make genotype-phenotype correlations possible for complex traits (Abecasis et al., 2010). In contrast to the human genome, much less is known about the genetic variation of the zebrafish genome, despite its similar size. The zebrafish (Danio rerio) has emerged as one of the most popular model organisms for phenotype-based forward-genetic mutagenesis screens owing to its optical clarity, rapid development and high fecundity (Patton and Zon, 2001). Unlike other vertebrate model organisms, most of the common laboratory zebrafish strains were established relatively recently from single pairs of founder fish caught in the wild or purchased from pet stores, with progeny selected for lack of embryonic lethality and developmental anomalies (Trevarrow and Robison, 2004). Generally, subsequent propagation of these wild-type strains has sought to avoid loss of genetic variability due to bottlenecking and genetic drift by avoiding inbreeding, maintaining a maximum effective population size and importing individuals of the same strain from other populations (Harper and Lawrence, 2011). However, most labs maintain their own small, closed populations of each wild-type strain, and adoption of the aforementioned measures varies greatly among fish facilities (D. Castranova, personal communication). Consequently, the genetic variability between and within wild-type strains probably varies considerably among different population isolates.
Only a handful of studies have attempted to quantify the genetic variability of laboratory zebrafish strains. Empirical measures of genetic variability in wild-type zebrafish strains have suggested high levels of both intra- and interstrain variation. Initial studies using simple sequence length polymorphism markers (SSLPs) were limited by either the number of markers used or by the number of individuals of a strain tested (Knapik et al., 1998; Nechiporuk et al., 1999; Coe et al., 2009). Similarly, subsequent studies using single nucleotide polymorphisms (SNPs) greatly expanded the number of markers tested, but few individuals of each strain were genotyped (Stickney et al., 2002; Guryev et al., 2006; Bradley et al., 2007; Whiteley et al., 2011). Recently, whole-genome sequencing (WGS) has been used to identify SNP and insertion/deletion (Indel) variation in wild-type and chemically mutagenized zebrafish (Bowen et al., 2012; Obholzer et al., 2012; Howe et al., 2013; Patowary et al., 2013). Many of the NGS positional cloning strategies described for zebrafish rely heavily on the ability to filter out normal variants and/or track strain-specific SNPs (Bowen et al., 2012; Leshchiner et al., 2012; Obholzer et al., 2012; Voz et al., 2012; Miller et al., 2013). Therefore, NGS positional cloning strategies would be enhanced by increased information on normal genetic variants found within the zebrafish genome. This would facilitate differentiation between neutral polymorphisms and phenotype-causing mutations, particularly when no ‘smoking gun’ mutations are identified in the critical regions.
In this study, WGS was employed to find SNP and Indel variant positions in three populations of commonly used laboratory zebrafish strains (EK, TL and WIK). An average of 5.04 million SNPs was identified in each strain surveyed. Each SNP was categorized according to its strain distribution, frequency and position (exon, intron, etc.) in the genome. The resulting SNP variation dataset was then organized into a user-friendly suite of tools named ‘SNPfisher’. SNP variants were also formatted for display in the UCSC genome browser for integrated genomic displays (Kent et al., 2002).
RESULTS
WGS of three common laboratory zebrafish
WGS was performed to identify both SNP and Indel variation in three common laboratory zebrafish strains, including Tüpfel long fin (TL), WIK and Tg(fli1a-eGFPy1). TL was originally purchased in a pet shop and derives its name from the recessive leopard phenotype (tüpfel translates to ‘dot’ in English) and dominant long fin mutant phenotypes (Tresnak, 1981; Haffter et al., 1996). WIK, formerly WIK11, originated from a single pair of wild-type zebrafish caught in India exhibiting the absence of embryonic and larval lethality (Rauch et al., 1997). The Tg(fli1a-eGFPy1) transgenic strain, popular in vascular biology and henceforth referred to as FLI, was originally generated from and has been subsequently back crossed to the EkkWill (EK) strain [originally derived from fish obtained from EkkWill Tropical Fish Farm (Gibsonton, FL, USA)] and therefore represents an isolate of EK (Lawson and Weinstein, 2002). Sequencing libraries were prepared from genomic DNA isolated from each of the above strains separately. Each strain library was generated from caudal fin clips from 18 individual adult zebrafish (nine females and nine males), pooled into a single sample for library construction. Illumina HiSeq sequencing reads were aligned to the Zv9 reference genome sequence, a genome sequence mainly generated from the Tübingen strain (TÜ) (Howe et al., 2013). An alternative allele was considered to be anything that differed from the Zv9 reference sequence at a given position. Evaluation of the resulting alignments demonstrated a mean of 16.1× genome coverage for each strain surveyed. With genome coverage of 16×, we expect to detect 80% of alternate alleles with frequencies equal to 0.1 assuming random sampling (supplementary material Fig. S1). However, a minimum read depth of three reads was required for inclusion in our dataset, which drops the expected detection to 78% for alternate alleles with frequencies equal to 0.4 (supplementary material Fig. S1). Prior to applying filters, Genome Annotation Toolkit (GATK) marked 35.4±0.2 million reads as containing a variant per strain (supplementary material Fig. S2). The total number of variant reads was reduced to 10.7±0.2 million (∼30% of total) once quality control filters (described in the Materials and Methods) were applied (supplementary material Fig. S2). The remaining variants were then further filtered by requiring a 3-read minimum in each strain and by removing any variants contained in repetitive sequence as annotated by Repeatmasker. After this filtering, the surveyed strains averaged 5.04±0.22 million variants per strain. This equates to a SNP ratio of seven SNPs per 1 kb of unique sequence in the zebrafish reference genome. In order to put the amount of variant positions in laboratory zebrafish into perspective, a scripted calculation was used to determine the total number of SNP variants in a single, ethnically defined human population when 18 individuals were randomly chosen from the 1000 genomes samples (Abecasis et al., 2012). Surprisingly, laboratory zebrafish ranged from 6.3 to 4.4 SNPs per 1 kb, which is a higher ratio than any ethnically defined human population (2.6-4.4 SNPs per 1 kb) measured (supplementary material Fig. S3). This suggests that either the human genome reference sequence is more representative of the consensus human genome among various populations or there is more genetic variation in the wild zebrafish populations that were used to establish laboratory strains.
Strain-specific and shared genetic variation in laboratory zebrafish
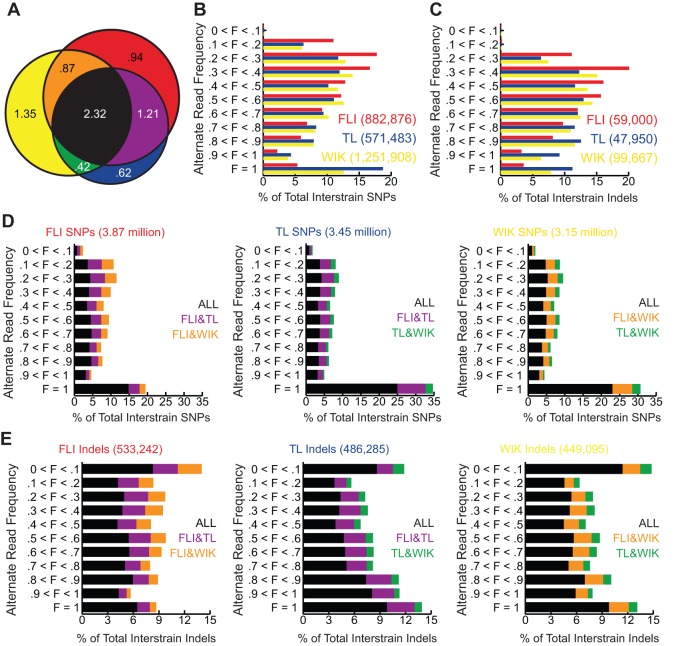
For many applications, especially genetic mapping, the strain specificity and allele frequency of a polymorphism will determine its utility. Therefore, SNPs and Indels that passed each of the filter criteria were classified according to their distribution among each strain (Fig. 1A). For purposes of our analysis, strain-specific variants were defined as alternate alleles observed in only one of the strains. Strain-specific SNP and Indel variation was measured by counting positions in which an alternate allele, considered any base differing from the Zv9 reference sequence, was present in only one of the three sampled strains (red, yellow and blue quadrants in Fig. 1A). The number of strain-specific variants ranged from a maximum of 1.35 million in WIK to a minimum of 0.62 million in TL, with a mean of 0.99±0.21 million SNPs (average strain-specific SNP ratio=1.34 SNPs/1 kb). The observed amount of relative strain-specific variation is similar to estimates determined with limited SSLP markers (Coe et al., 2009). In contrast to strain-specific variants, we defined shared variants as any alternate alleles differing from the Zv9 reference sequence and that were observed in more than one of the strains surveyed. The majority of the shared variants we identified (2.32 million) were observed in all three strains (Fig. 1A, black), suggesting that these genomic positions are commonly polymorphic in parental wild zebrafish populations from which the laboratory strains were derived. Some of the other shared variants demonstrated some strain specificity being present in two of the three surveyed strains (Fig. 1A, purple, orange and green quadrants). Of these, FLI (derived from EK) and TL have the highest number of shared variants (1.21 million), whereas WIK and TL contain the lowest (0.42 million).
Fig. 1.
Natural SNP and Indel variation in laboratory zebrafish. (A) Venn diagram demonstrating the number of positions in the zebrafish genome with alternate alleles specific to or shared between the FLI, TL and WIK strains as compared with the Zv9 reference genome. Numbers are in millions. See below for key to color scheme. (B,C) The numbers of strain-specific SNP (B) and Indel (C) variants (red, blue and yellow color-coded categories of variants shown in A) as percentages of total number of strain-specific variants (on the x-axis) and grouped by alternate allele read frequency (on the y-axis). The total number of strain-specific variants for each strain is shown in parentheses to the right of the name of each strain. (D,E) The numbers of shared SNP (D) and Indel (E) variants (purple, green, orange and black color-coded categories of variants shown in A), as percentages of total number of shared variants (on the x-axis) and grouped by observed alternate allele read frequency (on the y-axis). The total number of variants is shown above each graph and the color scheme is shown to the right. The color code in all panels denotes which strain or strains contain(s) a variant at a given position (at any frequency) relative to the Zv9 reference: Red, present in only FLI; blue, present in only TL; yellow, present in only WIK; purple, present in FLI and TL, but not in WIK; green, present in TL and WIK, but not in FLI; orange, present in FLI and WIK, but not in TL; black, present in all three strains. F, alternate allele read frequency.
As mentioned above, the frequency of a polymorphism often dictates its utility. In genetic mapping, the likelihood of the presence (high-frequency variants) or absence (low-frequency variants) of a polymorphism will determine its value as a genetic marker. Therefore, once grouped by strain specificity, variants were classified by alternate allele read frequency into 10% intervals (Fig. 1B-E). Alternate allele read frequencies (F) were simply calculated by dividing the number of alternate allele reads by the total number of sequence reads at a given position. As our sequencing libraries for each strain were generated from a pool of 18 individuals, the alternate allele read frequencies reflect estimates of the frequencies within the strain population rather than allele frequencies calculated from individual genotyping. In both the TL and WIK strains, the majority of strain-specific SNPs (60 and 55%, respectively) had F≥0.5, whereas only 29% of FLI-specific SNPs reached the same frequency (Fig. 1B). The higher number of fixed (F=1) alternate alleles in the TL and WIK strains (19 and 13%, respectively) compared with the FLI strain (5%) accounts for most of the observed differences. The difference in the number of fixed alleles might reflect a difference in population management, as FLI has been propagated at the National Institutes of Health (NIH, Bethesda, MD, USA) for 15 generations, whereas the TL and WIK strains, surveyed in this study, were propagated for two generations since acquisition from the Zebrafish International Resource Center (ZIRC, Eugene, OR, USA). Strain-specific Indels had a similar distribution when compared with strain-specific SNPs except that Indels at F<0.3 were underrepresented (Fig. 1C).
For shared variants, alternate alleles that were fixed in all three populations (Fig. 1D, black) accounted for the largest group in each of the strains surveyed, ranging from 25% (TL) to 15% (FLI). This observation suggests that the alleles found at these positions in the Zv9 reference genome sequence are probably not the true ‘consensus’ laboratory zebrafish alleles, but rather TÜ-specific variants. Unlike shared SNPs, shared Indels did not show a bias toward fixed alleles, but were more evenly distributed among the frequency intervals in each strain (Fig. 1E). Again, most Indels (59-70%) were present at some frequency in all three strains, suggesting that these were probably common among the wild zebrafish used to establish the laboratory strains.
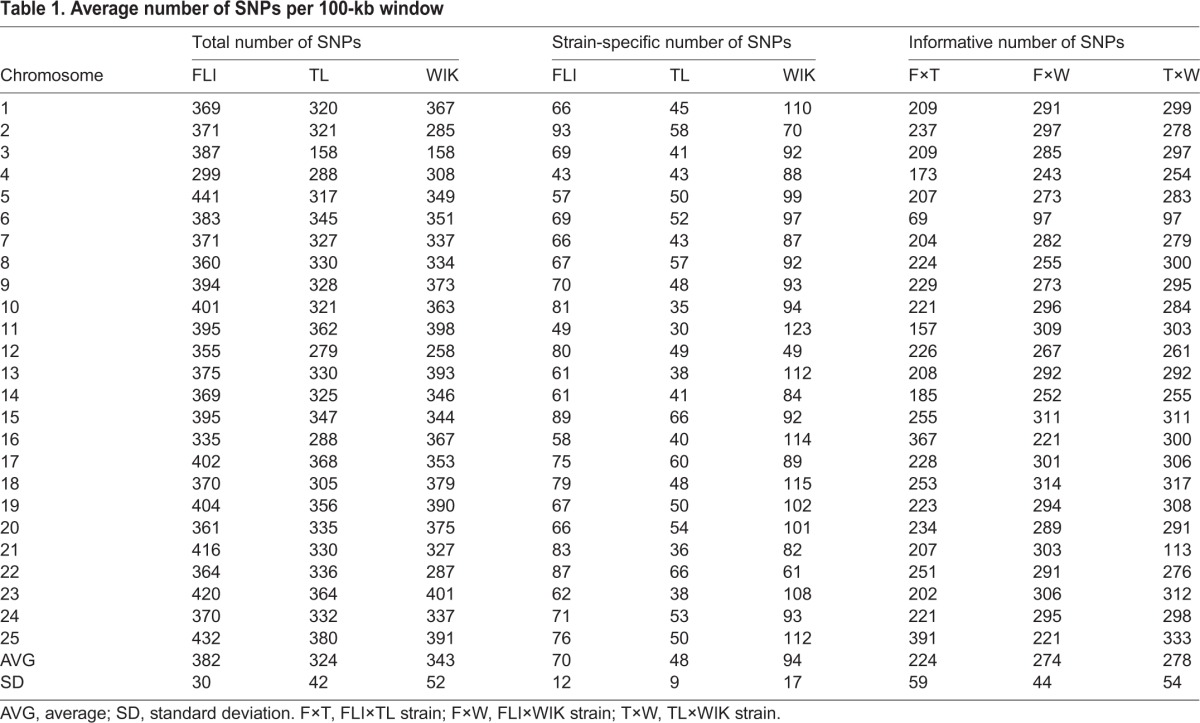
In order to evaluate the spatial density of the identified variants, the zebrafish genome was divided into 100-kb non-overlapping bins and the number of SNPs per each bin (regardless of alternate allele frequency) was measured. The resultant 100-kb bins were then averaged by chromosome (Table 1). Genome-wide calculations (average of each chromosome) of the total number of SNPs per 100 kb ranged from 382 (FLI) to 324 (TL), whereas the average number of strain-specific SNPs was between 94 (WIK) and 47 (TL). The ability to discriminate strains by SNP content (Table 1, informative SNPs) was determined by counting the number of differential SNPs expected from pairwise crosses ignoring allele frequency. With the exception of three chromosomes (16, 21 and 25), crosses between FLI and TL would yield the lowest number of polymorphisms compared with crosses between FLI and WIK or between TL and WIK. To provide a broader view of SNP density in the zebrafish genome, the number of SNPs per 100-kb non-overlapping bin was plotted along the length of each chromosome (supplementary material Fig. S4).
Table 1.
Average number of SNPs per 100-kb window
Gene-associated variation in the zebrafish genome
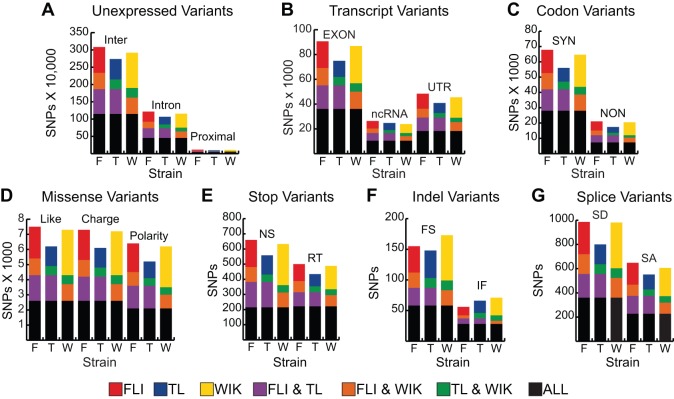
A great advantage of the zebrafish model is the ability to conduct large-scale forward-genetic screens. Most of these screens rely on chemical mutagenesis with N-ethyl-nitrosourea (ENU) (Solnica-Krezel et al., 1994). With advances in NGS technologies, WGS has been employed to find ENU-induced mutations responsible for phenotypes of interest (Bowen et al., 2012; Obholzer et al., 2012; Voz et al., 2012; Miller et al., 2013). However, to recognize a causative mutation, other mutation candidates need to be eliminated from consideration. We examined our natural SNP and Indel variants for their presence in coding regions of the genome and their putative effect, if any, on the encoded peptides. Natural SNP and Indel variation was categorized using Annovar (Fig. 2A-G) (Wang et al., 2010). As expected, the vast majority of SNP variants were located in either intergenic or intronic sequence (Fig. 2A). A mean of 8.4×104 (s.e.m.=0.5×104) SNP positions were located in the exons for each strain (Fig. 2B, EXON). Of the exonic variations, about one quarter (23.4%) caused non-synonymous amino acid changes in each strain (Fig. 2C). Many of these variants would be considered strong mutation candidates if located in crucial regions identified by meiotic mapping, as they include changes in polarity or charge of amino acid side chains (Fig. 2D), nonsense and read-through changes (Fig. 2E), and frameshift and non-frameshift insertions/deletions (Fig. 2F). Similarly, some of the SNP variants were located in splice acceptor and donor sites, resulting in potential aberrant splicing (Fig. 2G). In order to determine the distribution of non-synonymous mutations among genes, the number of non-synonymous changes per kb of exonic sequence was calculated for each Refseq gene annotation in the zebrafish genome. When all 14,484 genes are considered, an average of 56% of the total genes showed ≥1 non-synonymous change per kb of exonic sequence among the surveyed strains (supplementary material Fig. S5).
Fig. 2.
Spectrum of natural genetic variation in laboratory zebrafish. The number of SNPs or Indels per category. (A) Unexpressed variants, including intergenic (Inter), intronic (Intron) and within 5 kb of a transcript (Proximal) variants. (B) Transcript variants, including those falling in exons, non-coding RNA (ncRNA), and untranslated regions (UTR). (C) Codon variants, divided into synonymous (SYN) and non-synonymous (NON) changes. (D) Missense variants, divided into conservative changes, e.g. nonpolar-to-nonpolar (Like), charge changes, e.g. polar-to-positive (Charge) and polarity changes, e.g. polar-to-nonpolar (Polarity). (E) Stop codon variants, divided into nonsense (NS) and read-through (RT) alleles. (F) Indel variants within coding sequences, divided by whether they result in frameshift (FS) or in-frame (IF) alleles. (G) Splice site-altering variants, including those that alter splice donor dinucleotides (SD) and those that alter splice acceptor dinucleotides (SA). Color scheme used is as in Fig. 1, and is also noted at the bottom of this figure.
Alternate alleles are present in independent population isolates
The relative amount of genetic drift from the founders of a given strain will vary among different isolates maintained in independent laboratories. To determine the utility of our variation data for the larger zebrafish community, we compared our variation data (populations maintained by the Weinstein laboratory at the NIH) with independent variation data obtained from populations maintained by the Granato laboratory [University of Pennsylvania (UPenn), Philadelphia, PA, USA]. The NIH populations of TL and WIK were both acquired from the ZIRC and maintained at NIH for two generations. The UPenn and ZIRC populations of TL and WIK were derived from the same parental populations kept by the Max Planck Institute for Developmental Biology (Tübingen, Germany), but have been propagated independently for ∼10-15 generations. This comparison was restricted to SNP positions that had at least three reads in both the NIH and UPenn data. In this comparison, the TL and WIK strains shared 3.22 and 2.70 million SNPs (61% and 48% of total), respectively (Fig. 3A,B). The shared SNP positions were then quantified and grouped based on variant frequency in our dataset, as shown in Fig. 3C. Over 99% and 76% of the fixed alleles (1.0 value on y-axis) in the TL and WIK strains, respectively, were also detected in the UPenn dataset, suggesting that most of the SNP data at higher frequencies will be common among different isolates of zebrafish for both the TL and WIK strains (Fig. 3). The higher percentage of SNPs detected in the TL strain suggests a lower level of genetic diversity in this strain compared with the WIK strain.
Fig. 3.
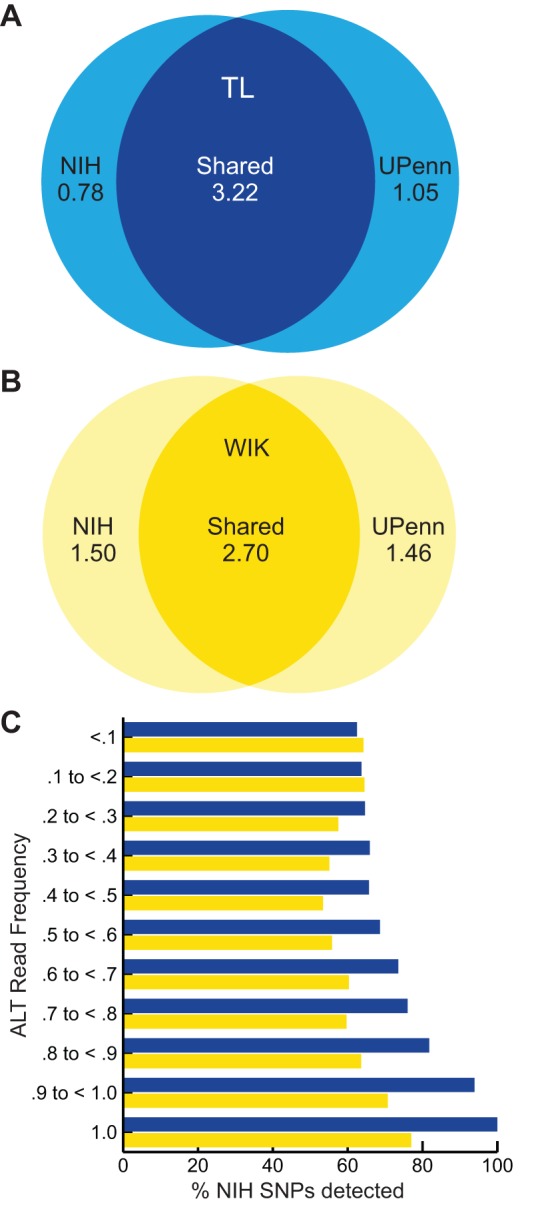
Variant sharing between independent populations of TL and WIK. (A,B) Venn diagrams showing the number of SNPs shared (overlapping areas) or not shared (non-overlapping areas) between independent isolates of the TL (A) and WIK (B) strains maintained at the NIH (leftmost circle) and UPenn (rightmost circle). (C) A stacked bar graph showing the percentage of total NIH SNPs also detected in the UPenn SNP dataset (x-axis), grouped by frequency category (y-axis). Frequency is calculated by dividing the number of alternate allele reads per total reads from the NIH strains. Values for the TL and WIK strains are shown in blue and yellow, respectively.
Zebrafish variant report compiler tool
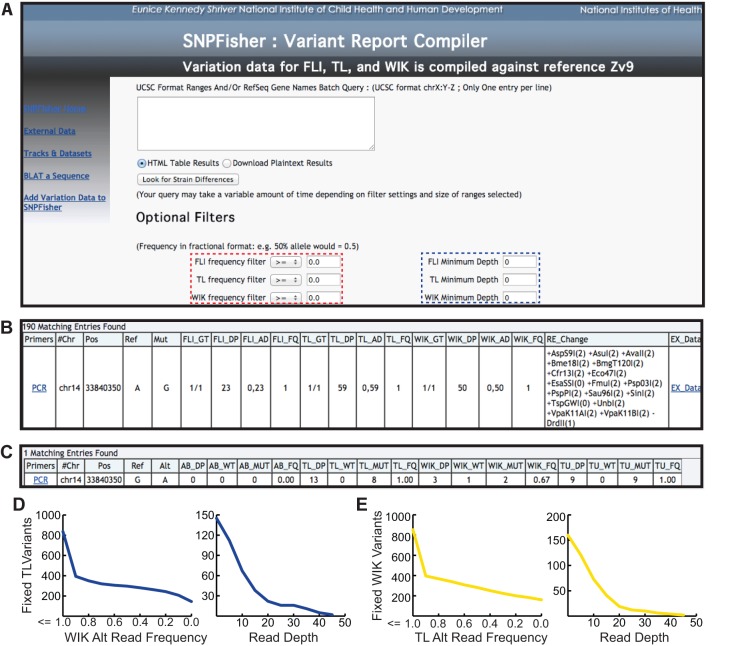
Our data on genetic variation in laboratory zebrafish were organized into an easy-to-use website termed ‘SNPfisher’ (snpfisher.nichd.nih.gov). For acquiring and filtering variants in a gene or genomic region of interest, the SNPfisher variant report compiler was developed (Fig. 4A). Users define a genomic region of interest using the UCSC genome browser coordinate format or provide a Refseq gene name in the query box (Fig. 4A). Optional filters located below the main query box allow user-defined criteria for the variant frequency and the minimum read depth (Fig. 4A, dashed red and blue boxes, respectively). An example of the SNPfisher variant report compiler annotation is shown in Fig. 4B. This report includes the genotype of the population (GT), read depth at the variant position (DP), the allele distribution (AD) and the alternate allele read frequency (FQ=alternate allele reads divided by the total number of reads) for each of the three surveyed strains (FLI, TL, WIK) (Fig. 4B). To expedite the use of these variants, each was provided a link to the ‘Primer3’ web interface to generate primers for PCR amplification (Rozen and Skaletsky, 2000). To further facilitate the use of these variants in applications like allele-specific PCR or genetic fine mapping, each variant was evaluated for restriction fragment length polymorphisms (Fig. 4B). Likewise, to aid in the selection of variants as molecular markers, an external data hyperlink (EX_Data; Fig. 4B) was built into the SNPfisher variation report to assess the presence or absence of variants in other laboratory populations (Fig. 4C). Additionally, external variant annotations can be queried directly by launching the ‘External Data’ hyperlink at the SNPfisher homepage. This link opens a query box to retrieve the desired external variation data by refseq gene name or genomic coordinates.
Fig. 4.
SNPfisher homepage and variant report compiler tool. (A) Screen shot of the SNPfisher homepage featuring the SNPfisher Variant Report Compiler. Users can define frequency values (dashed red box) and minimum read depth values (dashed blue box) for each strain in any gene or genomic region entered into the query box (white box in center). The drop-down filtering menus include ‘=’, ‘<’, ‘>’, ‘≤’ and ‘≥’ to adjust variant frequencies. (B) A variant annotation generated by the SNPfisher Variant Report Compiler homepage. The Primers column contains links to the ‘Primer3’ web interface to generate primers for amplification of the variant. The genotype (GT) value corresponds to genotype of the population of fish surveyed (0/0=ref. allele homozygote, 0/1=alt. allele heterozygote, 1/1=alt. allele homozygote). The allele distribution (AD) is composed of the number of ref. allele reads and then alt. allele reads separated by commas, respectively. Alt. allele read frequencies (FQ) were calculated by dividing the alt. allele reads by the total number of reads at a given position. Variants encoding RFLPs are documented in the RE_Changes column. The creation (+) or destruction (−) of a site is indicated prior to the restriction enzyme name. The number in parentheses following the restriction enzyme name indicates the number of restrictions recognition sites in the 800 bp surrounding the variant (400 bp upstream and downstream) in the Zv9 reference genome sequence. (C) An example of the variant annotation provided by the Ex_Data link in the SNPfisher report. The total read depth (DP), ref. allele read depth (WT), alt. allele read depth (MUT) and alt. allele read frequency (FQ) values represent the cumulative read sums calculated from published datasets. (D,E) Scatterplots demonstrate TL and WIK variant filtration in the cloche crucial region. The number of fixed TL variants (y-axis in left plot of D) decreases by filtering for lower alternate (Alt) allele read frequencies in WIK (x-axis). Increasing read depth (x-axis in right plot of D) decreases fixed TL variants absent in WIK (x-axis). Similar results are obtained for WIK variants in the cloche region (E).
In order to demonstrate the ease and utility of the SNPfisher variant report compiler, variants were evaluated for their informativeness in the ∼800 kb crucial region that has been reported for the cloche mutation (Leshchiner et al., 2012). Hypothetically, if recombination in the cloche region were to be determined by segregation of TL versus WIK alleles, the allele frequency and depth filters could quickly prioritize variants to serve as molecular markers. An unfiltered search of the region returns 4683 variants. Requiring fixation of variant alleles (alt. read frequency=1.0) in TL or WIK decreases the number of variants to 837 and 853, respectively. Requiring absence of TL variants in WIK and vice versa drops the number of variants to 160 and 145 for TL and WIK variants, respectively (left plots in Fig. 4D,E). Additional filtering by read depth, which is proportional to the accuracy of the alternate allele read frequency estimate, quickly reduces the number of candidate variant markers (right plots in Fig. 4D,E). Thus, the SNPfisher variant report compiler not only documents variants, but also prioritizes them to suit a user's needs.
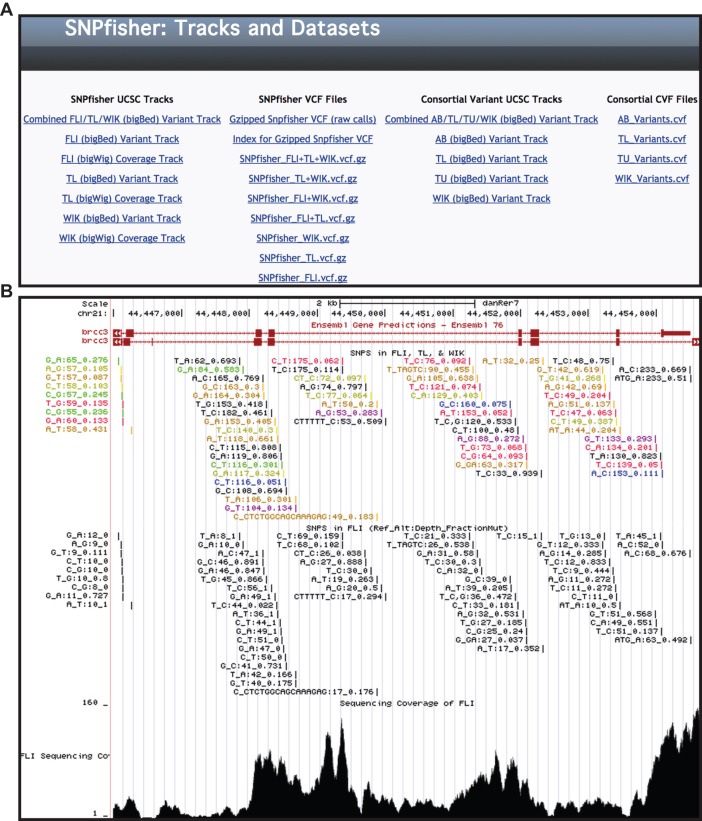
Genome-wide variant resources
To facilitate genome-wide approaches, like filtering strain variants from NGS-based positional cloning projects, the SNPfisher site provides our datasets in variant call format files (VCF). These files are available from the ‘Tracks and Datasets’ page of the SNPfisher website (Fig. 5A, column 2). The SNPfisher VCF contains 7.86 million variants. Additionally, VCFs partitioned by strain distribution are also provided for download. To demonstrate its filtering capability, the SNPfisher dataset was used to filter the linkage region of the published ca1 mutant (Leshchiner et al., 2012). The SNPTrack pipeline, after applying its own inherent variant filtration, identifies ten missense mutations as the most likely causative mutations. SNPfisher filtration reduces the number of candidates to six, having four annotated as natural variants.
Fig. 5.
SNPfisher tracks and dataset page. (A) Screen shot of SNPfisher ‘Tracks and Datasets’ page containing UCSC genome track links (columns 1 and 3) and downloadable datasets in VCF (column 2) and CVF format (column 4). The bigBed file hyperlinks (Combined FLI/TL/WIK, FLI, TL, WIK) display variation data and the bigWig files (FLI, TL and WIK) show base-by-base sequencing coverage for each strain in the UCSC genome browser. CVF variants were also formatted for display in the UCSC genome browser (column 3). (B) Screen shot of the brcc3 locus (Miskinyte et al., 2011) in the UCSC genome browser, in which SNP and Indel variation is demonstrated with the Combined FLI/TL/WIK Variant Track (top), the FLI Variant Track (middle) and the FLI Coverage Track (bottom). Variant annotations are formatted by Ref. Allele_Alt. Allele:Read Depth_Alt. Allele Read Frequency. In the Combined FLI/TL/WIK Variant Track, the strain distribution is annotated by color (FLI, red; TL, blue; WIK, yellow; FLI&TL, purple; FLI&WIK, orange; TL&WIK, green; ALL, black), and depth and alt allele read frequency are calculated from reads from all three strains.
In order to enhance genome-wide variant lists, we combined our variants along with previously published variants (Bowen et al., 2012; Obholzer et al., 2012). Our dataset, as well as the others, were generated from sequencing libraries of pooled genomic DNA samples. Consequently, the alternate allele read frequency is an estimate of the alternate allele frequency in the population of fish represented in the pooled genomic sample. Therefore, the available datasets were combined with our variants in a format we have named consortial variant format (CVF). CVFs contain the chromosome or contig (CHROM), position (POS), reference allele (REF), alternate allele (ALT), reference read depth (REF_DP), alternate allele read depth (ALT_DP), total read depth (DP) and alternate allele read frequency (AF). All of the depth values are sums calculated from the collective datasets available. CVFs for the AB, TL, TÜ and WIK strains, containing 5064481, 6994338, 4094439 and 9661402 variants, respectively, are available for download from the SNPfisher ‘Tracks and Datasets’ page (Fig. 5A, column 4).
SNPfisher genome browser displays
For integrated inquiries with other genome annotations, SNPfisher variants were formatted for display in the UCSC genome browser (Kent et al., 2002) and accessed through the ‘Tracks and Datasets’ link at the SNPfisher homepage (Fig. 5A, column 1). SNPfisher variants were organized into bigBed files and the sequencing coverage of each base of the zebrafish genome was provided in bigWig files. Variant annotation consists of the Zv9 reference allele, the alternate allele, the read depth and the frequency of the alternate allele per total alleles, as demonstrated in Fig. 5B. The frequency information for the Combined FLI/TL/WIK track is calculated from the sum of the alternate allele reads divided by the total number of reads from all strains. For the Combined track, the strains are represented by color, as shown in Fig. 5B (top). SNPfisher also provides a convenient means to scan short oligos for SNPs and Indels. Users must first launch the links for the strain variation of interest (bigBeds) on the ‘Tracks and Dataset’ page. Then, from the SNPfisher homepage, launching the ‘Blat a Sequence’ link will provide a query box for the UCSC Blat tool to search any oligo sequence (Fig. 4A, left) (Kent, 2002). To show the utility of using this tool, a genome-wide survey of splice morpholino (spMO) target site and predicted CRISPR site variation was conducted. Using the SNPfisher variation dataset alone, at least one variant was identified in 48% and 21% of spMO (498,064) and predicted CRISPR (4.28 million) target sites, respectively.
DISCUSSION
Historically, the low number of identified SNPs has hampered their use in the zebrafish field (Stickney et al., 2002; Guryev et al., 2006; Bradley et al., 2007). The recent use of WGS and RNA-seq to clone zebrafish mutations has generated a substantial amount of SNP data in the last few years (Bowen et al., 2012; Leshchiner et al., 2012; Obholzer et al., 2012; Voz et al., 2012; Miller et al., 2013). However, the annotation and relative ease of accessing SNP variation data remain scant and insufficient, respectively. Similarly, the utility of the known SNPs is often limited by the absence of annotations for strain specificity and allele frequency.
In this study, we sampled the natural genetic variation within and between three laboratory zebrafish strains by WGS of pooled DNA samples, encompassing 18 individuals per strain. Each sample was sequenced to ∼16× coverage, yielding an average of 5.04 million SNPs per strain when compared with the Zv9 reference strain. One drawback of using pooled DNA samples is the inability to directly measure allele frequencies for reference and alternate alleles. However, we assumed that each of the 36 alleles in our samples had an equal probability of being sequenced. Consequently, we can estimate the allele frequency from the ratio of alternate allele reads to reference allele reads at a given position. Clearly, the accuracy of our estimated allele frequencies will be proportional to the total read depth per position. A similar proportional relationship will exist between the observed strain specificity and total read depth, as alternate or reference alleles are less likely to be missed at higher read depths. Importantly, the SNPfisher reporter tool enables researchers to easily filter by minimum read depth, allowing each researcher to choose their confidence threshold.
For both the TL and WIK strains, we directly compared the variation in our populations to independent populations maintained in the Granato laboratory at the University of Pennsylvania to address the applicability of using our SNP data in other populations. When alternate allele frequency is considered, almost all (>99%) of the TL variant positions fixed in the NIH population (1.25 million) are detected in the UPenn population, suggesting that our variation data represent the TL strain well. At the same alternate allele frequency, fewer WIK variant positions in the NIH population (0.90 million; 77%) are detected in the UPenn population. Still, the vast majority of fixed variants are detected, suggesting that the variation observed in the WIK population at NIH can be applied reasonably well to other WIK isolates.
In order to facilitate access and use of our dataset, we generated the SNPfisher tool and formatted our data for display in the UCSC genome browser. SNPfisher provides a user-friendly, web-based interface for the acquisition of SNPs and Indels in any region of the zebrafish genome. Users can supply minimums for read depth and allele frequency in any or all strains surveyed (EK, TL or WIK) to quickly ascertain SNPs of interest. Additionally, the display of our SNP data in the UCSC genome browser facilitates integrated inquiries of SNP variation to augment primer selection and target site selection for TALENs, CRISPRs or morpholinos (Nasevicius and Ekker, 2000; Huang et al., 2011; Sander et al., 2011; Chang et al., 2013; Hwang et al., 2013). Variation in intended target sites can be a significant issue in the design of effective tools for site-directed mutation or gene knockdown. For example, unique CRISPR target sites (for the Schizosaccharomyces pombe Cas9 system) have been predicted for the entire zebrafish genome (http://www.genome-engineering.org/crispr/?page_id=41). Examination of the 4.28 million predicted CRISPR target sites revealed that 21% contained SNPs annotated by SNPfisher. Similarly, we evaluated over 498,000 splice morpholino target sites and determined that 48% had at least 1 annotated variant in SNPfisher. Taken together, these observations argue for the incorporation of variation data into sequence-based molecular biology reagent design, which is now easy with SNPfisher.
The SNP variation that we report in this study will enhance the identification of genetic lesions in forward-genetic approaches. Our data will improve the discrimination between causative mutations versus neutral polymorphisms. In addition, the identification of SNP variation in studies like ours will enable genome-wide association studies to tackle complex traits in the zebrafish. Hypothetically, an ENU-mutagenesis screen could be conducted to identify a family with an extreme spectral phenotype that exhibits non-Mendelian inheritance. Given the fecundity of zebrafish, a genome-wide association scan could be performed within the family of interest comparing affected to wild-type siblings, eliminating signals from population stratification without compromising sample size.
MATERIALS AND METHODS
Ethical statement
Zebrafish work was performed in accordance with NIH policy and approved by the Institutional Animal Care and Use Committee.
Zebrafish strains and gDNA isolation
NIH isolates. Tüpfel long fin (TL) and WIK (Rauch et al., 1997) zebrafish strains were purchased from the ZIRC and propagated for two generations. The Tg(fli1a-eGFP)y1 strain (Lawson and Weinstein, 2002) is derived from a transgenic insertion into the EkkWill (EK) strain and has been maintained at NIH for 14 generations, including several backcrosses to wild-type EKs. For each strain, the caudal fins were clipped from 18 individual adult zebrafish (nine female and nine male), and the clips were pooled into a single sample from which gDNA was isolated using standard techniques (Strauss, 2001).
UPenn isolates. TL and WIK samples contained gDNA isolated and pooled from the somatic tissue of 26 and 27 ENU-mutagenized adult males, respectively. Males were selected prior to mutagenesis based on the absence of startle response, performance and habituation behaviors.
Whole-genome small insert library method and sequencing
The NIH Intramural Sequencing Center (NISC) performed library preparation and WGS. Five micrograms (μg) of genomic DNA was sheared to ∼500 bp using a Covaris E210 with the following settings: duty cycle 20%; intensity 4; cycles 200. The DNA was size-selected for a range of 300-600 bp on a Pippin Prep (Sage Science) 1.5% cassette. The library was constructed on a SPRI-TE Robot (Beckman Coulter) using SPRI-TE reagents and Illumina Paired-End DNA Sample Prep Oligo Only Kit. The resulting library was size-selected on a Pippin Prep with a 1.5% cassette selecting for 440-560 bp. To ensure that the library was not overamplified, test amplification was performed in which aliquots of a PCR reaction were removed every two cycles from 4-16. These aliquots were evaluated on a 2% agarose gel and an optimal cycle number was selected for subsequent large-scale amplification in which unique barcodes were added to each library. For these libraries 12-14 cycles were selected. Amplification reactions were cleaned up using two rounds of Agencourt AMPure Beads. Library aliquots were pooled and the pool was evaluated on a MiSeq indexed run. The de-multiplexed read numbers were used to produce a normalized pool for sequencing on a HiSeq2000. The five-member pool was run in one lane to generate paired-end 101 base reads. Additional reads were generated for three of the libraries (FLI, WIK and TL). These three were run in individual lanes under the same run conditions. Data were processed using RTA1.12.4.2 or 1.13.48 and CASAVA 1.8.2.
WGS alignment and post-processing
Reads in FASTQ format obtained from Illumina HiSeq sequencing were aligned to the Zv9 assembly (Ensembl) for each sample using Bowtie2 (Langmead and Salzberg, 2012). Following alignment, the BAM files were treated according to the GATK best practices workflow (https://www.broadinstitute.org/gatk/guide/best-practices) (McKenna et al., 2010; DePristo et al., 2011). Read clipping was performed and duplicates were marked and removed with Picard (http://picard.sourceforge.net). In addition, reads with mapping quality under 30 were discarded using Samtools and a custom Perl script (Li et al., 2009). Finally, SNPs and Indels were called across all datasets in parallel using the GATK unified genotyper (McKenna et al., 2010; DePristo et al., 2011).
Filtering and annotation of SNPs and Indels
SNPs and Indels were split and flagged according to manually defined filters as follows (FLAG: Filter logic): InDel: SNP mutations occurring within 10 bp of Indels; MapQuality: Mapping Quality <40; LowQual: QUAL <30; HighDepth: Depth >120; LowDepth: Depth <3; SNPCluster: ≥3 SNPs in a 10-bp window. Indels were flagged according to the following: LowQD: Quality depth ratio (QD) <2. Flagged mutations were discarded from consideration. Additionally, called mutations were required to pass data for all three datasets; a minimum of three reads must exist in each of the three strains for any single mutation to be considered. Mutations mapped to repetitive elements and low-complexity regions were removed using a Repeatmasker annotation file of the Zv9 sequence (http://repeatmasker.org). The SNPs were categorized by location and consequence using Annovar (Wang et al., 2010). Basic statistics regarding gene position and resulting mutation changes were compiled using a custom Perl script. SNPs introducing or eliminating restriction enzyme recognition sites were predicted using a custom Perl script, using restriction enzyme mutation patterns for the list of enzymes available on the commercial website of New England Biolabs (NEB).
SNP density calculations
The size of the zebrafish genome (Zv9) is reported to be 1.506 Gb (Howe et al., 2013). Repeatmasker masked 769 Mb of the Zv9 genome as repetitive sequence (51%), leaving 737 Mb of unique sequence. The size of the human genome is 3.14 Gb, with Repeatmasker marking 1.47 Gb as repetitive sequence. The remaining 1.66 Gb of unique sequence was used for density calculations.
ca1 Mutant filtration
SRR453533 and SRR453534 were downloaded from the Sequence Read Archive (http://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?view=search_obj) and converted to FASTQ format with the fastq-dump tool in the SRA Toolkit (Wheeler et al., 2008). The resultant FASTQs were uploaded to SNPTrack for linkage analysis and mutation screening (Leshchiner et al., 2012). SNPs from the linked region (chr3:21.4-33.2 Mb), as indicated by the HMM score, were downloaded from SNPTrack if they met each of the following criteria: (1) homozygous in the mutant pool; (2) heterozygous in the wild-type pool; (3) not annotated in the SNPTrack SNP database; and (4) not repetitive. The most likely causative mutations (ten missense mutations) of these SNPs were evaluated for presence in SNPfisher.
Splice morpholino and CRISPR target site polymorphisms
CRISPR target sites were downloaded from the CRISPR Genome Engineering Resources website (http://www.genome-engineering.org/crispr/) via the UCSC genome browser. All annotated exon start and stop coordinates in Zv9 were extracted from Ensembl gene annotations downloaded from the UCSC genome browser (http://hgdownload.soe.ucsc.edu/goldenPath/danRer7/database/ensGene.txt.gz). spMO target sites were defined as 50 bp (25 upstream, 25 downstream) surrounding the last base of the exon of splice donors and the first base of the exon of splice acceptors. A custom script was written to generate a non-redundant bed of spMO target sites from the extracted exon target sites. The intersect function of bedtools was used to identify spMO and CRISPR target sites that contain SNPfisher variants (Quinlan and Hall, 2010).
SNPfisher tool
All SNPs and Indels passing all filtering criteria along with predicted restriction enzyme changes were compiled in a simple database accessible using a web interface (snpfisher.nichd.nih.gov). Simple filtering criteria are implemented in the submission form, including restriction digest requirements, allele frequencies and associated read depths for each or any of the strains. Sequencing data and mutation calls were compiled into UCSC genome browser tracks for easy viewing of both mutations and sequencing coverage for each genetic background.
Consortial variant format
Binary alignment maps (BAMs) for each wild-type strain sequenced by Bowen et al. were downloaded from the Resource page of the Harris Lab website (Boston Children's Hospital, Boston, MA, USA; http://fishbonelab.org/harris/Resources.html) (Bowen et al., 2012). BAMs divided by chromosome were concatenated into a single BAM per each strain. Variants were recalled and filtered independently, not with other strains, according to the criteria described by Bowen et al., with the exception that a low-depth filter was removed due to low genome coverage.
VCF files for the wild-type strains sequenced by Obholzer et al. were downloaded from the MegaMapper page of the Megason Lab website (https://wiki.med.harvard.edu/SysBio/Megason/MegaMapper; Obholzer et al., 2012). A Repeatmasker annotation file for the Zv9 assembly was downloaded from http://hgdownload.soe.ucsc.edu/goldenPath/danRer7/database/rmsk.txt.gz and re-ordered to match the contig order in the Zv9 fasta downloaded from the Wellcome Trust Sanger Institute website (ftp://ftp.sanger.ac.uk/pub/zfish/assembly/Zv9/Zv9_toplevel.fa.gz). The reordered Repeatmasker file was used as a mask with the GATK VariantFiltration tool to flag and then remove repeatmasked variants from the original VCFs created by Obholzer et al. A bash script was then written to calculate the QD values for each variant and remove any variants with a QD<2.0.
CVFs were constructed from VCFs from each dataset. CVFs are composed of the chromosome or contig (CHROM), position in the Zv9 genome reference assembly (POS), Zv9 reference allele (REF), alternate allele (ALT), cumulative Zv9 reference allele read depth (REF_DP), cumulative alternate allele read depth (ALT_DP), cumulative total read depth (DP) and the cumulative alternate allele read depth (AF). The allele read depths and allele read frequencies are referred to as cumulative, as they are the sum of the values from each independent dataset. Inclusion in the CVFs was restricted to biallelic variants, as REF_DP and ALT_DP values could not be calculated from DP4 values in the INFO fields of polyallelic variants in the datasets of Bowen et al. (2012) and Obholzer et al. (2012).
Data access
All SNP and Indel variants that passed all of the filters imposed by the authors are available at the snpfisher.nichd.nih.gov website. Raw seq files will be submitted to Sequence Read Archive (SRA) at http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi upon acceptance under the accession numbers SRS863087, SRS863089, SRS863090.
Supplementary Material
Acknowledgements
We would like to thank Alice Young (NISC) for providing the methods for genomic library construction and Illumina sequencing performed in this study. We would also like to thank Matthew Breymaier for providing website and server expertise. This study utilized the high-performance computational capabilities of the Helix Systems and Biowulf cluster at the NIH (http://helix.nih.gov).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
This work was supported by the intramural program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) and by the Fondation Leducq (Transatlantic Network of Excellence for the Identification of Novel Genetic Targets in Hemorrhagic Stroke) to B.M.W. This work was also supported by a National Institutes of Health grant [MH092257] to M.G. Deposited in PMC for release after 12 months.
Author contributions
M.G.B. and B.M.W. conceived and designed the experiments. M.G.B. performed the experiments. Data analysis was conducted by M.G.B., J.R.I. and J.A.E. M.G.B. and B.M.W. developed the SNPfisher website. J.R.I. wrote the code for the SNPfisher website. K.C.M. and M.G. provided WGS data for comparison. M.G.B. and B.M.W. wrote the paper.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.118786/-/DC1
References
- Abecasis G. R., Altshuler D., Auton A., Brooks L. D., Durbin R. M., Gibbs R. A., Hurles M. E. and McVean G. A. (2010). A map of human genome variation from population-scale sequencing. Nature 467, 1061-1073 10.1038/nature09534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis G. R., Auton A., Brooks L. D., DePristo M. A., Durbin R. M., Handsaker R. E., Kang H. M., Marth G. T. and McVean G. A. (2012). An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56-65 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen M. E., Henke K., Siegfried K. R., Warman M. L. and Harris M. P. (2012). Efficient mapping and cloning of mutations in zebrafish by low-coverage whole-genome sequencing. Genetics 190, 1017-1024 10.1534/genetics.111.136069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley K. M., Elmore J. B., Breyer J. P., Yaspan B. L., Jessen J. R., Knapik E. W. and Smith J. R. (2007). A major zebrafish polymorphism resource for genetic mapping. Genome Biol. 8, R55 10.1186/gb-2007-8-4-r55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang N., Sun C., Gao L., Zhu D., Xu X., Zhu X., Xiong J.-W. and Xi J. J. (2013). Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 23, 465-472 10.1038/cr.2013.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe T. S., Hamilton P. B., Griffiths A. M., Hodgson D. J., Wahab M. A. and Tyler C. R. (2009). Genetic variation in strains of zebrafish (Danio rerio) and the implications for ecotoxicology studies. Ecotoxicology 18, 144-150 10.1007/s10646-008-0267-0 [DOI] [PubMed] [Google Scholar]
- DePristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R., Hartl C., Philippakis A. A., del Angel G., Rivas M. A., Hanna M. et al. (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491-498 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guryev V., Koudijs M. J., Berezikov E., Johnson S. L., Plasterk R. H. A., van Eeden F. J. M. and Cuppen E. (2006). Genetic variation in the zebrafish. Genome Res. 16, 491-497 10.1101/gr.4791006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P., Odenthal J., Mullins M. C., Lin S., Farrell M. J., Vogelsang E., Haas F., Brand M., van Eeden F. J. M., Furutani-Seiki M. et al. (1996). Mutations affecting pigmentation and shape of the adult zebrafish. Dev. Genes Evol. 206, 260-276 10.1007/s004270050051 [DOI] [PubMed] [Google Scholar]
- Harper C. and Lawrence C. (2011). The Laboratory Zebrafish. Boca Raton, FL: CRC Press. [Google Scholar]
- Howe K., Clark M. D., Torroja C. F., Torrance J., Berthelot C., Muffato M., Collins J. E., Humphray S., McLaren K., Matthews L., et al. (2013). The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498-503 10.1038/nature12111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Xiao A., Zhou M., Zhu Z., Lin S. and Zhang B. (2011). Heritable gene targeting in zebrafish using customized TALENs. Nat. Biotechnol. 29, 699-700 10.1038/nbt.1939 [DOI] [PubMed] [Google Scholar]
- Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Tsai S. Q., Sander J. D., Peterson R. T., Yeh J.-R. J. and Joung J. K. (2013). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227-229 10.1038/nbt.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. J. (2002). BLAT--the BLAST-like alignment tool. Genome Res. 12, 656-664 10.1101/gr.229202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M. and Haussler D. (2002). The human genome browser at UCSC. Genome Res. 12, 996-1006 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapik E. W., Goodman A., Ekker M., Chevrette M., Delgado J., Neuhauss S., Shimoda N., Driever W., Fishman M. C. and Jacob H. J. (1998). A microsatellite genetic linkage map for zebrafish (Danio rerio). Nat. Genet. 18, 338-343 10.1038/ng0498-338 [DOI] [PubMed] [Google Scholar]
- Langmead B. and Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357-359 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson N. D. and Weinstein B. M. (2002). In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 248, 307-318 10.1006/dbio.2002.0711 [DOI] [PubMed] [Google Scholar]
- Leshchiner I., Alexa K., Kelsey P., Adzhubei I., Austin-Tse C. A., Cooney J. D., Anderson H., King M. J., Stottmann R. W., Garnaas M. K. et al. (2012). Mutation mapping and identification by whole-genome sequencing. Genome Res. 22, 1541-1548 10.1101/gr.135541.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G. and Durbin R. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078-2079 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M. et al. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297-1303 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A. C., Obholzer N. D., Shah A. N., Megason S. G. and Moens C. B. (2013). RNA-seq-based mapping and candidate identification of mutations from forward genetic screens. Genome Res. 23, 679-686 10.1101/gr.147322.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miskinyte S., Butler M. G., Hervé D., Sarret C., Nicolino M., Petralia J. D., Bergametti F., Arnould M., Pham V. N., Gore A. V. et al. (2011). Loss of BRCC3 deubiquitinating enzyme leads to abnormal angiogenesis and is associated with syndromic moyamoya. Am. J. Hum. Genet. 88, 718-728 10.1016/j.ajhg.2011.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasevicius A. and Ekker S. C. (2000). Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet. 26, 216-220 10.1038/79951 [DOI] [PubMed] [Google Scholar]
- Nechiporuk A., Finney J. E., Keating M. T. and Johnson S. L. (1999). Assessment of polymorphism in zebrafish mapping strains. Genome Res. 9, 1231-1238 10.1101/gr.9.12.1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obholzer N., Swinburne I. A., Schwab E., Nechiporuk A. V., Nicolson T. and Megason S. G. (2012). Rapid positional cloning of zebrafish mutations by linkage and homozygosity mapping using whole-genome sequencing. Development 139, 4280-4290 10.1242/dev.083931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patowary A., Purkanti R., Singh M., Chauhan R., Singh A. R., Swarnkar M., Singh N., Pandey V., Torroja C., Clark M. D. et al. (2013). A sequence-based variation map of zebrafish. Zebrafish 10, 15-20 10.1089/zeb.2012.0848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton E. E. and Zon L. I. (2001). The art and design of genetic screens: zebrafish. Nat. Rev. Genet. 2, 956-966 10.1038/35103567 [DOI] [PubMed] [Google Scholar]
- Quinlan A. R. and Hall I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841-842 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch G.-J., Granato M. and Haffter P. (1997). A polymorphic zebrafish line for genetic mapping using SSLPs on high-percentage agarose gels. In Technical Tips Online: BioMedNet. [Google Scholar]
- Rozen S. and Skaletsky H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365-386. [DOI] [PubMed] [Google Scholar]
- Sander J. D., Cade L., Khayter C., Reyon D., Peterson R. T., Joung J. K. and Yeh J.-R. J. (2011). Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat. Biotechnol. 29, 697-698 10.1038/nbt.1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solnica-Krezel L., Schier A. F. and Driever W. (1994). Efficient recovery of ENU-induced mutations from the zebrafish germline. Genetics 136, 1401-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickney H. L., Schmutz J., Woods I. G., Holtzer C. C., Dickson M. C., Kelly P. D., Myers R. M. and Talbot W. S. (2002). Rapid mapping of zebrafish mutations with SNPs and oligonucleotide microarrays. Genome Res. 12, 1929-1934 10.1101/gr.777302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss W. M. (2001). Preparation of genomic DNA from mammalian tissue. Curr. Protoc. Mol. Biol. 42, 2.2.1-2.2.3 10.1002/0471142727.mb0202s42 [DOI] [PubMed] [Google Scholar]
- Tresnak I. (1981). The long-finned zebra Danio. Trop. Fish Hobby 29, 43-56. [Google Scholar]
- Trevarrow B. and Robison B. (2004). Genetic backgrounds, standard lines, and husbandry of zebrafish. Methods Cell Biol. 77, 599-616 10.1016/S0091-679X(04)77032-6 [DOI] [PubMed] [Google Scholar]
- Turner D. J., Keane T. M., Sudbery I. and Adams D. J. (2009). Next-generation sequencing of vertebrate experimental organisms. Mamm. Genome 20, 327-338 10.1007/s00335-009-9187-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voz M. L., Coppieters W., Manfroid I., Baudhuin A., Von Berg V., Charlier C., Meyer D., Driever W., Martial J. A. and Peers B. (2012). Fast homozygosity mapping and identification of a zebrafish ENU-induced mutation by whole-genome sequencing. PLoS ONE 7, e34671 10.1371/journal.pone.0034671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li M. and Hakonarson H. (2010). ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler D. L., Barrett T., Benson D. A., Bryant S. H., Canese K., Chetvernin V., Church D. M., DiCuccio M., Edgar R., Federhen S. et al. (2008). Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 36Suppl. 1, D13-D21 10.1093/nar/gkm1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley A. R., Bhat A., Martins E. P., Mayden R. L., Arunachalam M., Uusi-Heikkila S., Ahmed A. T., Shrestha J., Clark M., Stemple D. et al. (2011). Population genomics of wild and laboratory zebrafish (Danio rerio). Mol. Ecol. 20, 4259-4276 10.1111/j.1365-294X.2011.05272.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.